Abstract
We present a microfluidic immunoassay platform based on the use of linear microretroreflectors embedded in a transparent polymer layer as an optical sensing surface, and micron-sized magnetic particles as light-blocking labels. Retroreflectors return light directly to its source and are highly detectable using inexpensive optics. The analyte is immuno-magnetically pre-concentrated from a sample and then captured on an antibody-modified microfluidic substrate comprised of embedded microretroreflectors, thereby blocking reflected light. Fluidic force discrimination is used to increase specificity of the assay, following which a difference imaging algorithm that can see single 3 μm magnetic particles without optical calibration is used to detect and quantify signal intensity from each sub-array of retroreflectors. We demonstrate the utility of embedded microretroreflectors as a new sensing modality through a proof-of-concept immunoassay for a small, obligate intracellular bacterial pathogen, Rickettsia conorii, the causative agent of Mediterranean Spotted Fever. The combination of large sensing area, optimized surface chemistry and microfluidic protocols, automated image capture and analysis, and high sensitivity of the difference imaging results in a sensitive immunoassay with a limit of detection of roughly 4000 R. conorii per mL.
Introduction
Immunoassays, widely used since the development of the radioimmunoassay by Yalow and Berson,1 have traditionally used macroscopic liquid handling systems such as microtiter plates. In recent years, immunoassays are increasingly being implemented in microfluidic formats that provide several advantages: smaller device footprints and sample/reagent quantities; shorter times to result; well-defined and precisely controllable flow; robust statistics from redundant, miniaturized detection regions; faster mass transfer; and the dominance of surface-mediated over volumetric phenomena.2 At the broadest level, microfluidic assay platforms comprise two components: (i) flow control by pressure-driven,3 electroosmotic,4 electrowetting,5 capillary,6 or centrifugal7 approaches, and (ii) a readout strategy, which may be label-free or label-dependent. Label-free readout strategies include the use of field effect transistors,8,9 cantilevers,10 or resonant optical waveguides,11,12 and the exploitation of phenomena such as surface plasmon resonance13 and surface acoustic waves.14 Fluorescence is commonly used for label detection in microfluidic immunoassays despite requiring complex, expensive hardware. The analyte is labeled using a detection antibody carrying small organic fluors, fluorescent proteins, or fluorescent nanoparticles, and is read out using CCD cameras and magnifying optics that have direct or engineered optical paths for signal enhancement.6,15-17 Other label-based readout techniques used in microfluidic assays include chemiluminescence,18 enzymatic electrochemistry,19 and surface-enhanced Raman spectroscopy.20
Superparamagnetic particles of nanometer to micrometer diameters are commercially available with well-characterized diameters, compositions and surfaces, and are widely used as substrates and/or labels in microfluidic biosensors.21 Magnetic beads have been used as labels in readout strategies including electrochemical detection,22 electrochemiluminescence,23,24 mass spectrometry,25 and agglutination-monitoring assays;20,26 they have also been commercialized in the Luminex xMAP platform that uses fluorescent detection.27 “Surface coverage” assays, in which analytes are affinity-sandwiched between magnetic beads and a detection surface, use the number of particles bound to the sensor surface as the readout. The techniques used to count beads in magnetic bead-based surface coverage assays include direct inspection and counting using an optical microscope,28-30 surface plasmon resonance to quantify binding-induced refractive index changes,31,32 magnetoresistive sensing technologies, such as the compact bead array sensor system (cBASS)30 and the bead array counter (BARC),33,34 mass-based detection using measurements of the shift in resonant frequency of a MEMS resonator,35 and nuclear magnetic resonance.36 In some cases, after surface capture, nonspecifically bound beads are selectively removed using fluidic drag forces to reduce the “background” bead count and therefore improve the limit of detection.28-30,34,37 A number of microfluidic assay platforms, based on magnetic bead labels and otherwise, have excellent limits of detection, but require significant user training, and are inherently difficult to automate. For example, when an optical microscope is used to directly obtain bead counts in a surface coverage assay, the actual enumeration of beads is potentially easy to automate, but an experienced operator would still need to fine-tune the focus for each assay area and/or microfluidic channel to obtain quality data. For use in a field-deployable manner by minimally trained personnel, easy-to-automate sensing modalities capable of excellent analytical sensitivities are highly desirable.
In this paper, we introduce a microfluidic surface coverage sandwich immunoassay using magnetic beads as light-blocking labels and photo-lithographically fabricated linear microretroreflectors, embedded in a transparent polymer layer, as the optical sensing surface. Retroreflectors are highly detectable structures that efficiently reflect light back to its source over a broad range of angles. Arrays of spherical (transparent high-refractive index spheres partially coated with a reflective surface) or corner-cube retroreflectors (with three mutually perpendicular reflective surfaces) with dimensions on the order of 100 μm to several millimeters find applications in road markings and personnel or vehicle conspicuity (as retroreflective tape or paint),38,39 remote sensing of air pollutants,40 lunar laser ranging,41 and as components of laser interferometers.42 We have previously reported the fabrication of suspended 5 μm corner cube retroreflectors43 and their use as optical immunoassay labels.44 In this work, we use densely packed arrays of thousands of micron-scale linear retroreflectors (dimensions 100 μm length × 3 μm width × 5 μm height), fabricated with precise positioning on a microfluidic substrate and embedded in a bio-functionalizable transparent polymer, as a microfluidic optical sensing substrate. Linear retroreflectors have one less reflective surface than corner cube reflectors, and are brightest at one azimuth. As shown in Fig. 1(A), embedded linear microretroreflectors present as a machine-registerable pattern of bright lines when viewed through an in-lens illuminated objective connected to a CMOS camera. The detection principle involves the analyte-dependent blocking of light reflected from the retroreflectors through a sandwich immunoassay, as shown in Fig. 1(B). The precise arrangement of retroreflectors over large areas allows automated image capture, and the difference between images captured before and after the assay is completed is used to determine the particle counts. We demonstrate the utility of this new sensing modality through a proof-of-concept immunoassay for the sensitive detection of the bacterium Rickettsia conorii. R. conorii is a Gram-negative, obligate intracellular bacterium that is the causative agent of the tick-transmitted Mediterranean Spotted Fever.45 An NIAID Category C priority pathogen, R. conorii is a potential bioterror agent due to the possibility of aerosolized release causing severe disease (infectious dose less than 10 organisms) and prior attempts to weaponize Rickettsiae.46 In this work, 3.0 μm magnetic beads modified with polyclonal anti-R. conorii antibodies were used to capture R. conorii spiked into buffer, and introduced after pre-concentration into a microfluidic channel with anti-R. conorii functionalized embedded microretroreflectors as the surface to complete the immuno-sandwich. Non-specifically bound beads were removed by fluidic force discrimination, and the remaining beads were counted using an automated image capture and image differencing approach. This assay was determined to have a limit of detection of around 4000 R. conorii/mL. Whereas the embedded microretroreflector-based assay platform is demonstrated here for a model analyte, it can be extended to the development of assays that rapidly and sensitively detect other biomolecules (e.g., proteins, viruses, DNA) with minimal user intervention, with the only pre-requisite being the availability of specific affinity reagents.
Figure 1.
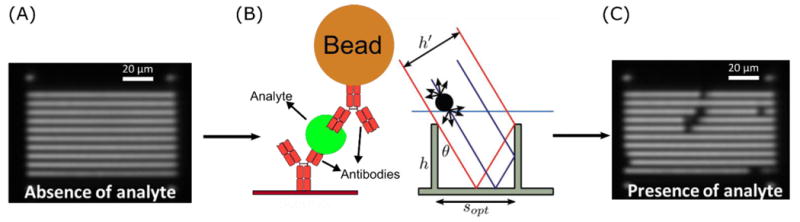
Detection principle. (A) Antibody-modified embedded linear microretroreflectors appear as bright white lines when imaged through an in-lens illuminated CMOS camera. (B) Analyte is sandwiched between embedded microretroreflector surface and antibody-modified magnetic beads, which block light. (C) Beads appear as dark spots on the retroreflector pattern; bead count correlates with analyte concentration.
Methods and Experiments
Bacteria and antibodies
R. conorii (Malish strain) was grown in Vero cell monolayers (ATCC, Manassas, VA) in 150 cm2 tissue culture flasks, cultured in Modified Eagle Medium (Gibco, Paisley, UK) and supplemented with 4% fetal calf serum and 2 mM L-glutamine. Five days after inoculation, infected cells were harvested with sterile glass beads and pelleted by centrifugation at 10,000 × g for 15 minutes. Bacteria were purified using sucrose gradients, and heat-inactivated by heating at 60°C for 30 minutes to downgrade biosafety requirement to BSL-2. An Izon qNano tunable resistive pulse sensing device (Izon Science, New Zealand) was used to confirm the concentration and structural integrity of heat-inactivated bacteria. To raise polyclonal rabbit anti-R. conorii antibodies, New Zealand rabbits, housed in an ABSL-3 facility at University of Texas Medical Branch-Galveston, were immunized with 106 R. conorii intravenously every 3 weeks until antibody titers by IFA reached 1:2096. The rabbits were humanely sacrificed according to IACUC protocols and blood was harvested. The serum was affinity purified using Protein A columns and re-suspended in phosphate buffered saline (PBS; Clontech Laboratories, Mountain View, CA) to give an antibody concentration near 1 mg/mL.
Embedded microretroreflector fabrication
Embedded microretroreflectors were fabricated on 100 mm diameter silicon wafers using a scalable photolithographic technique [Fig. 2]. A 5 μm thick layer of SU-8 (SU-8 2005, MicroChem Corp., Westborough, MA) was first spin-cast (Cee 200X, Brewer Science, Rolla, MO) onto an ozone-cleaned 100 mm silicon wafer and pre-baked at 95°C for 4 minutes. This was followed by UV exposure through a photomask using a mask aligner (Model TTMA, ABM, Inc., Scotts Valley, CA) and a post-exposure bake at 95°C for 3 minutes. The photomask was designed to allow the simultaneous formation of five 38×25mm2 chip patterns, each consisting of 15 channels. The pattern was developed in 1-methoxy-2-propanol acetate for 90 seconds, rinsed in isopropyl alcohol, and dried using nitrogen. Next, silver was evaporated using an electron-beam evaporator (3 kW e-Gun, Thermionic Vacuum Products, Hayward, CA) onto the structures at a 30° angle to the horizontal from both sides to a thickness of 120 nm, and at a 90° angle to a thickness of 60 nm, which ensured that the base and walls of the structures were reflective. The linear retroreflectors manufactured using this method were 5 μm tall, 3 μm wide and 100 μm long, and were spaced by an 8 μm pitch. To create a planar surface onto which the particles could be captured, the wafer bearing the retroreflectors was coated with a 6 μm thick layer of SU-8 2005 by spin-casting at 800 rpm for 1 minute, followed by a pre-exposure bake at 95°C for 4.5 minutes on a hot plate, a blanket UV exposure for 10 minutes (CL-1000 Ultraviolet Crosslinker, UVP, LLC, Upland, CA), and a post-exposure bake at 95°C for 3 minutes. Finally, a 2 μm thick layer of poly(methyl methacrylate) (PMMA C9, MicroChem Corp., Westborough, MA) was deposited by spin-coating at 2000 rpm for 30 seconds and baking at 180°C for 10 minutes, resulting in an overall planarization layer thickness of 8 μm. The silicon wafers containing the embedded microretroreflectors were cleaved, using a diamond-tipped scriber, into 38×25mm2 chips that would eventually form microfluidic substrates, and stored in a clean environment prior to antibody-modification.
Figure 2.
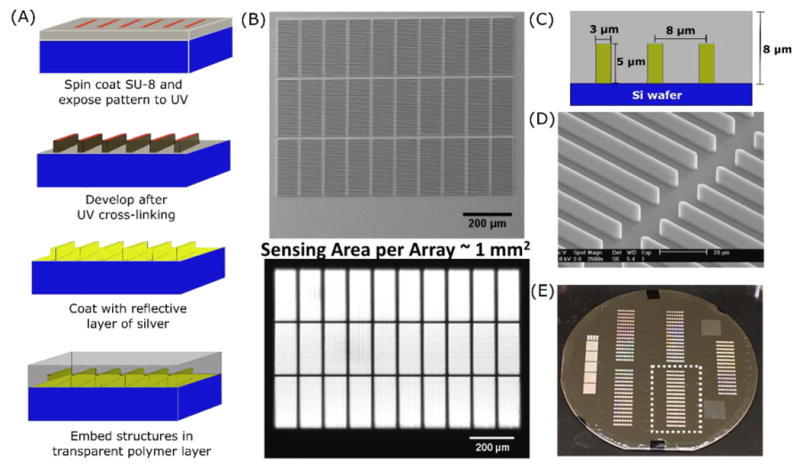
(A) Schematic of embedded microretroreflector fabrication. (B) (top) Scanning electron microscope (SEM) image of a single linear microretroreflector “array” – an arrangement of 30 “tiles” in three rows of 10 tiles each, with each tile consisting of 30 linear microretroreflectors; (bottom) image of linear microretroreflector array obtained using an in-lens illuminated CMOS camera with a 4× objective (middle row in focus). (C) Dimensions and intra-tile spacing of embedded linear microretroreflectors. (D) Close-up SEM image showing retroreflector structures from two adjacent tiles. (E) Picture of a 100 mm silicon wafer with five 38×25mm2 regions of embedded microretroreflectors that eventually form microfluidic substrates; one such region is indicated by the dotted rectangle.
Functionalization of microfluidic substrates and magnetic beads with anti-R. conorii antibodies
Two strategies were used to covalently immobilize anti-R. conorii on the embedded retroreflector surface (PMMA) of the microfluidic channels and the surface of the magnetic beads.
Strategy 1: Fc-mediated immobilization following oxidation of polysaccharides on antibodies
The presence of glycosylated residues in the Fc portion of rabbit IgGs47 enabled their immobilization on sensing surfaces in a favorably directed manner. A previously described method48 was adapted to oxidize the oligosaccharide moieties in the antibodies' Fc region and create amine-reactive aldehyde groups. Briefly, 20 μL of 0.2 M sodium (meta)periodate (Sigma-Aldrich) in water was added to 200 μL of 1 mg/mL anti-R. conorii antibody (in PBS) and allowed to react in the dark for 2 h at 4°C. The resulting solution of Fc-oxidized antibodies was purified from the reaction mixture using Zeba Spin Desalting Columns (7K MWCO, Thermo Scientific) and re-suspended in PBS to a concentration of 1 mg/mL (verified by absorbance at 280 nm).
Microfluidic substrate
Embedded microretroreflectors were covalently antibody-functionalized by first introducing primary amines on the PMMA surface and linking them to the aldehydes on the Fc-oxidized anti-R. conorii antibodies via reductive amination. Briefly, the substrates were cleaned by bath sonication (5 min) in 200-proof ethanol followed by thorough rinsing with MilliQ water, dried under a nitrogen stream, and immersed in a solution of 10% hexamethylenediamine (Sigma-Aldrich) in 100 mM borate buffer, pH 11.5, for 2 hours at room temperature to introduce primary amines.49 After washing with MilliQ water, the chips were dried at 40°C for 10 min, following which 6 μL of a 140 μg/mL PBS solution of Fc-oxidized anti-Rc antibodies was manually spotted on each six-array row of embedded microretroreflectors. Following incubation in a humidified chamber at room temperature for 4 hours, the substrates were quickly rinsed using 10 mM 2-(N-morpholino)ethanesulfonic acid (MES; Sigma-Aldrich) buffer, pH 6.0, and immediately immersed in a 50 mM solution of sodium cyanoborohydride (NaCNBH3; Sigma-Aldrich) in 10 mM sodium phosphate, pH 8.0, for 1 hour to reduce the unstable Schiff bases formed by the reaction of primary amines and aldehydes to stable secondary amines. The chips were then washed thoroughly with PBS and passivated by immersion in 20 mg/mL BSA in PBS for 2 hours at room temperature. They were then rinsed with MilliQ water, dried under a nitrogen stream, and stored at 4°C until further use (usually within a few hours).
Beads
3 μm NH2-modified magnetic beads (Bangs Laboratories, Fishers, IN) were covalently modified with Fc-oxidized anti-R. conorii antibodies via reductive amination. Briefly, 100 μL of beads, as purchased, were washed once with 1 mL 10 mM sodium phosphate, pH 8.0, using a magnetic separation stand (Thermo Scientific). They were then incubated with 400 μL of a 100 μg/mL solution of Fc-oxidized anti-R. conorii antibodies in 10 mM sodium phosphate, pH 8.0 for 4 hours at room temperature. After two washes with 10 mM MES, pH 6.0 the beads were incubated for 30 minutes at 37 °C, in 1 mL of a 250 mM NaCNBH3 solution in 10 mM sodium phosphate, pH 8.0. The antibody-modified beads were then washed twice with PBS and passivated by incubating with 15 mg/mL BSA in PBS for 2 hours at 37°C. They were then washed thrice and stored suspended in PBS at 4°C.
Strategy 2: Amine-mediated immobilization with PEG spacers
Microfluidic substrate
The PMMA surfaces of embedded retroreflectors were first PEGylated by nucleophilic aminolysis using a homobifunctional PEG molecule. After initial cleaning by 10 min immersion in isopropyl alcohol followed by rinsing with water, a 100 mg/mL solution of PEG bis(amine) (MW 3000; Sigma-Aldrich) in 100 mM borate buffer, pH 11.9, was spotted over the entire surface of the microfluidic substrates and incubated for six hours at room temperature. The substrates, now displaying primary amines at the end of PEG spacers, were immersed in 2.5% glutaraldehyde (by volume, in PBS) at room temperature for 2 hours. After rinsing with PBS, 6 μL anti-Rc antibody at 140 μg/mL in PBS (also containing 50 mM NaCNBH3), was spotted manually on the sensing areas of each microfluidic channel and incubated in a humidified environment for 4 hours. After removing the unreacted antibodies, passivation was performed using a solution of 500 mM hydroxylamine and 20 mg/mL BSA in PBS containing 50 mM NaCNBH3 for 2 hours at room temperature. This was followed by a final wash step, after which the substrates were dried under a nitrogen stream and stored at 4°C until use.
Beads
50 μL as-purchased aliquots of 3.0 μm streptavidin-modified magnetic beads (Bangs Laboratories, Fishers, IN) were washed twice with PBS, then PEGylated using a 2 mg/mL solution of biotin-PEG-COOH (MW 3400; Nanocs, New York, NY) in PBS for 2 hours. This was followed by two washes using 50 mM MES buffer, pH 5.5, after which the beads were resuspended in 200 μL of a solution containing 10 mg/mL 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and 5 mg/mL N-hydroxysuccinimide in MES buffer, pH 5.5, for 15 minutes at room temperature to activate terminal carboxyl groups. After two washes with 500 μL PBS, 50 μg anti-R. conorii antibodies (in 200 μL PBS) were added and allowed to react at room temperature for 4 hours. The beads were passivated after removing unreacted anti-R. conorii antibodies by incubating them with a mixture of 500 mM NH2OH and 15 mg/mL BSA in PBS for 2 hours at 37°C. After passivation, the beads were washed thrice and stored suspended in PBS at 4°C.
Microfluidic device and assembly
A photograph of the assay setup is shown in Fig. 3(A). Pressure-driven laminar flow in the channels was precisely controlled through a 10-channel syringe pump (Chemyx, Stafford, TX), capable of both infusion and withdrawal, using 1 mL plastic BD syringes (Beckton Dickinson, East Rutherford, NJ). The syringe pump was operated either manually, through its touch interface, or through a LabVIEW program. Microfluidic channels (7 channels per chip; 15 mm L × 1.5 mm W) were laser-cut into double-sided adhesive tape (300 LSE; 3M, Saint Paul, MN), following which the tape was cut into strips of the same dimension as the microfluidic substrate. Each microfluidic chip was assembled by sandwiching these laser-cut adhesive strips between a substrate containing embedded linear microretroreflectors (38 mm L × 25 mm W) and a transparent optical lid (0.2 mm thick TOPAS cyclic olefin copolymer) with inlet and outlet via holes (0.8 mm diameter) that allowed fluid flow [Fig. 3(C)]. As shown in Fig. 3(B), a re-usable microfluidic manifold, into which the chip was assembled by compression using fasteners, was used to interface fluid flow in the flow channels to the syringe pump using standard components including plastic ferrules and adapters, and polyetheretherketone (PEEK) tubing obtained from IDEX Corporation (Lake Forest, IL). The height of the microfluidic channels was defined by the thickness of the double-sided adhesive tape. In this case, the manufacturer's specifications for the triple-layer adhesive tape (two layers of 300LSE adhesive and an ultrathin, stabilizing polyester carrier sandwiched between them) added up to 175 μm. The actual height of the channels was lower due to the adhesive layers being compressed during assembly, and was measured using electronic calipers to be 150 ± 1 μm.
Figure 3.
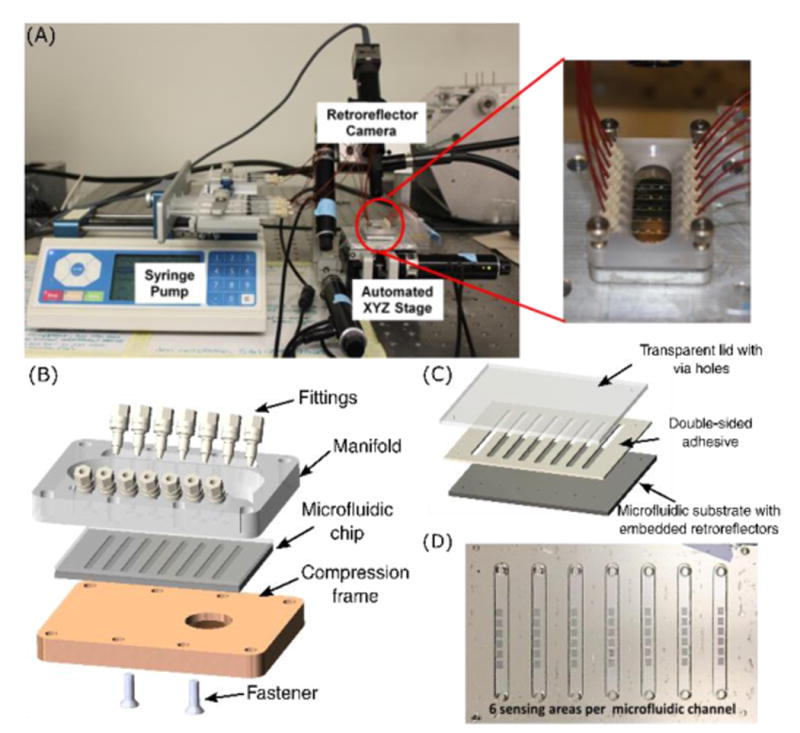
(A) Pictures of (left) assay setup and (right) microfluidic manifold. (B) Components of the microfluidic manifold. The microfluidic chip, held in place by the frame and fasteners, interfaces with the syringe pump through ferrules (fittings) and O-rings (not shown). (C) Components of the microfluidic chip. (D) Picture of an assembled microfluidic chip showing inlet and outlet via holes, and 6 embedded microretroreflector “arrays” (roughly 1 sq. mm each) per channel.
Optics and imaging
The optics used for imaging comprised a Dolan-Jenner MI-150 Fiber Optic Illuminator as the light source and an EO-5012M ½″ Monochrome Camera (CMOS; USB 2.0) mounted on an Infinitube FM-200 in-line assembly system (with 4× internal magnification) that ended in a 1× objective lens and interfaced, on the side, with the illuminator through a 48″ long fiber optic light guide. The ultimate field-of-view (FOV) of the acquired image, for retroreflective imaging, was 1.6 mm × 1 mm at a working distance of 44 mm. The Infinitube system was mounted on a machined structure that held it at a constant 35° angle to the vertical. All optical components were obtained from Edmund Optics (Barrington, NJ). An in-house LabVIEW program was used to control the automated XYZ-stage (Zaber Technologies, Vancouver, Canada) onto which the microfluidic manifold was placed, at the same location for each experiment, using pre-drilled alignment holes and fastening screws. The first sensing region in the first microfluidic channel was manually positioned before each experiment, following which a script was used to manipulate the XYZ-stage and USB-camera and capture images of all retroreflector arrays on the chip as required. Three images (with 0.15 mm differences in z-focus) were acquired for each 30-tile array (one image for each 10-tile row). The image acquisition process was rapid – for example, in experiments involving 42 sensing regions in 7 microfluidic channels, 126 total images were acquired and saved as PNG files in less than 150 seconds.
Assay procedure
R. conorii was spiked into PBS at various concentrations for assay testing. 250 μL samples were incubated using end-over-end mixing for 2 hours with 2 × 106 anti-R. conorii modified 3 μm magnetic beads. A magnetic separation stand (Thermo Scientific) was then used to concentrate the beads – the original sample was replaced with 20 μL assay buffer (PBS, 0.01% Tween-20), thereby concentrating the beads (and captured bacteria) by roughly an order of magnitude. In all microfluidic experiments, the channels were initially washed with assay buffer for 2 min at 100 μL/min after assembling the chip onto the manifold and verifying proper flow. A set of “before” images of all sensing regions on the chip were then automatically acquired using the image capture script. The magnetically concentrated assay samples were loaded at the end of the microfluidic channel not connected to the syringe pump, typically by directly inserting a micropipette tip into the ferrules of the microfluidic manifold. These samples, which contained either bead-R. conorii complexes or “blanks” (anti-R. conorii beads with no bacteria), were then introduced into the microfluidic channel (syringe pump in withdrawal mode) at 15 μL/min for 30 s and beads allowed to settle for 2 minutes. This initial settling period was followed by three withdrawal cycles on the syringe pump, each of which resulted in the flow of a 4 μL sample volume at 15 μL/min followed by 30 s of no flow. The syringe pump was then switched to infusion mode, following which four more flow-stop cycles (4 μL infusion at 15 μL/min followed by 30 s of no flow) were performed. This “flow-stop-drop” loading pattern was adopted to maximize bead interaction with the sensing surface. Wash buffer (PBS, 1% Tween-20) was then introduced into the channels, using the syringe pump in infusion mode, at a flow rate of 60 μL/min for ten minutes. It should be noted that fluidic force discrimination was likely completed in the first 30 seconds to 1 minute, but additional washing was required to clear out all residual beads. Following this step, the same image capture script as used for capturing “before” images was used to automatically capture “after” images from all sensing regions.
Difference imaging to extract bead count
To determine the location of the particles on the surface, two images of the retroreflector array were captured, one before the assay was run and one after the assay was completed. A Python-based script was then used to load each image and identify the location of the retroreflector tiles. This was accomplished by down-sampling the image, creating an idealized, low-resolution template of the tile pattern, and computing the cross-correlation of the template and the image. The maxima of the cross-correlation determined the location of the array of tiles and each tile could be saved as a separate image of the ‘before’ and ‘after’ images. Once each tile was identified in the two images, the offset between each ‘before’ and ‘after’ tile was determined by computing the cross-correlation function and using a cubic interpolation function to find the location of the maxima with sub-pixel resolution. By aligning the tiles individually, as opposed to attempting to align the full images, we were able to avoid correcting for small rotational alignment errors as the width of each tile is just 100 μm. Once the offset between each tile was computed, one of the images was shifted to compute a difference image for each tile. A user-specified background threshold, or one computed by the Otsu method,50 was used to isolate the beads from the background and a standard labeling algorithm was used to determine the location of the particles or particle clusters. The difference imaging algorithm typically took less than ten seconds to generate data from each 1 mm2 array. The bead counts per tile were exported and analyzed using Microsoft Excel or Igor. Fig. 4(B) shows an example of difference imaging output.
Figure 4. Difference Imaging.
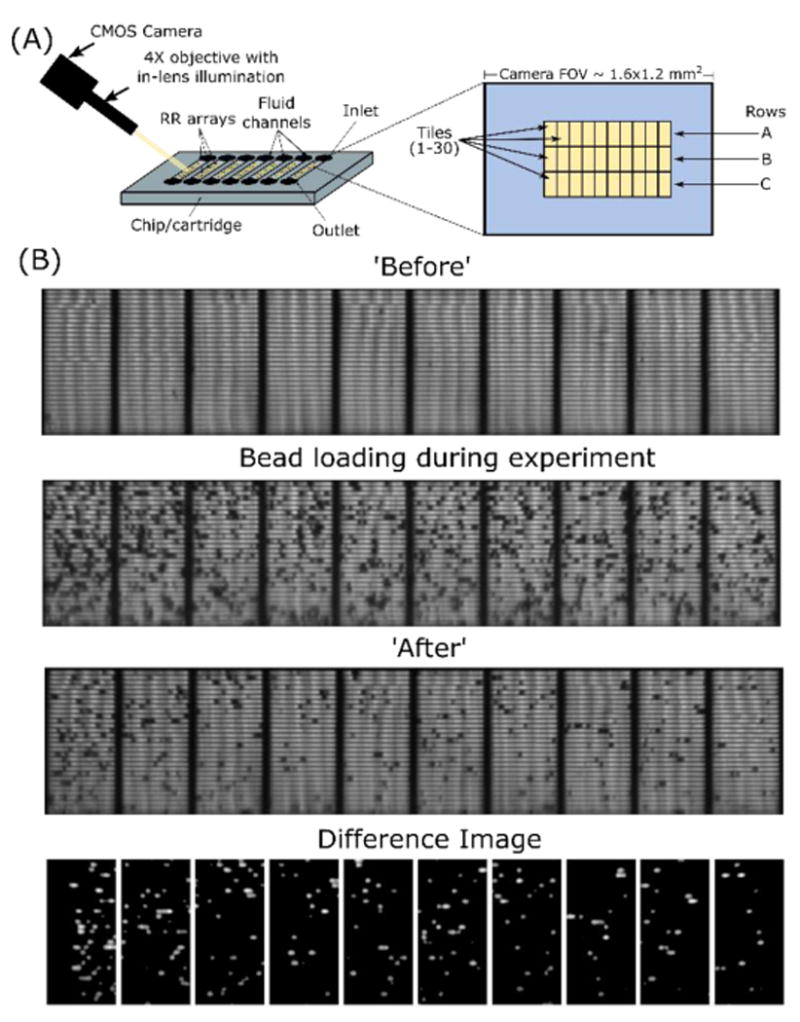
(A) Schematic showing image acquisition and tiling. (B) Representative example of difference imaging from a 10-tile row of embedded microretroreflectors. Particle count per tile as calculated by the difference imaging algorithm: mean = 21, median = 19.
Results and Discussion
qNano characterization of R. conorii
R. conorii was first quantified by an in-house real-time PCR method using 16S rDNA primers51 to obtain a concentration of 3.09 × 109 genome-equivalents/mL. The qNano, which uses tunable resistive pulse sensing to characterize particles with single-particle resolution, was used to verify this concentration and the size distribution, and therefore the structural integrity post-inactivation, of different aliquots of the bacteria. The qNano instrument consists of two fluid cells separated by an elastomeric polyurethane membrane into which a conical nanopore has been mechanically punched. A voltage is applied across the membrane, and the increase in electrical resistance from particles electrophoretically traversing the nanopore is observed as a decrease in current. The magnitude of these ‘blockade’ events is compared to that of known size standards to obtain an absolute size distribution. Concentration is determined by measuring samples at different external applied pressures and calculating the ratio of the slopes of pressure vs. translocation rate plots for the unknown sample and known concentration standards.52-55 The qNano-obtained equivalent spherical diameter distribution of heat-inactivated R. conorii bacteria [Fig. 5], with a mode of 678 nm, was in good agreement with the size of whole rickettsiae, which are rod-like with a diameter between 0.3 and 0.5 μm and length between 0.9 and 1.6 μm.56 The concentration of R. conorii aliquots was measured to be 3.56 ± 0.15 × 109/mL (n = 3), which was in good agreement with that measured by qPCR (3.09 × 109/mL), and therefore further confirmed the structural integrity of the heat-killed bacteria.
Figure 5.
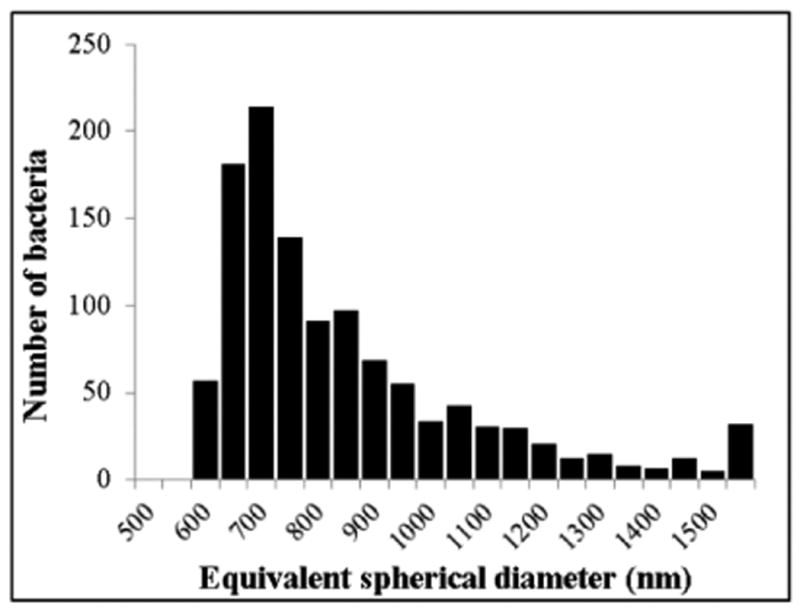
Equivalent spherical diameter distribution, with single-particle resolution, of heat-inactivated R. conorii measured using the qNano. 1,155 bacteria were measured. Mode = 678 nm.
Mass transport and fluidic force discrimination
The choice of 3 μm beads (over magnetic beads with smaller radii) as assay labels was dictated by a combination of mass transport (faster settling time), fluidic force discrimination (greater drag forces), and imaging (greater sensitivity of difference imaging using low numerical aperture optics). Irrespective of the sensing modality used, the performance of microfluidic surface-based biosensors can be significantly affected by convection and diffusion limitations that impair interaction of assay analytes with surface-immobilized receptors.57,58 The effects of these mass transport limitations were minimized in our assay through an optimized sample loading protocol consisting of the following steps: (i) the use of a magnetic separator to preconcentrate the analyte by roughly an order of magnitude,59 (ii) sample introduction by withdrawal, which eliminated dead volumes due to the syringe pump, and (iii) the adoption of a cyclical “flow-stop-drop” approach, which helped maximize the interaction of bacteria-laden beads with the embedded microretroreflector surface. Seven flow-stop-drop cycles, each of which lasted 45 seconds, were implemented after initial sample introduction; each cycle exchanged the entire volume of the microfluidic channel for a new volume of sample. The 3 μm beads, which have a Stokes settling velocity of roughly 2 μm/s, require 75 seconds to sediment from the top to the bottom of the 150 μm deep flow channels. The stop-drop portion of all seven cycles after the initial one (which had a 2 minute settling time) lasted 30 seconds to allow the beads to settle and interact with the antibody-functionalized surface. Further optimization of the flow-stop-drop loading protocol using a sample delivery mechanism with greater precision than a syringe pump conceivably could improve assay performance, as could changing the dimensions of the channel such that the microretroreflector pattern occupies a greater percentage of the channel's surface area.
Almost all solid-phase immunoassays include a washing step after analyte binding to reduce the background signal from nonspecifically-bound labels. Ideally, the wash step should expose the assay labels to forces stronger than the strongest non-specific interaction, but weaker than the weakest specific interaction. Atomic Force Microscopy (AFM) measurements provide rough estimates of the strengths of specific and non-specific interactions; widely cited examples for specific interactions include the rupture forces of covalent bonds (ca. 2 nN) and sulfur-gold anchors (ca. 1.4 nN),60 DNA duplexes (ca. 1 nN),61 biotin-streptavidin interactions (ca. 1 nN)62 and antibody-antigen interactions (ca. 250 pN).63 The strength of non-specific bonds in biological ligand-receptor systems has been demonstrated to be at least an order of magnitude weaker than specific antibody-antigen bonds,61,64 but hasn't been studied widely in AFM-based studies. Based on related work in non-biological systems, including the measurement of hydrophobic bond strength (18 pN)65 by Ray et al. and the calculation of van der Waals rupture forces by Lee et al. (12 pN),62 non-specific interactions in biological systems are estimated to have rupture forces of 0.1-10 pN.30 Forces between 10-100 pN, when applied to the magnetic beads, can therefore be expected to be sufficient to remove non-specifically bound beads. Fluidic force discrimination was first described by Rife and Whitman,66 and later demonstrated by Mulvaney et al. for magnetic bead-based surface coverage immunoassays.30,34,37 According to the Chang and Hammer bead detachment model, the tension force on a molecular tether holding beads to a microfluidic channel's surface is much greater than Stokes drag due to the lever-like effect created by the offset between the bead-surface contact point and the bead-tether contact point.67 By combining this model with the exact solutions for the force and torque on a stationary bead in laminar flow at a wall,68 the tension on the tethers holding 3 μm beads to the embedded microretroreflector surface can be derived to be T = 31.07 * ηa3/2vL−1/2, where η is the buffer viscosity, a the bead radius, v the fluid velocity at the center of the bead (calculated by assuming a parabolic velocity profile in our 150 μm high, 1.5 mm wide flow channel), and L the length of the molecular tether holding the bead to the surface. Surface roughness, a parameter in the Chang and Hammer model, was ignored due to the negligible roughness of spin-cast PMMA, and a tether length of 10 nm was used. It was observed that washing flow rates lower than 35 μL/min (T < 30 pN) resulted in poor signal-to-noise ratios, while flow rates 80 μL/min (T > 69 pN) resulted in scouring-off of specifically bound beads. We attribute the former to multiple weak interactions in the 0.1-10 pN range holding the beads down and the latter to our polyclonal anti-R. conorii antibodies binding to R. conorii with weaker affinities than some antibody-antigen systems reported in literature.63 A washing flow rate of 60 μL/min (T=52 pN) was ultimately used for fluidic force discrimination; this was sufficient to preferentially detach most 3 μm beads that were not non-specifically bound by more than a few weak interactions in our chosen antibody-antigen system; other systems would need to be calibrated appropriately. It should also be noted that the theoretical model for tension force estimation proposed by Mulvaney et al. can probably only be relied upon for order-of-magnitude approximations.
Assay performance
Poor signal-to-noise ratios in immunoassay-based biosensors are typically attributed to non-specific adsorption of proteins and other molecules to the sensing layers, which contributes to poor selectivity and therefore high background noise,69,70 or to the random orientation of capture antibodies on the sensor surface, which reduces target capture activity.71,72 Chemistries enabling favorable antibody orientations have been shown to improve assay performance,48,73 as has coating anti-fouling polymers between substrates and capture antibodies.74 In our assay for R. conorii, we explored two such chemistries on both the bead and PMMA surfaces: specific oxidation of the Fc region of anti-R. conorii antibodies, followed by covalent coupling to primary amine-modified surfaces; and the addition of layers of poly(ethylene) glycol (PEG) as spacers between the anchoring surfaces and antibodies.75
The use of bead count per tile as readout, rather than overall bead count per channel, resulted in robust statistics (180 tiles per channel); however, irregular bead counts were occasionally observed from a few tiles in certain channels. These outlier bead counts, which would significantly skew a conventional averaging approach, arose from local flow abnormalities due to the manual assembly of microfluidic chips, bubbles in the microchannels, and spatial irregularities in the density of immobilized antibody. Also, as a “yes/no” assay, the need for quantitation or precise dose-response measurements over a large dynamic range was obviated; rather, a clear distinction between a “blank” sample and a sample containing a target analyte was deemed sufficient. With these considerations in mind, bead counts per tile for different R. conorii sample concentrations were plotted and visualized as notched box plots. Notched box plots display the variation in a statistical population without making any assumptions about the underlying statistical distribution, and provide a rough guide to significance of difference of medians – notches on the box represent the equivalent of a confidence interval (for normal distributions) about the median value. For the data shown in Fig. 6, the displayed notches extend to 1.58 (IQR)/√n on either side of the median, where IQR is the interquartile range and n is the number of tiles from which bead counts were obtained. This value, based on the asymptotic normality of the median and roughly equal sample sizes for the two medians being compared, is insensitive to the underlying distributions of the samples. Non-overlapping notches, therefore, imply a significant difference in the medians of data represented by the relevant data sets with a 95% confidence level.76,77
Figure 6.
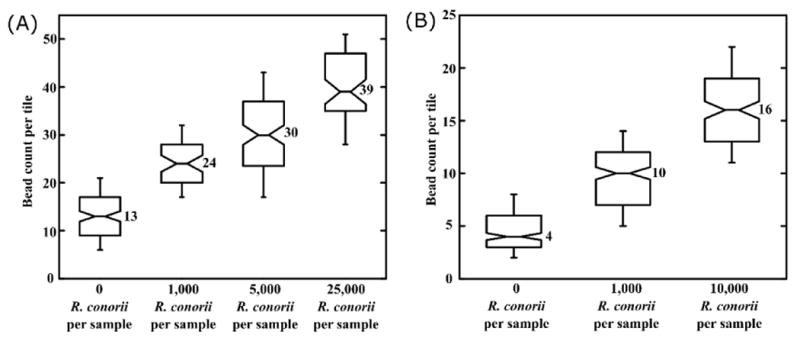
Assay results for R. conorii detection. (A) Notched box plots showing bead counts per tile for R. conorii detection using the Fc-mediated antibody immobilization strategy. (B) Notched box plots showing bead counts per tile for R. conorii detection using the PEGylation and amine-mediated antibody immobilization strategy. Horizontal lines on each box plot, from bottom to top beginning with the bottom whisker: 10th percentile, 25th percentile, median, 75th percentile, and 90th percentile. Non-overlapping notches imply significantly differing median values with 95% confidence. Box plots were generated using bead count per tile data from at least 3 independent experiments, with each experiment accounting for bead counts from 180 tiles.
Figure 6(A) shows the assay results for R. conorii detection using Fc-mediated anti-R. conorii immobilization on both beads and PMMA surfaces. The lowest detectable R. conorii concentration was 1000 per 250 μL sample, or 4000 rickettsiae per mL. Figure 6(B) shows the assay results for R. conorii detection using primary amine-mediated anti-R. conorii antibody immobilization on PEGylated beads and PMMA surfaces. The background bead count was substantially lowered by PEGylation (median bead count per tile = 4, compared to 13 using Fc-mediated anti-R. conorii immobilization), and the “positive” samples had proportionately higher numbers in terms of the signal-to-noise ratio. However, the analytical sensitivity was not improved, potentially indicating that the mass transport limit of affinity recognition during homogeneous mixing and/or solid-phase capture for the current assay configuration may have been reached. The current limit of detection is in the clinically relevant range for several pathogenic bacteria including R. rickettsii E. chaffeensis, E. ewingii, P. aeruginosa, and N. meningitidis.78
It should be noted that the sensing areas of each microfluidic channel, which currently total 180 tiles, only constituted 27% of the total available channel surface area – the assay's limit of detection might be improved by increasing this number. The limit of detection may also be improved by further optimizing antibody densities and immobilization/passivation chemistries on both surfaces. The bead counts per tile at the lowest and highest extremes of the data sets in Fig. 6 mostly arose from air bubbles in the microfluidic channel; some high bead counts were observed near the edges of the antibody spots due to coffee-ring effects. Depending on their size, position, and tendency to move during the washing step, air bubbles that occupied tiny fractions of the channel either caused beads to accumulate around them, or resulted in specifically bound beads being swept away by surface tension.79 The coffee-ring effect can be overcome by inducing Marangoni flow in spotted antibody droplets during functionalization,80 while the use of a gas-permeable material to form the microfluidic channels could minimize abnormal bead counts due to air bubbles.81
The ultimate goal of this work is to develop a benchtop device capable of multiplexed, high-throughput “yes/no” screening for pathogens and other analytes, with applications in areas including biodefense, food safety, and environmental monitoring. For multiplexed detection, we envision the use of a bead ‘cocktail’, comprising populations of magnetic beads against different analytes, for sample capture, and the antibody-modification of separate channels with secondary antibodies to individual analytes. There is also the possibility of modifying some sensing regions with other affinity reagents to serve as positive or negative controls. The density and fractional coverage of embedded microretroreflectors on each microfluidic chip can be easily increased by the current fabrication process to support high-throughput sample processing. For example, our current photomask design already includes enough microretroreflector arrays to support 15 flow channels on each 38 mm × 25 mm microfluidic chip. The sample-to-result time for the assay in its current form is dominated by the off-chip sample capture step, while the overall setup's portability is limited by the bulky syringe pump. The use of battery-powered electrokinetic pumps to drive flow, and the integration of analyte capture and magnetic concentration onto a second microfluidic chip upstream of the present one, may lead to a more rapid and portable assay platform.
Conclusion
In summary, a novel optical microfluidic immunoassay platform has been demonstrated through a proof-of-concept assay for detection of Rickettsia conorii. The platform is based on using embedded linear microretroreflectors, fabricated using easily scalable photo-lithographic techniques, as the microfluidic sensing surface, and micron-sized magnetic beads as light-blocking labels. The assay was implemented in a semi-homogeneous format – bacteria were captured by antibody-modified magnetic beads, following which they were introduced into antibody-modified microfluidic channels. A flow-stop-drop approach was adopted for optimal bead interaction with the sensing surface, and fluidic force discrimination was used to remove non-specifically bound beads and complete the assay. An automated XYZ stage and image capture script were used for the automatic imaging of all sensing regions before and after each experiment, and analyzed using a difference imaging algorithm to provide bead counts, which represented specifically-bound beads, as the readout. Two different strategies were used for the antibody-modification of beads and embedded microretroreflector surfaces; both of them resulted in a limit of detection of 4000 R. conorii/mL in buffer, a clinically relevant value for several pathogenic bacteria.
Acknowledgments
This work was supported by the NIAID/NIH (U54 AI057156). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the RCE Programs Office, NIAID, or NIH. Support was also provided by the Welch Foundation (E-1264), NSF (CBET-1438204) and Cancer Prevention & Research Institute of Texas (RP150343). The authors would also like to thank the University of Houston Nanofabrication Facility for providing excellent facilities and support. The authors also acknowledge Tecnológico de Monterrey Research chair (Grant CAT161).
Notes and references
- 1.Yalow RS, Berson SA. The Journal of clinical investigation. 1960;39:1157–1175. doi: 10.1172/JCI104130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mark D, Haeberle S, Roth G, von Stetten F, Zengerle R. Chemical Society reviews. 2010;39:1153–1182. doi: 10.1039/b820557b. [DOI] [PubMed] [Google Scholar]
- 3.Yang SY, Lien KY, Huang KJ, Lei HY, Lee GB. Biosensors & bioelectronics. 2008;24:861–868. doi: 10.1016/j.bios.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Gao Y, Hu G, Lin FY, Sherman PM, Li D. Biomedical microdevices. 2005;7:301–312. doi: 10.1007/s10544-005-6072-0. [DOI] [PubMed] [Google Scholar]
- 5.Sista R, Hua Z, Thwar P, Sudarsan A, Srinivasan V, Eckhardt A, Pollack M, Pamula V. Lab on a chip. 2008;8:2091–2104. doi: 10.1039/b814922d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chabinyc ML, Chiu DT, McDonald JC, Stroock AD, Christian JF, Karger AM, Whitesides GM. Analytical chemistry. 2001;73:4491–4498. doi: 10.1021/ac010423z. [DOI] [PubMed] [Google Scholar]
- 7.Gorkin R, Park J, Siegrist J, Amasia M, Lee BS, Park JM, Kim J, Kim H, Madou M, Cho YK. Lab on a chip. 2010;10:1758–1773. doi: 10.1039/b924109d. [DOI] [PubMed] [Google Scholar]
- 8.Uno T, Tabata H, Kawai T. Analytical chemistry. 2007;79:52–59. doi: 10.1021/ac060273y. [DOI] [PubMed] [Google Scholar]
- 9.Park HJ, Kim SK, Park K, Lyu HK, Lee CS, Chung SJ, Yun WS, Kim M, Chung BH. FEBS letters. 2009;583:157–162. doi: 10.1016/j.febslet.2008.11.039. [DOI] [PubMed] [Google Scholar]
- 10.McKendry R, Zhang J, Arntz Y, Strunz T, Hegner M, Lang HP, Baller MK, Certa U, Meyer E, Guntherodt HJ, Gerber C. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9783–9788. doi: 10.1073/pnas.152330199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voros J, Ramsden JJ, Csucs G, Szendro I, De Paul SM, Textor M, Spencer ND. Biomaterials. 2002;23:3699–3710. doi: 10.1016/s0142-9612(02)00103-5. [DOI] [PubMed] [Google Scholar]
- 12.Lai Z, Wang Y, Allbritton N, Li GP, Bachman M. Optics letters. 2008;33:1735–1737. doi: 10.1364/ol.33.001735. [DOI] [PubMed] [Google Scholar]
- 13.Hoa XD, Kirk AG, Tabrizian M. Biosensors & bioelectronics. 2007;23:151–160. doi: 10.1016/j.bios.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Lange K, Rapp BE, Rapp M. Analytical and bioanalytical chemistry. 2008;391:1509–1519. doi: 10.1007/s00216-008-1911-5. [DOI] [PubMed] [Google Scholar]
- 15.Bernard A, Michel B, Delamarche E. Analytical chemistry. 2001;73:8–12. doi: 10.1021/ac0008845. [DOI] [PubMed] [Google Scholar]
- 16.Cesaro-Tadic S, Dernick G, Juncker D, Buurman G, Kropshofer H, Michel B, Fattinger C, Delamarche E. Lab on a chip. 2004;4:563–569. doi: 10.1039/b408964b. [DOI] [PubMed] [Google Scholar]
- 17.Gervais L, Delamarche E. Lab on a chip. 2009;9:3330–3337. doi: 10.1039/b906523g. [DOI] [PubMed] [Google Scholar]
- 18.Yakovleva J, Davidsson R, Lobanova A, Bengtsson M, Eremin S, Laurell T, Emneus J. Analytical chemistry. 2002;74:2994–3004. doi: 10.1021/ac015645b. [DOI] [PubMed] [Google Scholar]
- 19.Tang CK, Vaze A, Rusling JF. Lab on a chip. 2012;12:281–286. doi: 10.1039/c1lc20833k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chon H, Lim C, Ha SM, Ahn Y, Lee EK, Chang SI, Seong GH, Choo J. Analytical chemistry. 2010;82:5290–5295. doi: 10.1021/ac100736t. [DOI] [PubMed] [Google Scholar]
- 21.Tekin HC, Gijs MAM. Lab on a chip. 2013;13:4711–4739. doi: 10.1039/c3lc50477h. [DOI] [PubMed] [Google Scholar]
- 22.Proczek G, Gassner AL, Busnel JM, Girault HH. Analytical and bioanalytical chemistry. 2012;402:2645–2653. doi: 10.1007/s00216-011-5495-0. [DOI] [PubMed] [Google Scholar]
- 23.Fu Z, Shao G, Wang J, Lu D, Wang W, Lin Y. Analytical chemistry. 2011;83:2685–2690. doi: 10.1021/ac1032116. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Ge S, Wang S, Yan M, Yu J, Song X, Liu W. The Analyst. 2012;137:2176–2182. doi: 10.1039/c2an16170b. [DOI] [PubMed] [Google Scholar]
- 25.Neubert H, Gale J, Muirhead D. Clinical chemistry. 2010;56:1413–1423. doi: 10.1373/clinchem.2010.144576. [DOI] [PubMed] [Google Scholar]
- 26.Teste B, Ali-Cherif A, Viovy JL, Malaquin L. Lab on a chip. 2013;13:2344–2349. doi: 10.1039/c3lc50353d. [DOI] [PubMed] [Google Scholar]
- 27.Sasso L, Johnston I, Zheng M, Gupte R, Ündar A, Zahn J. Microfluid Nanofluid. 2012;13:603–612. doi: 10.1007/s10404-012-0980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morozov VN, Shlyapnikov YM, Kidd J, Morozova TY, Shlyapnikova EA. Analytical chemistry. 2011;83:5548–5555. doi: 10.1021/ac201146w. [DOI] [PubMed] [Google Scholar]
- 29.Tekin HC, Cornaglia M, Gijs MA. Lab on a chip. 2013;13:1053–1059. doi: 10.1039/c3lc41285g. [DOI] [PubMed] [Google Scholar]
- 30.Mulvaney SP, Cole CL, Kniller MD, Malito M, Tamanaha CR, Rife JC, Stanton MW, Whitman LJ. Biosensors & bioelectronics. 2007;23:191–200. doi: 10.1016/j.bios.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Dostalek J, Knoll W. Analytical chemistry. 2011;83:6202–6207. doi: 10.1021/ac200751s. [DOI] [PubMed] [Google Scholar]
- 32.Krishnan S, Mani V, Wasalathanthri D, Kumar CV, Rusling JF. Angewandte Chemie (International ed in English) 2011;50:1175–1178. doi: 10.1002/anie.201005607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edelstein RL, Tamanaha CR, Sheehan PE, Miller MM, Baselt DR, Whitman LJ, Colton RJ. Biosensors & bioelectronics. 2000;14:805–813. doi: 10.1016/s0956-5663(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 34.Mulvaney SP, Ibe CN, Tamanaha CR, Whitman LJ. Analytical biochemistry. 2009;392:139–144. doi: 10.1016/j.ab.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 35.Waggoner PS, Varshney M, Craighead HG. Lab on a chip. 2009;9:3095–3099. doi: 10.1039/b907309b. [DOI] [PubMed] [Google Scholar]
- 36.Lee H, Yoon TJ, Weissleder R. Angewandte Chemie (International ed in English) 2009;48:5657–5660. doi: 10.1002/anie.200901791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulvaney SP, Myers KM, Sheehan PE, Whitman LJ. Biosensors & bioelectronics. 2009;24:1109–1115. doi: 10.1016/j.bios.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Luoma J, Schumann J, Traube EC. Accident; analysis and prevention. 1996;28:377–383. doi: 10.1016/0001-4575(96)00004-8. [DOI] [PubMed] [Google Scholar]
- 39.Schultz P, Cumby B, Heikenfeld J. Applied optics. 2012;51:3744–3754. doi: 10.1364/AO.51.003744. [DOI] [PubMed] [Google Scholar]
- 40.Switkes M, Ervin BL, Kingsborough RP, Rothschild M, Sworin M. Sensors and Actuators B: Chemical. 2011;160:1244–1249. [Google Scholar]
- 41.Bender PL, Currie DG, Poultney SK, Alley CO, Dicke RH, Wilkinson DT, Eckhardt DH, Faller JE, Kaula WM, Mulholland JD, Plotkin HH, Silverberg EC, Williams JG. Science (New York, NY) 1973;182:229–238. doi: 10.1126/science.182.4109.229. [DOI] [PubMed] [Google Scholar]
- 42.Bohling M, Seiler T, Wdowiak B, Jahns J, Mohr J, Borner M. Applied optics. 2012;51:5989–5995. doi: 10.1364/AO.51.005989. [DOI] [PubMed] [Google Scholar]
- 43.Sherlock T, Nasrullah A, Litvinov J, Cacao E, Knoop J, Kemper S, Kourentzi K, Kar A, Ruchhoeft P, Willson R. Journal of Vacuum Science & Technology B. 2011;29:06FA01. doi: 10.1116/1.3656801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garvey G, Shakarisaz D, Ruiz-Ruiz F, Hagström AEV, Raja B, Pascente C, Kar A, Kourentzi K, Rito-Palomares M, Ruchhoeft P, Willson RC. Analytical chemistry. 2014;86:9029–9035. doi: 10.1021/ac501491t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyka W. The Journal of infectious diseases. 1950;86:81–87. doi: 10.1093/infdis/86.1.81. illust. [DOI] [PubMed] [Google Scholar]
- 46.Azad AF. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007;45(Suppl 1):S52–55. doi: 10.1086/518147. [DOI] [PubMed] [Google Scholar]
- 47.Girardi E, Holdom MD, Davies AM, Sutton BJ, Beavil AJ. The Biochemical journal. 2009;417:77–83. doi: 10.1042/BJ20081355. [DOI] [PubMed] [Google Scholar]
- 48.Puertas S, Moros M, Fernández-Pacheco R, Ibarra MR, Grazú V, Fuente JMdl. Journal of Physics D: Applied Physics. 2010;43:474012. [Google Scholar]
- 49.Fixe F, Dufva M, Telleman P, Christensen CB. Nucleic acids research. 2004;32:e9. doi: 10.1093/nar/gng157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otsu N. Automatica. 1975;11:23–27. [Google Scholar]
- 51.Woods ME, Wen G, Olano JP. Annals of the New York Academy of Sciences. 2005;1063:239–245. doi: 10.1196/annals.1355.037. [DOI] [PubMed] [Google Scholar]
- 52.Willmott GR, Vogel R, Yu SS, Groenewegen LG, Roberts GS, Kozak D, Anderson W, Trau M. Journal of physics Condensed matter : an Institute of Physics journal. 2010;22:454116. doi: 10.1088/0953-8984/22/45/454116. [DOI] [PubMed] [Google Scholar]
- 53.Vogel R, Willmott G, Kozak D, Roberts GS, Anderson W, Groenewegen L, Glossop B, Barnett A, Turner A, Trau M. Analytical chemistry. 2011;83:3499–3506. doi: 10.1021/ac200195n. [DOI] [PubMed] [Google Scholar]
- 54.Vogel R, Anderson W, Eldridge J, Glossop B, Willmott G. Analytical chemistry. 2012;84:3125–3131. doi: 10.1021/ac2030915. [DOI] [PubMed] [Google Scholar]
- 55.Kozak D, Anderson W, Vogel R, Chen S, Antaw F, Trau M. ACS nano. 2012;6:6990–6997. doi: 10.1021/nn3020322. [DOI] [PubMed] [Google Scholar]
- 56.Rovery C, Raoult D. In: Rickettsial Diseases. Raoult D, Parola P, editors. ch. 10. CRC Press; Boca Raton, FL: 2007. pp. 125–138. [Google Scholar]
- 57.Squires TM, Messinger RJ, Manalis SR. Nat Biotech. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Dostalek J, Knoll W. Analytical chemistry. 2011;83:6202–6207. doi: 10.1021/ac200751s. [DOI] [PubMed] [Google Scholar]
- 59.Parsa H, Chin CD, Mongkolwisetwara P, Lee BW, Wang JJ, Sia SK. Lab on a chip. 2008;8:2062–2070. doi: 10.1039/b813350f. [DOI] [PubMed] [Google Scholar]
- 60.Grandbois M, Beyer M, Rief M, Clausen-Schaumann H, Gaub HE. Science (New York, NY) 1999;283:1727–1730. doi: 10.1126/science.283.5408.1727. [DOI] [PubMed] [Google Scholar]
- 61.Lee GU, Chrisey LA, Colton RJ. Science (New York, NY) 1994;266:771–773. doi: 10.1126/science.7973628. [DOI] [PubMed] [Google Scholar]
- 62.Lee GU, Kidwell DA, Colton RJ. Langmuir. 1994;10:354–357. [Google Scholar]
- 63.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metzger SW, Natesan M, Yanavich C, Schneider J, Lee GU. Journal of Vacuum Science & Technology A. 1999;17:2623–2628. [Google Scholar]
- 65.Ray C, Brown JR, Akhremitchev BB. The journal of physical chemistry B. 2006;110:17578–17583. doi: 10.1021/jp063517r. [DOI] [PubMed] [Google Scholar]
- 66.7,736,889. USA Pat. 2010
- 67.Chang KC, Hammer DA. Langmuir. 1996;12:2271–2282. [Google Scholar]
- 68.Goldman AJ, Cox RG, Brenner H. Chemical Engineering Science. 1967;22:653–660. [Google Scholar]
- 69.Ahluwalia A, Giusto G, De Rossi D. Materials Science and Engineering: C. 1995;3:267–271. [Google Scholar]
- 70.Wisniewski N, Reichert M. Colloids and Surfaces B: Biointerfaces. 2000;18:197–219. doi: 10.1016/s0927-7765(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 71.Kausaite-Minkstimiene A, Ramanaviciene A, Kirlyte J, Ramanavicius A. Analytical chemistry. 2010;82:6401–6408. doi: 10.1021/ac100468k. [DOI] [PubMed] [Google Scholar]
- 72.Saerens D, Huang L, Bonroy K, Muyldermans S. Sensors. 2008;8:4669–4686. doi: 10.3390/s8084669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cho IH, Seo SM, Jeon JW, Paek SH. Journal of Biomedicine and Biotechnology. 2009;2009:9. doi: 10.1155/2009/104094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen A, Kozak D, Battersby BJ, Forrest RM, Scholler N, Urban N, Trau M. Langmuir. 2009;25:13510–13515. doi: 10.1021/la903148n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hucknall A, Rangarajan S, Chilkoti A. Advanced Materials. 2009;21:2441–2446. doi: 10.1002/adma.200803125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chambers JM, Cleveland WS, Kleiner B, Tukey PA, editors. Graphical Methods for Data Analysis. Wadsworth & Brooks/Cole; 1983. [Google Scholar]
- 77.McGill R, Tukey JW, Larsen WA. The American Statistician. 1978;32:12–16. [Google Scholar]
- 78.Eshoo MW, Crowder CD, Li H, Matthews HE, Meng S, Sefers SE, Sampath R, Stratton CW, Blyn LB, Ecker DJ, Tang YW. Journal of clinical microbiology. 2010;48:472–478. doi: 10.1128/JCM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skottrup PD, Hansen MF, Lange JM, Deryabina M, Svendsen WE, Jakobsen MH, Dufva M. Acta biomaterialia. 2010;6:3936–3946. doi: 10.1016/j.actbio.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 80.Majumder M, Rendall CS, Eukel JA, Wang JYL, Behabtu N, Pint CL, Liu TY, Orbaek AW, Mirri F, Nam J, Barron AR, Hauge RH, Schmidt HK, Pasquali M. The Journal of Physical Chemistry B. 2012;116:6536–6542. doi: 10.1021/jp3009628. [DOI] [PubMed] [Google Scholar]
- 81.Kang JH, Kim YC, Park JK. Lab on a chip. 2008;8:176–178. doi: 10.1039/b712672g. [DOI] [PubMed] [Google Scholar]


