Abstract
Insect species in the Auchenorrhyncha suborder (Hemiptera) maintain ancient obligate symbioses with bacteria that provide essential amino acids (EAAs) deficient in their plant-sap diets. Molecular studies have revealed that two complementary symbiont lineages, “Candidatus Sulcia muelleri” and a betaproteobacterium (“Ca. Zinderia insecticola” in spittlebugs [Cercopoidea] and “Ca. Nasuia deltocephalinicola” in leafhoppers [Cicadellidae]) may have persisted in the suborder since its origin ∼300 Ma. However, investigation of how this pair has co-evolved on a genomic level is limited to only a few host lineages. We sequenced the complete genomes of Sulcia and a betaproteobacterium from the treehopper, Entylia carinata (Membracidae: ENCA), as the first representative from this species-rich group. It also offers the opportunity to compare symbiont evolution across a major insect group, the Membracoidea (leafhoppers + treehoppers). Genomic analyses show that the betaproteobacteria in ENCA is a member of the Nasuia lineage. Both symbionts have larger genomes (Sulcia = 218 kb and Nasuia = 144 kb) than related lineages in Deltocephalinae leafhoppers, retaining genes involved in basic cellular functions and information processing. Nasuia-ENCA further exhibits few unique gene losses, suggesting that its parent lineage in the common ancestor to the Membracoidea was already highly reduced. Sulcia-ENCA has lost the abilities to synthesize menaquinone cofactor and to complete the synthesis of the branched-chain EAAs. Both capabilities are conserved in other Sulcia lineages sequenced from across the Auchenorrhyncha. Finally, metagenomic sequencing recovered the partial genome of an Arsenophonus symbiont, although it infects only 20% of individuals indicating a facultative role.
Keywords: genome evolution, bacteria, aminoacyl tRNA synthetases, gene loss, nutritional symbioses, facultative symbiont
Introduction
At least 10% of all insect species harbor obligate symbionts that supply essential nutrients and other beneficial traits to their hosts (Buchner 1965; Baumann 2005; Moran et al. 2008; Sudakaran et al. 2017). Symbionts that provide essential services are generally maintained intracellularly within specialized organs and cells (bacteriomes and bacteriocytes, respectively) that regulate symbiont functions and ensure vertical transmission to subsequent generations (Nakabachi et al. 2005; Hansen and Moran 2012; Koga et al. 2012). As a result, many obligate symbionts have co-evolved with their hosts for 10’s to 100’s of millions of years (Moran et al. 2005; McCutcheon and Moran 2012). This arrangement has lead to the dramatic loss of genes from symbiont genomes due to genetic redundancy with their hosts, reduced selection, small population sizes, and frequent population bottlenecks (Moran et al. 1995; McCutcheon and Moran 2012; Wernegreen 2015). This process has produced the smallest known bacterial genomes (Moran and Bennett 2014). Nonetheless, while the relative ease of whole genomic sequencing has expanded our knowledge of genomic co-evolution in these symbioses (Wernegreen 2015; Wilson and Duncan 2015), the vast majority of insect symbiont lineages still remain unexplored.
Nutritional symbiotic relationships have played a fundamental role in the diversification of the hemipteran suborder, the Auchenorrhyncha, which contains several iconic insect groups: e.g., leafhoppers, spittlebugs, and cicadas (Moran et al. 2003; McCutcheon and Moran 2010; Urban and Cryan 2012; Koga et al. 2013; Bennett and Moran 2015; Douglas 2016; Sudakaran et al. 2017). Like other Hemiptera, species in this group have specialized piercing-sucking mouthparts adapted for feeding on phloem or xylem saps, or in some cases, plant parenchyma. Plant saps are depleted in essential amino acids (EAAS) and other nutrients that animals cannot make and must acquire from alternative sources (Sandström and Moran 1999; Douglas 2006). To overcome these dietary challenges, many species in the Auchenorrhyncha (Hemiptera) ally with at least two complimentary bacterial symbionts. Recent molecular and genomic work have shown that the common ancestor of the suborder was likely associated with “Candidatus Sulcia muelleri” (hereafter Sulcia) since its origin ∼300 Ma (Moran et al. 2005; McCutcheon and Moran 2010). Sulcia may have had a betaproteobacterial partner since very early in its association with the Auchenorrhyncha (Bennett and Moran 2013; Koga et al. 2013). This symbiont may still persist in several host lineages, although under several different names, including “Ca. Zinderia insecticola” in spittlebugs (Cercopoidea) (McCutcheon and Moran 2010), “Ca. Nasuia deltocephalinicola” in leafhoppers (Cicadellidae) (Noda et al. 2012), and “Ca. Vidania fulgoroideae” in planthoppers (Fulgoroidea) (Gonella et al. 2011) (note: were hereafter refer to symbionts by their italicized provisional genus name).
Whole genomic sequencing provides the opportunity to investigate the general biology, genomic evolution, and potential origins of unculturable symbionts. To date, the complete genomes of Sulcia have been sequenced from a range of hosts, revealing that it is generally responsible for eight of the ten essential amino acids (EAAs), but provides seven in the spittlebugs (McCutcheon and Moran 2010). Its partner symbiont complements the remaining two or three EAA pathways (Wu et al. 2006; McCutcheon et al. 2009; McCutcheon and Moran 2010; Bennett and Moran 2013). While Sulcia is rarely lost, the betaproteobacterium appears to have been replaced multiple times (e.g., “Ca. Baumannia cicadellinicola” in sharpshooter leafhoppers; Wu et al., 2006; Koga et al., 2013). In host lineages where both ancestral symbionts are retained, their genomes are the smallest known from any biological system aside from organelles. For example, the leafhopper host Macrosteles quadrilineatus (ALF) retains Nasuia-ALF (112 kb) and Sulcia-ALF (190 kb) (Bennett and Moran 2013). However, only a few representative symbiont genomes are available for the more than 42,000 recognized Auchenorrhyncha species. Furthermore, the co-primary betaproteobacterial lineage has been sequenced from only two insect lineages, spittlebugs, and leafhoppers, despite potentially being widespread among other major auchenorrhynchan groups.
To better understand the origins and genomic evolution of Sulcia and its betaproteobacterial partner, we sequenced their complete genomes from the phloem-feeding keeled treehopper, Entylia carinata (fig. 1A; Membracidae: ENCA). This species is widely distributed across the North American continent, feeding on a variety of herbaceous plant species, particularly in the family Asteraceae (McKamey 1998; Godoy et al. 2006; Deitz and Wallace 2010). The membracids are a diverse family, comprising over 400 genera and 3,500 described species, which exhibit several remarkable life-history traits including brood care and ant-tending (Buchner 1965; Cryan et al. 2004; Deitz and Wallace 2010; Deitz and Wallace 2012). Treehoppers are close relatives of leafhoppers and collectively comprise the large insect group, the Membracoidea. They maintain complex interactions with microbial symbionts with some species reported to harbor up to six partners (Buchner 1965; Dietrich et al. 2001). Recent microscopy and systematic work revealed that several lineages retain the ancient Sulcia symbiont and also a betaproteobacterium, which provide a simplified study system to investigate symbiont evolution in these lineages (Buchner 1965; Moran et al. 2005; Koga et al. 2013). Results from our study unequivocally confirm that the Betaproteobacteria in ENCA, and likely in other treehoppers, is a member of the Nasuia lineage found broadly in leafhoppers. We hereafter refer to it as Nasuia–ENCA. Our results further reveal that in contrast to deltocephaline leafhoppers, Sulcia-ENCA and Nasuia-ENCA retain larger genomes with expanded functional capacities. However, Sulcia-ENCA uniquely lost the capability to synthesize the branch-chain EAAs (leucine, isoleucine, and valine)—a first for this characteristically stable symbiont. Although we detected a possible facultative symbiont that could potentially help fill in these gaps, it does not appear to infect all members of the ENCA population sampled.
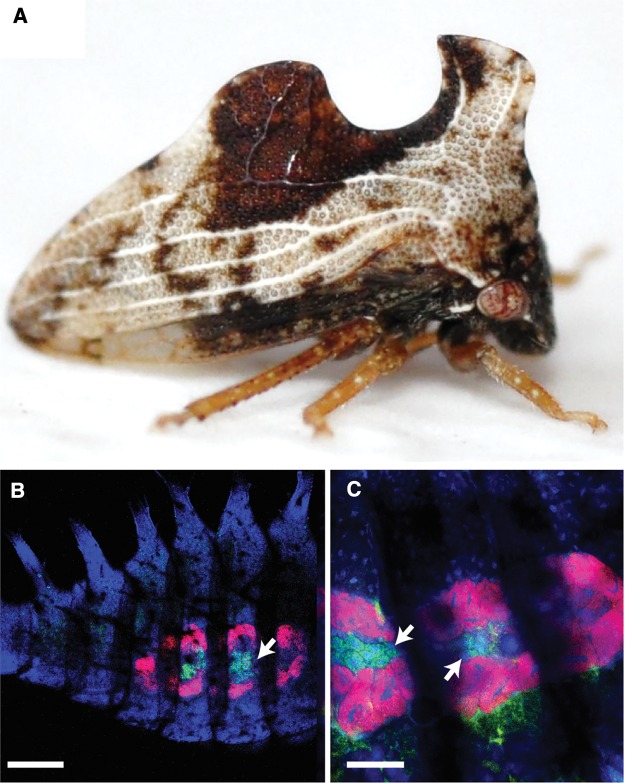
Fig. 1.
—Entylia carinata (Membracidae) host and fluorescence in situ hybridization of bacteriome organs containing the bacterial symbionts, “Ca. Sulcia muelleri” (red; Bacteroidetes) and “Ca. Nasuia koganicola” (green; Betaproteobacteria). DNA is counterstained with DAPI (blue) primarily showing host DNA. (A) Image showing E. carinata adult. (B) Lateral habitus of E. carinata nymph abdomen and (C) showing bacteriocytes containing bacteria (white arrows illustrate Nasuia containing bacteriocytes). White scale bars represent 200 µm (B) and 100 µm (C). Note: Green autofluorescence observed in abdominal cavity (without arrows) for Image (C). Photograph of E. carinata adult (A) provided by Kyle Kittelberger.
Materials and Methods
Fluorescence In Situ Hybridization Microscopy
Whole-mount fluorescence in situ hybridization was performed to confirm the bacteriome association of Nasuia and Sulcia in the ENCA host following Koga et al. (2009). Specimens of ENCA were field collected in 2013 from Yale West Campus, Orange, Connecticut, USA (41°15'16.3”N, 72°59'33.4”W). A total of 14 nymphs were examined; nymphs were chosen due to their pale coloration in contrast to the heavily pigmented adults (fig. 1A). Fluorochrome-labeled oligo probes that target bacterial 16S rRNA were adapted from Bennett and Moran (2013). Genomic sequences were used to verify the binding specificity of BET940-AL555 (5′-TTAATCCACATCATCCACCG-3′) and Sulc664R-AL647 (5′-CCMCACATTCCAGMTACTCC-3′) probes. Briefly, acetone preserved specimens were partially dissected, fixed in Carnoy’s solution, and bleached in a 6% hydrogen peroxide and 80% ethanol solution for 2 weeks. They were then rehydrated in PBSTx and washed with hybridization buffer. Material was finally incubated in hybridization buffer containing 10–50 µM of oligo-probes and DAPI counterstain. For Nasuia, multiple probe concentrations were tested due to low signal intensity and considerable autofluorescence in nymphs. Specimens were visualized using an Olympus FV1000 Confocal system at the Univ. Hawaii Mānoa, Biological Electron Microscope Facility.
Material Acquisition and Genome Sequencing
Whole abdomens of ten individuals were dissected and pooled for total genomic DNA (gDNA) extraction with DNeasy kit (Qiagen). gDNA was fragmented into a 550 bp insert size and sequenced on an Illumina MiSeq platform (2 × 300 bp paired-end [PE] reads). Library preparation and genomic sequencing were performed at the University of Texas, Austin Genomic Sequencing and Analysis Facility. Reads were de-multiplexed, adapter trimmed, and quality filtered with Trimmomatic v0.32 (program settings: ILLUMINACLIP:2:20:10:1 LEADING:20 TRAILING:20 SLIDINGWINDOW:4:20 MINLEN:50) (Bolger et al. 2014).
Genome Assembly, Verification, and Annotation
Sequencing and quality filtering yielded ∼5 million PE reads. Initial genome assembly was done from a subsampled read set (∼1 million pairs), since a large number of reads can result in highly fragmented contigs (Sloan et al. 2013). Assembly was performed with SPAdes V3.6.2 with multiple kmer lengths (program settings: -k 55,127 –only-assembler) (Bankevich et al. 2012). Assembly yielded two large contigs 218 and 144 kb in length that were identified with initial BLASTN V2.2.30+ searches as close matches to Sulcia and Nasuia lineages from leafhoppers, respectively. In order to estimate average coverage and to verify assembly quality (e.g., consistent coverage and no coverage breaks), all reads were mapped to the contigs with Bowtie2 V2.2.7 (default settings, Langmead and Salzberg 2012). Two A + T rich regions (within the gltX gene, and between the hisG and gltX genes, respectively) had broken coverage and were verified with Sanger sequencing. Finally, complete circular chromosomes were verified by breaking contigs at a known gene and remapping reads to assess coverage across joined ends.
Initial gene predictions and annotations were done with RAST (Aziz et al. 2008) and manually verified with Glimmer3 in Geneious 9.1.5 (Delcher et al. 2007; Kearse et al. 2012). Similar to Nasuia-ALF and Zinderia, Nasuia-ENCA was found to have an alternate genetic code (UGA Stop Codon → Trp); open reading frames were adjusted accordingly. Gene functions and genetic pathways were determined using the EcoCyc and KEGG databases (Kanehisa et al. 2004; Keseler et al. 2011). RNA-encoding genes were further verified with tRNAscan-SE and RNAmmer with the Rfam database (Lowe and Eddy 1997; Griffiths-Jones et al. 2005; Lagesen et al. 2007). Finally, Nasuia-ENCA and Sulcia-ENCA were aligned to related ALF genomes to verify and compare gene predictions and losses.
During the assembly process, we detected large contigs belonging to the facultative symbiont, Arsenophonus (Gammaproteobacteria). In order to test whether it is a permanent resident of our sampled population of ENCA, we performed a multi-locus sequence typing (MLST) screen of ten field-collected individuals. Adult specimens were collected from Orange, Connecticut, USA. High quality DNA was verified with Qubit 3.0 fluorometer (ThermoFisher). Diagnostic PCR was performed using primers designed for two protein coding genes: yaeT (primers: F-GCATACGGTTCAGACGGGTTTG and R-GCYGAAACGCCTTCAGAAAAG), and ftsK (Primers: F-GTTGTYATGGTYGATGAATTTGC and R-GCTCTTCATCACYATCAWAACC) (Duron et al. 2010). PCRs were carried out in a 25 µl reaction volume with standard NEB Taq reaction protocol (New England Biolabs Inc., Ipswich, MA). Thermo-cycling parameters were as follows: 95 °C for 2 min, 30 cycles of 95 °C for 15 s + 52 °C annealing 30 s + 68 °C for 1 min, and a final extension at 68 °C for 5 min. Taxonomic verification of the host was also done with Cytochrome Oxidase I barcoding from the completely sequenced mitochondrion (Mao et al. 2016).
Phylogenomic Reconstruction
The phylogenetic relationship of Sulcia lineages have been well illustrated by several recent molecular studies (Moran et al. 2005; Takiya et al. 2006; Noda et al. 2012; Nishino et al. 2016). The relationships and origins of Nasuia and Zinderia are less clear; and, the relationship between the ENCA betaproteobacterium and other leafhoppers has not been tested in a genomic framework. Thus, all currently available genomes were included in a phylum-level phylogenomic analysis. A total of 42 orthologous genes from 104 bacterial lineages and four outgroups from the other major proteobacteria groups (supplementary table 1, Supplementary material online) were determined using Phyla_AMPHORA (Wang and Wu 2013). Amino acid sequences for each gene were extracted and aligned with MAFFT with the L-INS-i model (Katoh et al. 2002). Ambiguously aligned regions for which homology was questionable were removed. Likelihood substitution models for each gene were estimated with ProtTest3 (Darriba et al. 2011). The final concatenated gene matrices were partitioned by gene and assigned independent substitution models. Phylogenetic reconstructions were inferred with maximum-likelihood using RAxML-HPC2 via the CIPRES server (Miller et al. 2010; Stamatakis 2014). A total of 1000 bootstrap replicates were performed with the GTR substitution model.
Results and Discussion
Localization of ENCA Symbionts
In order to verify the bacteriome association of Sulcia-ENCA and Nasuia-ENCA, we performed whole-mount FISH microscopy. Genome matched oligo-probes were hybridized to expressed bacterial 16S rRNA. Results demonstrate that Nasuia and Sulcia are co-localized in paired, nested bacteriomes located along the lateral sides of the abdomen (fig. 1B). This association is similar to that found in other Membracoidea, including other treehopper species and Deltocephalinae leafhoppers (Buchner 1965; Noda et al. 2012; Koga et al. 2013). We note, however, that a weak fluorescence signal was detected for Nasuia possibly due to the age of the samples (collected in acetone in 2013) and its deeply nested association within the Sulcia bacteriome (fig. 1C). Whole-mounted nymphs also exhibited considerable autofluorescence and internal structural complexity, which appears to be due to inter-instar nymphal development and molting fluid in the exuvial space (i.e. cavity between old and newly developing insect exoskeletons). Nevertheless, Sulcia and Nasuia were observable.
Basic Genome Features of ENCA Symbionts
The complete genomes of Sulcia and Nasuia from ENCA were sequenced to investigate the patterns of genomic evolution in these ancient symbionts. Both were fully assembled into single contigs, requiring Sanger verification of two low coverage regions in Nasuia. The complete Sulcia-ENCA genome consists of a single circular chromosome of 218,034 bp with a 23.0% G + C content (fig. 2; average coverage = 952×). It encodes 201 predicted protein-coding genes, 6 pseudogenes, a single rRNA operon, and 29 tRNAs capable of recognizing all amino acid codons. The genes that are highly truncated (<30% of their predicted lengths in close lineages, such as Sulcia-ALF and Sulcia-GWSS) or harbor internal stop codon were annotated as predicted pseudogenes. Predictions are based on in silico determination and have not been verified with gene expression experiments (e.g., RNA or proteomic expression). The Sulcia-ENCA genome is structurally conserved and, excluding gene deletions, perfectly syntenic compared with those sequenced from other auchenorrhynchan hosts. The Nasuia-ENCA genome is 144,602 bp long with a 15.2% G + C content (fig. 2; average coverage = 432×). Mapping revealed two regions with low or broken coverage (<50×) that were further verified with Sanger sequencing. The genome encodes 159 protein-coding genes, a single rRNA operon, and 29 tRNAs. A large noncoding region, including two tandem repeats and one A + T rich region (98.3% A + T content), was identified between the hisG and gltX genes. This region has also been identified in other Nasuia genomes (Bennett and Moran 2013; Bennett et al. 2016a). The functional significance of this feature is currently unknown. The Nasuia-ENCA genome is otherwise perfectly syntenic with that of the Nasuia genomes sequenced from Macrosteles leafhopper hosts, confirming the identity of the betaproteobacterium in ENCA.
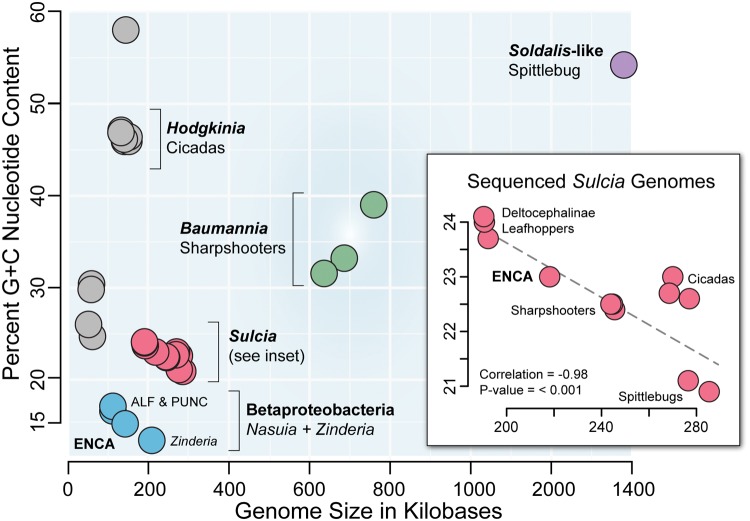
Fig. 2.
—The relationship between percent G + C nucleotide content and genome size of the major co-primary bacterial symbiont lineages in the Auchenorrhyncha (Hemiptera). Symbionts are color-coded by their lineage of origin (discussed in text; see labels, e.g., Hodgkinia = grey). Inset graph shows that G + C content is significantly correlated with genome size (Phylogenetic Generalized Lease Squares [PGLS]: correlation = −0.98 and P-value = <0.001) in sequenced Sulcia lineages. Sulcia phylogeny reconstructed with 16S rRNA using ML criteria in RAxML (Stamatakis 2014) and PGLS performed in R with nlme and ape (Paradis et al. 2004; Pinheiro et al. 2017). Representative host insects are abbreviated as follows: ALF = Macrosteles quadrilineatus, PUNC = M. quadripunctulatus, and ENCA = Entylia carinata.
While the general patterns of Nasuia genome evolution are only starting to emerge from increasing numbers of available genomes, Sulcia is known to exhibit unusual genomic stability. Aside from gene losses in different host groups, it exhibits highly conserved genomic synteny, an enigmatically depressed nucleotide substitution rate, and a higher A + T nucleotide bias relative to its partner symbionts (McCutcheon and Moran 2010; Bennett et al. 2014). The genomic stability of Sulcia remains something of a mystery. Within the Cicadomorpha, it retains a structurally conserved genome relative to its betaproteobacterial partner, Zinderia + Nasuia, which are highly rearranged. Both co-primary symbionts also have a G + C content ∼13 and ∼15%, respectively in contrast to their Sulcia partners that range between ∼21and 24%. The difference is rather striking considering that genetic drift should continue driving G + C downward and that both partner symbionts are among the most ancient, having evolved highly reduced genomes millions of years ago (fig. 2; Moran 1996). Bacteria generally show A + T biased DNA mutations, which is counterbalanced by natural selection or biased gene conversion that likely maintains a higher G + C content (Hershberg and Petrov 2010; Lassalle et al. 2015). In symbionts, the loss of DNA repair genes and ratcheting of slightly deleterious mutations offset these forces (Moran 1996; Moran et al. 2008). In Sulcia lineages, the elevated A + T content is actually higher in smaller genomes (fig. 2). A PGLS (phylogenetic generalized least squares) correlation analysis performed in R with nlme and ape shows that the GC content and genome size in Sulcia lineages is significantly correlated (correlation = −0.98 and P-value = <0.001) (fig. 2 Inset) (Paradis et al. 2004; Pinheiro et al. 2017). The same correlation was also observed in Betaproteobacteria. However, the same analysis was not undertaken for this symbiont lineage due to limited taxon sampling from closely related host lineages (n = 4; PUNC and ALF are from the same host genus, Macrosteles). This correlation raises the intriguing possibility that some selective constraints are maintaining nucleotide identity that could be further correlated with its extremely depressed mutation rates (Bennett et al. 2014).
Detection of Arsenophonus
Our metagenomic approach also detected the presence of a potential facultative symbiont, Arsenophonus sp. (Gammaproteobacteria). Assembly and reciprocal BLAST searches against a curated Arsenophonus and the NR databases recovered 818 contigs (>500 bp in length) assigned to Arsenophonus with an average coverage of 51×. From these contigs, RAST identified 4,080 protein-coding genes (1,208 hypothetical protein genes), 8 rRNAs, and 48 tRNAs, suggesting a relatively complete, but highly fragmented genome (Darby et al. 2010). To test the infection frequency of Arsenophonus sp., we screened ten field-collected individuals with PCR for the yaeT and ftsK loci. Our MLST screen revealed a 20% infection rate among individuals (both loci amplified in positive samples), supporting a possible facultative association; however, the effect of this symbiont on ENCA fitness remains to be tested. 16S rRNA BLAST searches further revealed that the Arsenophonus detected in ENCA is a close match to lineages found in other sap-feeding hemipterans (e.g., aphids [Aphis spp.] and psyllids [Glycaspis spp.]). Arsenophonus is also commonly found in other Auchenorrhyncha species, including planthoppers and leafhoppers, where it can have high infection rates (Xue et al. 2014; Kobiałka et al. 2016). Arsenophonus is among the most widespread insect symbionts known; however, its function in hosts is not well understood (Nováková et al. 2009). It has been predicted to provide B-vitamins to the host, contribute to virus transmission in plants, or protect hosts against parasitoids (Hansen et al. 2007; Rana et al. 2012; Xue et al. 2014). The genomes of Sulcia-, Nasuia-ENCA and Arsenophonus sp. are available on GenBank under accessions CP021172–CP021173 and NHNG00000000.
Phylogenomics and the Evolution of Auchenorrhyncha and Its Betaproteobacteria Symbiont
To verify the phylogenetic relationship between Nasuia lineages from the Membracoidea, and Zinderia from spittlebugs, a maximum likelihood phylogenomic tree was reconstructed for representative bacteria. Results confirm the close relationship between the Nasuia-ENCA and Nasuia from leafhoppers (Bootstrap Support [BS] = 100). Both are placed sister to Zinderia (BS = 100) and within the Oxalobacteriaceae (BS = 49; fig. 3). Our results support several previous molecular and genomic analyses (McCutcheon and Moran 2010; Noda et al. 2012; Bennett and Moran 2013; Koga et al. 2013). While the relationship between Nasuia and Zinderia could be an artifact of ancient A + T biased evolution and homoplasy, other biological features support this finding. Both lineages use the direct sulfhydrylation methionine synthesis pathway and an alternate genetic code (UGA Stop → Trp; McCutcheon and Moran 2010; Bennett and Moran 2013). The latter feature is a rare evolutionary event that is documented in only a handful of genomes across life. These results, along with shared synteny between Nasuia-ALF and Nasuia-ENCA, confirm an origin for the betaproteobacterial co-primary lineage at least as early as in the common ancestor of the Membracoidea >200 Ma and possibly the Cicadomorpha >270 Ma (Shcherbakov 2002; Cryan and Svenson 2010). It is further possible that Nasuia and Zinderia represent extant members of a Betaproteobacteria symbiont that originated with the establishment of Sulcia in the common ancestor of the Auchenorrhyncha ∼300 Ma (Bennett and Moran 2013). This hypothesis suggests that Vidania in planthoppers is also a member of this clade, which is supported in 16S rRNA phylogenetic analyses (Koga et al. 2013). Unfortunately, no genomes are currently available for Vidania to thoroughly test this hypothesis.
Fig. 3.
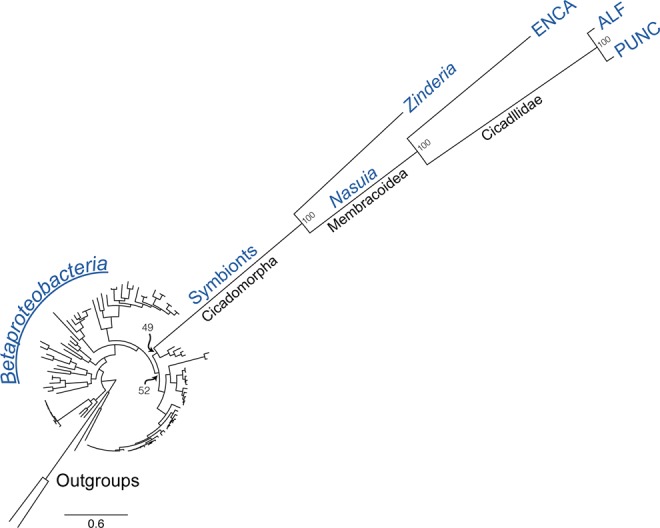
—Maximum likelihood phylogeny of Nasuia and other Betaproteobacteria based on 42 protein-coding genes for 108 taxa (104 ingroup taxa + 4 outgroup taxa). Relevant bootstrap support values are shown at each inter-node. Host lineages/names in Cicadomorpha are given beneath the branches. Representative host insects are abbreviated as follows: ALF = Macrosteles quadrilineatus, PUNC = M. quadripunctulatus, and ENCA = Entylia carinata.
Genomic Capabilities of Nasuia-ENCA
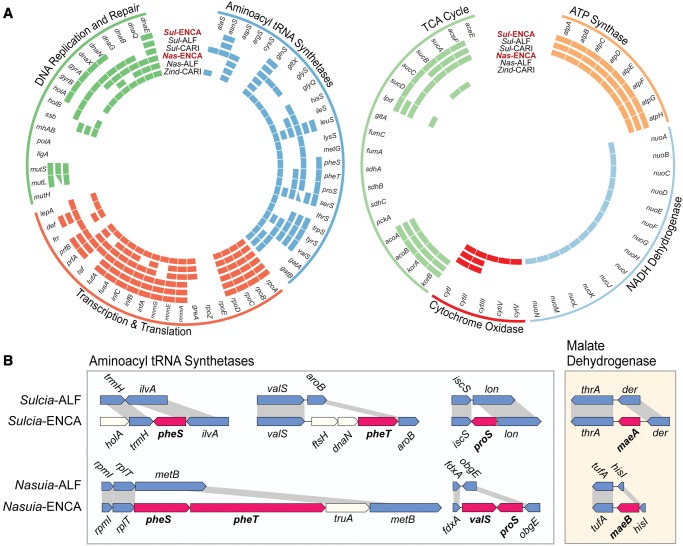
In comparison to Nasuia-ALF, the Nasuia-ENCA genome retains 26 unique genes (and four hypothetical genes) (fig. 4). The majority of genes are involved in cellular information replication, transcription, and translation (fig. 5A). Nasuia-ENCA retains two DNA replication genes, dnaG and dnaX, which are essential in the initiation and priming of DNA replication (reviewed by Baker and Wickner 1992). Nasuia-ENCA also encodes an expanded set of genes involved in translation (see below) and ribosome formation (e.g., rlmB, rpmCE, rsmA, rpsP, rplAIS)—a shared feature with Sulcia-ENCA (see discussion below). Nasuia-ENCA encodes two translation factors involved in initiation and elongation (infB and tsf, respectively), which are considered to be essential in Escherichia coli and may play a role in symbiont cell growth (Caldas et al. 2000; Gromadski et al. 2002). Nasuia-ENCA further encodes a larger, albeit incomplete, set of aminoacyl tRNA synthetase genes (13 instead of 7 found in Nasuia-ALF). Intriguingly, three of these genes, pheST and proS, are also retained in the genome of Sulcia-ENCA (fig. 5B). The concerted loss of these genes from Sulcia and Nasuia genomes in the ALF host suggests they may have lost these genes simultaneously as their host evolved mechanisms to supplant their impaired gene functions. The retention of tRNA synthetases is further known to vary among other co-primary symbionts. For example, while Zinderia retains a complete set, Hodgkinia encodes only ten, indicating that there is flexibility in the requirement for these genes across hosts (McCutcheon et al. 2009; McCutcheon and Moran 2010; Van Leuven et al. 2014).
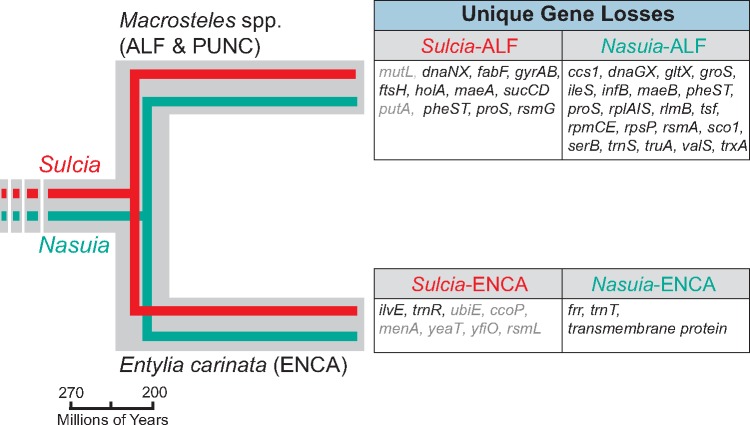
Fig. 4.
—Summary of the lineage-specific gene losses (excluding hypothetical genes) between obligate symbionts from the treehopper Entylia carinata (Sulcia- and Nasuia-ENCA) and leafhopper Macrosteles spp. (Sulcia- and Nauia-ALF & PUNC). The hypothetical establishment date of obligate symbionts in the common ancestor of the Auchenorrhyncha and the hypothetical diversification date of leafhoppers and treehoppers are based on previous studies (Shcherbakov 2002; Moran et al. 2005; Cryan and Svenson 2010; Koga et al. 2013). Evolutionary events of Sulcia and Nasuia are colored in red and green, respectively. Pseudogenes are shaded in gray.
Fig. 5.
—(A) Gene retention and losses involved in DNA replication and repair, transcription, translation and energy metabolisms in obligate symbionts from the treehopper Entylia carinata (Sul- and Nas-ENCA), leafhopper Macrosteles quadrilineatus (Sul- and Nas-ALF), and spittlebug Clastoptera arizonana (Sul- and Zind-CARI). (B) Retention of genes with the same predicted function in both Nasuia–ENCA and Sulcia-ENCA (genes shaded red) relative to their related lineages in the ALF leafhopper. Conserved orthologs are shaded blue and unique genes lost involved in other cellular functions are shaded white. (B) Note: tRNA genes omitted.
Nasuia-ENCA also uniquely retains malate dehydrogenase (maeB) that is important in bacterial energy and cellular metabolism by synthesizing pyruvate and NADPH (fig. 5B). The maeB gene functions as a major contributor of NADPH, which plays key roles in detoxifying and oxidative defense mechanisms, and acts as a universal electron donor to feed reductive biosynthesis (Agledal et al. 2010; Spaans et al. 2015). NADPH can be obtained by alternative sources, such as Ferredoxin-NADP+ reductase (fpr), which is also retained in other sequenced Nasuia genomes (Bennett and Moran 2013; Bennett et al. 2016a). Pyruvate is also an essential metabolite involved in a wide range of essential reactions, including the synthesis of energy, several EAAs (e.g., alanine and branch-chain), and acetyl-CoA (Shigenobu et al. 2000). Nasuia-ENCA is otherwise incapable of producing pyruvate. Due to its limited genome size, and the potential for the host to provide this resource, it is unclear what the pyruvate needs of Nasuia-ENCA are.
Finally, it is notable that Nasuia-ENCA has uniquely lost only a few genes in contrast to the dozens lost in the sequenced representative of Macrosteles leafhoppers. Nasuia-ENCA gene losses include ribosome recycling factor (frr), a predicted transmembrane protein, and a hypothetical protein. While the functions of the latter two are unknown, frr is essential for protein synthesis in bacteria (Janosi et al. 1994). The unique loss of so few genes in Nasuia-ENCA suggests that the genomic capabilities of Nasuia in the common ancestor to the Membracoidea may have either been very similar to that observed today in ENCA, or that there was extensive parallel gene loss. The former scenario is the most parsimonious expectation and suggests that Nasuia achieved a highly compact genome before the diversification of leafhoppers and treehoppers ∼200 Ma (Shcherbakov 2002). Recent evolution experiment of E. coli indicates that the obligate symbionts lost genes very quickly at the beginning of the symbiosis (Sabater-Muñoz et al. 2017). The factors contributing to the dramatic genomic differences observed in these symbiont lineages are unclear. However, they appear to have equally affected both symbiont partners in some fashion as to permit their genomes to shrink to the smallest known in any bacteria. Sulcia-ALF also exhibits extensive gene losses (see below).
Genomic Capabilities of Sulcia-ENCA
Relative to Sulcia in sharpshooter and Deltocephalinae leafhoppers, Sulcia-ENCA’s genome is roughly an intermediate size. In contrast to Sulcia-GWSS that has a larger genome (245 kb) (McCutcheon and Moran 2007), it has lost 16 genes mainly involved in oxidative phosphorylation and metabolite transport. Relative to Sulcia-ALF, Sulcia-ENCA encodes 14 additional protein-coding genes (and two hypothetical genes) that are absent in Sulcia-ALF (fig. 4). We focus most of our discussion on this comparison as it better highlights the capabilities of this symbiont; and, recent work has compared the genomes of ALF and GWSS (Bennett and Moran 2013).
Sulcia-ENCA’s expanded gene content broadly includes genes involved in DNA replication and repair, transcription, and translation (fig. 5A). Six of these genes (dnaNX, gyrAB, holA, and mutL) have important roles in DNA replication and repair in free-living bacteria (Moran et al. 2008; McCutcheon and Moran 2012). As discussed above, Sulcia-ENCA retains the aminoacyl tRNA synthetases, pheST and proS, also found in Nasuia-ENCA’s genome (fig. 5B). The incomplete set of the tRNA synthetases is commonly found in the Sulcia genomes, while the number of genes retained is variable among different lineages (Wu et al. 2006; McCutcheon and Moran 2007; McCutcheon et al. 2009; McCutcheon and Moran 2010; Bennett and Moran 2013; Bennett et al. 2014; Chang et al. 2015). It is notable that Sulcia-ENCA has the same tRNA synthetases as found in some lineages from xylem-feeding sharpshooter leafhoppers (Cicadellinae) (McCutcheon and Moran 2007). However, similar to Sulcia in Deltocephalinae leafhoppers (i.e., ALF), Sulcia from the Blue Green Sharpshooter (Graphocephala atropunctata) has also independently lost the pheS synthetase (Bennett et al. 2014). Additional losses in the deltocephaline lineages, and at least one sharpshooter, appear to be convergent evolutionary events. Their loss likely requires the incorporation of novel functions in existing genes, or some level of host intervention (McCutcheon et al. 2009).
Similar to Sulcia-ALF, Sulcia-ENCA is missing many of the genes involved in energy synthesis (fig. 5A). However, Sulcia-ENCA does retain several additional capabilities that are part of the TCA cycle, which includes the two-subunit Succinyl-CoA synthetase (sucCD). Since Sulcia lineages generally have an incomplete TCA cycle pathway, it is unclear to what extent it is able to generate energy in either lineage. Similarly, both Sulcia-ALF and Sulcia-ENCA have lost many genes involved in the formation of major oxidative phosphorylation (OXPHOS) protein complexes, including cytochrome C oxidase and NADH dehydrogenase. Although, both lineages do retain the subunits required for ATP synthase formation and is also conserved in all other sequenced Sulcia genomes (McCutcheon and Moran 2010; Koga and Moran 2014; Van Leuven et al. 2014; Chang et al. 2015). We were further able to identify a truncated gene fragment for the Cbb3-type cytochrome c oxidase subunit III (ccoP), which is ∼10% (105 bp) of its predicted length in E. coli. Cbb3-type cytochrome c oxidases (subunit I, II, III, and IV) are required for OXPHOS energy metabolism and were previously reported to be completely lost from Sulcia-ALF genome but incompletely remained in other Sulcia lineages in leafhoppers (McCutcheon and Moran 2007; Bennett and Moran 2013; Bennett et al. 2014), suggesting a more recent degradation of these genes in the ENCA lineage.
The apparent loss of ubiquinone and menaquinone cofactor biosynthesis, which supplies quinones as electron carriers for respiration (Fujimoto et al. 2012), is unique to Sulcia-ALF and Sulcia-ENCA. Both menA and ubiE are still identifiable as highly truncated fragments in the Sulcia-ENCA genome (12% [141 bp] and 8% [63 bp], respectively), which indicates, that like ccoP, these genes may have been recently degraded in the ENCA lineage. Ubiquinone and menaquinone cofactor are retained in all other sequenced Sulcia lineages, including those of sharpshooters, cicadas, and spittlebugs (Wu et al. 2006; McCutcheon and Moran 2007; McCutcheon et al. 2009; McCutcheon and Moran 2010; Woyke et al. 2010; Bennett et al. 2014; Koga and Moran 2014; Van Leuven et al. 2014). Thus, it seems likely that the host, or other elements of the symbiosis, requires these cellular products; however, it is unclear from where they are sourced in the ENCA system. One possibility is that the facultative symbiont Arsenophonus, which does retain the menA and ubiE genes, complements Sulcia and Nasuia. However, it is not ubiquitously infecting and it is uncertain if it plays any essential role in ENCA.
Essential Amino Acid Synthesis and Gene Loss in ENCA Symbionts
Both Sulcia and Nasuia from ENCA retain the pathways for synthesis of eight and two EAAs, respectively. Nasuia-ENCA retains the same pathways in the production of histidine and methionine as is found in the genomes of other Deltocephalinae leafhopper hosts (Bennett and Moran 2013; Bennett et al. 2016a). The histidine pathway is complete, while the methionine biosynthesis uses the direct sulfhydrylation pathway that appears to require several precursor metabolites, homoserine and coenzyme-A (CoA), which may be derived from Sulcia or the host (Hacham et al. 2003; McCutcheon and Moran 2010). Sulcia-ENCA is capable to make homoserine from aspartate and has been proposed to contribute this precursor to its partner (McCutcheon and Moran 2007; Douglas 2016). The obligate symbionts with extreme genomic degeneracy have lost many independent cellular activities, including the synthesis of EAAs. The most interpretable mechanism to complete the lost functions is to require metabolites from their partners (Douglas 2016). However, whether the metabolites are exchanged between symbionts housed in discrete bacteriome compartments has not been investigated.
Although Sulcia-ENCA retains most of the genes for EAA synthesis as has been reported in all other Sulcia lineages (McCutcheon and Moran 2007, 2010; Bennett and Moran 2013), it appears to require direct genetic input from the host or other local source. In general, Sulcia-ENCA has an expanded ability to synthesize EAAs relative to Sulcia-ALF. For example, it is capable of synthesizing pyruvate from malate using NAD-dependent malate dehydrogenase maeA (fig. 5B). However, remarkably, Sulcia-ENCA has uniquely lost the ilvE gene encoding branched-chain EAA transaminase (BCA) that carries out the terminal step in the synthesis of each of the BCAs (leucine, isoleucine, and valine; fig. 6). It is the first observed loss of an EAA synthesis capability from Sulcia, which is striking given that Sulcia is known for its unusually stable genome across host lineages that diverged several 100 Ma (McCutcheon and Moran 2010; Bennett et al. 2014). Furthermore, the symbionts of the Auchenorrhyncha generally retain complete, independent EAA pathways, requiring little or no input from their hosts or partners (McCutcheon and Moran 2010). The only other known loss of an EAA pathway in this insect group is that of the entire methionine synthesis pathway in Sulcia’s companion symbiont, Baumannia, from the Green sharpshooter leafhopper (Draeculacephala minerva); the loss may be related to a dietary and host-plant shift (Bennett et al. 2016b).
Fig. 6.
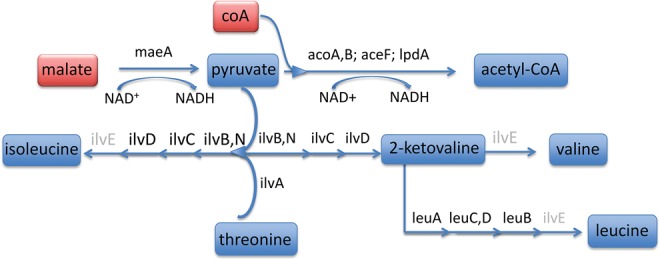
—Predicted branched-chain amino acid pathways in Sulcia-ENCA. The missing ilvE (branched-chain EAA transaminase) gene is shaded in gray. Substrates required for pathway completion and predicted to be imported by the host are shown in red box. Metabolites synthesized in Sulcia-ENCA are shown in blue box.
The complete synthesis of BCAs by Sulcia-ENCA may occur through contributions from either the host or additional bacterial symbionts. Although we assembled the partial genome of an Arsenophonus symbiont, its infection rate is not absolute, which would be required to permanently supplant missing obligate symbiont functions. Moreover, we were only able to identify a truncated ilvE gene in the Arsenophonus contigs, which is ∼14% (130 bp) of its predicted length in other Arsenophonus genomes (Darby et al. 2010; Xue et al. 2014) . This raises an intriguing possibility that this symbiont could also take advantage of the more plausible scenario that the ENCA host contributes a BCA transaminase to its symbionts. The loss of the ilvE gene is known to have occurred in several other obligate symbioses, including Buchnera aphidicola in pea aphids and “Ca. Tremblaya princeps” in the citrus mealybug. Gene expression analyses have shown that the host BCA gene is more highly expressed in bacteriocytes of these insects, indicating a direct host role in completing the synthesis of these particular EAAs (Hansen and Moran 2011; Husnik et al. 2013). Thus, we predict that the metabolic collaboration in BCA synthesis is likely to exist in the treehopper–Sulcia system.
Conclusion
Genome reduction is an inevitable fate of obligate nutritional symbionts in insects (McCutcheon and Moran 2012). However, gene loss can vary widely among different symbiont species and even between lineages of the same species (Koga and Moran 2014; Van Leuven et al. 2014; Bennett et al. 2016b). In this study, we sequenced the genomes of the two obligate symbionts Sulcia and the Betaproteobacteria from the first treehopper host, Entylia carinata (ENCA). Our genomic analyses confirm that the betaproteobacterial symbiont is a member of the Nasuia lineage found widely across leafhoppers and possibly the whole Auchenorrhyncha suborder (Iasur-Kruh et al. 2013; Ishii et al. 2013; Kobiałka et al. 2015, 2016; Szklarzewicz et al. 2016). Both symbionts were found to encode larger, more capable genomes than related lineages in deltocephaline leafhoppers (Bennett and Moran 2013; Chang et al. 2015). However, Sulcia-ENCA exhibits extensive gene losses relative to Sulcia in sharpshooter leafhoppers and all other lineages sequenced. In contrast, fewer Nasuia genomes are currently available to compare how this symbiont has evolved across hosts. Nevertheless, results from our study suggest that Nasuia likely already had a highly reduced genome in the common ancestor to the Membracoidea. Since it exhibits few unique losses, its encoded functional content was probably very similar to that observed in the Nasuia-ENCA today. Additional gene losses suggest that symbiont lineages with already highly degenerate genomes are generally restricted to DNA repair, transcription, and translation (Bennett and Moran 2013; Chang et al. 2015). ENCA further demonstrates that isolated loss of genes that provide essential functions such as EAA synthesis genes (e.g., ilvE) is also possible (Shigenobu et al. 2000; McCutcheon and von Dohlen 2011; Bennett et al. 2016b). Such additional gene losses are likely to impose a metabolic crisis for the host, requiring compensatory evolution to shore up losses as has been repeatedly observed in other highly divergent hemipteran host lineages (Hansen and Moran 2011; Husnik et al. 2013). Furthermore, the concerted loss of particular genes between companion symbionts in the same host (e.g., tRNA synthetases and malate dehydrogenases ALF symbionts) suggests that some feature of the symbiosis equally permits gene losses in bacterial partners, allowing their genomes to jointly shrink.
Finally, the presence of some symbionts in widely disparate host lineages raises some important implications for naming conventions. With regard to the betaproteobacteria symbiont in E. carinata, we propose to keep provisional generic name, “Ca. Nasuia”, given its syntenic genomic arrangement, shared origin, and phylogenetic relationships. However, the species moniker, deltocephalinicola, is not appropriate since it distinctly refers to the Cicadellidae subfamily from which this symbiont was originally described (Noda et al. 2012). Thus, we propose a new provisional Candidatus species name for the Nasuia lineage found in the Membracidae, which comprises a monophyletic insect group (Cryan et al. 2000). In honor of Ryuchi Koga (AIST), a positive mentor of the senior author in symbiont research, we propose the name—“Ca. Nasuia koganicola”—to refer to the Betaproteobacteria symbiont of treehoppers.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for their insightful and helpful comments. We thank Dr Nancy Moran (University of Texas at Austin) and Ryuchi Koga (AIST, Japan) for help finding populations of Entylia carianata, Tina Carvalho and Marilyn Dunlap (Biological Electron Miscroscope Facility, University of Hawaii Manoa) for assistance with FISH microscopy and Kyle Kittelberger (NC Museum of Natural Sciences, NRC) for photography of E. carinata adult. We thank UT Austin Genomics and Analysis Facility for assistance with sequencing. This work was supported by the National Science Foundation Award [IOS1347116] and new lab startup provided by UH Manoa, College of Tropical Agriculture and Dept. of Plan and Environmental Protection Sciences.
Literature Cited
- Agledal L, Niere M, Ziegler M.. 2010. The phosphate makes a difference: cellular functions of NADP. Redox Rep. 15:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Wickner SH.. 1992. Genetics and enzymology of DNA replication in Escherichia coli. Annu Rev Genet. 26:447–477. [DOI] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 59:155–189. [DOI] [PubMed] [Google Scholar]
- Bennett GM, Abbà S, Kube M, Marzachì C.. 2016a. Complete genome sequences of the obligate symbionts “Candidatus Sulcia muelleri” and “Ca. Nasuia deltocephalinicola” from the pestiferous leafhopper Macrosteles quadripunctulatus (Hemiptera: Cicadellidae). Genome Announc. 4:e01604–e01615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, et al. 2014. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. MBio 5:e01697–e01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, McCutcheon JP, McDonald BR, Moran NA.. 2016b. Lineage-specific patterns of genome deterioration in obligate symbionts of sharpshooter leafhoppers. Genome Biol Evol. 8:296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Moran NA.. 2015. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 112:10169–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Moran NA.. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 5:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. 1965. Endosymbiosis of animals with plant microorganims. New York (NY): Interscience. [Google Scholar]
- Caldas T, Laalami S, Richarme G.. 2000. Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J Biol Chem. 275:855–860. [DOI] [PubMed] [Google Scholar]
- Chang H-H, et al. 2015. Complete genome sequence of “Candidatus Sulcia muelleri” ML, an obligate nutritional symbiont of maize leafhopper (Dalbulus maidis). Genome Announc. 3:e01483–e01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JR, Svenson GJ.. 2010. Family‐level relationships of the spittlebugs and froghoppers (Hemiptera: Cicadomorpha: Cercopoidea). Syst Entomol. 35:393–415. [Google Scholar]
- Cryan JR, Wiegmann BM, Deitz LL, Dietrich CH.. 2000. Phylogeny of the treehoppers (Insecta: Hemiptera: Membracidae): evidence from two nuclear genes. Mol Phylogenet Evol. 17:317–334. [DOI] [PubMed] [Google Scholar]
- Cryan JR, et al. 2004. Treehopper trees: phylogeny of Membracidae (Hemiptera: Cicadomorpha: Membracoidea) based on molecules and morphology. Syst Entomol. 29:441–454. [Google Scholar]
- Darby A, et al. 2010. Characteristics of the genome of Arsenophonus nasoniae, son‐killer bacterium of the wasp Nasonia. Insect Mol Biol. 19:75–89. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitz L, Wallace M.. 2010. Treehoppers: Aetalionidae, Melizoderidae, and Membracidae (Hemiptera). In: The National Science Foundation, NCSU Insect Museum, and East Stroudsburg University of Pennsylvania, USA (updated August 2011).
- Deitz LL, Wallace MS.. 2012. Richness of the Nearctic treehopper fauna (Hemiptera: Aetalionidae and Membracidae). Zootaxa 3423:1–26. [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL.. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C, Rakitov R, Holmes J, Black W.. 2001. Phylogeny of the major lineages of Membracoidea (Insecta: Hemiptera: Cicadomorpha) based on 28S rDNA sequences. Mol Phylogenet Evol. 18:293–305. [DOI] [PubMed] [Google Scholar]
- Douglas A. 2006. Phloem-sap feeding by animals: problems and solutions. J Exp Bot. 57:747–754. [DOI] [PubMed] [Google Scholar]
- Douglas AE. 2016. How multi-partner endosymbioses function. Nat Rev Microbiol. 14:731–743. [DOI] [PubMed] [Google Scholar]
- Duron O, Wilkes TE, Hurst GD.. 2010. Interspecific transmission of a male‐killing bacterium on an ecological timescale. Ecol Lett. 13:1139–1148. [DOI] [PubMed] [Google Scholar]
- Fujimoto N, Kosaka T, Yamada M.. 2012. Menaquinone as well as ubiquinone as a crucial component in the Escherichia coli respiratory chain In: Ekinci D, editor. Chemical biology: InTech; p. 187–208. [Google Scholar]
- Godoy C, Miranda X, Nishida K.. 2006. Treehoppers of America Tropical. Costa Rica: INBio. [Google Scholar]
- Gonella E, et al. 2011. Bacterial endosymbiont localization in Hyalesthes obsoletus, the insect vector of Bois noir in Vitis vinifera. Appl Environ Microbiol. 77:1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, et al. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33:D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Wieden H-J, Rodnina MV.. 2002. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41:162–169. [DOI] [PubMed] [Google Scholar]
- Hacham Y, Gophna U, Amir R.. 2003. In vivo analysis of various substrates utilized by cystathionine γ-synthase and O-acetylhomoserine sulfhydrylase in methionine biosynthesis. Mol Biol Evol. 20:1513–1520. [DOI] [PubMed] [Google Scholar]
- Hansen A, Jeong G, Paine T, Stouthamer R.. 2007. Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Appl Environ Microbiol. 73:7531–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA.. 2012. Altered tRNA characteristics and 3′ maturation in bacterial symbionts with reduced genomes. Nucleic Acids Res. 40:7870–7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA.. 2011. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 108:2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA.. 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6:e1001115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F, et al. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153:1567–1578. [DOI] [PubMed] [Google Scholar]
- Iasur-Kruh L, et al. 2013. Novel Rickettsiella bacterium in the leafhopper Orosius albicinctus (Hemiptera: Cicadellidae). Appl Environ Microbiol. 79:4246–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, et al. 2013. Diversity of bacterial endosymbionts associated with Macrosteles leafhoppers vectoring phytopathogenic phytoplasmas. Appl Environ Microbiol. 79:5013–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L, Shimizu I, Kaji A.. 1994. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc Natl Acad Sci USA. 91:4249–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32:D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-i, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, et al. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39:D583–D590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiałka M, et al. 2016. Sulcia symbiont of the leafhopper Macrosteles laevis (Ribaut, 1927)(Insecta, Hemiptera, Cicadellidae: Deltocephalinae) harbors Arsenophonus bacteria. Protoplasma 253:903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiałka M, et al. 2015. Symbiotic microorganisms of the leafhopper Deltocephalus pulicaris (Fallén, 1806)(Insecta, Hemiptera, Cicadellidae: Deltocephalinae): molecular characterization, ultrastructure and transovarial transmission. Polish J Entomol. 84:289–304. [Google Scholar]
- Koga R, Bennett GM, Cryan JR, Moran NA.. 2013. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol. 15:2073–2081. [DOI] [PubMed] [Google Scholar]
- Koga R, Meng X-Y, Tsuchida T, Fukatsu T.. 2012. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte–embryo interface. Proc Natl Acad Sci USA. 109:E1230–E1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Moran NA.. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J. 8:1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Tsuchida T, Fukatsu T.. 2009. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl Entomol Zool. 44:281–291. [Google Scholar]
- Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F, et al. 2015. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet. 11:e1004941.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M, Yang X, Bennett G.. 2016. The complete mitochondrial genome of Entylia carinata (Hemiptera: Membracidae). Mitochondrial DNA Part B 1:662–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA.. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 106:15394–15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10:13–26. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 104:19392–19397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, von Dohlen CD.. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 21:1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKamey SH. 1998. General catalogue of the Hemiptera. Gainesville (FL: ): American Entomological Institute. [Google Scholar]
- Miller MA, et al. 2010. editors. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA.
- Moran NA. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 93:2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Bennett GM.. 2014. The tiniest tiny genomes. Annu Rev Microbiol. 68:195–215. [DOI] [PubMed] [Google Scholar]
- Moran NA, et al. 2003. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ Microbiol. 5:116–126. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A.. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 42:165–190. [DOI] [PubMed] [Google Scholar]
- Moran NA, Tran P, Gerardo NM.. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 71:8802–8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, von Dohlen CD, Baumann P.. 1995. Faster evolutionary rates in endosymbiotic bacteria than in cospeciating insect hosts. J Mol Evol. 41:727–731. [Google Scholar]
- Nakabachi A, et al. 2005. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 102:5477–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, et al. 2016. Fungal and bacterial endosymbionts of eared leafhoppers of the subfamily Ledrinae (Hemiptera: Cicadellidae). Appl Entomol Zool. 51:465–477. [Google Scholar]
- Noda H, et al. 2012. Bacteriome-associated endosymbionts of the green rice leafhopper Nephotettix cincticeps (Hemiptera: Cicadellidae). Appl Entomol Zool. 47:217–225. [Google Scholar]
- Nováková E, Hypša V, Moran NA.. 2009. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2017. nlme: linear and nonlinear mixed effects models. R Package Version 3:1–131. https://CRAN.R-project.org/package=nlme. [Google Scholar]
- Rana VS, et al. 2012. Arsenophonus GroEL interacts with CLCuV and is localized in midgut and salivary gland of whitefly B. tabaci. PLoS One 7:e42168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabater-Muñoz B, Toft C, Alvarez-Ponce D, Fares MA.. 2017. Chance and necessity in the genome evolution of endosymbiotic bacteria of insects. ISME J. 11(6):1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström J, Moran N.. 1999. How nutritionally imbalanced is phloem sap for aphids?. Entomol Exp Appl. 91:203–210. [Google Scholar]
- Shcherbakov D. 2002. The 270 million year history of Auchenorrhyncha (Homoptera). Denisia 4:29–36. [Google Scholar]
- Shigenobu S, et al. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86. [DOI] [PubMed] [Google Scholar]
- Sloan DB, et al. 2013. Disentangling associated genomes. Method Enzymol. 531:445–464. [DOI] [PubMed] [Google Scholar]
- Spaans SK, Weusthuis RA, Van Der Oost J, Kengen SW.. 2015. NADPH-generating systems in bacteria and archaea. Front Microbiol. 6:742.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran S, Kost C, Kaltenpoth M.. 2017. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 27:375–390. [DOI] [PubMed] [Google Scholar]
- Szklarzewicz T, Grzywacz B, Szwedo J, Michalik A.. 2016. Bacterial symbionts of the leafhopper Evacanthus interruptus (Linnaeus, 1758) (Insecta, Hemiptera, Cicadellidae: Evacanthinae). Protoplasma 253:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiya DM, Tran PL, Dietrich CH, Moran NA.. 2006. Co‐cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol Ecol. 15:4175–4191. [DOI] [PubMed] [Google Scholar]
- Urban JM, Cryan JR.. 2012. Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea). BMC Evol Biol. 12:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leuven JT, Meister RC, Simon C, McCutcheon JP.. 2014. Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one. Cell 158:1270–1280. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu M.. 2013. A phylum-level bacterial phylogenetic marker database. Mol Biol Evol. 30:1258–1262. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. 2015. Endosymbiont evolution: predictions from theory and surprises from genomes. Ann N Y Acad Sci. 1360:16–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Duncan RP.. 2015. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc Natl Acad Sci USA. 112:10255–10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, et al. 2010. One bacterial cell, one complete genome. PLoS One 5:e10314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, et al. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4:e188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, et al. 2014. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15:521.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.