Abstract
Antibiotic treatments have been used to modulate intestinal bacteria and investigate the role of intestinal bacteria on bile acid (BA) homeostasis. However, knowledge on which intestinal bacteria and bile acids are modified by antibiotics is limited. In the present study, mice were administered various antibiotics, 47 of the most abundant bacterial species in intestine, as well as individual BAs in plasma, liver, and intestine were quantified. Compared to the two antibiotic combinations (vancomycin+imipenem and cephalothin+neomycin), the three single antibiotics (metronidazole, ciprofloxacin and aztreonam) have less effect on intestinal bacterial profiles, and thus on host BA profiles and mRNA expression of genes that are important for BA homeostasis. The two antibiotic combinations decreased the ratio of Firmicutes to Bacteroidetes in intestine, as well as most secondary BAs in serum, liver and intestine. Additionally, the two antibiotic combinations significantly increased mRNA of the hepatic BA uptake transporters (Ntcp and Oatp1b2) and canalicular BA efflux transporters (Bsep and Mrp2), but decreased mRNA of the hepatic BA synthetic enzyme Cyp8b1, suggesting an elevated enterohepatic circulation of BAs. Interestingly, the two antibiotic combinations tended to have opposite effect on the mRNAs of most intestinal genes, which tended to be inhibited by vacomycin+imipenem but stimulated by cephalothin+neomycin. To conclude, the present study clearly shows that various antibiotics have distinct effects on modulating intestinal bacteria and host BA metabolism.
Introduction
Primary bile acids (BAs) are synthesized from cholesterol in hepatocytes. The two primary BAs synthesized in human liver are cholic acid (CA) and chenodeoxycholic acid (CDCA). Rodent livers can convert CDCA to form α-muricholic acid (αMCA) and βMCA (Botham and Boyd, 1983). Therefore, in addition to CA and CDCA, α- and β-MCA are also primary BAs in rodents. Primary BAs are then conjugated with glycine or taurine before secretion into bile and small intestine. In intestine, bacterial enzymes modify primary BAs to produce secondary BAs, which are reabsorbed and returned to the liver for further processing (Hofmann and Hagey, 2008). Deoxycholic acid (DCA) and lithocholic acid (LCA) are the two major secondary BAs in humans. The structure and nomenclature of various BAs are shown in Supplemental Figure 1.
The BA profile is important not only to BA signaling pathways but also to BA-induced toxicity. BAs are endogenous ligands for farnesoid × receptor (FXR) (Makishima et al., 1999; Parks et al., 1999; Wang et al., 1999) and the plasma membrane-bound G-protein-coupled BA receptor 1 (TGR5 or GPBAR1) (Kawamata et al., 2003). Individual BAs differ markedly in their potency to activate BA receptors; the potency of BAs to activate FXR is CDCA>DCA>LCA>CA (Parks et al., 1999), and to activate TGR5 is LCA>DCA>CDCA>CA (Sato et al., 2008). However, these data are based on in vitro non-hepatocyte-derived cell cultures and do not necessarily reflect the in vivo BA potencies in liver. Increased BA concentrations may cause toxicities to liver and intestine due to their detergent properties. Secondary BAs are more hydrophobic than their corresponding primary BAs, and thus are thought to be more toxic. Our laboratory has shown that individual BAs produce hepatotoxicity with different potencies when fed to mice: LCA>DCA>CDCA>CA (Song et al., 2011).
The BA profile is largely dependent on activities of intestinal bacterial enzymes that mediate deconjugation, dehydrogenation, dehydroxylation and epimerization of primary BAs in the distal small intestine as well as large intestine (Ridlon et al., 2006). Bacterial bile salt hydrolases (BSH) mediate the deconjugation of BAs, whereas bacterial hydroxysteroid dehydrogenases (HSDH) mediate the oxidation and epimerization of the 3-, 7-, and 12-hydroxy groups of BAs. Despite trillions of bacteria in the intestine, only a few species have been examined for their ability to metabolize primary BAs, due to the limitations of cultivating anaerobic bacteria that make up the predominant intestinal microbiota (Ridlon et al., 2006; Ridlon and Hylemon, 2011).
Antibiotics can alter the intestinal bacterial composition, and thus they represent one strategy to investigate the role of intestinal bacteria on BA metabolism. Swann et al. (2010) treated rats with streptomycin and penicillin to assess the role of intestinal bacteria on the BA profile. Antunes et al. (2011) demonstrated that streptomycin markedly affects intestinal BA metabolism in mice. Kuribayashi et al. (2012) showed that ampicillin treatment increases total BA concentrations in intestine of mice. Recently, Sayin et al. (2013) showed that a cocktail of antibiotics (bacitracin, neomycin and streptomycin) increases primary BAs in gallbladder and decreases secondary BAs in serum of mice. However, little is known which antibiotics alter intestinal microbiota and thus bile acids.
In the present study, several popular antibiotics were selected to test our hypothesis that antibiotics have different effect on bile acid metabolism due to their different ability to modify intestinal bacteria. Mice were treated with either single antibiotics (metronidazole, ciprofloxacin and aztreonam) or antibiotic combinations (vancomycin+imipenem and cephalothin+neomycin) that are expected to selectively eliminate intestinal bacterial genera. These antibiotics belong to various classes with different spectrums of activity and intestinal absorption (Table 1). The purpose of the present study was to systematically explore the impact of intestinal bacteria on BA profiles by modulating intestinal bacteria with various antibiotics.
Table 1.
Properties of antibiotics used in present study
| Antibiotics | Category | Mechanism | Spectrum | Intestinal Absorption |
|---|---|---|---|---|
| Metronidazole | nitroimidazole | deactivation of critical enzymes | narrow spectrum; anaerobic bacteria | complete (>90%) |
| Ciprofloxacin | fluoroquinolone | inhibition of topoisomerase II and IV | broad spectrum; gram-positive and negative | well (>70%) |
| Aztreonam | β-lactam | inhibition of cell wall | narrow spectrum; gram-negative | poorly |
| Vancomycin | glycopeptide | inhibition of cell wall synthesis | narrow spectrum; gram-positive | poorly |
| Imipenem | β-lactam | inhibition of bacterial cell wall formation | broad spectrum; gram-positive and negative | poorly |
| Cephalothin | β-lactam | inhibition of bacterial cell wall formation | broad spectrum; gram-positive and negative | poorly |
| Neomycin | aminoglycosides | inhibition of protein synthesis in bacteria | broad spectrum; excellent for gram-negative | poorly |
Materials and Methods
Chemicals and Reagents
Aztreonam, cephalothin, ciprofloxacin, imipenem, metronidazole, neomycin, and vancomycin were obtained from Sigma-Aldrich (St. Louis, MO). CA, CDCA, αMCA, βMCA, DCA, LCA, and murideoxycholic acid (MDCA) were purchased from Steraloids, Inc. (Newport, RI) and Sigma-Aldrich (St Louis, MO). Other BA standards were generous gifts from Dr Alan F Hofmann (University of California, San Diego, CA). The structure and abbreviations of various BAs can be found in our previous manuscript (Zhang et al., 2012a).
Animals and Chemical Treatments
Eight week-old male C57BL/6 mice were purchased from Charles River Laboratories, Inc. (Wilmington, MA). All the animal maintenance and treatment protocols were in compliance with the Guide for Care and Use of Laboratory animals as adopted and promulgated by the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee. Mice were singly housed in the facility with a 14-h light/10-h dark-cycle, temperature-, and humidity-controlled environment and given ad libitum access to water and rodent chow (Harlan-Teklad 8064, Madison, WI). Mice were acclimated for one week before starting treatment. Mice (N=4-5/group, singly housed) received either normal drinking water or drinking water supplemented with: metronidazole (0.1 mg/ml for 14 days), ciprofloxacin (30 mg/ml for 14 days), aztreonam (0.1 mg/ml for 7 days), vancomycin + imipenem (0.1 mg/ml of each for 14 days), or cephalothin + neomycin (2 mg/ml of each for 7 days). The dosages of antibiotics were according to previous studies (Goris et al., 1986; Hoentjen et al., 2003), and were found to have little effect on water and food consumption or body weight of mice in the present study (data not shown). Drinking water with and without antibiotics was prepared freshly and replaced daily. At the end of the treatment period, mice were anesthetized between 8:00 AM and 11:45 AM. Blood was collected by orbital sinus bleeding, and serum was obtained by centrifuging blood at 6,000 g for 15 min. Livers were harvested after the gallbladders were removed from the same animals, washed with saline, frozen in liquid nitrogen, and stored at −80°C. Intestinal contents were flushed using PBS containing 10 mM dithiothreitol (DTT) as the mucolytic agent. Intestinal segments were frozen in liquid nitrogen. All samples were stored at −80°C until further analysis.
Bacterial DNA Extraction
Luminal contents of intestine were collected in PBS containing 10 mM DTT and were centrifuged at 20,000×g for 30 min at 4°C. Total genomic bacterial DNA was extracted from the pellet using QIAmp DNA® stool kit (Qiagen, Valencia, CA) following the manufacturer’s instructions with slight modification where the entire volume of the lysate was processed instead of using only 200 μl as suggested by the manufacturer. The volumes needed for further processing were adjusted accordingly. The integrity, concentration, and quality of the total DNA were assessed by agarose gel electrophoresis, and determined by absorption at A260, and A260 to A280 ratio, respectively. DNA solutions were stored at −20°C until further analysis.
Quantification of Bacteria
The bacteria quantified in the present study were chosen based on a previous publication on the major intestinal microbiota in mice (Salzman et al., 2002). A branched DNA (bDNA) assay based on the 16S rDNA gene was developed for 47 individual bacteria (Supplemental Table 1) belonging to Firmicutes (total 22 species), Bacteroidetes (total 14 species), and other bacteria (total 11 species). Compared with real-time qPCR and gene sequencing methodologies, the method developed in the present study is much simpler and more rapid. The 16S rDNA gene assay was performed using Quantigene 2.0 Reagent System (Panomics/Affymetrix, Fremont, CA) according to the manufacturer’s protocol. Because the majority of the bacteria residing in the intestine are not easily cultivable, the bacterial sequences are defined as the closest known relative in the phylogenic tree (Salzman et al. 2002). Accession numbers for the 16S-rRNA genes of the corresponding bacteria are given in Supplementary Table 1. Total bacterial DNA (1 ng/μl, 20 μl) was added to each well containing 80 μl of lysis buffer containing blocking reagent and each probe set. Sample DNA was allowed to hybridize to each probe set overnight at 55°C. Subsequently, the plate was washed with washing buffer 3 times. Samples were hybridized with the amplification reagent (100 μl/well) in the amplifier/label probe buffer for 1 h at 55°C. The plate was washed 3 times with wash buffer. Label probe diluted in amplifier/label probe buffer was added to each well (100 μl/well), and the alkaline phosphatase-conjugated label probe was allowed to hybridize to the bDNA-DNA complex for 1 h at 50°C. The plate was washed with wash buffer 3 times. The enzyme reaction was triggered by the addition of substrate solution (100 μl/well) and incubated for 5 min. The resulting luminescence was quantified using a luminometer set at an integration time of 0.2 sec.
BA Extraction and Analysis
BA extraction and analysis of liver, serum, and intestinal contents were according to previously published methods (Zhang et al., 2010; Zhang et al., 2011; Zhang et al., 2012b).
RNA Extraction and mRNA Quantification by Multiplex Suspension Array
Total RNA was isolated using RNA-Bee reagent (Tel-Test Inc., Friendswood, TX) according to the manufacturer’s protocol. Total RNA concentrations were quantified spectrophotometrically at 260 nm. Integrity of RNA samples was determined by formaldehyde-agarose gel electrophoresis with visualization by ethidium bromide fluorescence under ultraviolet light. The mRNA expression was quantified by mutiplex suspension array (Panomics-Affymetrix, Inc., Fremont, CA). Individual gene accession numbers can be accessed at www.panomics.com (panels #21021 and #21151). Assays were performed according to each manufactures’ protocol (Zhang et al., 2012b). RNA data were normalized to Gapdh mRNA.
Statistical Analysis
Statistical differences in BAs as well as mRNAs between control and antibiotic-treated mice were determined using a two-tailed Student’s t-test. Comparisons resulting with p<0.05 were considered statistically significant. Heatmaps were generated in R Bioconductor using the heatmap.2 function (omitting row and column dendrograms) of the gplots package (http://cran.r-project.org/web/packages/gplots/index.html). Data points statistically different (p<0.05) from control have the percent of control indicated within the heatmap square (control=100%).
Results
Antibiotics produced marked alterations in the relative abundance of various bacterial species
To examine the effects of antibiotics on the composition of bacterial communities in intestinal contents, a simple and rapid multiplex assay was developed to quantify the 47 most abundant bacterial species in the intestine of mice treated with metronidazole, ciprofloxacin, aztreonam, vancomycin+imipenem, or cephalothin+neomycin (Figure 1) (see also Supplemental Figures 2–6).
Figure 1.
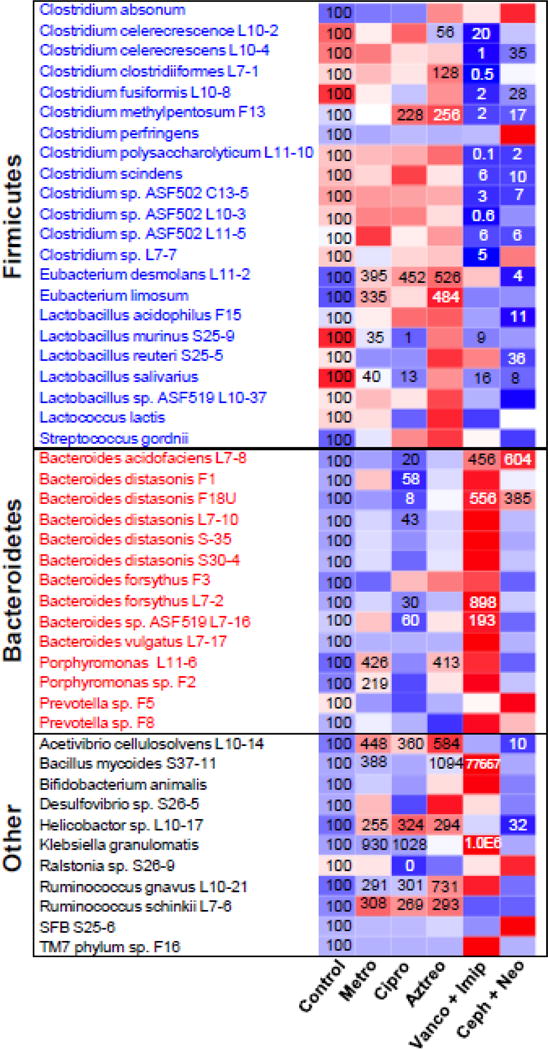
Heatmap presentation of antibiotic modulation of the intestinal microbiome. The heatmap was generated in R Bioconductor using the heatmap.2 function (omitting row and column dendrograms) of the gplots package. Data points statistically different (p<0.05) from control have the percent of control indicated within the heatmap square (control=100%). Red indicates an increased value, whereas blue indicates a decreased value.
Metronidazole tended to increase most of the Firmicutes with a significant increase in Eubacterium desmolans L11-2 (295%↑) and Eubacterium limosum (235%↑), whereas it significantly decreased Lactobacillus murinus S25-9 (65%↓) and Lacobacillus salivarius (60%↓). In contrast, metronidazole tended to suppress a majority of the Bacteroidetes, though it significantly increased Porphyromonas L11-6 (326%↑) and Porphyomonas sp. F2 (119%↑). In addition, metronidazole significantly increased six other bacterial species [Acetivibrio cellulosolvens L10-14 (348%↑), Bacillus mycoides S37-11 (290%↑), Helicobactor sp. L10-17 (155%↑), Klebsiella granulomatis (830%↑), Ruminococcus gnavus L10-21 (190%↑), and Ruminococcus schinkii L7-6 (208%↑)]. Taken together, metronidazole had limited ability to suppress intestinal bacteria, but increased several bacterial species.
Ciprofloxacin significantly increased two Firmicutes [Clostridium methylpentosum F13 (128%↑) and Eubacterium desmolans L11-2 (352%↑)] whereas it significantly decreased two Firmicutes [Lactobacillus murinus S25-9 (99%↓) and Lactobacillus salivarius (87%↓)] and six Bacteroidetes [Bacteroides acidofaciens L7-8 (80%↓), Bacteroides distasonis F1 (42%↓), Bacteroides distasonis F18U (92%↓), Bateroides distasonis L7-10 (57%↓), Bacteroides forsythus L7-2 (70%↓), and Bacteroides sp. ASF519 L7-16 (40%↓)]. In addition, ciprofloxacin significantly increased several other bacterial species [Acetivibrio cellulosolvens L10-14 (260%↑), Helicobactor sp. L10-17 (224%↑), Klebsiella granulomatis (928%↑), Ruminococcus gnavus L10-21 (201%↑), and Ruminococcus schii L7-6 (169%↑)]. Interestingly, ciprofloxacin eliminated Ralstonia sp. S26-9 to an undetectable level. Taken together, ciprofloxacin treatment resulted in a marked suppression of Bacteroidetes, whereas it increased many other bacterial species.
Aztreonam showed a strong stimulatory effect toward Firmicutes species with a significant increase in Clostridium clostridiiformes L7-1 (28%↑), Clostridium methylpentosum F13 (156%↑), Eubacterium desmolans L11-2 (426%↑), and Eubacterium limosum (384%↑). Aztreonam suppressed only one Firmicutes [Clostridium celerecrescence L10-2 (44%↓)]. In addition, aztreonam significantly increased one Bacteroidetes [Porphyromonas L11-6 (313%↑)] and five other bacterial species [Acetivibrio cellulosolvens L10-14 (484%↑), Bacillus mycoides S37-11 (994%↑), Helicobactor sp. L10-17 (194%↑), Ruminococcus gnavus L10-21 (631%↑), and Ruminococcus schinkii L7-6 (193%↑)]. Taken together, aztreonam had little suppressive effect on intestinal bacteria, but increased many bacterial species.
Vancomycin+imipenem significantly suppressed 13 out of 22 Firmicutes [Clostridium celerecrescence L10-2 (80%↓), Clostridium celerecrescens L10-4 (99%↓), Clostridium clostridiiformes L7-1 (99.5%↓), Clostridium fusiformis L10-8 (98%↓), Clostridium methylpentosum F13 (98%↓), Clostridium polysaccharolyticum L11-10 (99.9%↓), Clostridium scindens (94%↓), Clostridium sp. ASF502 C13-5 (97%↓), Clostridium sp. ASF 502 L10-3 (99.4%↓), Clostridium sp. ASF502 L11-5 (94%↓), Clostridium sp. L7-7 (95%), Lactobacillus murinus S25-9 (91%↓), and Lactobacillus salivarius (87%↓)]. In contrast, vancomycin+imipenem significantly increased 4 Bacteroidetes [Bacterides acidofaciens L7-8 (356%↑), Bacteroides distasonis F18U (456%↑), Bacteroides forsythus L7-2 (798%↑), and Bacteroides sp. ASF519 L7-16 (93%↑)] and 2 other bacterial species [Bacillus mycoides S37-11 (77,500%↑) and Klebsiella granulomatis (990,000%↑) ]. Taken together, vancomycin+imipenem treatment produced a selective suppressive effect on Firmicutes.
Cephalothin+neomycin significantly suppressed 11 out of 22 Firmicutes [Clostridium celerescrescens L10-4 (65%↓), Clostridium fusiformis L10-8 (72%↓), Clostridium methylpentosum F13 (83%↓), Clostridium polysaccharolyicum L11-10 (98%↓), Clostridium scindens (90%↓), Clostridium sp. ASF502 C13-5 (93%↓), Clostridium sp. ASF502 L11-5 (94%↓), Eubacterium desmolans L11-2 (96%↓), Lactobacillus acidophilus F15 (89%↓), Lactobacillus reuteri S25-5 (64%↓), and Lactobacillus salivarius (92%↓)], and two other bacterial species [Acetivibrio cellulosolvens L10-14 (90%↓) and Helicobactor sp. L10-17 (68%↓)]. In contrast, cephalothin+neomycin significantly increased two Bacteroidetes [Bacteroides acidofaciens L7-8 (504%↑) and Bateroides distasonis F18U (285%↑)]. Taken together, cephalothin+neomycin treatment showed a suppressive effect on most of the bacterial species, in particular Firmicutes.
Antibiotics treatment restructured bacterial community
The intestinal microbiota in both humans and mice consist mainly of species belonging to the Firmicutes and Bacteroidetes phyla (Ley et al., 2008; Salzman et al., 2002). In general terms, the Firmicutes to Bacteroidetes ratio is regarded as having significant relevance to the composition of intestinal microbiota. Figure 2 summarizes the effect of the five antibiotic treatments on the total Firmicutes and Bacteroidetes species investigated in the present study. Metronidazole slightly decreased the Firmicutes to Bacteroidetes ratio, mainly due to the increase in Bacteroidetes. Ciprofloxacin increased the Firmicutes to Bacteroidetes ratio mainly due to its suppression of Bacteroidetes. Unlike metronidazole and ciprofloxacin, aztreonam had little effect on the Firmicutes to Bacteroidetes ratio although it increased both of them. Vancomycin+imipenem decreased Firmicutes but increased Bacteroidetes, resulting in a prominent decrease in the Firmicutes to Bacteroidetes ratio. Cephalothin+neomycin decreased both Firmicutes and Bacteroidetes, leading to a slight decrease in the Firmicutes to Bacteroidetes ratio. Taken together, the Firmicutes to Bacteroidetes ratio was decreased by metronidazole, vancomycin+imipenem and cephalothin+neomycin, whereas it was increased by ciprofloxacin.
Figure 2.
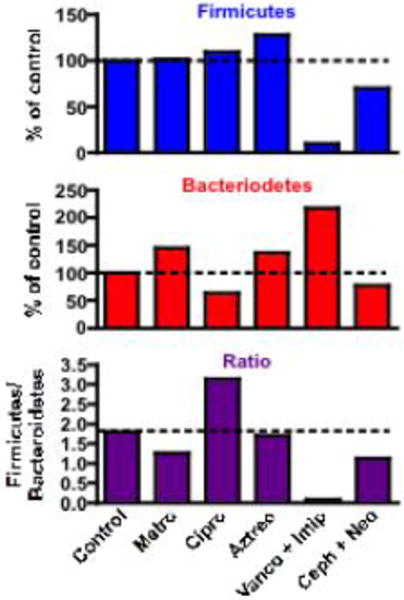
Bar graph representation of the effect of antibiotics on total intestinal Firmicutes species(top panel), Bacteroidetes species (middle panel), and the ratio of total Firmicutes/Bacteroidetes species (bottom panel).
Single antibiotics had little effect on intestinal BA profile, whereas antibiotic combinations markedly inhibited secondary BA formation in the large intestine
Contents of large intestines were collected and BAs were quantified to evaluate the effect of antibiotics on BA metabolism (Figure 3) (see also Supplemental Figures 2–6). Surprisingly, single antibiotics had little effect on BA composition in the large intestine, except that they tended to suppress secondary BA synthesis (7-dehydroxylation, 6β to 6α epimerization and 3α to 3β epimerization). Several BAs were significantly altered after single antibiotic treatments, including metronidazole-mediated decrease in UDCA (55%↓), TLCA (72%↓) and TUCA (83%↓), ciprofloxacin-mediated increase in TUCA (354%↑), as well as aztreonam-mediated decrease in TLCA (96%↓) and TUCA (90%↓). In contrast, combination antibiotic treatments markedly increased conjugated primary BAs but decreased secondary BAs in the large intestine. Vancomycin+imipenem significantly increased TCA (2800%↑), TαMCA (3800%↑), TβMCA (6100%↑), TUDCA (1800%↑), TUCA (2400%↑), and TωMCA (460%↑), whereas it significantly decreased αMCA (91%↓), TDCA (67%↓), TLCA (79%↓), DCA (99.8%↓), LCA (99%↓), MDCA (97%↓), HDCA (98%↓), isoDCA (99.7%↓), and isoLCA (97%↓). Cephalothin+neomycin significantly increased TCA (1900%↑), TCDCA (310%↑), TαMCA (301%↑), TUDCA (793%↑), TUCA (246%↑), and TωMCA (588%↑), whereas it significantly decreased αMCA (84% ↓), TLCA (43%↓), DCA (93%↓), LCA (88%↓), MDCA (92%↓), HDCA (95%↓), ωMCA (95%↓), isoDCA (97%↓), and isoLCA (94%↓). Taken together, single antibiotics treatments had little effect on intestinal BA composition, whereas antibiotic combinations treatments markedly inhibited bacteria-mediated secondary BA formation.
Figure 3.
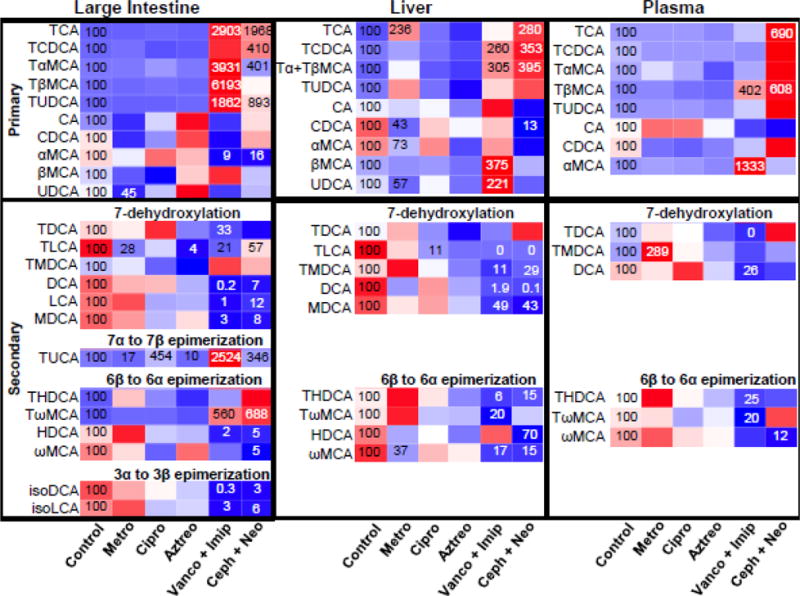
Heatmap presentation of the effect of antibiotics on host BA profiles in large intestine, liver, and plasma. The heatmap was generated in R Bioconductor using the heatmap.2 function (omitting row and column dendrograms) of the gplots package. Data points statistically different (p<0.05) from control have the percent of control indicated within the heatmap square (control=100%), Red indicates an increased value, whereas blue indicates a decreased value.
Metronidazole, vancomycin+imipenem and cephalothin+neomycin increased the total BA concentrations in liver due to the increase in conjugated primary BAs
The concentrations of individual BAs in livers of mice treated with single or antibiotic combinations were quantified to determine whether antibiotics-induced modulation of intestinal bacteria had an effect on liver BA profiles (Figure 3) (see also Supplemental Figures 2–6). Metronidazole significantly increased TCA (136%↑), but it significantly decreased CDCA (57%↓), αMCA (27%↓), UDCA (43%↓), and ωMCA (63%↓). In addition, metronidazole also tended to increase other conjugated BAs, such as TCDCA, Tα/βMCA, TUDCA, TDCA, TMDCA, THDCA, and TωMCA. Unlike metronidazole, ciprofloxacin and aztreonam had little effect on BAs in liver, except that ciprofloxacin significantly decreased TLCA (89%↓). In general, vancomycin+imipenem increased primary BAs but decreased secondary BAs in the liver. Vancomycin+imipenem significantly increased TCDCA (160%↑), Tα/βMCA (205%↑), βMCA (275%↑), and UDCA (121%↑), but significantly decreased TLCA (100%↓), TMDCA (89%↓), DCA (92%↓), MDCA (51%↓), THDCA (94%↓), TωMCA (80%↓), and ωMCA (83%↓). Similarly, cephalothin+neomycin significantly increased TCA (180%↑), TCDCA (253%↑), and Tα/βMCA (295%↑), but significantly decreased CDCA (87%↓), TLCA (100%↓), TMDCA (71%↓), DCA (99.9%↓), MDCA (57%↓), THDCA (85%↓), HDCA (30%↓), and ωMCA (85%↓). Taken together, metronidazole, vancomycin+imipenem, and cephalothin+neomycin increased total liver BAs (Supplemental Figure 7) due to elevated conjugated primary BAs, in particular TCA and Tα/βMCA.
Vancomycin+imipenem and cephalothin+neomycin markedly altered serum BA profiles
Serum BA profiles of antibiotic-treated mice revealed that single antibiotics had little effect on individual BA concentrations in the serum, except that metronidazole significantly increased serum TMDCA (Figure 3) (see also Supplemental Figures 2–6). In contrast, antibiotic combinations had more prominent effects on serum BA profiles. Vancomycin+imipenem increased serum TβMCA (302%↑) and αMCA (1200%↑), but decreased TDCA (100%↓), DCA (74%↓), THDCA (75%↓), and TωMCA (80%). Cephalothin+neomycin increased TCA (590%↑) and TβMCA (508%↑), but decreased ωMCA (88%↓). Taken together, single antibiotics had little effect on serum BA profiles, whereas antibiotic combinations increased primary BAs (in particular TβMCA) and decreased secondary BAs in serum.
Antibiotics alter the mRNA expression of genes in hepatic BA synthesis, transport, and signaling
The mRNA expression of genes involved in liver BA synthesis, transport, and signaling were quantified to evaluate whether the altered BA profile in liver was associated with antibiotic-mediated regulation of genes involved in BA homeostasis. In agreement with their minimal effect on liver BA concentrations, single antibiotics had little effect on the mRNA expression of genes that regulate BA homeostasis in liver. The significant changes included an increase in Oatp2b1 (24%↑) and Abcg8 (14%↑) by metronidazole, an increase of Oatp1b2 (22%↑) and SHP (48%↑) by ciprofloxacin, as well as an increase of Ntcp (14%↑), Mrp2 (9%↑), LRH-1 (11%↑), Abcg5 (43%↑) and Abcg8 (27%↑) by aztreonam. Vancomycin+imipenem produced a prominent effect on gene expression in liver, including a significant decrease in Cyp7a1 (69%↓), Cyp8b1 (46%↓), Baat (9%↓), Oatp1a4 (29%↓), Mrp3 (27%↓), as well as a significant increase in BAL (7%↑), Ntcp (18%↑), Oatp1a1(20%↑), Oatp1b2 (23%↑), Oatp2b1 (31%↑), Bsep (23%↑), Mrp2(8%↑), SHP (53%↑), LRH-1 (23%↑), Abcg5 (66%↑) and Abcg8 (60%↑). Cephalothin+neomycin significantly decreased Cyp8b1 (64%↓), but increased BA uptake transporters Ntcp (26%↑) and Oatp1b2 (28%↑). In addition, cephalothin+neomycin tended to increase the mRNA of many genes, such as BA conjugation enzymes Baat and BAL, as well as BA efflux transporters Mrp4 and Ostβ. Taken together, in agreement with the alterations in liver BA profile, antibiotic combinations showed a more prominent effect on liver BA gene expression profile than did single antibiotics.
Antibiotics altered mRNA expression of intestinal BA transporters and genes in BA signaling pathway
Because intestine is the primary site of the antibiotic effects, the mRNA expression of intestinal BA transporters, cholesterol transporters, and genes in BA signaling were quantified to further evaluate the effect of antibiotics on BA homeostasis. In general, single antibiotics had less effect on intestinal gene expression than did antibiotic combinations. Compared to other BA transporters, efflux transporters such as Mrp2, Mrp3, and Ostβ were relatively more susceptible to antibiotic treatments. Mrp2 was significantly decreased by metronidazole (23%↓), ciprofloxacin (13%↓) and vancomycin+imipenem (33%↓), whereas it was significantly increased by cephalothin+neomycin (43%↑). Mrp3 was significantly increased by metronidazole (26%↑), ciprofloxacin (29%↑) and cephalothin+neomycin (27%↑). Ostβ was significantly decreased by metronidazole (21%↓), ciprofloxacin (18%↓), aztreonam (27%↓), and cephalothin+neomycin (23%↓). In addition, the significant changes in BA transporters also included a decrease of Asbt by aztreonam and a decrease of Mdr1a and Ostα by vancomycin+imipenem. Single antibiotics had little effect on BA signaling genes and cholesterol transporters, except that ciprofloxacin decreased Fgf15 (48%↓) and azteronam decreased Npc1l1 (24%↓). Vancomycin+imipenem had little effect on BA signaling genes but significantly decreased cholesterol transporters Abcg5 (16%↓), Abcg8 (20%↓), and Npc1l1 (44%↓) in the intestine. Cephalothin+neomycin increased BA signaling genes LRH-1 (89%↑), Fgf15 (292%↑), and VDR (42%↑), as well as cholesterol transporter Npc1l1 (30%↑). Taken together, antibiotics had a major effect on intestinal BA efflux transporters, and cephalothin+neomycin had prominent effects in activating genes in the intestinal BA signaling pathway.
Discussion
Although oral antibiotics have been used in animal models during the last few years to explore the function of intestinal bacteria in BA metabolism, knowledge about which intestinal bacteria and host bile acids are modified by antibiotics is limited. In the present study, we developed a simple and rapid multiplex bDNA assay to quantify the 47 most abundant bacterial species in the intestine of mice treated with various antibiotics. It was expected that metronidazole, ciprofloxacin and aztreonam treatment would lead to distinct intestinal bacterial profiles, due to different molecular structures, intestinal absorption abilities as well as bactericidal mechanisms and spectrums (Table 1). Consistently, these single antibiotics resulted in different ratios of intestinal Firmicutes to Bacteroidetes (decreased by metronidazole, increased by ciprofloxacin and unaltered by aztreonam) (Figure 2). In addition, they all significantly increased Eubacterium desmolans, Acetivibrio cellulosolvens, Helicobactor sp., Ruminococcus gnavus, and Ruminococcus schinkii (Figure 1). Antibiotic combinations had a larger impact on intestinal bacterial profile than single antibiotics, which is not surprising because they possess more broad-spectrum activities.
Analysis of intestinal bacteria and BA composition in antibiotic-treated mice reveals a potential correlation between intestinal Clostridia and intestinal formation of secondary BAs. The three single antibiotics (metronidazole, ciprofloxacin and aztreonam) had little effect on intestinal BA composition (Figure 3), which is in agreement with the minimal alterations they exerted on bacterial profile in the intestine. Although the two antibiotic combinations demonstrated distinct effects on intestinal BA communities (e.g. vancomycin+imipenem increased Bacteroidetes, whereas cephalothin+neomycin tended to decrease Bacteroidetes), both vancomycin+imipenem and cephalothin+neomycin significantly decreased the majority of intestinal Clostridia (Figure 1). In addition, both vancomycin+imipenem and cephalothin+neomycin significantly increased conjugated primary BAs and decreased secondary BAs, in particular, those formed by 7-dehydroxylation of primary BAs (Figure 3). Therefore, a positive correlation exists between intestinal secondary BAs and intestinal Clostridia, which have been suggested to possess critical activity for BA deconjugation and 7-dehydroxylation (Ridlon et al., 2006; Ridlon and Hylemon, 2011). Vancomycin+imipenem was more effective in increasing primary BAs in the intestine than was cephalothin+neomycin, probably due to its more prominent effect in suppressing intestinal Clostridia. However, little is known on the role the Clostridia species listed in the present study have in regulating BA metabolism. Future studies, such as in vitro studies of Clostridia incubation with bile acids and in vivo Clostridia transplantation to germ-free mice, are required to determine the effect of Clostridia on intestinal primary conjugated BAs after antibiotic treatments.
The present study suggests a potential beneficial outcome of antibiotic combinations to the host to protect the liver and intestine from injuries induced by toxic BAs. Decades of research has strongly suggested that secondary BAs are involved in many human liver diseases as well as liver and colon cancer (Chaplin, 1998; Bayerdorffer et al., 1995; Changbumrung et al., 1990). LCA causes segmental bile-duct obstruction, destructive cholangitis, and periductal fibrosis (Benedetti et al., 1997). LCA concentrations are also linked to primary biliary cirrhosis (PBC) and primary sclerosing cholangitis (PSC) (Sinakos et al., 2010). Toxicity of BAs is attributed partially to their detergent properties, which is dependent on their hydrophilicity. The combinaton antibiotics increased the hydrophilicity of BAs in both the intestine and liver, especially by suppressing the de-conjugation of primary BAs. In addition, the antibiotic combinations tended to induce the synthesis of UDCA, a therapeutic BA used in many liver diseases. UDCA has been considered a secondary BA in humans and rodents, and can be synthesized by Bacteroides distasonis (Takamine et al., 1985). However, the increase of total UDCA (TUDCA+UDCA) in both large intestine and livers of mice treated with the antibiotic combinations suggests that UDCA is not likely a synthetic product by intestinal bacteria. This is further confirmed by our findings in germfree animals (data not shown) and consistent with a recent finding (Sayin et al., 2013). Thus, in contrast to humans (Russell, 2003), UDCA appears to be a primary BA in rodents. A previous clinical study reported that the use of oral vancomycin in pediatric PSC patients resulted in favorable changes in liver tests and clinical symptoms, whereas metronidazole treatment failed to prevent disease progression (Davies et al., 2008). This could be explained by the current finding that vancomycin but not metronidazole increases liver BA hydrophilicity and thereby improves liver function.
The current findings lead to our hypothesis that intestinal bacteria are not only involved in the formation of secondary BAs, but also important for liver and serum BA pool size by regulating the expression of intestinal BA transporters as well as the expression of the intestinal enzymes critical for BA homeostasis. Metronidazole increased total BA concentrations in liver, whereas it had very little effect on BA profile in the intestine. It cannot be excluded that metronidazole affects liver BA metabolism by another mechanism in addition to modulating intestinal bacteria, because metronidazole is almost completely absorbed from the intestine. To simplify, in the current study, only those antibiotics with poor intestinal absorption (aztreonam, vancomycin+imipenem and cephalothin+neomycin) are used to assess the effect of intestinal bacteria on liver and serum BA profiles. A positive correlation exists between intestinal BA profiles and alterations of BAs in liver and serum (Figure 3). The two antibiotic combinations with prominent effects in suppressing the formation of secondary BAs, significantly increased the total BA concentrations in liver, mainly due to the increase in conjugated primary BAs. This may reflect increased BA reabsorption in the intestine of mice treated with antibiotic combinations, and also suggests the intestinal transport system (Asbt and Ostα/β) is very efficient for conjugated BAs. Although the increased expression of intestinal BA transporter Asbt was observed in germ-free mice (Sayin et al., 2013), the current study showed that neither of the two antibiotic combinations had any effect on Asbt mRNA expression. In addition, both of the two antibiotic combinations significantly increased mRNA expression of uptake BA transporters, Ntcp and Oatp1b2, in liver. Two canalicular BA efflux transporters, Bsep and Mrp2, were also increased or tended to be increased by antibiotic combinations. This suggests that the liver mayadapt to the increased conjugated BAs by boosting its BA transport system to facilitate the enterohepatic circulation of BAs. The increased BA concentrations in liver may suppress hepatic BA biosynthesis, as evidenced by a significant decrease in Cyp7a1 and 8b1 after vancomycin+imipenem treatment, as well as a significant decrease in Cyp8b1 mRNA after cephalothin+neomycin treatment. However, additional experiments are required to obtain a defined effect of antibiotics on liver BA metabolism.
Identification of the BA receptor FXR has facilitated mechanistic investigations on BA-mediated physiological and toxic responses (Kawamata et al., 2003; Makishima et al., 1999). FXR is highly expressed in liver and ileum, and regulates BA synthesis via the liver FXR-SHP pathway and the intestinal FXR-Fgf15 pathway (Goodwin et al., 2000). It was initially recognized that the liver FXR-SHP pathway suppresses hepatic Cyp7a1 mRNA as a negative feedback mechanism, however, recent studies suggest that this liver FXR-SHP pathway has a minor role in the regulation of Cyp7a1, but is more important for Cyp8b1 regulation (Kim et al., 2007; Kong et al., 2012). In the present study, both vancomycin+imipenem and cephalothin+neomycin significantly suppressed Cyp8b1 mRNA, which is likely due to activation of the liver FXR-SHP pathway, as indicated by the increased mRNA of SHP. Interestingly, vancomycin+imipenem appeared to have less of an effect on the intestinal FXR-Fgf5 pathway than did cephalothin+neomycin, which significantly increased mRNA of Fgf15 in the intestine and tended to increase mRNA of Fgfr4 in the liver, the receptor for Fgf15. Recently, Sayin et al. (2013) identified TαMCA and TβMCA as FXR antagonists, and the ratio of TMCA/TCA ratio was negatively correlated with FXR activity. This may explain the different effects of the two antibiotic combinations on intestinal FXR-Fgf15 pathway, because the intestinal TMCA/TCA ratio was increased by vancomycin+imipenem but decreased by cephalothin+neomycin. However, this might not explain why vancomycin+imipenem significantly decreased hepatic Cyp7a1 mRNA, because intestinal FXR-Fgf15 pathway has been shown to play a major role in Cyp7a1 suppression. Further studies using whole body FXR-null, liver-specific and intestine-specific FXR-null mouse models are expected to elucidate the role of FXR in antibiotic-mediated BA regulation.
In summary, we have demonstrated that antibiotic treatments result in not only marked changes in intestinal bacterial communities and diversity, but also a profound systemic effect on BA metabolism and signaling pathways. Antibiotic combinations are more efficient in suppressing secondary BA formation in the intestine, resulting in increased conjugated primary BAs in plasma, liver, and intestine of mice. The abundance of Clostridia appears to be critical for the formation of secondary BAs in the intestine of mice. However, the present study did not exclude the possibility that antibiotics might have a direct effect on bile acid metabolism. Therefore, future studies, such as antibiotic treatments of germ-free mice are required to elucidate the role of intestinal microbiota in antibiotic-mediated regulation of BA metabolism. Overall, the findings from this study will provide guidance in selection of antibiotics for BA-associated studies by modulation of intestinal microbiota.
Supplementary Material
Figure 4.
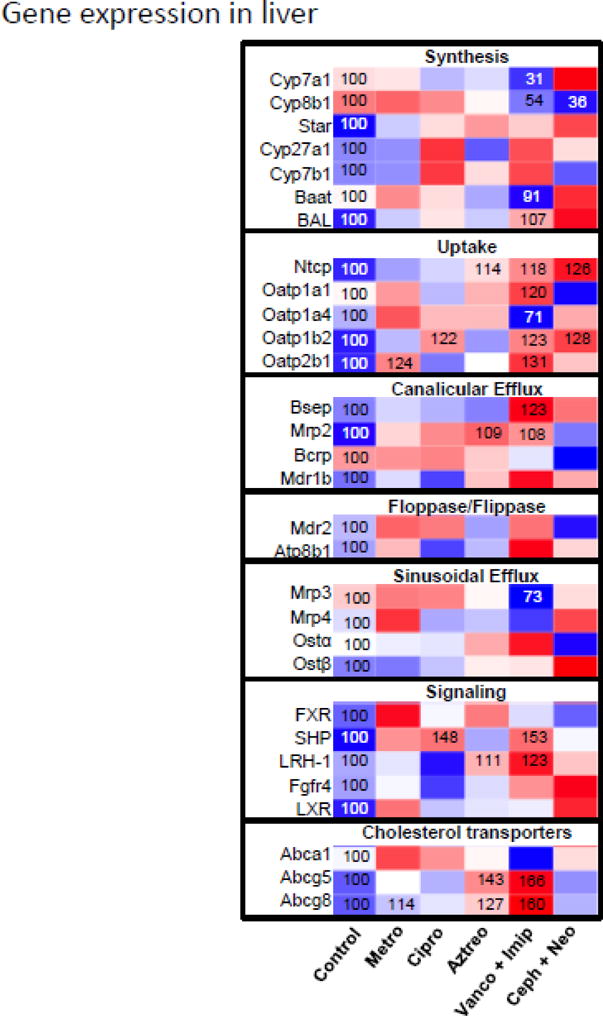
Heatmap presentation of the effect of antibiotics on mRNA expression of genes involved in liver BA homestasis. The heatmap was generated in R Bioconductor using the heatmap.2 function (omitting row and column dendrograms) of the gplots package. Data points statistically different (p<0.05) from control have the percent of control indicated within the heatmap square (control=100%). Red indicates an increased value, whereas blue indicates a decreased value.
Figure 5.
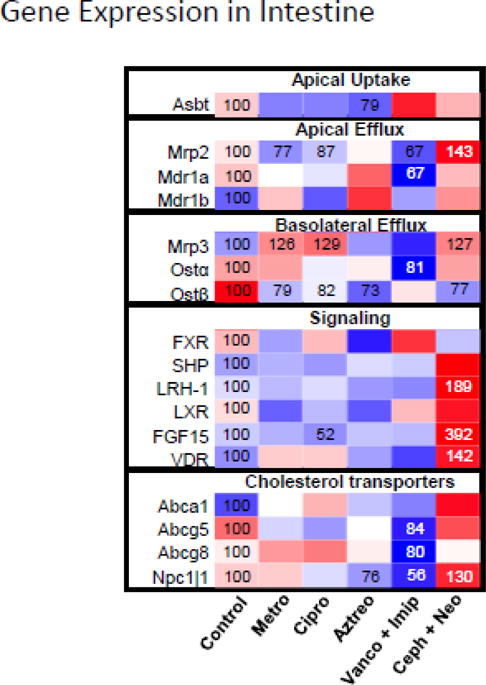
Heatmap presentation of the effect of antibiotics on mRNA expression of genes involved in intestinal BA homeostasis. The heatmap was generated in R Bioconductor using the heatmap.2 function (omitting row and column dendrograms) of the gplots package. Data points statistically different (p<0.05) from control have the percent of control indicated within the heatmap square (control=100%). Red indicates an increased value, whereas blue indicates a decreased value.
Research highlights.
Various antibiotics have different effect on intestinal bacteria.
Antibiotics alter bile acid composition in mouse liver and intestine.
Antibiotics influence genes involved in bile acid homeostasis.
Clostridia appear to be important for secondary bile acid formation.
Acknowledgments
The authors would like to thank Dr. Cheryl Rockwell for optimizing the multiplex suspension mRNA array and Felcy Selwyn for her assistance in mRNA extraction. The authors also thank Dr. Julia Yue Cui and the rest of the members of Dr Klaassen’s laboratory for their help in tissue collection and manuscript editing. This study was funded by NIH grants: ES-009649.
Abbreviations
- Abca1
ATP-binding cassette transporter a1
- BA
bile acid
- CA
cholic acid
- Bsep
bile salt-export pump
- CDCA
chenodeoxycholic acid
- Cyp
cytochrome P450
- DCA
deoxycholic acid
- Fxr
farnesoid × receptor
- Gapdh
glyceraldehyde 3-phosphate dehydrogenase
- HDCA
hyodeoxycholic acid
- IS
internal standard
- LCA
lithocholic acid
- MCA
muricholic acid
- MDCA
murideoxycholic acid
- Mrp
multidrug resistance-associated protein
- Ntcp
sodium taurocholate cotransporting polypeptide
- Oatp/OATP
organic anion transporting polypeptide
- Ost
organic solute transporter
- 7-oxoDCA
7-oxo-deoxycholic acid
- Shp
small heterodimer partner
- TCA
tauro-cholic acid
- T-12-epiDCA
tauro-12-epi deoxycholic acid
- UDCA
ursodeoxycholic acid
- UPLC
ultra performance liquid chromatography
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship Contributions
Conceived and designed the experiments: PL, YZ, and CDK. Performed the experiments: PL and YZ. Analyzed the data: PL, YZ, HR, and CDK. Wrote the paper: YZ, PL, and CDK.
References
- Antunes LC, Han J, Ferreira RB, Lolic P, Borchers CH, Finlay BB. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayerdorffer E, Mannes GA, Ochsenkuhn T, Dirschedl P, Wiebecke B, Paumgartner G. Unconjugated secondary bile acids in the serum of patients with colorectal adenomas. Gut. 1995;36:268–273. doi: 10.1136/gut.36.2.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti A, Alvaro D, Bassotti C, Gigliozzi A, Ferretti G, La Rosa T, Di Sario A, Baiocchi L, Jezequel AM. Cytotoxicity of bile salts against biliary epithelium: a study in isolated bile ductule fragments and isolated perfused rat liver. Hepatology. 1997;26:9–21. doi: 10.1002/hep.510260102. [DOI] [PubMed] [Google Scholar]
- Botham KM, Boyd GS. The metabolism of chenodeoxycholic acid to beta-muricholic acid in rat liver. Eur J Biochem. 1983;134:191–196. doi: 10.1111/j.1432-1033.1983.tb07550.x. [DOI] [PubMed] [Google Scholar]
- Changbumrung S, Tungtrongchitr R, Migasena P, Chamroenngan S. Serum unconjugated primary and secondary bile acids in patients with cholangiocarcinoma and hepatocellular carcinoma. J Med Assoc Thai. 1990;73:81–90. [PubMed] [Google Scholar]
- Chaplin MF. Bile acids, fibre and colon cancer: the story unfolds. J R Soc Health. 1998;118:53–61. doi: 10.1177/146642409811800111. [DOI] [PubMed] [Google Scholar]
- Davies YK, Cox KM, Abdullah BA, Safta A, Terry AB, Cox KL. Long-term treatment of primary sclerosing cholangitis in children with oral vancomycin: an immunomodulating antibiotic. J Pediatr Gastroenterol Nutr. 2008;47:61–67. doi: 10.1097/MPG.0b013e31816fee95. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Goris H, de Boer F, van der Waaij D. Oral administration of antibiotics and intestinal flora associated endotoxin in mice. Scand J Infect Dis. 1986;18:55–63. doi: 10.3109/00365548609032307. [DOI] [PubMed] [Google Scholar]
- Hoentjen F, Harmsen HJM, Braat H, Torrice CD, Mann BA, Sartor RB, Dieleman LA. Antibiotics with a selective aerobic or anaerobic spectrum have different therapeutic activities in various regions of the colon in interleukin 10 gene deficient mice. Gut. 2003;52:1721–1727. doi: 10.1136/gut.52.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol Life Sci. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ. Differential regulation of bile acid homeostasis by the farnesoid × receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid × receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuribayashi H, Miyata M, Yamakawa H, Yoshinari K, Yamazoe Y. Enterobacteria-mediated deconjugation of taurocholic acid enhances ileal farnesoid × receptor signaling. Eur J Pharmacol. 2012;697:132–138. doi: 10.1016/j.ejphar.2012.09.048. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Hylemon PB. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7alpha-dehydroxylating intestinal bacterium. J Lipid Res. 2011;53:66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Salzman NH, de Jong H, Paterson Y, Harmsen HJ, Welling GW, Bos NA. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology. 2002;148:3651–3660. doi: 10.1099/00221287-148-11-3651. [DOI] [PubMed] [Google Scholar]
- Sato H, Macchiarulo A, Thomas C, Gioiello A, Une M, Hofmann AF, Saladin R, Schoonjans K, Pellicciari R, Auwerx J. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J Med Chem. 2008;51:1831–1841. doi: 10.1021/jm7015864. [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Sinakos E, Marschall HU, Kowdley KV, Befeler A, Keach J, Lindor K. Bile acid changes after high-dose ursodeoxycholic acid treatment in primary sclerosing cholangitis: Relation to disease progression. Hepatology. 2010;52:197–203. doi: 10.1002/hep.23631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Zhang Y, Klaassen CD. Dose-response of five bile acids on serum and liver bile Acid concentrations and hepatotoxicty in mice. Toxicol Sci. 2011;123:359–367. doi: 10.1093/toxsci/kfr177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2010;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamine F, Imamura T. 7 beta-dehydroxylation of 3,7-dihydroxy bile acids by a Eubacterium species strain C-25 and stimulation of 7 beta-dehydroxylation by Bacteroides distasonis strain K-5. Microbiol Immunol. 1985;29:1247–1252. doi: 10.1111/j.1348-0421.1985.tb00915.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Csanaky IL, Cheng X, Lehman-McKeeman LD, Klaassen CD. Organic anion transporting polypeptide 1a1 null mice are sensitive to cholestatic liver injury. Toxicol Sci. 2012a;127:451–462. doi: 10.1093/toxsci/kfs123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hong JY, Rockwell CE, Copple BL, Jaeschke H, Klaassen CD. Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver Int. 2011;32:58–69. doi: 10.1111/j.1478-3231.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Klaassen CD. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J Lipid Res. 2010;51:3230–3242. doi: 10.1194/jlr.M007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Limaye PB, Lehman-McKeeman LD, Klaassen CD. Dysfunction of organic anion transporting polypeptide 1a1 alters intestinal bacteria and bile acid metabolism in mice. PLoS One. 2012b;7:e34522. doi: 10.1371/journal.pone.0034522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


