Abstract
Rationale
Discerning cardiac myocyte cell cycle behavior is challenging owing to commingled cell types with higher proliferative activity.
Objective
To investigate cardiac myocyte cell cycle activity in development and the early postnatal period.
Methods and Results
To facilitate studies of cell type–specific proliferation, we have generated tissue-specific cell cycle indicator BAC transgenic mouse lines. Experiments using embryonic fibroblasts from CyclinA2-LacZ-floxed-EGFP, or CyclinA2-EGFP mice, demonstrated that CyclinA2-βgal and CyclinA2-EGFP were expressed from mid-G1 to mid-M phase. Using Troponin T-Cre;CyclinA2-LacZ-EGFP mice, we examined cardiac myocyte cell cycle activity during embryogenesis and in the early postnatal period. Our data demonstrated that right ventricular cardiac myocytes exhibited reduced cell cycle activity relative to left ventricular cardiac myocytes in the immediate perinatal period. Additionally, in contrast to a recent report, we could find no evidence to support a burst of cardiac myocyte cell cycle activity at postnatal day 15.
Conclusions
Our data highlight advantages of a cardiac myocyte–specific cell cycle reporter for studies of cardiac myocyte cell cycle regulation.
Keywords: cardiac myocyte, cell cycle, cyclins, heart development, proliferation
Cell cycle regulation is a key factor in disease and regeneration. 1–4 Healing responses to injury often require cell cycle re-entry of tissue parenchyma and cognate vascular stro-mal fraction, comprising endothelial cells, vascular mural cells, and fibroblasts.1,3 Tissues that heal well in response to injury are able to replenish both tissue parenchymal cells and vascular stroma because of the ability of these cells to re-enter the cell cycle, or to be provided by tissue-resident stem cells.1,4,5 Some tissues, however, are composed of parenchymal cells that are refractory to cell cycle re-entry, and a major goal of regenerative medicine is to understand factors regulating cell cycle within these cell types toward manipulations that may promote cell cycle re-entry. One such tissue is the heart, where adult cardiac myocytes are withdrawn from the cell cycle.6,7
Understanding cardiac myocyte cell cycle regulation lies at the heart of regenerative therapies for cardiac diseases and has been the subject of intensive study. One of the great challenges to this field is to clearly define the myocyte identity of proliferating cells.8 This is particularly challenging because myocytes comprise less than one half of all cell types within the heart9 and are closely intermingled with multiple other cell types of smaller size, including fibroblasts, pericytes, vascular smooth muscle cells, and endothelial cells.
Studies of cell cycle regulation can be facilitated by cell cycle reporter transgenes. Previous cell cycle reporter mice have been generated by in-frame fusion of fluorescent reporter transgenes to genes encoding protein fragments that are destabilized at discrete stages of the cell cycle.10 Initial iterations of this kind of reporter were generated using a cytomegalo-virus-based enhancer, were subject to issues with transgene expression, and were not cell type–specific, although in more recent iterations, tissue conditional expression is possible.11,12 Investigation of cardiac myocyte proliferation with these reporters was recently reported.13 Here, we undertook a distinct approach and generated a CyclinA2-reporter fusion protein under the control of the endogenous CyclinA2 locus. We adopted this strategy because CyclinA2 is an essential regulator of cell cycle expressed in all cycling cells from S to M phase,14 reasoning that this approach would give us a robust and sensitive reporter of cell cycle activity in any cell type of interest.
To facilitate studies of cell type–specific cell cycle behavior, we generated a BAC transgenic line, CyclinA2-lacZ-EGFP, that contained a transgene encoding a lacZ fusion in frame to the C-terminal coding sequence of cyclinA2. In these mice, the CyclinA2-lacZ fusion reporter is expressed under control of the cyclinA2 locus within BAC sequences and, on Cre-mediated excision, is converted to a cyclinA2-EGFP fusion gene, expressed under the control of the cyclinA2 locus only in the cell type that expressed Cre.
In this report, we demonstrate and characterize cell cycle–specific expression of cyclinA2-lacZ and cyclinA2-EGFP fusion reporter transgenes and use this new tool, in concert with a cardiac myocyte–specific Cre, Troponin T (TnT)-Cre,15 to uncover unexpected findings concerning cardiac myocyte cell cycle regulation.
Methods
Transgenic Animals
All animals were maintained and experiments performed in accordance with institutional guidelines at University of California, San Diego. Protamine-Cre16 and TnT-Cre15 were purchased from Jackson Laboratories. CyclinA2-EGFP allele was produced by crossing CyclinA2-LacZ-EGFP with Protamine-Cre line. CyclinA2-LacZ-EGFP transgenics have been bred into a Black Swiss outbred background for >15 generations and has not exhibited gene silencing. Genotyping for transgene was performed by PCR, using following primers, forward primer: 5′-CAGCCACAACGTCTATATCATGGC-3′, reverse primer: 5′-TTGTCTGTGGCTATACCATC-3′, giving rise to 405bp amplicon.
Generation of BAC Transgenic Mice
BAC clone RP23-297G4 containing cyclinA2 was purchased from BACPAC Resources Center. BAC recombineering was performed with galK selection, and recombineering was performed as detailed (https://ncifrederick.cancer.gov/research/brb/protocol.aspx). After linearization with AscI, BAC DNA was purified by Sepharose column fractionation. DNA purity was examined by pulse field gel electrophoresis, and a fraction with highest purity was used for pronuclear injection.
Southern Blotting
Genomic DNA was purified from tail tip biopsies, followed by digestion with NcoI. Digested genomic DNA was separated by agarose gel electrophoresis. Short sequence of vector backbone adjacent to linearized AscI site was used as a probe (Figure 1A, pink bar). The probe was amplified by PCR using following primers, forward primer: 5′-TAAAGTAGTGGTAATACTCCTGCTTAC-3′, reverse primer: 5′-TCTATCTGCTACATAACTTACTT-3′.
Figure 1. A, Diagram of CyclinA2 indicator bacterial artificial chromosome (BAC) transgene.
A floxed lacZ cassette and an enhanced green fluorescent protein (EGFP) cassette were inserted in frame immediately before the cyclinA2 stop codon. B, CyclinA2-LacZ will be expressed under control of cyclinA2 sequences on the BAC. TnT-Cre;CyclinA2-LacZ-EGFP mice express CyclinA2-EGFP selectively in cardiac myocytes. C, Southern blot confirming transgene integration.
Cell Culture and Cell Synchronization
Mouse embryonic fibroblasts (MEFs) were isolated by trypsinization of skin tissues of embryonic day 14.5 mouse embryos. MEFs were maintained in DMEM (Gibco), 10% fetal bovine serum at 37°C in 5% CO2. MEFs were synchronized in G0 phase by culturing in DMEM, 0.1% fetal bovine serum at 37°C in 5% CO2 for 72 hours.
Immunofluorescence
Embryos were fixed in 4% paraformaldehyde and embedded in Tissue-Tek OCT after sucrose gradient treatment. After denaturation with 1% SDS for 5 minutes, frozen sections were blocked with 10% donkey serum, 3% skim milk, 0.1% Triton, donkey anti-mouse IgG Fab fragment (715-007-003, Jackson Immunoresearch, 1:100), followed by incubation with following primary antibodies. Primary antibodies used for immunohistochemistry were anti-TnT mouse monoclonal (13-11, Thermo Scientific, 1:200), anti-GFP rabbit polyclonal (ab290, Abcam, 1:400), anti-Serine28 phosphoHistone H3 rat monoclonal (HTA28, BD Pharmingen, 1:25), anti-Serine 10 phosphoHistone H3 rabbit polyclonal (06-570, Millipore, 1:200), anti-α-actinin (Sarcomeric) mouse monoclonal (EA53, Sigma, 1:100), anti-PDGFRα goat poly-clonal (AF1062, R&D Systems, 1:100), anti-CD31 rat monoclonal (ME13.3, BD Pharmingen, 1:50), anti-CD45 rat monoclonal (30-F11, eBioscience, 1:50), or anti-CD146 rat monoclonal (ME-9F1, BioLegend, 1:50). Secondary antibodies used were Alexa 488, 555, or 647 anti-rabbit, mouse, rat, or goat IgG (Life Technologies), followed by nuclear staining with DAPI. Stained sections were mounted with Dako fluorescence mounting medium and visualized using an Olympus confocal microscope (FV1000). 5-Ethynyl-2′-deoxyuridine (EdU; 30 μg/g, dose per mouse body weight; Life Technologies) was injected 2 to 10.5 hours before dissection and detected with the Click-iT EdU Alexa Fluor 647 Imaging Kit (Life Technologies). EdU was intraperitoneally injected in pregnant dams and adult mice or subcutaneously injected in the back of postnatal day (PN) 0 and PN1 mice.
Quantitative Analysis
Quantitative analysis was performed using Image J software or Volocity software. To determine the number of myocardial nuclei of postnatal heart tissue sections, nuclei surrounded by staining with anti-PDGFRα, anti-CD31, anti-CD45, or anti-CD146 antibody were subtracted from total number of nuclei.
Results
Using BAC recombineering technology with galK selection,17 a BAC plasmid containing 83.2 kbp upstream and 87.4 kbp downstream of the cyclin A2 transcription start site was engineered to insert a lacZ cassette flanked by loxP sites, followed by an EGFP cassette in frame immediately before the stop codon of cyclin A2 (Figure 1A). Thus, the cyclinA2-lacZ fusion gene would be expressed under control of the cyclin A2 locus as contained within BAC sequences. On Cre-mediated excision, CyclinA2-EGFP would be expressed under control of the cyclinA2 locus only in lineages that had expressed Cre (Figure 1B). After linearization, recombineered BAC DNA was injected into pronuclei (Online Figure IA). Southern blot screening confirmed 2 independent mouse lines (CyclinA2-lacZ-EGFP; Figure 1C).
To define phases of cell cycle in which CyclinA2-EGFP was expressed, a CyclinA2-EGFP mouse line was generated by germline ablation of the lacZ cassette in cyclinA2-lacZ-EGFP, using Protamine Cre.16 In this line, CyclinA2-EGFP should be expressed under control of the cyclinA2 locus within BAC sequences. MEFs were harvested from CyclinA2-EGFP embryos. MEFs were synchronized in G0 by serum starvation,18 then induced to re-enter cell cycle by addition of 10% fetal bovine serum in the presence of EdU (12 μM; Figure 2A). Cultures were fixed with 4% paraformaldehyde every 6 hours, then immunostained with antibodies to GFP, the M phase–specific marker, Serine 28 phosphohistone 3,19 and stained for EdU, with DAPI nuclear staining. Attempts to visualize CyclinA2-EGFP expression by live imaging were not successful, for reasons which are not clear, but may be owing to low levels of transgene expression or quenching of EGFP within the context of the CyclinA2 fusion protein.
Figure 2.
A, Diagram of time-course experiment shown in B. B, Time-course experiment after synchronization of CyclinA2-enhanced green fluorescent protein (EGFP) mouse embryonic fibroblasts (MEFs). Cells were immunostained with antibodies to green fluorescent protein (GFP), Serine 28 phosphohistone 3 (S28pH3), and stained for 5-ethynyl-2′-deoxyuridine (EdU) and with DAPI. Scale bar: 100 μm. C, Fluorescence microscope images of CyclinA2-EGFP MEFs in mid G1 (a1–4), S (b1–4), metaphase (c1–4), and telophase (d1–4). Note cytosolic dispersion of CyclinA2-EGFP in metaphase (c2) and its disappearance in telophase (d2). Scale bar: 50 μm. D, Quantitative analysis of CyclinA2-EGFP or EdU-positive cells at each time point. E, Quantitative analysis of CyclinA2-EGFP/EdU double- or single-positive cells at each time point. F, Schematic diagram of CyclinA2-EGFP expression and EdU incorporation throughout cell cycle.
As shown in Figure 2B, MEFs initiated expression of CyclinA2-EGFP by 18 hours after serum induction (Figure 2B, c2), at mid-G1 when no cells yet demonstrated EdU incorporation (Figure 2B, c3). EdU incorporation, marking S phase, was evident by 24 hours after induction, and all cells marked by EdU were also positive for CyclinA2-EGFP (Figure 2B, d2, d3). By 30 hours after induction, the M phase marker, Serine 28 phosphohistone 3, was observed (eg, in cells undergoing telophase as shown in Figure 2B, e4, yellow ellipses). CyclinA2-EGFP expression was no longer evident in cells at telophase (Figure 2B, e2, yellow ellipses). This observation was consistent with CyclinA2-EGFP dispersion throughout the cytosol during metaphase (Figure 2C, c2) and its absence at late telophase (Figure 2C, d2). Altogether, the foregoing indicated that CyclinA2-EGFP was expressed from mid-G1 to mid-M phase.
Quantitative analysis revealed dynamics of CyclinA2 and EdU labeling (Figure 2D and 2E). CyclinA2(+)/EdU(−) cells reached a peak at mid-G1 phase, followed by a peak of CyclinA2(+)/EdU(+) cells at S phase. At M/G0, CyclinA2(+)/ EdU(−) cells began to disappear, whereas CyclinA2(−)/EdU(+) cells began to appear. From foregoing data, as schematically diagrammed in Figure 2F, CyclinA2-EGFP appeared somewhat earlier than EdU and disappeared at telophase, whereas, as expected, EdU label remained after cell division, indicating that CyclinA2-EGFP provided a more sensitive and accurate read-out for current cell cycle activity than EdU.
To examine dynamics of CyclinA2-β-galactosidase expression, MEFs were harvested from CyclinA2-lacZ-EGFP embryos and used for a time-course experiment, performed as described earlier. After fixation, cultures of MEFs were stained with X-gal and immunostained with Ser10 anti-phosphohistone 3 antibody, marking G2 to M nuclei,20 and DAPI nuclear staining. As observed for CyclinA2-EGFP and as shown in Figure 3A, CyclinA2-β-galactosidase was first evident at mid-G1 (Figure 3A, c2) and remained evident through S/G2 phase (Figure 3A, d2, e2). At metaphase, CyclinA2-β-galactosidase was dispersed throughout the cytosol, (Figure 3A, e2, white circles and Figure 3B, c2; and Online Figure II, b2) and was no longer evident in telophase (Online Figure II, c2). Therefore, timing of CyclinA2-β-galactosidase expression during cell cycle mirrored that of CyclinA2-EGFP.
Figure 3. A, Time-course experiment after synchronization of CyclinA2-LacZ-enhanced green fluorescent protein (EGFP) mouse embryonic fibroblasts (MEFs).
MEFs were stained with X-gal, followed by immunostaining with anti-Serine10 phosphohistone 3 antibody (S10pH3) and staining with DAPI. Cells begin to express CyclinA2-βgal at mid G1 (c2). Scale bar: 50 μm. B, Images of CyclinA2-LacZ-EGFP MEFs at G0 (a1–3), S (b1–3), and M phase (c1–3). Cells were stained with X-gal, followed by immunostaining for S10pH3 and staining with DAPI. CyclinA2-β-galactosidase (β-gal) was expressed during S phase (b2) and dispersed within cytosol at M phase (c2). C, Diagram of time-course experiment shown in D. D, Time-course experiment after serum starvation of CyclinA2-EGFP MEFs. MEF cells were stained for CyclinA2-EGFP, S28pH3, and DAPI. CyclinA2-EGFP disappeared at telophase after serum starvation (b1–3, white circles), then completely disappeared (c1–3, d1–3). Scale bar: 100 μm.
Next, to further verify that CyclinA2-EGFP expression was not carried over from one cell cycle to the next, MEFs were synchronized in G0 by serum starvation, then induced to re-enter cell cycle by addition of 10% fetal bovine serum. At 24 hours after stimulation (t=0, Figure 3D, a1–3), when most cells were in S phase, MEFs were again subjected to serum starvation to arrest cell cycle (Figure 3C). At t=0, as expected, all cells expressed CyclinA2-EGFP (Figure 3D, a2). However, 6 hours after serum withdrawal, most cells had ceased CyclinA2-EGFP expression, reflecting entry into late M/G0 (Figure 3D, b1–3), although cells in M phase, including early telophase as marked by Serine 28 phosphohistone 3, still exhibited faint transgene expression (Figure 3D, b2, white ellipse). At 12 hours after serum withdrawal, CyclinA2-EGFP had disappeared completely (Figure 3D, c2, d2). Thus, CyclinA2-EGFP had disappeared by late telophase/G0.
To understand cardiac myocyte cell cycle regulation during heart development, CyclinA2-lacZ-EGFP mice were crossed with a cardiac myocyte–specific Cre mouse line, TnT-Cre.15 To compare in vivo expression of our cell cycle indicator with EdU labeling, pregnant dams were injected intraperitoneally with EdU (30 μg/g; dose per body weight) 2 hours before harvest. Embryos were harvested at embryonic day 10.5, 12.5, 14.5, and 18.5, fixed with 4% paraformaldehyde, and immunostained with antibodies to GFP, TnT, stained for EdU, and with DAPI (Figure 4A).
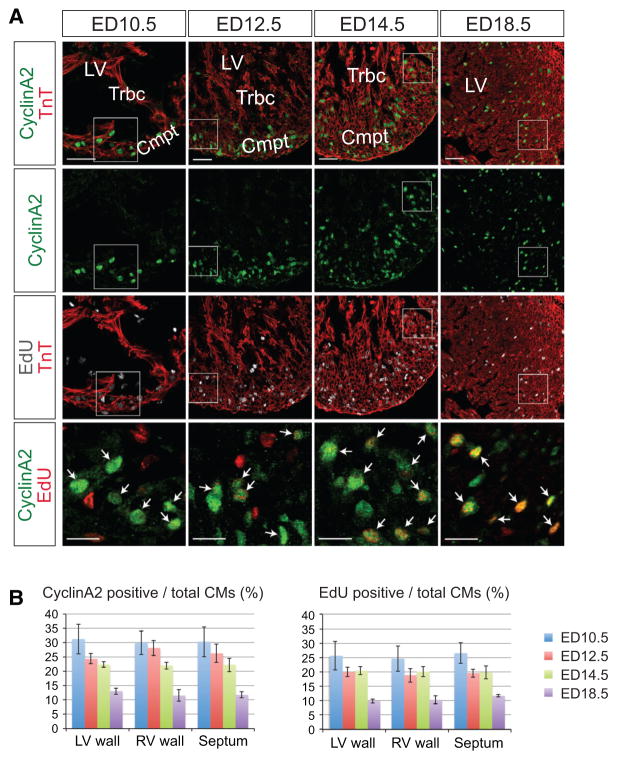
Figure 4. A, Fluorescence microscopy of embryonic day (ED) 10.5, ED12.5, ED14.5, and ED18.5 heart sections of Troponin T (TnT)-Cre;CyclinA2-LacZ-EGFP mice.
Sections were immunostained for CyclinA2-EGFP and Troponin T and stained for 5-ethynyl-2′-deoxyuridine (EdU) and with DAPI. CyclinA2-EGFP expression was largely confined to the compact layer. CyclinA2-EGFP/EdU double-positive cells are indicated by white arrows. For lower magnification (upper three panels), scale bar denotes 50 μm; for higher magnification images (lower panel), scale bar denotes 20 μm. B, Quantitative analysis of CyclinA2-EGFP and EdU-positive myocardial cells at ED10.5, ED12.5, ED14.5, and ED18.5. Myocardial cells were defined by Troponin T staining. Note gradual and steady decrease of CyclinA2-EGFP- and EdU-positive myocardial cells. Cmpt indicates compact layer; and Trbc, trabecula.
As expected,13,21,22 at embryonic day 10.5, 12.5, and 14.5, CyclinA2-EGFP was selectively expressed in the ventricular compact zone, not in trabeculae. Most CyclinA2-EGFP-positive cardiac myocytes overlapped with EdU staining (white arrows). Some CyclinA2-EGFP-positive cardiac my-ocytes were EdU-negative, as expected, because CyclinA2-EGFP is expressed from mid-G1 through M phase, whereas EdU is taken up only by cells undergoing S phase during the EdU pulse. From quantitative analysis, ≈81% of CyclinA2-EGFP-positive cardiac myocytes were also positive for EdU staining (Online Figure III). As expected, EdU staining was also observed in noncardiac myocyte lineages that were not stained for TnT. Thus, Cyclin A2-EGFP was expressed in a lineage-specific manner and allowed for sensitive detection of cycling cardiac myocytes.
Quantitative analysis of CyclinA2-EGFP expression during examined embryonic stages showed that, as previously found,6,21 the proliferation rate of cardiac myocytes gradually decreased with progressive development (Figure 4B, left panel). Quantitative analysis demonstrated a consistent profile of CyclinA2-EGFP expression and EdU labeling of cardiac myocytes, identified by TnT staining (Figure 4B, right panel). Overall, consistency of these results with previous studies of embryonic cardiac myocyte proliferation6,13 validated CyclinA2-EGFP as a cardiac myocyte–specific cell cycle indicator.
At birth, the circulatory system undergoes radical changes, profoundly affecting cardiac physiology.23 To examine proliferation of cardiac myocytes perinatally, EdU labeling was performed on TnT-Cre;CyclinA2-lacZ-EGFP mice by subcutaneous injection 4 hours before harvest. Hearts were harvested at postnatal day 0 (PN0), 1 (PN1), 5 (PN5), and 10 (PN10), fixed with 4% paraformaldehyde, sectioned, and immunostained with antibodies to GFP and stained for EdU with DAPI nuclear staining (Figure 5A). Identification of myocyte nuclei in the context of postnatal heart tissue is challenging.8 Because of the large area comprised by individual cardiomyocytes relative to other cardiac cell types, immunostaining for highly expressed myocyte cytoskeletal markers, such as TnT, can appear to encompass nuclei that are in fact nonmyocyte. Therefore, to identify myocyte nuclei, we used a negative staining protocol, using a cocktail of antibodies that comprehensively detected nonmyocyte cell types within the heart, including fibroblasts (PDGFRα), endothelial cells (CD31 and CD146), blood cells (CD45), and vascular support cells (CD146).24,25 Results of this analysis demonstrated that, as expected,6 the number of CyclinA2-EGFP-positive cardiac myocytes was reduced between PN0 and PN1 and further reduced by PN10. Intriguingly, during the immediate perinatal period (PN0-PN1), we noted fewer cardiac myocytes expressing CyclinA2-EGFP in right ventricle relative to left ventricle (LV) or interventricular septum. As seen during embryonic stages, from stages examined from PN1 to PN10, most CyclinA2-EGFP cardiac myocytes exhibited EdU staining. As expected, however, a majority of EdU staining at these stages was observed within noncardiac myocyte populations.6
Figure 5. A, Fluorescence microscopy of heart sections from postnatal day 0 (PN0), day 1 (PN1), day 5 (PN5), and day 10 (PN10) TnT-Cre;CyclinA2-LacZ-EGFP mice.
Sections were immunostained with antibodies for CyclinA2-EGFP, a cocktail of antibodies to PDGFRα, CD31, CD45, and CD146 stained for 5-ethynyl-2′-deoxyuridine (EdU) and DAPI. Images of left ventricle (LV), right ventricle (RV), and interventricular septum (IVS) are shown. CyclinA2-EGFP/EdU double-positive cells are indicated by white arrows. Note reduced number of CyclinA2-EGFP- or EdU-positive cardiac myocyte nuclei in RV relative to LV or IVS at PN0 and PN1. Scale bar: 50 μm. B, Quantitative analysis of CyclinA2-EGFP- and EdU-positive myocardial nuclei at PN0, PN1, PN5, and PN10. Myocardial cells were defined by excluding nonmyocardial cells labeled with either PDGFRα, CD31, CD45, or CD146. Note that all CyclinA2(−)/EdU(+) cells were labeled with either PDGFRα, CD31, CD45, or CD146.
Quantitative analysis of CyclinA2-EGFP and EdU labeling of cardiac myocyte nuclei during perinatal stages (Figure 5B) showed expected overall agreement between CyclinA2-EGFP and cardiac myocyte EdU labeling. Quantitative analyses confirmed lower rates of cell cycle activity of right ventricle cardiac myocytes relative to LV and interventricular septum cardiac myocytes at PN0 and PN1 (Figure 5B, right panel).
Binucleation of cardiac myocytes, where they undergo a round of cell cycle with karyokinesis but not cytokinesis, occurs from PN4 to PN7,6 suggesting that CyclinA2-EGFP expression at PN5 reflected cell cycle activity during binucleation. To address this, PN5 cardiac myocytes were isolated and plated on glass slides, immunostained with antibodies to GFP and sarcomeric-α-actinin, stained for EdU, and with DAPI. Quantitative analysis with isolated cardiac myocytes (n=451) demonstrated that ≈60% of myocytes at this stage were mononucleated and ≈35% were binucleated (Figure 6A). Approximately 14% of mono-nucleated cardiac myocytes expressed CyclinA2-EGFP, whereas only 2% of binucleated cardiac myocytes were positive for CyclinA2-EGFP (Figure 6B). The extent of EdU labeling of either mononucleated or binucleated cardiac myocytes was again in overall agreement with CyclinA2-EGFP expression (Figure 6B). Representative mononucleated and binucle-ated cardiac myocytes isolated at PN5 are shown (Figure 6C). Together, these results indicated that, as expected, CyclinA2-EGFP was expressed during cell cycle events required for bi-nucleation, but disappeared after binucleation.
Figure 6. A, Quantitative analysis of the fraction of mononucleated and binucleated cardiac myocytes from PN5 TnT-Cre;CyclinA2-LacZ-enhanced green fluorescent protein (EGFP) hearts.
A total of 451 cardiac myocytes were subjected to quantification. B, Quantitative analysis of the percentage of mononucleated or binucleated cardiac myocytes that were positive for CyclinA2-EGFP or 5-ethynyl-2′-deoxyuridine (EdU). C, Fluorescence microscopy of cardiac myocytes isolated from PN5 TnT-Cre;CyclinA2-LacZ-EGFP hearts. Cells were immunostained for CyclinA2-EGFP, sarcomeric-α–actinin, stained for EdU and DAPI. Scale bars: 20 μm.
Recent results suggested that a proliferative burst of ventricular cardiac myocytes occurs at PN15.26 If this were the case, our CyclinA2-EGFP should be expressed accordingly in cardiac myocytes at this stage. To investigate this, and guided by protocols used in the previous study,26 EdU (30 μg/g) was injected into TnT-Cre;CyclinA2-lacZ-EGFP mice at 11:30 am or 9:30 pm on PN14 or 11:30 am on PN15. Hearts were harvested 10.5 hours after each EdU injection, at 10 pm on PN14, 8 am on PN15, or 10 pm on PN15. Heart tissue sections were immunostained with antibodies to GFP, TnT, and stained for EdU, and with DAPI. Extremely rare Cyclin A2-EGFP cardiac myocytes were observed, some of which were also positive for EdU label (Figure 7A). As expected, the majority of EdU-positive cells were labeled by the cocktail of antibodies to noncardiac myocyte markers (Figure 7B, blue circles). Quantitative analysis revealed that the number of CyclinA2-EGFP-positive cells per total cardiac myocytes gradually decreased in the range from 0.025% to 0.010% throughout PN14 and PN15 (Figure 7C). These numbers are consistent with quantitative analysis of cardiac myocyte proliferation rates in young adult mice using stable isotope labeling.27 We further investigated rates of cardiac myocyte cell cycle activity from PN14 to PN20 by quantification of CyclinA2-EGFP- and EdU-labeled cells. As shown in Online Figure IV, rates of cardiac myocyte cell cycle activity gradually and steadily decreased from PN14 throughout PN20.
Figure 7. A, Fluorescence microscopy of heart sections from TnT-Cre;CyclinA2-LacZ-enhanced green fluorescent protein (EGFP) mice at PN14 and PN15 stained for CyclinA2-EGFP, Troponin T, 5-ethynyl-2′-deoxyuridine (EdU), and DAPI.
Mice were labeled with EdU during indicated time frames. CyclinA2-EGFP-positive cells were extremely rare. A single CyclinA2-EGFP-positive cardiac myocyte in the field is shown (white circle). CyclinA2-EGFP-positive but EdU-negative cardiac myocytes were also observed. Scale bar: 50 μm. B, Fluorescence microscopy of heart sections from TnT-Cre;CyclinA2-LacZ-EGFP mice at PN14 stained for CyclinA2-EGFP, PDGFRα, CD31, CD45, CD146, EdU, and DAPI. All EdU-positive/ CyclinA2-EGFP-negative cells were labeled with either anti-PDGFRα, anti-CD31, anti-CD45, or anti-CD146 antibody (blue circles). Scale bar: 50 μm. C, Quantitative analysis of CyclinA2-EGFP myocardial cells throughout PN14 and PN15. Note gradual and steady decrease.
To ensure that CyclinA2-lacZ-EGFP transgenes could be expressed at this stage, EdU labeling of Protamine-Cre;CyclinA2-EGFP mice was performed by peritoneal injection 6 hours before harvest, and hearts were harvested at PN15. CyclinA2-EGFP-positive cells that colocalized with EdU staining were observed within the LV wall at PN15 (Online Figure VA). Sections of PN15 hearts from CyclinA2-lacZ-EGFP mice were also stained for X-gal. CyclinA2-β-galactosidase cells were observed scattered throughout the heart (Online Figure VB). Thus, CyclinA2-LacZ and -EGFP transgenes could be expressed at these stages.
Discussion
In summary, we have generated a CyclinA2-EGFP transgene that reports cell cycle activity in a Cre-dependent manner, providing sensitive readout of cell cycle activity in cardiac myocytes or other cell types of interest. One advantage of our indicator relative to recently described tissue-specific FUCCI (fluorescent ubiquitination-based cell cycle indicator) indicators11,12 is that our indicator relies on a single fluorescence marker to report cell cycle activity, leaving other fluorescence channels available for additional markers. CyclinA2-EGFP was expressed from mid-G1 to mid-M phase, similar to endogenous cyclinA2,14,28 although slightly earlier and later, perhaps owing to enhanced sensitivity of detection or transgene stability. Using TnT-Cre, we compared cardiac myocyte–specific expression of CyclinA2-EGFP to results with EdU labeling. Owing to tissue complexity, attribution of cardiac myocyte–specific EdU staining is extremely challenging.8 In contrast, CyclinA2-EGFP provided a robust and sensitive readout of cardiac myocytes actively engaged in cell cycle activity. Additionally, the CyclinA2-EGFP reporter indicates cells that are actively engaged in cell cycle at the time of harvest, whereas EdU label can also be present in cells that have exited cell cycle. Another potential advantage to our reporters is that nucleosides and nucleoside analogs like thymidine and EdU are also incorporated during DNA repair consequent to DNA damage, whereas CyclinA2 is activated during bona fide cell cycle activity. This issue may be of particular relevance in the perinatal period, where cardiac myocytes exhibit increased DNA damage in response to increased oxidative stress, promoting cell cycle withdrawal.29
Using CyclinA2-EGFP, we discovered that rates of cell cycle activity in right ventricular cardiac myocytes at P0 and P1 were reduced relative to those of LV cardiac myocytes. This differential rate of cardiac myocyte cell cycle activity might, at least in part, contribute to differences in size between postnatal right ventricle and LV and may reflect relative increased pressures experienced by the LV and interventricular septum after birth.23
Utilization of the cardiac myocyte–specific cell cycle indicator allowed us to re-examine recent findings of a burst of myocyte cell cycle re-entry at PN15.26 Examination of CyclinA2-EGFP expression consequent to TnT-Cre activity, in concert with EdU labeling, demonstrated that cardiac myocytes do not exhibit a burst of proliferative activity at PN15. These findings serve to illustrate the challenges of attributing observed cell cycle activity to cardiac myocytes within complex cardiac tissue in the absence of a clear myocyte-specific indicator, as now provided by this new cell-specific cell cycle reporter. We also describe the development of a complementary method to negatively define cardiac myocyte nuclei by utilization of a cocktail of antibodies that comprehensively marks nonmyocyte populations within the heart.
In this report, we have focused on the utility of our reporter for studying cardiac myocyte cell cycle activity. It should be noted that the CyclinA2-lacZ-EGFP transgene will also have great utility in the context of other cell types during development and in the adult and in the context of both regenerative medicine and cancer.
Supplementary Material
Novelty and Significance.
What Is Known?
Understanding and promoting postnatal cardiac myocyte cell cycle regulation is a major goal of regenerative therapies for heart disease.
Studies of myocyte proliferation in postnatal heart are challenging owing to tissue complexity.
What New Information Does This Article Contribute?
We characterize a novel cell-type–specific cell-cycle reporter and use it to examine cardiac myocyte proliferation.
We found reduced proliferation of right ventricular cardiac myocytes relative to left ventricular cardiac myocytes in perinatal period.
In contrast to a recent study, there was no proliferative burst of cardiac myocytes at postnatal day (PN) 14 or PN15.
We have developed a new transgenic mouse line that enables visualization of cell cycle activity in specific cell types, including cardiac myocytes, consequent to Cre activity. Characterization of CyclinA2-floxed:LacZ-GFP mice demonstrated excellent agreement between pulse EdU labeling and CyclinA2-reporter expression. CyclinA2-fusion reporters were expressed from mid-G1 to late-M phase. Unexpectedly, studies using the Troponin T-Cre;CyclinA2-floxed:LacZ-GFP mice revealed reduced proliferation of right ventricular cardiac myocytes relative to left ventricular cardiac myocytes in the perinatal period. In contrast to a recent study, studies with Troponin T-Cre;CyclinA2-floxed;LacZ-GFP mice showed no proliferative burst of cardiac myocytes at PN14 or PN15. CyclinA2-floxed;LacZ-GFP reporter mice can be used to selectively visualize proliferating cardiac myocytes during embryonic development and in the postnatal period, addressing a current limitation in the field.
Acknowledgments
We thank L. Field, P. Cattaneo, N. Guimarães-Camboa for helpful discussions and D. Savic, M. Nobrega, T. Moore-Morris for technical advice.
Sources of Funding
This work was supported by grants from National Institutes of Health (NIH), DP1HL117649 and R01HL123747, to S.M. Evans, and Daiichi Sankyo Foundation of Life Science and Uehara Memorial Foundation to M. Hirai. Imaging was supported by UCSD Neuroscience Microscopy Shared Facility Grant P30 NS047101.
Nonstandard Abbreviations and Acronyms
- EdU
5-ethynyl-2′-deoxyuridine
- LV
left ventricle
- MEF
mouse embryonic fibroblast
- PN
postnatal day
- TnT
Troponin T
Footnotes
Disclosures
None.
References
- 1.DeLeve LD. Liver sinusoidal endothelial cells and liver regeneration. J Clin Invest. 2013;123:1861–1866. doi: 10.1172/JCI66025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chien KR, Domian IJ, Parker KK. Cardiogenesis and the complex biology of regenerative cardiovascular medicine. Science. 2008;322:1494–1497. doi: 10.1126/science.1163267. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J Clin Invest. 2010;120:11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiac myocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 7.Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90:1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- 8.Mark HS, Michael R, Loren JF. Challenges measuring cardiac myocyte renewal. Biochimica et Biophysica Acta (BBA) Molecular Cell Research. 2012:1833. [Google Scholar]
- 9.Senyo SE, Lee RT, Kühn B. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. 2014;13:532–541. doi: 10.1016/j.scr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2007;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 11.Takaya A, Asako S-S, Hiroshi K, Go S, Ken-ichi I, Toshitaka H, Kazuki N, Atsushi M, Shinichi A, Toshihiko F. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development. 2013;140:237–246. doi: 10.1242/dev.084111. [DOI] [PubMed] [Google Scholar]
- 12.Richard Lester M, Matthew Jonathan F, Asako S-S, Nils Olof L, Angela C, Adam Thomas D, Margaret Anne K, Peter H, Atsushi M, Ian James J. Fucci2a: a bicistronic cell cycle reporter that allows Cre-mediated tissue-specific expression in mice. Cell Cycle. 2014;13:2681–2696. doi: 10.4161/15384101.2015.945381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto H, Yuasa S, Tabata H, et al. Time-lapse imaging of cell cycle dynamics during development in living cardiomyocyte. J Mol Cell Cardiol. 2014;72:241–249. doi: 10.1016/j.yjmcc.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Helfrid H, Shunichi T, Tim H. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 15.Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BL. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson AM, White LS, Donovan PJ, Piwnica-Worms H. Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol Cell Biol. 2005;25:2853–2860. doi: 10.1128/MCB.25.7.2853-2860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J Cell Sci. 2003;116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- 21.de Boer BA, van den Berg G, de Boer PA, Moorman AF, Ruijter JM. Growth of the developing mouse heart: an interactive qualitative and quantitative 3D atlas. Dev Biol. 2012;368:203–213. doi: 10.1016/j.ydbio.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Chen H, Qu X, Chang CP, Shou W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC) Am J Med Genet C Semin Med Genet. 2013;163C:144–156. doi: 10.1002/ajmg.c.31369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudolph AM. The changes in the circulation after birth. Their importance in congenital heart disease. Circulation. 1970;41:343–359. doi: 10.1161/01.cir.41.2.343. [DOI] [PubMed] [Google Scholar]
- 24.Moore-Morris T, Guimarães-Camboa N, Banerjee I, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest. 2014;124:2921–2934. doi: 10.1172/JCI74783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Covas DT, Panepucci RA, Fontes AM, Silva WA, Jr, Orellana MD, Freitas MC, Neder L, Santos AR, Peres LC, Jamur MC, Zago MA. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Exp Hematol. 2008;36:642–654. doi: 10.1016/j.exphem.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Nawazish N, Ming L, John WC, et al. A proliferative burst during preadolescence establishes the final cardiac myocyte number. Cell. 2014;157:795–807. doi: 10.1016/j.cell.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erlandsson F, Linnman C, Ekholm S, Bengtsson E, Zetterberg A. A detailed analysis of cyclin A accumulation at the G(1)/S border in normal and transformed cells. Exp Cell Res. 2000;259:86–95. doi: 10.1006/excr.2000.4889. [DOI] [PubMed] [Google Scholar]
- 29.Puente BN, Kimura W, Muralidhar SA, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.