Abstract
The important role that cilia and flagella play in human disease creates an urgent need to identify genes involved in ciliary assembly and function. The strong and specific induction of flagellar-coding genes during flagellar regeneration in Chlamydomonas reinhardtii suggests that transcriptional profiling of such cells would reveal new flagella-related genes. We have conducted a genome-wide analysis of RNA transcript levels during flagellar regeneration in Chlamydomonas by using maskless photolithography method-produced DNA oligonucleotide microarrays with unique probe sequences for all exons of the 19,803 predicted genes. This analysis represents previously uncharacterized whole-genome transcriptional activity profiling study in this important model organism. Analysis of strongly induced genes reveals a large set of known flagellar components and also identifies a number of important disease-related proteins as being involved with cilia and flagella, including the zebrafish polycystic kidney genes Qilin, Reptin, and Pontin, as well as the testis-expressed tubby-like protein TULP2.
Keywords: cilia, flagella, polycystic kidney disease, RNA transcription
Cilia are sensory and motile structures that play critical roles in animal physiology and development. Ciliary defects have been implicated in several human genetic diseases, including polycystic kidney disease (PKD), Bardet-Biedl Syndrome (BBS), and primary ciliary dyskinesia (also known as Kartagener's Syndrome) (1). The symptoms of these diseases include physiological defects such as kidney failure, defective mucus clearance, neurosensory deficits, and obesity, as well as developmental defects such as situs inversus and polydactyly. To understand the role of cilia in development and physiology, it is essential to identify the gene products necessary for ciliary assembly and function. A proteomic analysis of human cilia has been presented (2), but such analyses are limited in their ability to detect low-abundance proteins. Two recent studies used comparative genomics to identify genes found only in the genomes of organisms that contain cilia and flagella (3, 4), and the set of genes thus identified is referred to as the flagellar and basal body proteome. Those bioinformatics studies detected several disease genes related to cilia, including several genes mutated in BBS patients. The success of the comparative genomics approach in identifying BBS genes suggests that analysis of the components of cilia and flagella would be an effective way to identify additional genes involved in ciliary diseases.
In this report, we take an alternative strategy for identifying ciliary components, by taking advantage of the fact that most known components of cilia and flagella are strongly induced during regeneration of flagella in Chlamydomonas reinhardtii (5). Chlamydomonas is a green alga with genetics similar to yeast, but unlike yeast, it has flagella that are virtually identical to human cilia. Most of the previously identified human ciliary disease genes have orthologs in Chlamydomonas that have been shown to be involved in flagellar assembly (1). Chlamydomonas cells can be induced to shed their flagella via katanin-mediated severing, after which the flagella immediately begin to regenerate. During this process, it has been shown that many known flagellar components are transcriptionally induced, with most transcripts reaching maximum accumulation between 30 and 45 min after deflagellation (5). In contrast, genes encoding components of other organelles do not show this induction in response to deflagellation. These observations, coupled with the recent sequencing of the Chlamydomonas genome (http://genome.jgi-psf.org/chlre2), suggest a systematic strategy for flagellar/ciliary gene identification. Specifically, we have conducted a genome-wide analysis of the RNA transcriptional profile observed during flagellar regeneration, to identify flagellar genes as those whose RNA transcription is activated in response to flagellar regeneration.
In addition to revealing genes that encode flagellar/ciliary components, we expect this approach to be able to identify genes whose products are involved in regulating flagellar assembly or function but that are not themselves components of cilia or flagella. In this sense, the RNA expression data provide information about potential cilia-related genes that is complementary to direct proteomic approaches, which can reveal only intrinsic components of the flagellum. The RNA transcriptional profiling approach is also complementary to the comparative genomics approaches referred to above, because it can reveal genes that are found in organisms lacking flagella but that nevertheless play important roles in flagellar assembly (e.g., tubulin). This type of approach is best suited to analyzing structures whose initial assembly is much faster than subsequent turnover, because such structures require a large initial burst of protein production. Flagellar turnover at steady state is ≈10 times slower than the rate of flagellar assembly during regeneration (6), suggesting that this is an ideal case for using transcriptional induction to identify organelle components. Gene function discovery by RNA transcriptional profiling tends to be most effective at identifying genes responsible for the development of new structures, such as in development (7), rather than identifying catalytic functions such as enzyme activities, which are typically not modulated in abundance by varying RNA transcription (8). In the case of flagella, even though turnover entails continuous assembly at the tip, the steady-state turnover is sufficiently smaller than the initial assembly rate. Therefore, we expect flagellar gene levels to be significantly higher during initial assembly compared with the subsequent steady state.
Materials and Methods
Design of Arrays. A set of 389,285 different 36-mer oligonucleotide probes were selected from the V2 genome sequence of C. reinhardtii that was available from the JGI web site (ftp://ftp.jgi-psf.org/pub/JGI_data/Chlamy/chlre2) in February 2004. The web site also listed 19,832 tentative predicted protein sequences generated by gene modeling software. The locations of these genes were identified by using both publicly available software [blast (9) and blat (10)] and software developed at the National Aeronautic and Space Administration (NASA) (available on request). Probes were selected for all exons of these proteins, and additional probes were chosen from the intergenic regions. The selected oligonucleotides were synthesized on a glass-based array by using an in situ maskless photolithographic synthesis device (11) and hybridized with mRNA extracted from C. reinhardtii.
Sample Labeling. RNA was extracted from the cells of wild-type (strain cc-124) C. reinhardtii cells grown 30, 45, or 120 min after removal of flagella by pH-mediated severing (12) as well as from normal cells that were not subjected to severing. Total RNA extraction was performed by lysing cells in TRIzol (Invitrogen) followed by phenol-chloroform extraction and ethanol precipitation. This RNA was converted to double-stranded cDNA by using a GIBCO/BRL SuperScript Choice System and mRNA was labeled by using an oligo(dT) primer containing the T7 RNA polymerase promoter (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-T24-3′). Briefly, 10 μg of total RNA was incubated with 1× first-strand buffer/10 mM DTT/500 μM dNTPs/5 pM primer for 60 min at room temperature. Second-strand synthesis was accomplished by incubation with 200 μM dNTPs/0.07 units/μl DNA ligase/0.27 units/μl DNA polymerase I/0.013 units/μl RNase/1× second-strand buffer/10 units of T4 DNA polymerase for 2 h. Double-stranded cDNA was purified by using phenol-chloroform extraction and Eppendorf Phase-Lock Gel tubes and ethanol precipitated, washed with 80% ethanol, and resuspended in 3 μl of water. In vitro transcription was used to produce biotin-labeled cRNA from the cDNA by using the Ambion (Austin, TX) MEGAscript T7 kit. Briefly, 1 μg of double-stranded cDNA was incubated with 7.5 mM ATP and GTP/5.6 mM UTP and CTP/1.9 mM bio-11-CTP and bio-16-UTP (Sigma-Aldrich) in 1× transcription buffer and 1× T7 enzyme mix for 5 h at 37°C. Before hybridization, cRNA was fragmented to an average size of 50-200 bp by incubation in 100 mM potassium acetate/30 mM magnesium acetate/40 mM Tris-acetate for 35 min at 94°C. For quality control at all steps, including input RNA quality, first- and second-strand cDNA synthesis, in vitro transcription, and fragmentation, assay performance was monitored by running small sample aliquots on the Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Hybridization and Washing. High-density 36-mer tiling arrays were hybridized with 12 μg of cRNA in 300 μl, in the presence of 50 mM Mes/0.5 M NaCl/10 mM EDTA/0.005% (vol/vol) Tween 20 for 16 h at 45°C. Before application, samples were heated to 95°C for 5 min, then 45°C for 5 min, and then clarified by centrifugation at 10,000 × g for 5 min. Hybridization was performed in a hybridization oven with continuous mixing. After hybridization, arrays were washed in nonstringent (NS) buffer [6× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA/0.01% Tween 20) for 5 min at room temperature, followed by washing in stringent buffer (100 mM Mes/0.01 M NaCl/0.01% Tween 20) for 30 min at 45°C. After washing, arrays were stained with streptavidin-Cy3 conjugate (Amersham Pharmacia) for 25 min at room temperature, followed by a 5-min wash in NS buffer, a 30-sec rinse in final rinse buffer, and a blow-dry step by using high-pressure grade 5 argon.
Array Scanning. Arrays were scanned on an Axon 4000B scanner and features were extracted by using nimblescan software (NimbleGen). Six data sets were generated for four time points, because the arrays for the normal state (initial state) and the state 120 min after pH shock (final state) were measured at two different photomultiplier tube settings. The raw data from this measurement have been deposited to the National Center for Biotechnology Information GEO database in the miame format (accession nos. GSE 2247; GSM 41324-41336; and GPL 1840-1842, 1844-1853).
Data Analysis. Before comparing data from multiple arrays, proper normalization was necessary to compensate for variations of average signal levels in different arrays. Probe signals were normalized by dividing them by the median intensity of all probes of the respective arrays. The logarithm (base 2) of the normalized numbers was used for subsequent analysis. For each protein-coding gene, the average expression level was computed by taking the median of log-normalized signals on all probes within the exon regions of the gene. Average expression levels for the genes are shown in Table 3, which is published as supporting information on the PNAS web site. Detailed plots of the expression levels for all of the genes are available upon request, where researchers can use our user-friendly graphics tools to plot signals at any region of interest. To decide whether a gene is expressed, we used signals on 244 nonmatching probes intentionally placed on our array. These probes did not match any region of the genome and therefore provided a reasonable estimate of the background level. A gene was considered expressed if its average expression level at any time point was >85% of the nonmatching probes. In addition, all genes that were >140% induced from normal level during flagellar regeneration were included in the list of expressed genes. Both of these groups had very large overlaps (>95%), and therefore second criterion added only a small fraction of the genes to the combined pool. In total, our study detected 8,078 genes (41% of all predicted genes) as expressed. This number lies between the number of verified genes of yeast Saccharomyces cerevisiae and of nematode Caenorhabditis elegans and is consistent with the genome complexity of the organism under study. Large numbers of undetected genes (11,725; 59%) can be explained by the limited accuracy of gene prediction methods for newly sequenced C. reinhardtii and are consistent with similar initial microarray study of other organisms such as C. elegans that detected only 56% of the predicted genes (13). It is probably not due to the sensitivity of our experimental procedure, because a similar technique has been used successfully to measure gene expressions for organisms of higher-complexity genomes (7, 14, 15), although it is possible that the unusually high GC content of the Chlamydomonas genome may result in decreased sensitivity.
Identification of Putative Consensus Motifs. Five hundred-base upstream regions of 122 most induced genes were considered in our search for putative promoter motifs using the standard programs mdscan (16) and meme (17). The number of considered genes (122 in total) was limited by the maximum file size limit specified by meme. For two pairs of genes (IDs 155875, 155853 and 170055, 169905) that had overlapping upstream regions, only one from each pair was taken. The search criteria used in mdscan were: motif width, 10; number of top sequences, 10; and number of candidate motifs for restsequence, 20. Search criteria used in meme were minimum width, 6; and maximum width, 15. Complete outputs from both programs are available upon request. It was also noticed that two of the most highly induced genes (protein IDs 155785 and 155853) shared the same 650-base upstream region. Therefore, all 8- to 12-base words present in either strand of this 650-bp region were compared against 500-base upstream regions of all genes by using a program developed by us (M.P.S., unpublished data). The top five words more likely to be present in the 147 most-induced genes than all other genes are given as the last set of entries in Table 2.
Table 2. Promoter motifs identified upstream of induced genes, using three different programs.
| Program | Consensus motif | International Union of Pure and Applied Chemistry |
|---|---|---|
| meme | CGCCCCACCCCGCCC | |
| ACATATCTTACTACA | ||
| TATCAACAGCA | ||
| CCCATGCA | ||
| ATTATATGACA | ||
| mdscan | CCCCCGCCCC | CCCSCGCCCC |
| GGGCGCGGGG | GGGCGSGGGS | |
| CGCCCGCGCC | CSBCCGCGSC | |
| TTCCAAGCCC | YYCCAAGCCC | |
| ATAGATATCA | ATASATATMA | |
| 650-bp region | TTCGGGAATACTC | |
| TAGAGCCAACGAG | ||
| GATTTTCCACGAT | ||
| ACTTTTAAAGCTA | ||
| GACGCTTCGAGAA |
Results
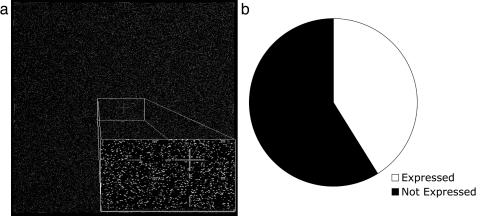
Genome-Wide Transcriptional Analysis of Flagellar Regeneration. Using maskless photolithographic DNA synthesis technology (7, 11, 18), we have constructed high-density DNA oligonucleotide microarrays to represent all exons from each strand of the Chlamydomonas genome. A total of 389,285 36-mer oligonucleotide probes, positioned within every exon, were selected to interrogate 19,803 genes at a feature density of ≈390,000 probes per array (Fig. 1). To measure transcriptional activity for all of the Chlamydomonas exons, the arrays were probed with fluorescence-labeled cDNA, reverse-transcribed from total RNA isolated from cells that were grown for 30, 45, and 120 min after deflagellation (12). A reference RNA sample was probed from the same culture before deflagellation.
Fig. 1.
Measurement of the transcriptional activity for all predicted C. reinhardtii.(a) Fluorescence micrograph of a maskless high-density oligonucleotide array. The array is designed to contain ≈390,000 36-mer features in a 14- × 17.4-mm area, with feature size of ≈15 μm. A control oligonucleotide probe is hybridized to an embedded set of features (Inset), which are used for array identification and extraction of experimental hybridization fluorescence data using software. (b) Analysis of raw data over all time points during flagellar regeneration shows that 41% of the predicted genes were expressed during this study. Median expression levels of all genes are provided in Table 3.
The reliability of the measured data was first verified (summarized in Table 1) by inspecting the RNA expression levels for 56 genes that were known to be induced according to previous studies (e.g., ref. 3). Thirty-five of those genes showed 175% or higher RNA expression levels during the period of flagellar regeneration compared with predeflagellation expression levels. In striking contrast, among another 54 genes previously shown not to be induced, not a single one had 175% or higher RNA expression levels relative to predeflagellation levels during the two intermediate time points (30 and 45 min) of flagellar regeneration. These data confirmed that the microarray method can reliably distinguish genes that are induced during flagellar regeneration experiments, with a very low false-positive rate. Gene lists used for this validation can be found in Tables 4, 5, 6, 7, 8, which are published as supporting information on the PNAS web site.
Table 1. Reference list for validation of known flagella-associated genes.
| Gene list | Induced genes | Total genes |
|---|---|---|
| Known induced | 35 | 56 |
| Known not induced | 0 | 54 |
| Known flagellar proteins | 33 | 61 |
| Known nonflagellar proteins | 0 | 20 |
| Random genes | 0 | 21 |
Results indicate the number of genes within each gene list that were induced by 175% or more relative to predeflagellation RNA expression levels. Gene lists and detailed results can be found in Table 3.
Transcriptional Induction Identifies Known Flagellar Genes. Having demonstrated that the Chlamydomonas microarray can reliably detect induced RNA transcripts, we next validated the strategy of identifying flagellar genes based on induction. As summarized in Table 1, we addressed RNA transcription levels of 61 known flagellar components, including genes encoding inner and outer dynein arms, radial spokes, intraflagellar transport components, and regulatory proteins. Of these 61 genes, 33 were found to be strongly induced (175% or higher RNA expression level during regeneration). In general, RNA expression levels of inner dynein arm components increased less dramatically than other flagellar components. By contrast, the ODA15 gene that codes for an outer-arm docking complex subunit was expressed up to seven times its normal level. As a general pattern, most of the induced flagellar genes had the strongest RNA expression level 30 min after the removal of flagella and then lapsed back to normal RNA expression levels 120 min after the removal of flagella. However, several genes still maintained a high level of RNA expression (although subdued from their peaks) at t = 120 min, indicating that, although flagellar regeneration was complete, the cell had not returned to steady-state conditions.
We also examined genes encoding 20 nonflagellar proteins, as well as 21 randomly chosen genes, and none of these genes showed induction, confirming the low false-positive rate in our analysis. Note that to guarantee a low rate of erroneous positive results, we have deliberately used a conservative criterion for RNA transcription level induction, which accounts for the high false-negative rate seen with known flagellar genes. Gene lists and detailed results can be found in Tables 4, 5, 6, 7, 8.
Having validated the strategy of identifying flagellar genes by microarray analysis of their induction during flagellar reassembly, we next explored the total set of genes within the Chlamydomonas genome that are induced in response to deflagellation. We classified any gene as “strongly induced” and hence potentially flagellar if it was induced at least 2-fold, which is an even more conservative criterion than that used in our initial validation experiments. An annotated list of all 220 strongly induced genes is given in Table 9, which is published as supporting information on the PNAS web site. Of the 220 strongly induced genes, 85 had previously been implicated with cilia and flagella, based either on localization studies, presence in the flagellar and basal body proteome, previous induction analysis, or proteomic analysis of human cilia (information about these classifications is given in Tables 4, 5, 6, 7, 8).
Among the genes whose RNA expression increased significantly during flagellar regeneration were cofactors of tubulin folding, including CPN2 and eight subunits of the T-complex protein 1 tubulin-folding factor (also known as CCT), previously shown to be involved with ciliary assembly (19). The elevated requirement for tubulin-folding chaperones in assembling a microtubule-based structure such as the flagellum likely explains the induction of these genes. These results support the idea that analysis of RNA transcription induction during flagellar assembly can identify genes involved in assembly that are not themselves flagellar components, which represents an important difference between our approach and proteomic analysis.
Heat-shock protein (HSP) molecular chaperones were also highly induced during flagellar regeneration. This is unlikely to be simply a generalized stress response, because several HSPs are known to localize within cilia and flagella. These include HSP70A∥ and HSP90A (2), both of which were strongly induced during flagellar regeneration in our experiment. It thus appears that molecular chaperones play important roles in flagellar and ciliary assembly, which is not surprising given the high complexity and continuous turnover of these large structures.
As indicated in Table 2, we have analyzed the upstream sequences corresponding to strongly induced genes. This analysis has yielded a set of putative regulatory motifs nonrandomly enriched upstream of flagellar genes, potentially indicating binding sites for transcription factors that regulate flagellar assembly.
For some genes, we detected an apparent repression of RNA transcription during flagellar regeneration. For instance, the RNA expression level of one G protein-coupled receptor was reduced almost 60% during flagellar regeneration but returned close to normal at t = 120 min, when flagellar regeneration was complete. Most control genes encoding chloroplast components showed similar down-regulation. Seven of the 10 genes encoding chloroplast-localized proteins, including rubisco and cytochrome components, showed >35% decrease in RNA expression levels, but none had a significant increase. This apparent repression of genes that are abundantly transcribed before deflagellation is not a statistical artifact and may represent a biologically significant activity that remains to be investigated.
Flagellar Induction of Zebrafish Polycystic Kidney Gene Orthologs. Having established that the transcriptional profiling strategy is effective in distinguishing known flagellar proteins from nonflagellar proteins, we next investigated whether any of the newly identified flagellar gene candidates might share sequence similarity with genes implicated in possible ciliary diseases. PKD is thought to result from defects in sensory cilia in the kidney, suggesting that PKD genes might be particularly likely to encode components of cilia and flagella. We found that our high-expression gene list (Table 9) contains the Chlamydomonas ortholog of the zebrafish gene qilin, which was cloned in a screen for zebrafish mutants that form kidney cysts similar to those seem in human patients with PKD (20). In contrast to another zebrafish cystic kidney gene scorpion (which we found to be moderately induced), zebrafish qilin mutants still formed cilia, raising doubts about whether the qilin gene product is actually involved with cilia, and whether the qilin mutation caused cystic kidneys by a cilia-independent mechanism. Because we found qilin to be strongly induced during flagellar regeneration, this suggests a connection with ciliary assembly or function. The RT-PCR analysis of Li et al. (3) also found this transcript to be induced during regeneration, although the identity of the gene as a polycystic kidney gene was not indicated. Similarly, a proteomic analysis of human cilia also identified a homolog of qilin, although this homology was not known when the cilia proteome was first annotated (2). These results strongly suggest that qilin is a ciliary protein. Given the lack of a ciliary assembly defect in qilin mutants in zebrafish, we propose that this gene might encode a protein that performs a sensory or regulatory function within cilia, without being required for ciliary assembly itself.
Two other potential PKD-related genes identified in zebrafish are pontin (20) and reptin (21), mutants in either of which cause cystic kidneys in fish. In mammals, pontin (also known as TIP49a or RuvBL1) is found in a complex with reptin (also known as TIP49b or RuvBL2), and the complex acts as a DNA helicase thought to be involved in gene regulation during development. Although both proteins are highly expressed in the testis (22), a major site of flagellar biosynthesis, the proteins themselves localize to the nucleus, and hence there had been no previous suggestion of an involvement with flagella or cilia. However, that mutation of pontin can cause cystic kidneys in zebrafish suggested a potential cilia-related function. This raised the question of whether reptin and pontin could have any functional role in ciliary assembly or function, even if they are not themselves found within cilia. Our analysis indicates that RNA transcription of Chlamydomonas homologs of both reptin (protein ID 156724, mutual best match in mouse genome with RuvBL2/reptin) and pontin (Chlamydomonas protein ID 169116, mutual best match in mouse genome with RuvBL1/pontin) is strongly induced during flagellar regeneration (Table 9). Because reptin and pontin proteins localize to the nucleus, but their RNA transcription levels are induced during flagellar regeneration, we propose that they act in the nucleus either to direct the transcriptional program of flagellar assembly or to mediate gene regulation in response to sensory cilia functions. It would be interesting to determine whether reptin/pontin activity is altered in mutants that fail to assemble cilia.
Flagellar Induced Genes with Potential Sensory Functions. Another interesting regulatory gene induced during regeneration was the Chlamydomonas homolog of Tubby-like protein 2 (TULP2). This predicted Chlamydomonas gene (protein ID 170392) is a mutual best match with mouse TULP2 and, like other members of the tubby superfamily, it contains a tubby-domain in its C terminus. The Tubby family of proteins (23) is named for the Tubby mouse mutant, which showed late-onset obesity and neurosensory defects, similar to the defects seen in BBS patients. Because BBS is now known to involve cilia and basal bodies, this suggests that Tubby genes may also be involved in ciliary assembly or function. Indeed, tubby mutant mice show defects in transport through the connecting cilium in retinal rod and cone cells similar to defects seen in intraflagellar transport gene deletions. Although the functional role of TULP2 is currently unknown, this particular member of the Tubby superfamily is particularly interesting, because in mouse it is expressed not only in the retina but also in the testis, consistent with a potential role in sperm flagella. Many of the genes in our high-expression list are also expressed in mouse testis. Despite these circumstantial connections between TULP2 and cilia, the protein was not identified in the flagellar and basal body proteome comparative genomics search and has not previously been detected as a flagellar protein. However, given its high level of RNA transcription induction during flagellar regeneration in Chlamydomonas, it seems likely that TULP2 may play a role in ciliary function. Tubby-like proteins are generally thought to play roles in signaling from G protein-coupled receptors (GPCRs). Interestingly, most cilia-localized receptors, such as the somatostatin receptor, 5-HT6 serotonin receptor, and olfactory odorant receptors, are GPCRs (24). We propose that TULP2, and possibly other Tubby family proteins, may function in signaling pathways downstream of GPCRs located in sensory cilia. As with reptin and pontin, this function need not involve localization within cilia or flagella and reinforces the value of our functional transcriptome approach for identifying genes that could not be found by direct proteomic analysis.
Finally, the set of induced genes also includes a Chlamydomonas homolog of dpy-30, a gene involved in dosage compensation in the worm C. elegans. This induction is unlikely to be coincidental, because a human orthologue of dpy-30 was found in a proteomic analysis of human cilia (2), and the dpy-30 gene product was identified in the expressed flagellar and basal body proteome comparative genomics analysis of Dutcher and co-workers (3). The connection between dpy-30 and cilia remains unclear; however, we note that in male worms, dpy-30 is required for proper mating behavior and tail development (25), both of which could be consistent with a role for dpy-30 in assembly or function of sensory cilia in the male ray sensilla. Based on the deflagellation-driven induction of dpy-30 in Chlamydomonas, a species that is evolutionarily highly diverged from C. elegans, we propose that dpy-30 homologs may play a conserved function related to assembly of cilia and flagella in all organisms.
Discussion
Genome-wide analysis of the transcriptional program of flagellar assembly provides a way to identify new components of cilia and flagella and hence to identify genes potentially involved in human ciliary diseases. These data are complementary to proteomic approaches because, in addition to revealing protein components of the flagellum, they should include genes whose products are needed for assembly or function of cilia/flagella but that do not themselves localize within the flagellum itself. This analysis also provides the basis for a future dissection of the regulatory pathways that drive flagellar assembly in response to deflagellation. We also note that the whole-genome microarray that we have developed for Chlamydomonas will allow genome-wide transcriptional analysis of other important phenomena in this organism, such as mating and photosynthesis.
Supplementary Material
Acknowledgments
We thank Himanshu Sethi for technical assistance and Mohua Bose, Elisa Kannegaard, Jessica Feldman, Kim Wemmer, Lani Keller, and Shigenori Nonaka for critical reading of the manuscript. This work was supported by grants (to V.S.) from the NASA Center for Nanotechnology; the NASA Fundamental Biology Program; the NASA Biomolecular Systems Research Program; the Computing, Information and Communications Technology Program (contract NAS2-99092); and National Science Foundation Grant 0416310 (to W.F.M.). M.P.S. has been supported by NASA contract to the NASA Ames University Affiliated Research Center.
Author contributions: V.S., M.P.S., W.T., and W.F.M. designed research; V.S., M.P.S., and W.F.M. performed research; V.S., M.P.S., W.T., and W.F.M. contributed new reagents/analytic tools; V.S., M.P.S., W.T., and W.F.M. analyzed data; and V.S., M.P.S., W.T., and W.F.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PKD, polycystic kidney disease; BBS, Bardet-Biedl Syndrome; NASA, National Aeronautic and Space Administration; TULP2, Tubby-like protein 2.
Footnotes
Shapiro, J. & Johnson, K. A. (1997) Mol. Biol. Cell 8, 53a (abstr.).
References
- 1.Pazour, G. J. & Rosenbaum, J. L. (2002) Trends Cell Biol. 12, 551-555. [DOI] [PubMed] [Google Scholar]
- 2.Ostrowski, L. E., Blackburn, K., Radde, K. M., Moyer, M. B., Schlatzer, D. M., Moseley, A. & Boucher, R. C. (2002) Mol. Cell. Proteomics 1, 451-465. [DOI] [PubMed] [Google Scholar]
- 3.Li, J. B., Gerdes, J. M., Haycraft, C. J., Fan, Y., Teslovich, T. M., May-Simera, H., Li, H., Blacque, O. E., Li, L., Leitch, C. C., et al. (2004) Cell 117, 541-552. [DOI] [PubMed] [Google Scholar]
- 4.Avidor-Reiss, T., Maer, A. M., Koundakjian, E., Polyanovsky, A., Keil, T., Subramaniam, S. & Zuker, C. S. (2004) Cell 117, 527-539. [DOI] [PubMed] [Google Scholar]
- 5.Lefebvre, P. A. & Rosenbaum, J. L. (1986) Annu. Rev. Cell Biol. 2, 517-546. [DOI] [PubMed] [Google Scholar]
- 6.Marshall, W. F. & Rosenbaum, J. L. (2001) J. Cell Biol. 155, 405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M. F., Rifkin, S. A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P. E., et al. (2004) Science 306, 655-660. [DOI] [PubMed] [Google Scholar]
- 8.Birrell, G. W., Brown, J. A., Wu, H. I., Giaever, G., Chu, A. M., Davis, R. W. & Brown, J. M. (2002) Proc. Natl. Acad. Sci. USA. 99, 8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent, W. J. (2002) Genome Res. 12, 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuwaysir, E. F., Huang, W., Albert, T. J., Singh, J., Nuwaysir, K., Pitas, A., Richmond, T., Gorski, T., Berg, J. P., Ballin, J., et al. (2002) Genome Res. 12, 1749-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witman, G. B. (1986) Methods Enzymol. 134, 280-290. [DOI] [PubMed] [Google Scholar]
- 13.Hill, A. A., Hunter, C. P., Tsung, B. T., Tucker-Kellogg, G. & Brown, E. L. (2000) Science 290, 809-812. [DOI] [PubMed] [Google Scholar]
- 14.Bertone, P., Stolc, V., Royce, T. E., Rozowsky, J. S., Urban, A. E., Zhu, X., Rinn, J. L., Tongprasit, W., Samanta, M., Weissman, S., et al. (2004) Science 306, 2242-2246. [DOI] [PubMed] [Google Scholar]
- 15.Stolc, V., Samanta, M. P., Tongprasit, W., Sethi, H., Liang, S., Hegeman, A., Nelson, C., Rancour, D., Bednarek, S., Ulrich, E., et al. (2005) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 16.Liu, X. S., Brutlag, D. L. & Liu, J. S. (2002) Nat. Biotechnol. 20, 835-839. [DOI] [PubMed] [Google Scholar]
- 17.Bailey, T. (1995) Ph.D. thesis (Univ. of California, San Diego).
- 18.Albert, T. J., Norton, J., Ott, M., Richmond, T., Nuwaysir, K., Nuwaysir, E. F., Stengele, K. P. & Green, R. D. (2003) Nucleic Acids Res. 31, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seixas, C., Casalou, C., Melo, L.V., Nolasco, S., Brogueira, P. & Soares, H. (2003) Exp. Cell Res. 290, 303-321. [DOI] [PubMed] [Google Scholar]
- 20.Sun, Z., Amsterdam, A., Pazour, G. J., Cole, D. G., Miller, M. S. & Hopkins, N. (2004) Development (Cambridge, U.K.) 131, 4085-4093. [DOI] [PubMed] [Google Scholar]
- 21.Amsterdam, A., Nissen, R. M., Sun, Z., Swindell, E. C., Farrington, S. & Hopkins, N. (2004) Proc. Natl. Acad. Sci. USA 101, 12792-12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanemaki, M., Kurokawa, Y., Matsu-ura, T., Makino, Y., Masani, A. Okazaki, K., Morishita, T. & Tamura, T. A. (1999) J. Biol. Chem. 274, 22437-22444. [DOI] [PubMed] [Google Scholar]
- 23.Carroll, K., Gomez, C. & Shapiro, L. (2004) Nat. Rev. Mol. Cell. Biol. 5, 55-63. [DOI] [PubMed] [Google Scholar]
- 24.Pazour, G. J. & Witman, G. B. (2003) Curr. Opin. Cell Biol. 15, 105-110. [DOI] [PubMed] [Google Scholar]
- 25.Hsu, D. R. & Meyer, B. J. (1994) Genetics 137, 999-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.