Abstract
The human PNPLA3 (patatin-like phospholipase domain-containing 3) gene codes for a protein which is highly expressed in adipose tissue and liver, and is implicated in lipid homeostasis. While PNPLA3 protein contains regions homologous to functional lipolytic proteins, the regulation of its tissue expression is reflective of lipogenic genes. A naturally occurring genetic variant of PNPLA3 in humans has been linked to increased susceptibility to alcoholic liver disease. We have examined the modulatory effect of alcohol on PNPLA3 protein and mRNA expression as well as the association of its gene promoter with acetylated histone H3K9 by chromatin immunoprecipitation (ChIP) assay in rat hepatocytes in vitro, and in vivo in mouse and rat models of acute binge, chronic, and chronic followed by acute binge ethanol administration. Protein expression of PNPLA3 was significantly increased by alcohol in all three models used. PNPLA3 mRNA also increased, albeit to a varying degree. ChIP assay using H3AcK9 antibody showed increased association with the promoter of PNPLA3 in hepatocytes and in mouse liver. This was less evident in rat livers in vivo except under chronic treatment. It is concluded for the first time that histone acetylation plays a role in the modulation of PNPLA3 levels in the liver exposed to binge ethanol both in vitro and in vivo.
Keywords: Binge ethanol, Chronic-binge ethanol, Epigenetics, Histone acetylation, PNPLA3
Introduction
Steatosis (fatty liver) is a common outcome of chronic alcohol consumption, and binge (acute) drinking in chronic alcohol users may predispose them to more severe alcoholic liver disease (ALD). Increased lipogenesis driven by the sterol response element binding protein (SREBP) transcription factors is essential for steatosis associated with chronic alcohol ingestion, but the mechanisms underlying steatosis following binge alcohol abuse are unknown. Fatty liver involves the accumulation of triglycerides and other lipids in hepatocytes as a result of defective fatty acid metabolism. PNPLA3 (also termed adiponutrin, or calcium-independent phospholipase A2 epsilon) is a nutritionally regulated protein belonging to the patatin-like phospholipase domain-containing family of lipid metabolizing proteins that is highly expressed in adipocytes and liver (Sookoian & Pirola, 2014). This enzyme was first discovered in potato, hence the term patatin. Genetic variations in the human PNPLA3 gene (e.g., the rs738409 C/G, M148I allele) have been associated with fatty liver disease (Romeo et al., 2008). Interestingly, the same genetic polymorphism in PNPLA3 has also been identified by several laboratories as a risk factor for alcohol-induced progression from fibrosis to cirrhosis and its related complication, i.e., hepatocarcinoma (HCC) in different populations (Buch et al., 2015; Nakaoka et al., 2015; Stickel, Hampe, Trépo, Datz, & Romeo, 2015; Tian, Stokowski, Kershenobich, Ballinger, & Hinds, 2010; Trépo et al., 2011; Valenti et al., 2013). Although human PNPLA3 has triacylglycerol (TG) hydrolase and transacylase activities in vitro, its in vivo function and physiological relevance remain uncertain. In vivo studies suggest that PNPLA3 activity is most consistent with lipogenesis (Huang et al., 2010; Kershaw et al., 2006). Equally unknown is the mechanisms for its expression. Any involvement of the chromatin modifications on the PNPLA3 transcriptional machinery that orchestrates the PNPLA3-related network has yet to be known (see Sookoian & Pirola, 2014). Histone H3K9 acetylation in chromatin has been shown to increase in vitro in a hepatocyte model and in animal models in vivo of ALD (Aroor, Restrepo, Kharbanda, & Shukla, 2014; Shukla, Aroor, Restrepo, Kharbanda, & Ibdah, 2015; Shukla, Restrepo, Fish, Lim, & Ibdah, 2015). Increases in TG levels after acute, chronic, and acute on chronic exposure to alcohol were reported in rats (Aroor, Jackson, & Shukla, 2011) and mice (Shukla, Aroor, et al., 2015). The present study examines the effects of alcohol on the expression of PNPLA3 and the potential epigenetic mechanism for its expression involving acetylation of histone H3K9.
Materials and methods
Hepatocyte culture methods
Hepatocyte preparation and treatments were as described previously (Shukla, Restrepo, et al., 2015). Briefly, hepatocytes from male Sprague-Dawley rats (250–300 g) were isolated using an in situ collagenase perfusion method. The viability of the hepatocytes was monitored by trypan blue dye exclusion and was routinely 90 ± 5%. Cells were treated with either saline (control) or ethanol, 100 mM in DMEM medium containing 1% fetal bovine serum, 2 mM L-glutamine, and 100 units/mL penicillin and 100 mg/mL streptomycin for 24 h in an incubator equilibrated with air/CO2 (95%/5%). Subsequently, the cells were processed for various analyses.
Mouse model
Diets, treatments, and sample processing have been previously described (Shukla, Aroor, et al., 2015). Briefly, 7-week-old C57BL/6J male mice housed under a 12-h/12-h light/dark cycle were allowed ad libitum consumption of standard laboratory chow for 1 week to acclimatize. This was followed by feeding animals a Lieber-DeCarli liquid diet (see Shukla, Aroor, et al., 2015; Dyets, Inc., Bethlehem, PA). Ethanol was progressively introduced into the liquid diet for 1 week up to 4% (wt/vol) then kept at this level up to 4 weeks. Pair-fed control mice were given a liquid diet in which ethanol was replaced by dextrin/maltose to maintain the isocaloric intake in the two groups. At the end of the 4-week feeding, mice were divided into four groups for administering binge ethanol (3 doses of ethanol at 3.5 g/kg, intragastric at 12-h intervals between each dose). For the binge period, the control group (C) received water, the chronic-ethanol group (E) received water, the control-binge group (CB) received three doses of ethanol, and the chronic ethanol-binge group (EB) received three binge doses of ethanol after chronic ethanol (see Shukla, Aroor, et al., 2015).
Rat model
Briefly, 8-week-old Sprague-Dawley rats were fed ethanol in a liquid diet (5%, wt/vol) for 4 weeks. Pair-fed control rats were given a liquid diet in which ethanol was replaced by dextrin/maltose to maintain the isocaloric intake in the two groups. Similar to the mouse study above, at the end of the 4-week feeding, rats were divided into four groups for ethanol binge (5.0 g/kg, intragastric) as follows: For the binge period, the control group (C) received water, the chronic-ethanol group (E) received water, the control-binge group (CB) received three doses of ethanol but no chronic ethanol, and the chronic ethanol-binge group (EB) received three binge doses of ethanol after chronic ethanol (Aroor et al., 2011). The animal care and protocol used in this study was approved by the University of Missouri Animal Care Committee (Protocol # 8175) and complied with the NIH guidelines for the care and use of laboratory animals.
Western blotting of PNPLA3
Western blotting of PNPLA3 for primary rat hepatocytes, mouse, and rat liver tissues was done in whole cell extracts of hepatocyte or liver tissue as described previously (Aroor et al., 2014; Shukla, Aroor, et al., 2015; Shukla, Restrepo, et al., 2015). An equal amount of protein (80 μg) was loaded in each gel lane for western blotting. After the transfer of proteins on blot, staining of the gel with Ponceau-S dye (Romero-Calvo et al., 2010) showed equal levels of transfer. Furthermore, this technique used in our laboratory has consistently shown the equal loading when actin or histone H3 were used as housekeeping proteins whose levels remain unaltered by ethanol exposure (Aroor, Jackson, & Shukla, 2012; Shukla, Aroor, et al., 2015). The blots showed a single band of PNPLA3 protein in the region of 52 KDa molecular weight, consistent with the data sheet provided for this antibody by the supplier (Pierce, product # PA5-18901 from Thermo Fisher Scientific, Rockford, Illinois).
Quantitative polymerase chain reaction (qtPCR) of cDNA from primary rat hepatocytes, mouse, and rat liver tissues
The protocols used for cDNA synthesis and mRNA amplification were the same as previously described (Shukla, Aroor, et al., 2015). Briefly, cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Primers specific for mouse and rat PNPLA3 were designed with the NCBI primer design tool. A CFX96 (Bio-Rad) qtPCR detection system was used for amplification. Primers for rat and mouse mRNA were obtained from Integrated DNA Technologies (Coralville, IA). Their sequences were as follows:
| Mouse | Fwd AGGACACCTTTGGTCCTTA | Rev GGATGCTCTCCCTACCTAATT |
| Rat | Fwd TCTCCCTCTCGATCACATCAT | Rev CCATCGGACACTCTGGTGAG |
Data for mRNA expression were calculated by the Delta-Delta method with GAPDH as the housekeeping gene (see Shukla, Aroor, et al., 2015; Shukla, Restrepo, et al., 2015). Cycle threshold (CT) values of less than 36 were used.
ChIP assay of mouse and rat liver tissues
Crosslinking and chromatin purification protocol
The protocols used for chromatin immunoprecipitation (ChIP) assay for mouse and rat liver tissues were the same as described in upstate Magna Chip A (catalog # 17-610) and were adapted for our assay. Briefly, 40 mg of liver sample were chopped in 2-mL centrifuge tubes containing PBS plus formaldehyde at 1% and allowed to crosslink at room temperature with rotation on an Adams Nutator (Clay Adams, Becton, Dickinson and Co., Sparks, MD) for 15 min. It was next stopped by the addition of 0.5-M glycine and rotated for an additional 5 min. Sample pellets were washed and homogenized in PBS in 7.5 mL Dounce homogenizer. Lysates were transferred to 1.5-mL tubes, pelleted by centrifugation, and washed once with 1X PBS. Chromatin washes and solutions were similar to those described in Millipore Salmon Sperm DNA/Protein A/G Agarose Application Protocol (see Browne, Harris, & Leir, 2014; Tomita, Buchholz, Obata, & Shi, 2003). Homogenates were resuspended in cell lysis buffer and kept at 4 °C for 15 min with 10-sec vortexing every 3 min, then centrifuged at 1000 rpm for 2 min at 4 °C. The resulting pellet was resuspended in nuclei lysis buffer and sonicated 3 times for 10 s in an ice-cold condition in a Vibra Cell model VC 600-1 sonicator (120 V, 600 W, 50/60 kHz; frequency 20 kHz; Sonics & Materials Inc., Danbury, CT). The sonicator used a 4-mm probe at a setting of 30% duty cycle. Samples were centrifuged at 13,000 rpm for 10 min at 4 °C to remove insoluble material. Sonication had been optimized to a chromatin size range of 200–800 bp. Supernatants were diluted 10-fold with ChIP dilution buffer, and 1 mL aliquots were stored at −80 °C for later use.
ChIP assay
One-mL chromatin-protein aliquots were precleared with 100 μL 50% DNA/Protein A/G slurry (Salmon Sperm DNA/Protein G Agar, catalog # 16-201; SSDNA/Protein A Agar, catalog # 16-157; Normal Mouse IGG, catalog # 12-371, EMD-Millipore Corporation, Billerica, MA). Precleared lysates were incubated overnight at 4 °C with rotation with 4 μg of anti-H3AcK9 (Anti-Acetyl Histidine H3-lys 9, catalog # 06-942 from Millipore) or 4 μg of normal mouse IgG (Normal Mouse IgG, catalog #12-371). Immunocomplexes were transferred to 1.5-mL tubes containing 100 μL of Salmon Sperm DNA/Protein A/G Agarose-50% slurry and incubated with rotation at 4 °C for 4 h. Agarose-immunocomplexes were pelleted by centrifugation at 750 rpm for 1 min at 4 °C. The supernatant was carefully removed and the agarose-immunocomplexes were subjected to two washes with low-salt buffer (150 mM NaCl, pH 8.0), two washes with high-salt buffer (500 mM NaCl, pH 8.0) and two washes with LiCl buffer (250 mM LiCl, pH 8.0), followed by one wash with Tris-EDTA (TE).
qtPCR of PNPLA3–H3AcK9-ChIP samples from mouse and rat liver tissues
H3AcK9/DNA and IgG/DNA complexes were eluted from the Protein A/G Agarose using two extractions with 250 μL SDS-NaHCO3 buffer. Twenty μL of 5 M NaCl were added to the eluates and incubated at 65 °C overnight with rotation to de-crosslink the H3AcK9/DNA or IgG/DNA complexes. After reverse crosslinking, 10 μL TE and 2 μL proteinase K (QIAGEN proteinase K; Valencia, CA) were added to the samples and incubated for 1 h at 56 °C. DNA was purified using QIAquick PCR Purification Kit (cat # 28104; QIAGEN, Valencia, CA) and resuspended in 100 μL elution buffer.
Primers for rat and mouse PNPLA3 promoters were obtained from Integrated DNA Technologies (Coralville, IA). A CFX96 (Bio-Rad) qtPCR detection system was used for amplification. Proximal promoter primer sequences relative to the TSS for mouse −158 to −23 and for rat −95 to +16 were designed using promoter information on EPD and/or NCBI (Promoter sequences −499 to +100 relative to the TSS). Their sequences are shown below. Data for analysis included CT values of less than 36. IgG/DNA samples produced no amplification.
| Fw1prPNPLA3mo | AGGATTTGGGAAAGCCCACG |
| Rv1prPNPLA3mo | TAAGCTGTCTTCGGGTGACG |
| Fw1prPNPLA3rat | ATGGTGGCATCTGTCGTGTAAT |
| Rv1prPNPLA3rat | CCTAATCGTAGGCTTGGATGCT |
Hepatocyte ChIP
Primary rat hepatocytes were collected and treated as described previously (Shukla, Restrepo, et al., 2015). Frozen cells were thawed on ice with the addition of 0.5 mL of a crosslinking solution consisting of 1X PBS/1% formaldehyde at room temperature with tube rotation for 12 min. Fixation was terminated by addition of 0.5 M glycine and rotated for an additional 5 min. Crosslinked cell pellets were washed with PBS and resuspended in 0.5 mL 1X cell lysis buffer with inhibitors and homogenized in 2 mL Dounce homogenizer. Tubes were centrifuged and supernatants removed and the remaining pellets resuspended in 0.5 mL of nuclear lysis buffer. DNA shearing by sonication was optimized to a chromatin fragment size range of 200–800 bp. Briefly, nuclear lysates were sheared with four 15-sec pulses with 60-sec intervals at 35% duty cycle using a Vibra Cell sonicator as detailed earlier. Sonicated samples were cleared by centrifugation at 12,000 × g for 15 min at 4 °C, and the supernatant was diluted 1:5 with ChIP dilution buffer, aliquoted into 1 mL tubes, snap-frozen in liquid nitrogen, and stored at −80 °C.
The protocol used for hepatocyte ChIP was adapted from earlier reports (Browne et al., 2014; Tomita et al., 2003). Magnetic beads from GenScript (Protein A/G MagBeads, GenScript catalog #L00277) were used. In addition to protease inhibitors (SIGMA cocktail P8340), histone deacetylase inhibitors TSA and Na-butyrate (at 20 nM and 5 mM, respectively) were added to all solutions. For complexing primary antibody and magnetic beads, 4 μg of anti-H3AcK9 (Anti-Acetyl Histone H3-lys 9, catalog # 06-942 from Millipore) or 4 μg of normal mouse IgG (Normal Mouse IgG, catalog #12-371 from Millipore) were added to 200 μL of a precleared magnetic bead slurry and incubated overnight at 4 °C with rotation. One mL of hepatocyte chromatin preparation was added to the magnetic bead-H3AcK9 or magnetic bead-IgG complexes and incubated overnight at 4 °C with rotation. Immunocomplexes were centrifuged at 750 rpm for 1 min, and the pellet was collected after discarding supernatant. The pellet was washed five times with 1 mL LiCl wash buffer (250 mM, pH 8.0), two times with 1 mL TE, and resuspended in 200 μL of elution buffer and incubated at 65 °C for 1 h with rotation. The supernatant containing ChIP DNA was incubated at 65 °C overnight to complete the reversal of the formaldehyde cross-links. ChIP DNA was purified using QIAquick PCR purification kit and eluted in a total of 50 μL elution buffer. Data for analysis included CT values of less than 36. IgG/DNA samples produced no amplification.
Results
Increase in PNPLA3 protein after ethanol treatment
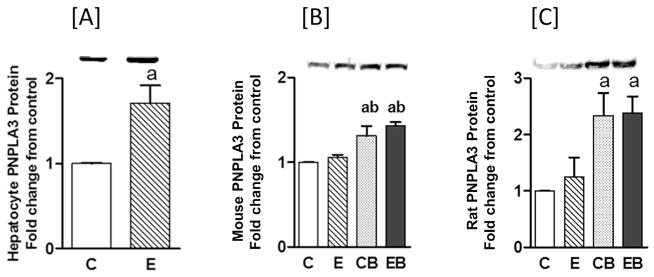
PNPLA3 protein levels increased almost 2-fold in rat hepatocytes treated with an acute alcohol dose of 100 mM for 24 h (Fig. 1A). The in vivo mouse and rat models exhibited responses to similar degrees in samples from acute (CB) and acute on chronic (EB) alcohol administration (Fig. 1B and C). In mice, PNPLA3 protein expression was increased to about 1.5-fold over controls for both acute binge and acute binge on chronic, while chronic ethanol alone did not alter PNPLA3 protein expression (Fig. 1B). A similar pattern was noted for rat liver, although the magnitude of the increase in PNPLA3 protein expression was greater than in mice, averaging about 2.5-fold over control (Fig. 1C).
Fig. 1. Western blot analysis of PNPLA3 in primary rat hepatocytes in vitro [A], and in mouse [B], and rat [C] liver in vivo.
Experimental details are described under Materials & Methods. Values are mean ± S.E.M. (n = 3 hepatocyte experiments; n = 4 mice; n = 4 rats). a: significant compared to control (p < 0.05); and b: significant compared to chronic (p < 0.05). In [A] hepatocyte in vitro, C: Control, E: Ethanol. For in vivo treatments of mouse [B] and rat [C], C: Control (pair fed); E: Chronic ethanol; CB: Control binge ethanol; EB: Chronic ethanol followed by binge ethanol.
qtPCR analysis of primary rat hepatocytes, mouse, and rat liver tissues
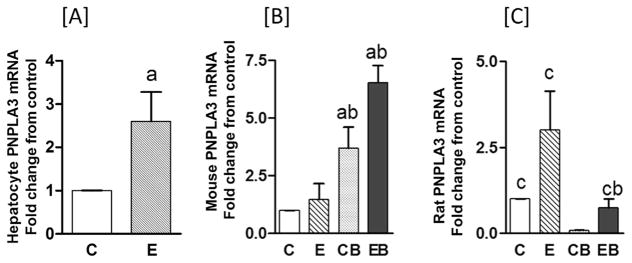
Our observations that PNPL3 protein expression increased after acute binge treatment and acute binge on chronic alcohol treatment led us to question whether or not increased gene expressions (i.e., mRNA levels) were affected by alcohol exposure. In cultured hepatocytes, PNPLA3 mRNA significantly increased in similar fashion as protein expression, with a 2.5-fold increase in mRNA (Fig. 2A) and 1.5-fold increase for protein (Fig. 1A) over controls.
Fig. 2. qtPCR analysis of PNPLA3 rat hepatocytes [A], mouse [B], and rat [C] liver tissues.
Experimental details were as described in Materials & Methods and in Fig. 1. Values are mean ± S.E.M. (n = 3 hepatocyte experiments; n = 4 mice; n = 4 rats). a: significant compared to control (p < 0.05); b: significant compared to chronic (p < 0.05); and c: significant compared to control binge (p < 0.05). In [A] hepatocyte in vitro, C: Control, E: Ethanol. For in vivo treatments of mouse [B] and rat [C], C: Control (pair fed); E: Chronic ethanol; CB: Control binge ethanol; EB: Chronic ethanol followed by binge ethanol.
In mice, chronic alcohol treatment appeared to increase PNPLA3 mRNA by about 2-fold over controls, although the difference was not significant statistically. In contrast, the data for acute binge and acute binge on chronic alcohol showed significant increases of approximately 4- and 6-fold over controls, respectively (Fig. 2B). Thus, for mice, the PNPLA3 data on protein and mRNA expression for acute binge and acute binge on chronic treatment followed a similar pattern of increase. Surprisingly, the data on rat PNPLA3 mRNA (Fig. 2C) were different from the results observed in cultured hepatocytes and mouse liver in vivo, in that there was no correlation with the protein levels. In the case of rat, PNPLA3 mRNA increased significantly (about 3-fold) only after chronic ethanol treatment, whereas binge ethanol, either in control animals or in animals after chronic ethanol exposure apparently led to a reduction of the mRNA compared to controls (Fig. 2C).
ChIP assay to monitor interaction between H3AcK9 and PNPLA3 promoter
Histone H3K9 acetylation, global as well as gene-specific, is one of the significant epigenetic effects of ethanol treatment in primary hepatocytes, and in mouse and rat livers in vivo (Shukla, Aroor, et al., 2015; Shukla, Restrepo, et al., 2015). It must be noted that the global histone H3K9 acetylation does not change markedly after 4 weeks of chronic ethanol treatment alone in rats (Park, Lim, & Shukla, 2012) and mice (Shukla, Aroor, et al., 2015; Table 1), despite the fact that ChIP analysis showed individual gene associations with acetylated histone H3 (Park et al., 2012). Ethanol therefore can increase association of H3AcK9 with gene promoter in the absence of detectable increase in the global acetylation of histone H3K9. Furthermore, ethanol can elevate expression of several genes, but the expression of only some of these genes are associated with histone H3 acetylation (Park et al., 2012; Shukla & Lim, 2013). The emerging pattern of PNPLA3 increases by ethanol led us to investigate whether or not histone H3K9 acetylation had any relationship to the expression of PNPLA3 mRNA. Various samples from each experimental model were therefore chromatin immuno-precipitated with H3AcK9 antibody and then subjected to qtPCR using the primers of PNPLA3 promoter (see Materials and Methods). Significant increases in the association between the PNPLA3 promoter and H3AcK9 were found in ChIP samples from primary hepatocytes (Fig. 3A), and from chronic, acute binge, and acute binge on chronic alcohol samples from mice compared to controls (Fig. 3B). Moreover, acute ethanol exposure after chronic treatment led to a further enhancement of H3AcK9 association over chronic treatment alone in mice (compare E & EB in Fig. 3B).
Table 1. Changes in global histone H3K9 acetylation by ethanol.
This table shows increases in the levels of H3AcK9 after ethanol treatment in different models. The values (mean ± SEM) represent fold-change over control (C). E: Chronic ethanol, CB: Control binge, EB: Chronic ethanol followed by binge (see text for details).
| In vitro | Reference | Fold-increase in H3AcK9 levels over control. | ||
|---|---|---|---|---|
| Rat hepatocyte | Shukla, Restrepo, et al., (2015) | 3.59 ± 1.16 | ||
| In vivo | E | CB | EB | |
| Rat | Aroor et al., (2014) | 1.63 ± 0.54 | 4.35 ± 1.26 | 5.46 ± 2.66 |
| Mouse | Shukla, Arror et al., (2015) | 0.97 ± 0.03 | 1.47 ± 0.09 | 0.91 ± 0.05 |
Fig. 3. ChIP assay for PNPLA3 promoter association with H3AcK9 in hepatocyte [A], mouse [B], and rat [C] liver tissues.
Experimental details were as described in the Materials & Methods. Values are mean ± S.E.M. (n = 4 mice; n = 4 rats). a: significant compared to control (p < 0.05); b: significant compared to chronic (p < 0.05). In [A] hepatocyte in vitro, C: Control, E: Ethanol. For in vivo treatments of mouse [B] and rat [C], C: Control (pair fed); E: Chronic ethanol; CB: Control binge ethanol; EB: Chronic ethanol followed by binge ethanol.
For rats, even though the mRNA expression pattern differed from that of mice, there was still some correlation between mRNA expression and ChIP profile. The values for the PNPLA3-H3AcK9 association for the chronic treatment were significantly higher (2-fold) over the controls in rats. In acute binge, and acute binge on chronic alcohol-treated rats, the increases in H3AcK9 association over control were small and statistically insignificant (Fig. 3C).
Discussion
This study demonstrated that ethanol increased PNPLA3 levels in liver in both in vitro and in vivo conditions. In rat hepatocytes in vitro, the expressions of mRNA and protein were elevated by alcohol exposure. Interestingly, ChIP analysis with H3AcK9 antibody provided evidence for increased association of the acetylated histone H3 with the promoter of PNPLA3 in cultured primary rat hepatocytes. This implies a role of histone acetylation in regulating its transcriptional expression. In vivo studies with rats and mice were also distinctive. In these in vivo models we utilized novel approaches that included chronic ethanol treatment, repeat three-binge (acute) ethanol exposure, and acute (three-repeat binge) on chronic ethanol treatments. In mice, binge ethanol and binge on chronic ethanol treatments increased PNPLA3 protein expression to a similar degree (Fig. 1B). In rats, the increases in PNPLA3 protein by binge alcohol and binge on chronic alcohol were also to a similar extent (Fig. 1C). The degree of increases by binge was greater in rats compared to mice (Fig. 1B vs. 1C). On the other hand, chronic ethanol treatment had negligible influence on the expression of this protein in both rats and mice. However, the pattern of PNPLA3 mRNA expression in rats and mice was different from that noted for protein expression for the three different alcohol-exposure protocols. In mice, the mRNA levels increased after binge alcohol treatment but increased even more after binge on chronic ethanol treatment. In rat liver, only chronic ethanol treatment produced increases in PNPLA3 mRNA, while acute or acute on chronic treatments exhibited an apparent decrease in mRNA and indicated that there may be species-related differences in PNPLA3 mRNA expression by alcohol. Despite this, the ChIP analysis after H3AcK9 antibody immunoprecipitation demonstrated that changes in PNPLA3 mRNA expression were consistent with the histone H3K9 acetylation in both animal species. The mRNA changes observed in mice appeared to more closely parallel the H3AcK9 changes compared to that observed in rats.
Steatosis is a common disorder of lipid metabolism produced by alcohol. Li et al. (2012) reported that a missense sequence variation in PNPLA3 (PNPLA3 I148M) was associated with increased hepatic TG content. While moderate drinking has very limited effects on triglycerides, binge drinking may cause an increase in the synthesis of large VLDL particles in the liver (Van de Wiel, 2012, pp. 1–4). A potential lipogenic role of PNPLA3 has also been suggested (Kershaw et al., 2006). Reducing PNPLA3 expression using specific antisense oligonucleotides prevented hepatic steatosis in rats fed a high-fat diet (Kumashiro et al., 2013). Liver PNPLA3 mRNA expression was strongly correlated with liver triglyceride and diglyceride content. The present study demonstrated increased PNPLA3 protein levels in rat hepatocytes, and in mouse and rat liver. Previously, we had reported increased TG levels after chronic, binge, and chronic plus binge alcohol treatments (Aroor et al., 2014; Shukla, Aroor, et al., 2015). For example, in mice 2-, 4-, and 13-fold increases in TG over control were noted for chronic, acute, and acute on chronic treatments, respectively (Shukla, Aroor, et al., 2015). Thus, ethanol-induced increases in PNPLA3 accompany increases in TG content.
PNPLA3 I148M induces a defect in TG remodeling, resulting in increased TG accumulation and more extensive association with LD (lipid droplets) surfaces than PNPLA3 WT (Ruhanen et al., 2014; Smagris et al., 2015). We infer that our alcohol treatments lead to a similar situation as the PNPLA3 I148M. The increased fatty liver observed with PNPLA3 I148M and alcohol treatment could be directly or secondarily correlated to altered PNPLA3 activity. Levels of mRNAs that are frequently increased in mouse models of hepatic steatosis are also increased in livers of PNPLA3 I148M transgenic mice (Li et al., 2012). Changes in the gene expression profile and fatty acid composition of hepatic TG in PNPLA3 I148M transgenic mice were found to be consistent with increased de novo fatty acid synthesis in these animals (Li et al., 2012). Altered lipogenesis by PNPLA3 due to alcohol treatment would produce a fatty acid profile prone to lipid peroxidation and liver injury. Regardless of whether the increase in PNPLA3 protein by ethanol is directly responsible for the increase in TG content, our data suggest that a similar increase in PNPLA3 protein is likely to occur in humans following repeated binge alcohol usage. This would certainly exacerbate the detrimental effect of the PNPLA3 M148I variant in humans carrying that mutation and may explain their increased susceptibility to advanced alcoholic liver disease.
In the mouse model, the increase in PNPLA3 protein correlates strongly with an increase in corresponding mRNA. ChIP assay and EMSA experiments indicated that SREBP-1 interacts directly with PNPLA3 via a response element (Huang et al., 2010). Our ChIP assay provides evidence for a close association of PNPLA3 promoter with H3AcK9. Our earlier experiments have shown that histone H3K9 acetylation increased in rat hepatocytes in vitro (Shukla, Restrepo, et al., 2015), and in vivo in rat and mouse models subjected to acute binge alcohol (Aroor et al., 2014; Shukla, Aroor, et al., 2015). We propose that H3AcK9 acetylation by alcohol directly increases the transcription-translation axis resulting in increased mRNA and protein levels of PNPLA3. While we cannot completely rule out stabilization of mRNA by ethanol, the ChIP data are more consistent with an increased transcription via histone acetylation. However, discordant mRNA and protein data in rats and mice may also suggest that ethanol may have species-specific post-translational effects. In summary, we conclude that histone H3 acetylation at lysine 9 is involved in the expression of PNPLA3 induced by ethanol in the liver. This epigenetic mechanism appears to be involved in vivo in rats and mice subjected to chronic and chronic-binge ethanol treatments, and also in vitro in rat primary hepatocytes.
Acknowledgments
This work was supported in part by Margaret Proctor Mulligan Endowment Funds to SDS and by the USA National Institutes of Alcohol Abuse & Alcoholism grant # AA-022108 to RJK.
References
- Aroor AR, Jackson DE, Shukla SD. Elevated activation of ERK1 and ERK2 accompany enhanced liver injury following alcohol binge in chronically ethanol-fed rats. Alcoholism: Clinical and Experimental Research. 2011;35:2128–2138. doi: 10.1111/j.1530-0277.2011.01577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroor AR, Jackson DE, Shukla SD. Dysregulated phosphorylation and nuclear translocation of cyclic AMP response element binding protein (CREB) in rat liver after chronic ethanol binge. European Journal of Pharmaceutics. 2012;679:101–108. doi: 10.1016/j.ejphar.2011.12.045. [DOI] [PubMed] [Google Scholar]
- Aroor AR, Restrepo RJ, Kharbanda KK, Shukla SD. Epigenetic histone modifications in a clinically relevant rat model of chronic ethanol-binge-mediated liver injury. Hepatology International. 2014;8(Suppl 2):S421–S430. doi: 10.1007/s12072-014-9546-4. [DOI] [PubMed] [Google Scholar]
- Browne JA, Harris A, Leir SH. An optimized protocol for isolating primary epithelial cell chromatin for ChIP. PLoS One. 2014;9(6):e100099. doi: 10.1371/journal.pone.0100099. http://dx.doi.org/10.1371/journal.pone.0100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch S, Stickel F, Trépo E, Way M, Herrmann H, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nature Genetics. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- Huang Y, He S, Li JZ, Seo YK, Osborne TF, Cohen CJ, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. PNAS. 2010;107(17):7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw EE, Hamm JK, Verhagen LA, Peroni O, Katic M, Flier JS. Adipose triglyceride lipase: Function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55:148–157. [PMC free article] [PubMed] [Google Scholar]
- Kumashiro N, Yoshimura T, Cantley JL, Majumdar SK, Guebre-Egziabher F, Kursawe R, et al. Role of patatin-like phospholipase domain-containing 3 on lipid-induced hepatic steatosis and insulin resistance in rats. Hepatology. 2013;57:1763–1772. doi: 10.1002/hep.26170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Huang Y, Karaman R, Ivanova PTH, Brown HA, Roddy T, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic stea-tosis. Journal of Clinical Investigation. 2012;122(11):4130–4144. doi: 10.1172/JCI65179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka K, Hashimoto S, Kawabe N, Nitta Y, Murao M, Nakano T, et al. PNPLA3 I148M associations with liver carcinogenesis in Japanese chronic hepatitis C patients. SpringerPlus. 2015;4(83):1–11. doi: 10.1186/s40064-015-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P, Lim RW, Shukla SD. Gene selective histone H3 acetylation in the absence of increase in global histone acetylation in liver in rats chronically fed alcohol. Alcohol and Alcoholism. 2012;47:233–239. doi: 10.1093/alcalc/ags004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Calvo I, Ocón B, Martínez-Moya P, Suárez MD, Zarzuelo A, Martínez-Augustin O, et al. Reversible Ponceau staining as a loading control alternative to actin in Western blots. Analytical Biochemistry. 2010;401:318–320. doi: 10.1016/j.ab.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Ruhanen H, Perttila J, Holtta-Vuori M, Zhou Y, Yki-Jarvinen H, Ikonen E, et al. PNPLA3 mediates hepatocyte triacylglycerol remodeling. The Journal of Lipid Research. 2014;55:739–746. doi: 10.1194/jlr.M046607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Aroor AR, Restrepo RJ, Kharbanda KK, Ibdah JA. In vivo acute on chronic ethanol effects in liver: A mouse model exhibiting exacerbated injury, altered metabolic and epigenetic responses. Biomolecules. 2015;5(4):3280–3294. doi: 10.3390/biom5043280. http://dx.doi.org/10.3390/biom5043280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Lim RW. Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Research: Current Reviews. 2013;35:47–55. [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Restrepo RJ, Fish P, Lim RW, Ibdah JA. Different mechanisms for histone acetylation by ethanol and its metabolite acetate in rat primary hepatocytes. Journal of Pharmacology Experimental Therapeutics. 2015b;354:18–23. doi: 10.1124/jpet.115.223867. [DOI] [PubMed] [Google Scholar]
- Smagris E, BasuRay S, Li J, Huang Y, Kaman V, et al. Pnpla3I148M Knockin mouse accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61:108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Pirola CJ. PNPLA3, the history of an orphan gene of the potato tuber protein family that found an organ: The liver. Hepatology. 2014;59:2068–2071. doi: 10.1002/hep.26895. [DOI] [PubMed] [Google Scholar]
- Stickel F, Hampe J, Trépo E, Datz C, Romeo S. PNPLA3 genetic variation in alcoholic steatosis and liver disease progression. Hepatobiliary Surgery Nutrition. 2015;4(3):152–160. doi: 10.3978/j.issn.2304-3881.2014.11.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nature Genetics. 2010;42:21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- Tomita A, Buchholz DR, Obata K, Shi YB. Fusion protein of retinoic acid receptor α with promyelocytic leukemia protein or promyelocytic leukemia zinc finger protein recruits N-CoR-TBLR1 corepressor complex to repress transcription in vivo. The Journal of Biological Chemistry. 2003;278:30788–30795. doi: 10.1074/jbc.M303309200. [DOI] [PubMed] [Google Scholar]
- Trépo E, Gustot T, Degre D, Lemmers A, Verset L, Demetter P, et al. Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. Journal of Hepatology. 2011;55:906–912. doi: 10.1016/j.jhep.2011.01.028. [DOI] [PubMed] [Google Scholar]
- Valenti L, Motta BM, Soardo G, Lavarone M, Donati B, Sangiovanni A, et al. PNPLA3 I148M polymorphism, clinical presentation, and survival in patients with hepatocellular carcinoma. PLoS One. 2013;8(10):e75982. doi: 10.1371/journal.pone.0075982. http://dx.doi.org/10.1371/journal.pone.0075982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wiel A. The effect of alcohol on postprandial and fasting tri-glycerides. International Journal of Vascular Medicine. 2012;2012 doi: 10.1155/2012/862504. http://dx.doi.org/10.1155/2012/862504. [DOI] [PMC free article] [PubMed] [Google Scholar]