ABSTRACT
Current methods for characterisation of extracellular vesicles (EVs) need further standardisation in order to obtain an acceptable level of data comparability. Size and concentration of EVs can be determined by nanoparticle tracking analysis (NTA). However, both the heterogeneity of EVs and the choice of instrument settings may cause an appreciable analytical variation. Intra-assay (within-day, n = 6) and inter-assay (day-to-day, n = 6) variations (coefficient of variation, % CV) of different preparations of EVs and artificial vesicles or beads were determined using two NanoSight NS500 instruments, located at different laboratories. All analyses were performed by the same operator. The effect of applying identical software settings or instrument-optimised settings for each sample type and instrument was also evaluated. Finally, the impact of different operators and the use of two different software versions were investigated. The intra-assay CVs were 1–12% for both EVs and artificial samples, measured on the same instrument. The overall day-to-day variation was similar for both instruments, ranging from 2% to 25%. However, significantly different results were observed between the two instruments using identical software settings. The effect of applying instrument-optimised settings reduced the mismatch between the instruments, resulting in little to no significant divergences. The impact of using different operators and software versions when analysing silica microspheres and microvesicles from monocytes using instrument-optimised settings on the same instrument did not contribute to significant variation compared to the overall day-to-day variation of one operator. Performance differences between two similar NTA instruments may display significant divergences in size and concentration measurements when analysing EVs, depending on applied instrument settings and technical conditions. The importance of developing a streamlined and standardised execution of analysis, as well as monitoring longitudinal variation parameters on both biological and synthetic samples, should be highlighted.
KEYWORDS: Exosomes, microvesicles, nanoparticle tracking analysis, repeatability, reproducibility
Introduction
Cells release different types of membrane-embedded extracellular vesicles (EVs) that, based on their mechanisms of release and size, are grouped into different classes. Exosomes are secreted after fusion of multivesicular bodies with the plasma membrane, whereas microvesicles are thought to bud directly from the plasma membrane. The majority of EVs occur within the sub-micron range (30–1000 nm) [1], where exosomes are believed to be most abundantly present in the lowest size range (30–150 nm). The potential of EVs for diagnosis and treatment of different diseases is being intensely investigated, as EV size, composition and concentration may give important clinical information. However, their small size and heterogeneity present challenges for their isolation, analysis and biomedical use. Methods for isolation, identification and quantification of EVs are far from standardised, and there is an on-going call for consensus in order to ensure an acceptable level of data comparability [2].
Nanoparticle tracking analysis (NTA) is a powerful characterisation technique that combines the properties of both laser light scattering microscopy and Brownian motion in order to obtain size distributions of particles in liquid suspension. NanoSight instruments (Malvern, UK), which currently are the most widely used instruments for NTA in the EV field, are equipped with one or more lasers and an optical microscope connected to a digital camera. According to the manufacturer, NanoSight enables characterisation of particles from 10–2000 nm in solution. Particles are visualised by the light they scatter upon laser illumination, and their Brownian motion is monitored. The NTA software enables sizing of single particles by tracking their mean squared displacement and thereby calculating their theoretical hydrodynamic diameter using the Stokes Einstein equation. On the basis of knowing the analysed sample volume, NTA also allows for an estimation of particle concentration [3]. More details on the principle of NTA have been described elsewhere [4,5].
Analysis of size distribution of homogeneous particle preparations of either same or mixed sizes by NanoSight has previously been shown to be accurate [6], whereas analysis of heterogeneous biological vesicles has proven to be more challenging. In 2013, a study showed that silica microspheres, with a refractive index close to EVs, could be used for calibration of NanoSight measurements [7]. Moreover, operator adjustments of capture and analysis settings, such as camera level and detection threshold, have been shown to strongly affect the NanoSight-based quantification of EVs [7,8]. Consequently, a standardised protocol for the best capture and analysis settings, both with respect to the type of sample being analysed and individual instrument could minimise the variability and imprecision associated with the technique. In addition, results from an inter-laboratory comparison of size measurements on synthetic, monodisperse nanoparticles [9], revealed the importance of a well-established procedure for highly reproducible and accurate NTA measurements. The study showed improved reproducibility of an average coefficient of variation (CV) from 38.5% down to 4.4% throughout four rounds of measurements, by introducing protocols with analysis guidelines to the laboratories involved. The importance of assessing measurement variation regarding size and concentration of EVs has been strongly emphasised in the literature, in order to increase sensitivity and reproducibility of particle measurement techniques [10–12]. To our knowledge, assessing measurement variation of EVs using NTA has otherwise been sparsely described in the literature, thus there still is a need to elucidate this issue.
In this study, we aimed to evaluate intra- and inter-assay variation of NanoSight measurements of both size and concentration of different preparations of EVs including microvesicles from human monocytes, exosomes from cell lines and outer membrane vesicles (OMVs) from Neisseria meningitidis. In addition, we performed variation analyses of different commercially available artificial vesicles/beads. The effect of applying identical vs. instrument-optimised measurement and analysis settings on two identical NanoSight instruments located separately, was also evaluated.
Methods
Sample preparation and isolation of EVs
Exosomes from cell cultures
PC-3 cells (ATCC) were grown in Ham’s F-12/DMEM (1:1) medium supplemented with 7% w/v foetal bovine serum (FBS), 100 units/mL penicillin and 100 units/mL streptomycin, in a humidified 5% CO2 atmosphere at 37°C. When cells were approximately 70% confluent, they were washed twice with FBS-free media, and incubated in this medium for approximately 24 h. Exosomes were then isolated from the culture medium by sequential ultracentrifugation [13]. Briefly, the supernatant was centrifuged at different speeds, 300g for 10 min, 1000g for 10 min and 10,000g for 30 min to remove cells, cell debris and microvesicles. Finally, the supernatant was ultracentrifuged at 100,000g for 70 min to pellet exosomes. The exosome pellet was washed with PBS and ultracentrifuged again at 100,000g for 70 min. Exosome pellets were resuspended in PBS, aliquoted and stored at −80°C until further use.
Jurkat cells (Clone E6-1, ATCC) were cultivated for 7 days in a CELLine bioreactor (CELLine CL 1000, Integra) with RPMI 1640 supplemented with 10% w/v FBS. The exosome-containing medium from the cell compartment was centrifuged stepwise: 200g for 10 min, 400g for 10 min, then 2000g for 30 min at room temperature. The exosomes were then enriched from the cell culture media using the Total Exosome Isolation Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
OMVs from N. meningitidis
The bacteria were grown on blood agar plates for 15 h at 37°C, harvested in STA buffer (0.2 M LiCl, 0.1 M NaAc, pH 5.8), and inactivated for 30 min at 60°C. Cells and debris were removed from the suspension by sequential centrifugation (4000g, 20 min and 18,000g, 15 min). The supernatant was further centrifuged at 140,000g for 90 min and the pellet resuspended in PBS, aliquoted and stored at −80°C.
Microvesicles from primary human monocytes
Microvesicles were harvested from supernatants (30 mL) of E. coli LPS (10 ng/mL)-stimulated primary human monocytes (1 × 108) [14] after incubation for 4 h in RPMI 1640 medium containing 5% v/v FBS depleted for EVs (ultracentrifuged at 113,000g for 16 h). Cells and debris were removed by centrifugation at 4500g for 5 min, and the microvesicles pelleted by centrifugation at 17,000g for 30 min. Pelleted microvesicles were dissolved in 200 µL medium (same as above), aliquoted and stored at −80°C.
Artificial vesicles/beads
Artificial vesicles (Invivofectamine® 2.0, Invitrogen), 100 nm polystyrene latex beads (calculated concentration 1.8 × 1013 particles/mL, given at a prediluted stock solution of 1:100, Duke Scientific Corp./NanoSight) and 150 nm silica microspheres (predicted concentration 3.0 × 1013 particles/mL, Polysciences, Inc.) were stored at +4°C. Predicted concentrations of 100 nm polystyrene beads were calculated as follows:
where W equals % mass of polymer (% solid), D is diameter in microns of latex particles and ρ is density of polymer in grams per millilitre (1.05 for polystyrene).
Transmission electron microscopy of EVs
To verify the presence of intact EVs, the preparations were analysed using transmission electron microscopy (TEM). Briefly, fixed (4% formaldehyde, 0.2% glutaraldehyde) EV samples were allowed to attach to Formvar/carbon-coated grids for 15–20 min. For labelling, PC-3-derived exosomes were first blocked with 0.5% BSA and then successively incubated with mouse anti-CD63 (H5C6) (Developmental Studies Hybridoma Bank, Iowa City, IA) followed by rabbit anti-mouse (Dako, Glostrup, Denmark) and then by 10 nm protein A-gold conjugates (Cell Microscopy Centre, Utrecht, Netherlands). Samples were then contrasted and embedded in a mixture of 0.4% uranyl acetate and 1.8% methyl cellulose. Exosomes from Jurkat cells were blocked with 0.5% BSA for 10 min, followed by labelling with anti-human CD81 (M38) (Invitrogen, Thermo Fisher Scientific) for 30 min, subsequent washing with PBS and addition of rabbit anti-mouse for 30 min. The EVs were then washed with PBS, incubated with 10 nm Protein A-gold particles for 15 min, washed again in PBS followed by distilled water and finally embedded in 0.3% uranyl acetate. For the other preparations, the grids were washed with PBS followed by double distilled water and stained with 0.4% uranyl acetate/1.8% methyl cellulose, and then dried. Exosomes from PC-3 cells were observed using a JEOL-JEM 1230 (JEOL Ltd., Tokyo, Japan) at 80 kV and images were acquired using a Morada digital camera and iTEM software (Olympus, Münster, Germany). The other EV preparations were observed using a FEI CM200 transmission electron microscope at 120 kV, and the images were recorded with a Quemesa CCD digital camera (Olympus Soft Imaging Solutions). The TEM images suggested that the preparations contained vesicles of various sizes (Figure 1).
Figure 1.
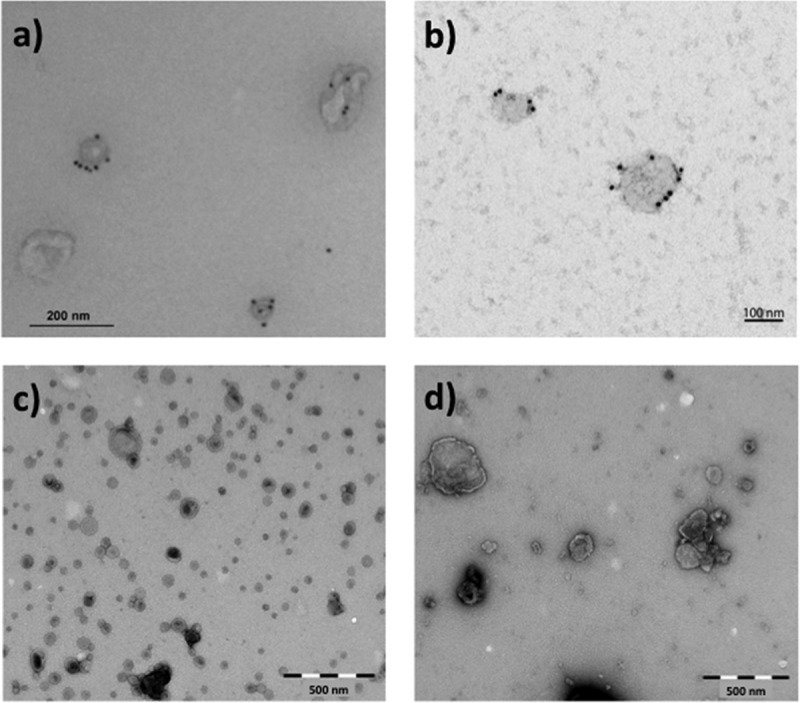
Representative transmission electron microscopy (TEM) images of negatively stained EVs used for NTA measurement variation assessment. The images show (a) exosomes derived from PC-3 cells labelled for CD63, (b) exosomes from Jurkat cells labelled for CD81, (c) OMVs derived from Neisseria meningitidis bacteria and (d) microvesicles from human monocytes. Scale bar sizes are 200, 100, 500 and 500 nm for (a), (b), (c) and (d), respectively. All images indicate the presence of intact vesicles of various sizes.
Intra/inter-assay variation of NanoSight measurements
Analyses were performed by the same operator on two different NanoSight NS500 instruments (Malvern Instruments, Amesbury, UK) located at different laboratories. The instruments were equipped with a 488 nm laser, a high sensitivity sCMOS camera and a syringe pump. The EVs and artificial vesicles/beads were mixed by vortexing, and subsequently diluted in particle-free PBS (0.02 µm filtered) to obtain a concentration within the recommended measurement range (1–10 × 108 particles/mL), corresponding to dilutions from 1:100 to 1:100,000 depending on the initial sample concentration (Tables 1 and 2). Experiment videos were analysed using NTA 2.3 build 17 or NTA 3.1 build 54 software (Malvern) (specified in Tables 1 and 2) after capture in script control mode (3 videos of 60 s per measurement) using syringe pump speed 20. A total of 1500 frames were examined per sample. Samples were captured and analysed by applying either identical or instrument-optimised (see “Inter-assay variation, instrument-optimised settings”) settings (Tables 1 and 2, respectively). Further settings, such as blur, minimum track length and minimum expected size were set to “automatic” and viscosity to “water” (0.909–0.90 cP).
Table 1.
Instrument settings for “identical settings” setup.
| “Identical settings” setup |
Instruments 1 and 2 |
|||
|---|---|---|---|---|
| Sample/vesicle type | Sample dilution | Camera level | Detection threshold | |
| EVs | Exosomes from PC-3 cells | 200 | 12 | 3 |
| Exosomes from Jurkat cells | 1000 | 13 | 4 | |
| OMV from Neisseria meningitidis | 10,000 | 12 | 3–4 | |
| Microvesicles from monocytes | 400 | 12 | 3–4 | |
| Artificial vesicles/beads | Artificial vesicles (Invivofectamine® 2.0) | 100,000 | 13–14 | 3–4 |
| Polystyrene latex beads 100 nm | 500 | 8 | 4 | |
| Silica microspheres 150 nm | 100,000 | 10 | 4 | |
Samples were analysed using NTA 2.3 build 17 software (Malvern).
Table 2.
Instrument settings for “instrument-optimised settings” setup.
| “Optimised settings” setup | Instrument 1 |
Instrument 2 |
||||
|---|---|---|---|---|---|---|
| Sample/vesicle type | Sample dilution | Camera level | Detection Threshold | Camera level | Detection Threshold | |
| EVs | Exosomes from PC-3 cells | 100 | 14–15 | 3 | 14–16 | 3 |
| Microvesicles from monocytes | 500 | 13–14 | 3 | 14–15 | 3 | |
| Beads | Polystyrene latex beads 100 nm | 1,000 | 11–12 | 3 | 12–13 | 3 |
Samples were analysed using NTA 3.1 build 54 software (Malvern).
Intra-assay variation, identical settings
Intra-assay variation of vesicle size and concentration measurements was determined by analysing the same samples sequentially within the same day (n = 6) on instrument 1. Consequently, % CVs of mean vesicle size (nm), mode size (nm) and concentration (particles/mL) were calculated (Table 3). Camera levels were determined for each sample type by scatter properties of the first preparation. All samples were measured at 23°C, as selected in the software script. Video analysis settings were determined by increasing the screen gain and adjusting the detection threshold for single particle tracking.
Table 3.
Intra-assay variation (within day, n = 6) of size and concentration measurements on instrument 1, using identical settings in NTA 2.3 build 17 software.
| Intra-assay (within day) | Mean size |
Mode size |
Mean concentration |
||||
|---|---|---|---|---|---|---|---|
| Sample/vesicle type | (nm) | % CV | (nm) | % CV | E8 particles/mL | % CV | |
| EVs | Exosomes from PC-3 cells | 128 | 2 | 94 | 5 | 5.8 | 5 |
| OMV from Neisseria meningitidis | 138 | 2 | 104 | 4 | 6.4 | 5 | |
| Microvesicles from monocytes | 141 | 1 | 103 | 4 | 6.8 | 7 | |
| Artificial vesicles/beads | Artificial vesicles (Invivofectamine® 2.0) | 87 | 2 | 81 | 4 | 5.4 | 12 |
| Polystyrene latex beads 100 nm | 102 | 2 | 94 | 3 | 4.9 | 8 | |
| Silica microspheres 150 nm | 140 | 2 | 132 | 2 | 7.8 | 8 | |
| Overall mean % CV | 2 | 3 | 8 | ||||
| Range % CV | 1–2 | 2–5 | 5–12 | ||||
Inter-assay variation, identical settings
Inter-assay variation was assessed by analysing the same sample aliquots with the same settings on two identical instruments located in two different laboratories on different days (n = 6) (Table 4). See also “Intra-assay variation, identical settings” section. Measurement curves from instrument 1 were generated (Supplementary Figure 1).
Table 4.
Inter-assay variation (day-to-day, n = 6) of size and concentration measurements on two NS500 instruments, using identical analysis settings in NTA 2.3 build 17 software.
| Mean size | Mean size | Mode size | Mode size | Mean concentration | Mean concentration | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Instrument 1 |
Instrument 2 |
Instrument 1 |
Instrument 2 |
Instrument 1 |
Instrument 2 |
|||||||||||
| Inter-assay (day-to-day)Sample/vesicle type | (nm) | % CV | (nm) | % CV | Per cent mismatch instrument 2 from 1 | (nm) | % CV | (nm) | % CV | Per cent mismatch instrument 2 from 1 | E8 particles/mL | % CV | E8 particles/mL | % CV | Per cent mismatch instrument 2 from 1 | |
| EVs | Exosomes from PC-3 cells | 128 | 3 | 142 | 2 | 11* | 94 | 2 | 113 | 3 | 20* | 6.3 | 18 | 10.8 | 10 | 72* |
| Exosomes from Jurkat cells | 130 | 4 | 144 | 6 | 11* | 91 | 6 | 99 | 6 | 9 | 5.6 | 12 | 5.1 | 7 | 8 | |
| OMV from Neisseria meningitidis | 139 | 2 | 156 | 4 | 12* | 102 | 5 | 122 | 6 | 20* | 5.8 | 16 | 8.9 | 8 | 54* | |
| Microvesicles from monocytes | 147 | 4 | 166 | 6 | 13* | 104 | 4 | 127 | 5 | 22* | 7.0 | 10 | 10.0 | 12 | 44* | |
| Artificial vesicles/beads | Artificial vesicles (Invivofectamine® 2.0) | 86 | 2 | 89 | 1 | 3* | 76 | 6 | 82 | 4 | 8* | 5.8 | 15 | 11.7 | 17 | 102* |
| Polystyrene latex beads 100 nm | 99 | 1 | 103 | 2 | 4* | 94 | 2 | 96 | 1 | 2* | 6.0 | 13 | 7.6 | 5 | 27* | |
| Silica microspheres 150 nm | 139 | 3 | 146 | 4 | 5 | 130 | 2 | 138 | 4 | 6* | 7.6 | 11 | 10.3 | 11 | 35* | |
| Overall mean % CV | 3 | 4 | 4 | 4 | 14 | 10 | ||||||||||
| Range % CV | 1–4 | 1–6 | 2–6 | 1–6 | 10–18 | 5–17 | ||||||||||
*Significant difference (paired t-test, p < 0.05).
Inter-assay variation, instrument-optimised settings
Inter-assay variation was assessed by analysing the same sample aliquots with instrument-optimised settings on different days (n = 3) (Table 5). The term “instrument-optimised” settings were in these experiments referred to as best visualisation of particles by applying software adjustments (camera level, focus and detection threshold) in order to optimise analysis results with respect to both sample type and individual instrument, without changing the settings within comparable sample series (Appendix 1). Samples were measured at 24°C. Automated image setup (camera level and focus) was chosen whenever applicable. See also the “Intra-assay variation, identical settings” section.
Table 5.
Inter-assay variation (day-to-day, n = 3) of two NS500 instruments, using instrument-optimised analysis settings in NTA 3.1 build 54 software.
| Mean size | Mean size | Mode size | Mode size | Mean concentration | Mean concentration | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inter-assay (day-to-day) |
Instrument 1 |
Instrument 2 |
Instrument 1 |
Instrument 2 |
Instrument 1 |
Instrument 2 |
||||||||||
| Sample/vesicle type | (nm) | % CV | (nm) | % CV | Per cent mismatch instrument 2 from 1 | (nm) | % CV | (nm) | % CV | Per cent mismatch instrument 2 from 1 | E8 particles/mL | % CV | E8 particles/mL | % CV | Per cent mismatch instrument 2 from 1 | |
| EVs | Exosomes from PC-3 cells | 125 | 2 | 135 | 12 | 8 | 89 | 8 | 92 | 16 | 3 | 3.1 | 15 | 2.9 | 25 | 8 |
| Microvesicles from monocytes | 171 | 4 | 167 | 2 | 2 | 127 | 3 | 119 | 10 | 6 | 4.3 | 4 | 3.9 | 13 | 8 | |
| Beads | Polystyrene latex beads 100 nm | 97 | 2 | 105 | 2 | 8* | 96 | 1 | 99 | 1 | 4* | 4.7 | 1 | 4.1 | 13 | 13 |
| Overall mean % CV | 3 | 5 | 4 | 9 | 7 | 17 | ||||||||||
| Range % CV | 2–4 | 2–12 | 1–8 | 1–16 | 1–15 | 13–25 | ||||||||||
*Significant difference (paired t-test, p < 0.05).
Inter-operator variation
Analyses were independently performed by four different operators on Instrument 1 by measurement of 150 nm silica microspheres and microvesicles from monocytes, using instrument-optimised settings. Additionally, all sample videos were acquired and analysed in software versions NTA 2.3 and NTA 3.1 (Table 6).
Table 6.
Inter-operator variation by measurement of 150 nm silica microspheres (1:100,000) and microvesicles from monocytes (1:400) in software versions NTA 2.3 and NTA 3.1, using instrument-optimised settings.
| Inter-operator |
Operator 1 |
Operator 2 |
Operator 3 |
Operator 4 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Instrument-optimised settings | NTA v2.3 | NTA v3.1 | NTA v2.3 | NTA v3.1 | NTA v2.3 | NTA v3.1 | NTA v2.3 | NTA v3.1 | |
| Silica microspheres 150 nm | Mean size (nm) | 142 | 137 | 136 | 135 | 129 | 136 | 130 | 136 |
| Mode size (nm) | 135 | 135 | 131 | 129 | 123 | 133 | 121 | 132 | |
| Concentration (E8 particles/mL) | 6.5 | 7.4 | 7.2 | 9.9 | 8.1 | 8.5 | 9.7 | 7.1 | |
| Camera level | 10 | 12 | 10 | 13 | 12 | 12 | 12 | 11 | |
| Detection threshold | 5 | 3 | 4 | 4 | 5 | 5 | 5 | 3 | |
| Microvesicles from monocytes | Mean size (nm) | 182 | 182 | 182 | 184 | 179 | 166 | 168 | 179 |
| Mode size (nm) | 138 | 148 | 128 | 134 | 134 | 123 | 125 | 138 | |
| Concentration (E8 particles/mL) | 4.6 | 3.9 | 5.1 | 5.5 | 4.9 | 2.9 | 3.9 | 3.1 | |
| Camera level | 14 | 14 | 14 | 15 | 14 | 15 | 15 | 14 | |
| Detection threshold | 3 | 3 | 4 | 4 | 4 | 3 | 5 | 3 | |
Statistical analysis
Calculation of % CVs of size and concentration measurements was performed using GraphPad PRISM® Version 6.03 (San Diego, CA, USA). Additionally, statistical significance in measurements between instruments (Tables 4 and 5) and between software versions (Supplementary Table 1) were determined using paired t-tests (p < 0.05). Observational errors for polystyrene beads and silica microspheres were calculated as % BIAS. Longitudinal measurements of 100 nm polystyrene beads were monitored in Levey Jennings diagrams for up to 2 SD of mean values.
Results
Intra-assay variation
In order to investigate the performance of one of the NanoSight instruments (Instrument 1), three different EV preparations (exosomes from PC-3 cells, microvesicles from monocytes and OMVs from N. meningitidis (Figure 1)) and three different preparations of artificial vesicles/beads (Invivofectamine® 2.0, 100 nm polystyrene latex beads, 150 nm silica microspheres) were measured. The intra-assay CVs were determined within the same day (n = 6) using identical settings within sample series, and ranged from 1–5% for size measurements and from 5–12% for concentration measurements (Table 3).
Inter-assay variation
Inter-assay CVs of four different preparations of EVs (same as in “Intra-assay variation” in addition to exosomes from Jurkat cells) and three different artificial vesicles or beads on two differently located NS500 instruments using identical settings were evaluated from day to day (n = 6). The inter-assay CVs ranged from 1–6% for size measurements, and from 5–18% for concentration measurements. Significantly different results on both size and concentrations were observed when comparing the two instruments (p < 0.05) (Table 4). Measurement curves illustrating inter-assay variation (day-to-day) from Instrument 1 are shown in Supplementary Figure 1. Longitudinal monitoring of 100 nm polystyrene latex beads measured on Instrument 1 over a period of 18 months is shown in Levey Jennings plots (Supplementary Figure 2), corresponding to inter-assay CVs of 3%, 4% and 9% on mean size, mode size and concentration, respectively.
To investigate the effect of applying instrument-optimised measurement settings (best visualisation of particles), three different sample preparations were measured on three different days on both instruments. The resulting inter-assay CVs ranged from 1–16% for size measurements, and from 1–25% for concentration measurements. Use of instrument-optimised settings on the two instruments resulted in no significant divergences considering EVs (Table 5). For polystyrene beads, observational errors (% BIAS) were calculated to be 158% in Instrument 1 and 128% in Instrument 2 (data not shown), reflecting predicted versus measured concentrations of 1.8 vs 4.7 × 108 particles/mL and 1.8 vs 4.1 × 108 particles/mL, respectively (Table 5). For silica microspheres, % BIAS was calculated to be 168% in Instrument 1 (data not shown), reflecting predicted versus mean measured concentration of 3.0 vs 8.0 × 108 particles/mL (Table 6).
Inter-operator variation
In order to investigate the influence of the operator on instrument-optimised settings, associated operator-dependent variations were also determined (Table 6 and Supplementary Table 1). For 150 nm silica microspheres and microvesicles from monocytes, the choice of camera levels ranged from 10 to 13 and 14 to 15, respectively. Detection threshold levels ranged from 3 to 5 for both sample types. There were no significant differences between the % CVs for either size or concentration measurements when comparing inter-operator variations in the two software versions NTA 2.3 and 3.1 (Supplementary Table 1).
Discussion
Standardisation of methodological platforms used for characterisation of EVs, both within and between laboratories, in order to compare research practice in the EV field, is of utmost importance [2,10]. The current study has evaluated single particle sizing and enumeration using NTA, for the primary purpose of obtaining standardised measurement practices across two laboratories. This was performed by analysing intra- and inter-assay variations on different biological and artificial vesicles or beads on two NanoSight NS500 instruments at different locations. The analyses were carried out by the same operator and by applying identical or instrument-optimised measurement settings. To elucidate the influence of the operator using instrument-optimised settings, four different operators performed measurements of silica microspheres and microvesicles from monocytes on the same instrument independently. Moreover, the sample videos were acquired and analysed in software versions NTA 2.3 and 3.1 to investigate potential software differences.
Repeatability and reproducibility of EV sizing and enumeration by NTA
Repeatability in particle sizing techniques may reflect the stability of a system when assessing precision of repeated consecutive measurements on a group of particles under identical conditions. The term reproducibility may be used when evaluating results obtained from measuring multiple aliquots of a bulk sample, but may also reflect variation occurring from inter-laboratory testing, such as instrument-to-instrument or day-to-day [15]. Evaluating repeatability and reproducibility in NTA measurements may be to consider intra- (within-day repeatability) and inter-assay (day-to-day reproducibility) variation of the measurements performed, as well as instrument-to-instrument reproducibility. A large number of variables may influence this assessment, either dependent on the operator or on the technical conditions of the instrument. Adjusting software settings, such as camera level (shutter and gain) and detection threshold may alone entail a noticeable variation when analysing samples. Various instrumental challenges such as air bubbles, temperature changes or laser misalignment may as well arise during use [7]. In addition, NTA detection limits may differ between various instrument versions depending on the hardware, such as illumination intensity, efficiency of the collection optics and quantum efficiency of the camera. Refractive index of the particles and the medium also influences NTA detection limits [16,17]. In our study, the two NTA instruments possessed identical hardware, and the sample aliquots were prepared uniformly in order to reduce pre-analytical variability. Variation originating from pre-analytical conditions, such as sample preparation, storage, freezing, thawing or precision of pipetting, may further contribute to the total variation of the samples to be compared. Pre-analytical issues are, however, beyond the scope of this paper and therefore have not been elucidated.
Different methodological platforms can be used for single particle sizing and enumeration of EVs. Recent studies using tuneable resistive pulse sensing (TRPS) have evaluated the reproducibility of size and concentration measurements on EVs using optimised operating procedures. These studies showed inter-assay CVs ranging from 5–15% on size measurement of urine vesicles [12], while CVs on the concentration measurements of vesicles isolated from citrated plasma and analysed in six different laboratories was up to 52.5% [18]. Most studies that have evaluated NTA variation measurements to date have utilised synthetic beads or artificial vesicles, such as liposomes [7,9]. Only a few studies have reported measurement variation in biological samples analysed by NTA, however, verification on the presence of EVs has largely lacked. Analyses of particles with a diameter less than 300 nm in human unprocessed urine showed a within-sample variation of 47% when measuring the area under the curve of particle size vs. concentration [19]. Recently, a study assessing analytical, pre-analytical and biological variation on particle levels by NTA and TRPS in blood plasma revealed a generally low (below 10%) analytical variation on samples [20]. However, a considerable intra- and inter-individual variation was demonstrated. In our investigation, the focus has been to include biological vesicles isolated from different sources, such as cell lines (PC-3 and Jurkat), primary human monocytes and N. meningitidis colonies. As suggested by TEM, these samples are all heterogeneous in terms of size, concentration and morphology of vesicles. Moreover, we included a selection of artificial vesicles or beads, representing homogenous particle sources of known sizes and predicted concentrations.
Reduced variation between NanoSight instruments using instrument-optimised software settings
In general, the obtained % CVs observed in our study were reasonable, ranging from 3% to 14% when measuring with identical settings (Table 4) and 3–17% when measuring with instrument-optimised settings (Table 5) in both biological and artificial vesicles or beads on both instruments. Compared to inter-assay, the intra-assay variation on one of the instruments (Table 3) resulted in overall lower mean % CVs (2–8%), as could be expected when making repeated measurements on the same aliquot rather than from different aliquots on different days.
When comparing results from the preparations analysed on two NanoSight instruments for size and concentration measurements using identical measurement settings (Table 4), a larger heterogeneous size distribution was observed for biological vesicles compared to artificial for both instruments. Furthermore, the measured mean and mode sizes showed significant divergences between the instruments. Also for the concentration measurements, the two instruments gave significantly different results. By applying instrument-optimised settings (Table 5), comparable inter-assay CVs were observed for both size and concentration measurements as for identical settings (Table 4). However, the divergences between the two instruments were greatly reduced for the observed sizes and concentrations of EVs to below the level of significance (Table 5).
Overall, higher camera levels were applied using instrument-optimised settings (Table 2) than for identical settings (Table 1). This may partly be explained by new aliquot series for EVs, explaining the changed sample dilution. Additionally, both instruments had undergone several service operations (laser realignments, optical configurations) between the measurement periods, which might explain the need for changed camera levels.
The overall mean % CVs were higher for concentration measurements than for size measurements, independently of whether identical or instrument-optimised settings were applied. Thus, we observed CVs of 10–14% compared to 3–4% for identical settings, and 7–17% compared to 3–9% for instrument-optimised settings for concentration and size measurements, respectively. This is likely due to the fact that concentration determination is more affected by camera level and detection threshold than size calculations, as explained by the Brownian motion and scatter properties of visible particles. An updated feature in newer software versions enables automated image setup (camera level and focus). However, we have found this to be most applicable for homogenous sample preparations, such as uniformly sized beads and less for heterogeneous samples, such as EVs.
The influence of different operators could lead to additional variation beyond the existing inter-assay and inter-instrument variations. In our study, the inter-operator CVs for both size and concentration measurements were comparable to the inter-assay (day-to-day) CVs for one operator (Tables 4 and 5 and Supplementary Table 1). These observations suggest that the influence of the operator when applying instrument-optimised settings is low.
As a bullet point in Appendix 2, we recommend to keep the software updated. However, one should take caution when comparing results measured by different software versions. Improved sizing algorithms and vibration correction in versions 3.X may result in improved measurement variation. Hence, this could have explained that our findings for instrument-optimised settings showed a lower per cent mismatch between the instruments (Tables 4 and 5). However, the overall % CVs per instrument were not markedly improved from software version 2.3 to version 3.1, also supported by the inter-operator variation results (Supplementary Table 1). It is therefore reasonable to infer that the instrument-to-instrument variation in our case was mainly reduced by applying instrument-optimised settings.
In Table 4, the mean and mode sizes measured by instrument 1 are generally slightly lower than for instrument 2 using identical settings. This could suggest that instrument 1 is more sensitive for the settings used, expecting to detect higher particle concentrations than instrument 2; however, this is not the case. We may speculate that, throughout the period of the measurements, there was a slight difference in microscope magnification between the instruments, possibly due to inconsistent microscope calibration. Microscope objectives may deviate a few per cent from their specified magnifications. If the magnification of instrument 1 is underestimated, the distances of particle tracks could be overestimated, leading to overestimation of diffusion coefficients and underestimation of particle sizes. Likewise, an underestimation of the magnification of instrument 1 could result in a smaller field-of-view than assumed, and thus an underestimation of the particle concentration.
Improved reproducibility of NTA measurements by combining the use of traceable standards with close monitoring of instrument plasticity
Applying traceable standards in the field of sizing and enumeration of EVs is of utmost importance. However, challenges may arise when transferring the calibration properties of homogeneous, synthetic standards onto heterogeneous, biological samples, such as EVs. Traceable standards, such as silica beads, obviously give substantial information of instrument performance. The use of a correction factor from measuring silica microspheres in order to calibrate results obtained from measuring EVs has been suggested [7]. Additionally, the settings used for measuring silica particle size could be more transmissible than for polystyrene beads, reasoned that they have a refractive index closer to EVs. An important challenge for a calibrator is to mimic the analyte as much as possible; however, there is still a gap in order to reach calibration properties that can be directly transferable to EVs. The use of nanoerythrosomes, possessing suitable EV properties, that are stable, safe and can be manufactured relatively cheaply, is currently being investigated [21]. However, the use of such reference materials still needs to be further validated and implemented. In line with others [8,20], we yet believe that the importance of relative comparisons of particle concentrations within experiments should therefore be underlined, rather than using mathematical corrections. The fact that we, in our experiments, observed little differences in variation when comparing heterogeneous biological EVs to homogeneous synthetic samples for both size and concentration measurements, could support the future applicability of biological reference materials for EV measurements by NTA.
When comparing our obtained numbers on synthetic beads from inter-assay (day-to-day) instrument-optimised settings, with the predicted concentrations (108 particles/mL), we found large observational errors (see Results section), suggesting quite unsatisfactory trueness of measurements. However, the predicted concentrations of these standard solutions are mathematically calculated from density, volume (% solids) and diameter, and could be largely inaccurate. We believe that, rather than reporting “true” concentrations of EV samples, the focus should be on relative comparisons of samples as well as monitoring measurement variability over time.
Taken together, our findings suggest that instrument-optimised settings provide improved reproducibility on both size and concentrations on biological vesicles when comparing NTA instruments. Consequently, the importance of monitoring “instrument plasticity” during measurement should be highlighted. Up until present, there has been much focus on the importance of reporting NTA settings in EV publications. This could “mislead” new operators to directly copy software settings from other groups, given that their sample type and instrument hardware are similar. We believe that reproducible NTA measurements can be obtained by both the use of traceable standards and the close monitoring of instrument plasticity. This might result in different software settings on two identical instruments when measuring the same sample. Capture optimisation might be done by providing the best visualisation of particles, meaning that they are adequately exposed and focused. Furthermore, keeping the balance between generating background exposure and reduced particle visibility, potentially leading to falsely elevated or low counts, should be emphasised. Nevertheless, comparable settings when handling samples of similar refractive index [16,17] and within measurement series on one instrument, are recommended. Technical conditions should, however, remain consistent during the measurement period.
Implementation of the NTA analysis in our lab has been time consuming. We have found it extremely important to have a dedicated person in the lab to develop and maintain a streamlined and standardised execution of analysis, thereby obtaining acquaintance with appropriate monitoring of the instrument and to assess any technical challenges related to the application used prior to evaluating results. Moreover, laboratories using NTA for EV research should be encouraged to monitor longitudinal variation parameters using both synthetic beads and biological samples, in order to validate the accuracy of their NTA measurements. Based on our own experience, we have listed several technical suggestions on how to reduce analytical variation when analysing EVs using NTA in Appendix 2.
In summary, the NTA measurements in the present study showed good repeatability, as evaluated by the good precision of repeated consecutive measurements on a NanoSight NS500 instrument. The results were further verified by the use of two NS500 instruments at different locations (Table 4), implying fair reproducibility from day to day and between the instruments. However, the overall variation assessment showed significant divergences on size and concentrations between the two instruments compared, suggesting there could be performance differences between same type of NTA instruments depending on applied instrument settings and technical conditions. Whereas the influence of the operator and software version may become a contributing factor of variation when measuring EVs by NTA, the close monitoring of instrument plasticity and longitudinal measurements could greatly improve the variation between instruments.
Supplementary Material
Acknowledgements
We thank The Norwegian Radium Hospital node of the electron microscopy core facility at Oslo University Hospital and the electron microscopy core facility at the University of Oslo. We would also like to thank Anne Marie Siebke Trøseid, Trude Aspelin and Tonje Bjørnetrø for contributing with NTA measurements. In addition, we thank Tone Tønjum and Håvard Homberset at The Genome Dynamics Group at the Department of Microbiology, OUH, Norway, for kindly providing OMVs from Neisseria meningitidis, Ketil Winther Pedersen (Thermo Fisher Scientific) for kind contribution of the TEM image of exosomes from Jurkat cells, and Nina Pettersen Hessvik at the Department of Molecular Cell Biology, Oslo University Hospital, Norway, for her expertise with NTA.
Appendix
Instrument-optimised software settings on NanoSight NS500
Video capture:
There should be approximately 20–60 particles in the field of view to reach acceptable statistics.
Increase the camera level (CL) to maximum level (usually 16).
Gradually decrease the CL until appropriate exposure is obtained, without losing visibility of particles (usually 12–15 for EVs). Overexposure of particles could cause the adjacent scatter noise falsely being tracked as particles.
Adjust focus according to the dimmest particles in the field of view. In heterogeneous samples, there will be a few particles out of focus. However, the software can in most cases still track these particles. For further visualisation on how to adjust focus, see the work by Gardiner et al from 2013 [7].
Start capture script (i.e. 3 × 60 s if using a syringe pump, or multiple shorter videos to reach a minimum of 1000 completed tracks per sample to get decent statistics).
Video analysis:
Before start, representative images from the video may be examined using the mouse cursor to make sure that the noise level is acceptable.
The detection threshold (DT) will be at default level of 7 in newer software versions. Decrease the DT as much as possible, to include all actual particles (particles to be tracked will be marked with red plus signs), while making sure there will be a minimum of particles marked with blue plus signs (noise detection). Additionally, visible scatter noise should not be marked with red plus signs, and thereby tracked, leading to falsely elevated particle counts. Further visualisation of correct DT adjustment can be obtained from the 2016 technical note by Malvern; “Measuring NIST Standards with NanoSight NTA”.
Appendix 1. Suggestions for applying instrument-optimised software settings after loading of EV samples into NanoSight NS500 (could also be applicable for other NanoSight instrument series). The settings should be identical for samples within comparable sample series.
Technical suggestions for reducing inter-assay variability for EV analysis using NTA (NanoSight NS500):
Make sure to install the updated version of the NTA software (http://www.malvern.com), as recent upgrades have increased the number of automated settings for capture and analysis, as well as improved vibration corrections and algorithms for calculation of size and concentration. Note that measurements made in some previous software versions, such as versions 2.X, may be reanalysed in newer versions (versions 3.X), while not the other way around.
Measure reference samples regularly, such as artificial beads and/or aliquots of EV samples, in order to monitor instrument performance over time.
Use freshly drawn particle-free water (MilliQ etc.) in a clean bottle, or from particle-free containers (such as a 50 mL Falcon tubes).
Dilute EV preparations with particle free PBS (0.02 µm filtered) right before analysis. All samples should be properly vortexed. Be aware that samples may “settle” quickly!
Dilute samples to reach the upper part of the measurement range (i.e. (5–9) × 108 particles/mL, equivalent to approximately 20–60 particles in the field of view), as statistical variation is more prevalent in the lower part (i.e. (1–4) × 108 particles/mL).
If available, use a syringe pump to achieve a more representative sample distribution and reduced variability between sample series. Adjust infusion rate of the syringe pump accordingly.
Use script control mode with repeated measurement videos, such as 3 × 60 s. If measuring too many videos or too long videos, particles may “settle” in the syringe, falsely underestimating particle concentration.
Right after sample load and video capture, stop the syringe pump (may also be set up as a part of the script, SYRINGESTOP) and flush the chamber and tubing system immediately with particle-free water to avoid salts from PBS building up in the system and reduce the need for mechanical cleaning of the chamber and gasket sheet.
Extensive measuring as well as heating/cooling of the laser over time may cause the laser module temperature to rise. If so, take small breaks, and open the hatch for ventilation. Temperature rise may cause shift or misalignment of the laser beam or zero point position.
Clean the gasket sheet when salts or other substances have accumulated in the gasket channel, or whenever particles are “stuck” or visible in zero point position. However, too extensive manual cleaning of the gasket sheet and chamber may cause abrasion. At the same time, unexpected values may signal the need for cleaning.
Regularly inspect the system for trapped air bubbles or leakage during use, especially when experiencing sample drift. Flushing the system often helps or emptying the fluidics then repriming with water. Leakage in the gasket may result from pressure build up or if the gasket sheet has not been allowed to settle for long enough to create a sealed channel.
Always check whether the zero point and scatter positions are correctly adjusted. At high camera levels, the scatter profile of water may reveal incorrect alignment of the laser beam and gasket. If the problem cannot be solved in the software, contact local Malvern technicians to help resolve the issue, as the laser module might need physical realignment.
Appendix 2. Technical suggestions based on our own practical experience to help reduce analytical variation when measuring EVs with NanoSight NS500.
Responsible Editor Eva-Maria Kramer-Albers, Johannes Gutenberg Universitat, Germany
Funding Statement
The work has received funding from the South-Eastern Norway Regional Health Authority.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental data
Supplemental data for this article can be accessed here.
References
- [1].Colombo M, Raposo G, Théry C.. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. [DOI] [PubMed] [Google Scholar]
- [2].Lotvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Carr B, Wright M. Nanoparticle tracking analysis; a review of applications and usage 2010-2012 amesbury. Wiltshire: NanoSight; 2013. [Google Scholar]
- [4].Dragovic RA, Gardiner C, Brooks AS, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011. December;7(6):780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Saveyn H, De Baets B, Thas O, et al. Accurate particle size distribution determination by nanoparticle tracking analysis based on 2-D Brownian dynamics simulation. J Colloid Interface Sci. 2010. December 15;352(2):593–600. [DOI] [PubMed] [Google Scholar]
- [6].Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010. May;27(5):796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gardiner C, Ferreira YJ, Dragovic RA, et al. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2 DOI: 10.3402/jev.v2i0.19671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Maas SL, De Vrij J, Van der Vlist EJ, et al. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J Control Release. 2015. February 28;200:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hole P, Sillence K, Hannell C, et al. Interlaboratory comparison of size measurements on nanoparticles using nanoparticle tracking analysis (NTA). J Nanopart Res. 2013;15:2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Witwer KW, Buzas EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Van der Pol E, Coumans FA, Grootemaat AE, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014. July;12(7):1182–1192. [DOI] [PubMed] [Google Scholar]
- [12].Coumans FA, Van der Pol E, Boing AN, et al. Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J Extracell Vesicles. 2014;3:25922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Phuyal S, Skotland T, Hessvik NP, et al. The ether lipid precursor hexadecylglycerol stimulates the release and changes the composition of exosomes derived from PC-3 cells. J Biol Chem. 2015. February 13;290(7):4225–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lund PK, Joø GB, Westvik AB, et al. Isolation of monocytes from whole blood by density gradient centrifugation and counter-current elutriation followed by cryopreservation: six years’ experience. Scand J Clin Lab Invest. 2000;60(5):357–365. [DOI] [PubMed] [Google Scholar]
- [15].ASTM E2834-12 Standard guide for measurement of particle size distribution of nanomaterials in suspension by nanoparticle tracking analysis (NTA). West Conshohocken (PA): ASTM International; 2012. Available from: www.astm.org. [Google Scholar]
- [16].Gardiner C, Shaw M, Hole P, et al. Measurement of refractive index by nanoparticle tracking analysis reveals heterogeneity in extracellular vesicles. J Extracell Vesicles. 2014;3:25361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Van der Pol E, Coumans FA, Sturk A, et al. Refractive index determination of nanoparticles in suspension using nanoparticle tracking analysis. Nano Lett. 2014. November 12;14(11):6195–6201. [DOI] [PubMed] [Google Scholar]
- [18].Vogel R, Coumans FA, Maltesen RG, et al. A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing. J Extracell Vesicles. 2016;5:31242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Oosthuyzen W, Sime NE, Ivy JR, et al. Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol. 2013. December 1;591(23):5833–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mork M, Pedersen S, Botha J, et al. Preanalytical, analytical, and biological variation of blood plasma submicron particle levels measured with nanoparticle tracking analysis and tunable resistive pulse sensing. Scand J Clin Lab Invest. 2016;19:1–12. [DOI] [PubMed] [Google Scholar]
- [21].Valkonen S, Van der Pol E, Boing A, et al. Biological reference materials for extracellular vesicle studies. Eur J Pharm Sci. 2017. February;15(98):4–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


