Abstract
Loss of imprinting (LOI) of the IGF2 gene (which encodes insulin-like growth factor II) is the most common genetic or epigenetic alteration in Wilms tumor; LOI involves aberrant activation of the normally repressed maternally inherited allele. We found previously that LOI of IGF2 occurs in approximately half of all Wilms tumors (i.e., those arising from lineage-committed nephrogenic progenitor cells). We investigated whether LOI of IGF2 is associated with relaxation of imprinting at loci other than IGF2 or with widespread alterations in DNA methylation. We stratified 59 Wilms tumor samples by IGF2 LOI status by use of hot-stop reverse transcription-polymerase chain reaction and/or methylation analysis of the differentially methylated region of the H19 gene and identified 31 samples with and 28 without LOI. We used quantitative allele-specific expression analysis to determine whether six other imprinted genes (i.e., H19, KCNQ1, LIT1, TSSC5, GRB10, and MEG3) had subtle LOI. No statistically significant difference in allele-specific expression between Wilms tumor with or without LOI was found for LIT1, TSSC5, GRB10, and MEG3. For the KCNQ1 gene there was a slight difference between the groups with (37.0%, 95% confidence interval [CI] = 31.8% to 42.2%) and without (27.7%, 95% CI = 21.8% to 33.5%) LOI (P = .02 for F test of group differences in a mixed-effects model). For H19, we also found a slight difference between the groups with [7.5%, 95% CI = 2.4% to 12.7%) and without (2.2%, 95% CI = −3.2% to 7.6%) LOI of IGF2 (P = .15 for F test). In 27 tumor samples, we also used a microarray technique to analyze methylation of 378 genes, 38 of which were suspected or confirmed imprinted genes. We found that statistically significant alterations in only the differentially methylated region of the H19 gene were associated with LOI of IGF2. Thus, epigenetic alterations in Wilms tumors are not widespread, supporting the gene and lineage specificity of LOI of IGF2.
Genomic imprinting is the differential expression of the two alleles of a gene that is dependent on the parent of origin of the allele (1). Loss of imprinting (LOI) of the IGF2 gene (encoding insulin-like growth factor II) involves aberrant activation of the normally repressed maternally inherited allele (1), related to biallelic hypermethylation of the H19 differentially methylated region. LOI of IGF2 was first described in Wilms tumor, the most common solid tumor during childhood (2,3), and has been found in the last decade in many human cancers, including other embryonic tumors (e.g., rhabdomyosarcoma) and adult tumors including colorectal, ovarian, and lung cancers (1,4).
Wilms tumor has two distinct histopathologic types (i.e., arising from perilobar nephrogenic rests or intralobar nephrogenic rests) that represent differing stages of renal development, with each group representing about half of all Wilms tumors (5). LOI is associated virtually uniformly with Wilms tumors that arise from perilobar nephrogenic rests, i.e., lineage-committed nephrogenic progenitor cells located in the periphery of the developing renal lobe (6,7). Perilobar nephrogenic rests also show LOI of IGF2, leading to expression from both alleles of the IGF2 gene and a double dose of IGF2 mRNA (6). Wilms tumors that arise from intralobar nephrogenic rests, which are not yet fully committed to renal development, do not have LOI of IGF2 (6), indicating the specificity of LOI for perilobar nephrogenic rest–like tumors. A previous study (8) found that TSSC3, TSSC5, KCNQ1, and ZNF195 genes were normally methylated, but the authors did not examine imprinting directly or by stratifying by IGF2 imprinting status. Another study (9) found rare (i.e., in four of 25 samples) hypomethylation of the LIT1 gene, also termed KCNQ1OT1, but no association of this change with LOI of IGF2. A recent study showed that genetic alterations at other loci are also uncommon in Wilms tumor with LOI (10). An exception appears to be chromosome 16q loss, which is associated with LOI of IGF2, possibly because of haploinsufficiency in the CTCF gene, which maps to that region and regulates IGF2 imprinting (11,12). As CTCF plays a role in the imprinting of multiple genes, this observation raises the question that imprinting alterations in Wilms tumor might be generalized.
Subtle changes in the proportion of alleles at single-nucleotide polymorphisms can now be measured by pyrosequencing (13), and microarray-based approaches allow methylation analysis of hundreds of target genes simultaneously (14). We therefore used these methods to determine whether LOI is specific to IGF2 or whether there is a generalized disruption of the epigenome in Wilms tumor with LOI.
Fifty-nine tissue samples were obtained from the National Wilms Tumor Study tissue bank (Edmonton, Alberta, Canada) and the Cooperative Human Tissue Network (Columbus, OH), under a local institutional review board–approved exemption. These samples had previously been examined by a pathologist to verify identity and homogeneity (6). A subset of these samples (n = 39) had available information regarding LOI status (6), and an additional 20 samples were typed for LOI status. Total cellular RNA was isolated by use of RNA STAT-60 (Tel-Test, Friendswood, TX). Genomic DNA was isolated from the organic–aqueous interphase of the RNA STAT-60 preparation with the protocol for TRIZOL Reagent (Invitrogen, Carlsbad, CA). RNA samples were treated with deoxyribonuclease by use of the TURBO DNA-free kit (Ambion, Austin, TX), and then reverse transcription (RT) was performed by use of the Superscript First-Strand Synthesis System for RT–polymerase chain reaction (PCR) (Invitrogen, Carlsbad, CA). We used the higher concentration of Superscript II (200 U/μL), which is available as a separate item but not as part of the Superscript First-Strand Synthesis System (50 U/μL). Primers were designed to amplify both genomic DNA and cDNA, so that genomic DNA results could be used to verify the linearity of results for each batch run, thereby acting as an intrinsic quality control measure. Therefore, a reaction mixture without RT was used as a negative control for each of the samples to exclude DNA contamination.
We assessed imprinting of IGF2 in all Wilms tumors examined, so that we would not be limited by the informativeness of an IGF2 single-nucleotide polymorphism for comparisons between IGF2 LOI status to the allele-specific expression results from the other genes. To increase the informativeness of IGF2, we took advantage of the fact that LOI of IGF2 in Wilms tumor occurs only if there is aberrant methylation of the differentially methylated region, which is upstream of the nearby imprinted gene H19 that regulates IGF2 imprinting (15,16). This observation is due to the fact that silencing of the maternal allele of IGF2 in the kidney requires that the maternal allele of H19 be unmethylated so that the insulator protein CTCF can bind to the differentially methylated region of the maternal allele and insulate IGF2 from downstream enhancers (17). LOI was assessed by methylation analysis (18) on all samples and by hot-stop RT–PCR (19) on the 26 samples that were heterozygous for a transcribed polymorphism in IGF2 (rs680). As expected, agreement was found between the two methods—11 samples had LOI and 15 did not have LOI.
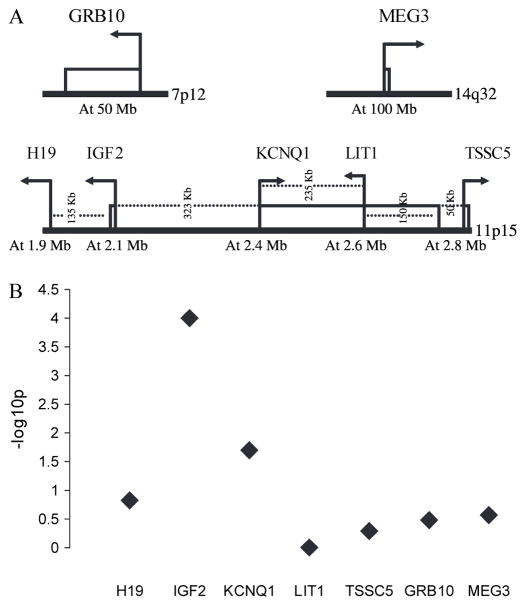
Using both hot-stop PCR of a transcribed polymorphism within IGF2 and Southern blot hybridization of the differentially methylated region of the H19 gene, 31 Wilms tumor samples could be classified as showing LOI of IGF2 and 28 Wilms tumor samples as showing normal imprinting (Supplementary Table 1, available online). We investigated whether there are subtle quantitative allele-specific expression differences in other imprinted genes in Wilms tumors with LOI of IGF2 by use of highly quantitative allele-specific expression assays for four genes located in the same chromosomal band as IGF2—i.e., H19, KCNQ1, LIT1, and TSSC5—and two genes outside of that band—i.e., GRB10 at chromosome 7p12 and MEG3 at chromosome 14q32 (Fig. 1, A; Supplementary Table 2, available online). Reconstitution experiments for LIT1, GRB10, and MEG3 DNA and for IGF2 RNA (Supplementary Fig. 1, available online) demonstrated the linearity of the method (13), indicating that the assays can accurately predict a wide range of allelic ratios.
Fig. 1.
Localization of genes of study and the strength of association for each of the genes for the allele-specific expression of differences between Wilms tumors with and without loss of imprinting (LOI) of IGF2 (encoding insulin-like growth factor II). A) Localization and orientation of genes selected for allele-specific expression analysis. For this analysis, we selected five genes on 11p15 (i.e., H19, IGF2, KCNQ1, LIT1, and TSSC5) and two genes at other locations (i.e., GRB10 at 7p12 and MEG3 at 14q32). The approximate distances in megabases (Mb) between genes and their approximate locations are according to coordinates in the Santa Cruz Genome Browser (http://genome.ucsc.edu/). B) Log10 (1/P) plot for the strength of association for each gene, comparing Wilms tumors with and without LOI of IGF2. The difference is represented by plotting the values in a log10(1/P) plot to visually demonstrate the difference in allele-specific expression for each gene, comparing Wilms tumors with and without LOI of IGF2.
As expected, for the 26 samples heterozygous for a transcribed polymorphism in IGF2 there was a marked difference in the average expression of the less abundantly expressed allele between the groups with (46.8%, 95% confidence interval [CI] = 44.4% to 49.2%) and without (5.6%, 95% CI = 3.5% to 7.6%) LOI of IGF2. This difference was statistically significant (P<.001 for F test comparing groups in a mixed-effects model; Table 1). We next assessed the H19, KCNQ1, LIT1, TSSC5, GRB10, and MEG3 genes, respectively, for heterozygosity of transcribed polymorphisms and identified 19, 18, 23, 23, 27, and 26 tumors with such heterozygosities. Allele-specific expression of these samples was then assessed by pyrosequencing analysis. For KCNQ1, we found a small but statistically significant difference between average expression of the lower allele among groups with (37.0%, 95% CI = 31.8% to 42.2%) and without (27.7%, 95% CI = 21.8% to 33.5%) LOI of IGF2 (difference = 9.2%; P = .02) (Table 1). For H19, we found a slight difference between groups with (7.5%, 95% CI = 2.4% to 12.7%) and without (2.2%, 95% CI = −3.2% to 7.6%) LOI of IGF2 (difference = 5.3%; P = .15) (Table 1), which could be related to the epigenetic silencing of the maternal allele of H19 in LOI (15,16) rather than activation of the paternal allele. No statistically significant differences in allele-specific expression of LIT1, TSSC5, GRB10, or MEG3 were found between groups with or without LOI of IGF2 (Table 1). As expected, IGF2 has the highest difference between the tumors with and without LOI of IGF2. Although of relatively lower magnitude, the second and third greatest P values were for genes on either side of IGF2, H19 and KCNQ1 (Fig. 1, B).
Table 1.
Allele-specific expression of imprinted genes in Wilms tumors with or without loss of imprinting of IGF2*
| Gene | Chromosomal location | % total expression for less abundant allele (95% CI) | F | P value | |
|---|---|---|---|---|---|
|
| |||||
| IGF2 LOI(+) tumors | IGF2 LOI(−) tumors | ||||
| H19 | 11p15 | 7.5 (2.4 to 12.7) | 2.2 (−3.2 to 7.6) | 2.3 | .15 |
| IGF2 | 11p15 | 46.8 (44.4 to 49.2) | 5.6 (3.5 to 7.6) | 741.2 | <.001 |
| KCNQ1 | 11p15 | 36.9 (31.8 to 42.2) | 27.7 (21.8 to 33.5) | 6.3 | .02 |
| LIT1 | 11p15 | 6.4 (4.1 to 8.8) | 6.4 (4.0 to 8.9) | 0.0 | .99 |
| TSSC5 | 11p15 | 40.9 (36.2 to 45.6) | 38.6 (33.1 to 43.9) | 0.45 | .51 |
| GRB10 | 7p12 | 26.3 (18.2 to 34.4) | 31.7 (24.1 to 39.2) | 1.0 | .33 |
| MEG3 | 14q32 | 2.1 (1.7 to 2.4) | 2.3 (2.0 to 2.7) | 1.27 | .27 |
To determine whether subtle loss of imprinting (LOI) of genes other than IGF2 (encoding insulin-like growth factor II) was associated with LOI of IGF2, we performed allele-specific expression analysis on a PSQ HS96 Pyrosequencer (Biotage, Uppsala, Sweden). The allele-specific expression results are given as expression of the lower (repressed) allele as a proportion of the total expression. Therefore, a gene with complete LOI (like IGF2 in LOI-positive tumors) would be expected to have a number close to 50, whereas a gene with strong imprinting (IGF2 in LOI-negative tumors) would be expected to be close to zero. The polymerase chain reaction (PCR) was performed on a GeneAmp PCR machine (Applied Biosystems, Foster City, CA), with the following conditions: 94 °C for 2 minutes; 40 cycles of 94 °C for 1 minute, 60 °C for 30 seconds, and 72 °C for 1 minute; and a primer extension at 70 °C for 9 minutes. The primer sequences for the seven assays are shown in Supplementary Table 1 (available online). All allele-specific expression assays were performed in triplicate. Genomic DNA heterozygotes were always analyzed concurrently. All statistical tests were two-sided and were performed with SAS version 9.1. CI = confidence interval. Genes with statistically significant P values are shown in boldface type.
Finally, we also performed pairwise comparisons of the allele-specific expression measurements across genes in tumors that were heterozygous for both genes. The neighboring genes LIT1 and TSSC5 demonstrated a statistically significant correlation between their respective allele-specific expression (R = 0.817 and P = .004; Supplementary Fig. 2, available online), which might indicate coregulation of these two genes at the locus, although allele-specific expression of these genes was not related to IGF2 imprinting. Thus, LOI of IGF2 appears to be a specific epigenetic event, in that there is not a generalized disruption of imprinting stringency associated with it.
DNA methylation analysis was also performed at 1178 sites in 378 genes, 38 of which were suspected or confirmed imprinted genes, with altered DNA methylation in other cancers, as well as the CTCF-binding sites in the H19 differentially methylated region (Supplementary Table 3, available online). We examined 14 Wilms tumors with and 13 without LOI of IGF2, by use of a modification of the Illumina Golden Gate genotyping assay as described (14). Briefly, DNA (1 μg) was treated with sodium bisulfite to convert unmethylated DNA to uracil while leaving methylated DNA intact, by use of the EZ DNA methylation kit (Zymo Research, Orange, CA). Therefore, methylated versus unmethylated cytosines were analyzed as single-nucleotide polymorphisms. Methylation status of CpG sites were quantified by use of a β value, which is defined as the ratio of the fluorescent signal of converted allele to the sum of the fluorescent signals from the converted and unconverted alleles (14). A difference of 0.17 between β values is the limit of detection of methylation changes (14). Data were analyzed with the Comparative Marker Selection algorithm in GenePattern Client software (Broad Institute, Boston, MA).
Using permutation analysis (20) and a cutoff of P value less than or equal to .01, 25 sites showed statistically significant methylation changes between Wilms tumors with and without LOI. Of these 25 sites, only six had a difference in β values greater than 0.17, the detection limit of the assay. All six sites were associated with the H19 gene, five within the differentially methylated region and one within the promoter (Supplementary Fig. 3, available online). Nineteen additional sites showed statistically significant methylation differences but did not have a difference in β values greater than 0.17. Eighteen of these sites were also associated with the H19 gene, one in the promoter and seventeen in the differentially methylated region. The remaining site was in the androgen receptor gene on the X chromosome (Supplementary Table 4, available online). This would suggest that methylation changes at known tumor-related sites appear specific to the H19 gene in Wilms tumors with LOI of IGF2.
In summary, using highly quantitative allele-specific expression assays, we have demonstrated that LOI of IGF2 in Wilms tumors appears epigenetically specific; i.e., LOI of IGF2 was not associated with a generalized disruption of imprinting. In addition, a microarray-based analysis of methylation changes in tumors with LOI of IGF2, compared with those with normal imprinting of IGF2, showed that methylation differences in Wilms tumors were not widespread and appeared to be confined to the differentially methylated region, which controls IGF2 imprinting. Thus, epigenetic changes in Wilms tumor with LOI appeared specific, just as the histopathology of Wilms tumors with LOI is specific for tumors of the perilobar nephrogenic rest type. Interestingly, there appeared to be regional specificity of epigenetic abnormalities, in that the genes that demonstrated subtle LOI were located on either side of IGF2 (Fig. 1, B).
Limitations of the study are that we have neither examined all imprinted genes nor analyzed all potentially methylated sites within the genome. However, given that the true number of imprinted genes is unknown, and the technology for gene-specific genome-wide methylation analysis is still in the “incubator” phase, the next level of resolution will require further technical development.
These results may have important therapeutic implications. Because of their potential reversibility, epigenetic modifications are under investigation as potential targets for the treatment of various common cancers (21). The epigenetic specificity of LOI of IGF2 in Wilms tumors indicates that IGF2 might be a good pharmacologic target for drug development, without the usual substantial concerns about other preexisting genetic or epigenetic changes that would be selected for by the treatment.
CONTEXT AND CAVEATS.
Prior knowledge
Loss of imprinting (LOI) of the IGF2 gene is the most common genetic or epigenetic alteration in Wilms tumor and involves aberrant activation of the normally repressed maternally inherited allele. LOI of IGF2 occurs in approximately half of all Wilms tumors.
Study design
Molecular biologic study of 59 Wilms tumor samples to determine whether LOI of IGF2 is associated with relaxation of imprinting at loci other than IGF2 or with widespread alterations in DNA methylation.
Contribution
LOI of IGF2 in Wilms tumors appears epigenetically specific and is not associated with a generalized disruption of imprinting.
Implications
Epigenetic alterations in Wilms tumors are not widespread, supporting the gene and lineage specificity of LOI of IGF2. Thus, IGF2 might be a good pharmacologic target for drug development, without the usual substantial concerns about other preexisting genetic or epigenetic changes that would be selected for by the treatment.
Limitations
Only 59 tumors were examined. Not all imprinted genes or all potentially methylated sites could be examined.
Acknowledgments
Funding
National Institutes of Health (CA65145 to A. P. Feinberg).
We thank Christine Ladd-Acosta for assistance with data analysis and Lotta M. Ellingsen for assistance with creating supplementary data figures. The authors were responsible for study design; data collection, analysis, and interpretation; and preparation and submission of the manuscript.
References
- 1.Feinberg AP. Genomic imprinting and gene activation in cancer. Nat Genet. 1993;4:110–3. doi: 10.1038/ng0693-110. [DOI] [PubMed] [Google Scholar]
- 2.Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–9. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 3.Ogawa O, Eccles MR, Szeto J, McNoe LA, Yun K, Maw MA, et al. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms’ tumour. Nature. 1993;362:749–51. doi: 10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14:427–32. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Beckwith JB, Kiviat NB, Bonadio JF. Nephrogenic rests, nephroblastomatosis, and the pathogenesis of Wilms’ tumor. Pediatr Pathol. 1990;10:1–36. doi: 10.3109/15513819009067094. [DOI] [PubMed] [Google Scholar]
- 6.Ravenel JD, Broman KW, Perlman EJ, Niemitz EL, Jayawardena TM, Bell DW, et al. Loss of imprinting of insulin-like growth factor-II (IGF2) gene in distinguishing specific biologic subtypes of Wilms tumor. J Natl Cancer Inst. 2001;93:1698–703. doi: 10.1093/jnci/93.22.1698. [DOI] [PubMed] [Google Scholar]
- 7.Fukuzawa R, Breslow NE, Morison IM, Dwyer P, Kusafuka T, Kobayashi Y, et al. Epigenetic differences between Wilms’ tumours in white and east-Asian children. Lancet. 2004;363:446–51. doi: 10.1016/S0140-6736(04)15491-3. [DOI] [PubMed] [Google Scholar]
- 8.Dao D, Walsh CP, Yuan L, Gorelov D, Feng L, Hensle T, et al. Multipoint analysis of human chromosome 11p15/mouse distal chromosome 7: inclusion of H19/IGF2 in the minimal WT2 region, gene specificity of H19 silencing in Wilms’ tumorigenesis and methylation hyper-dependence of H19 imprinting. Hum Mol Genet. 1999;8:1337–52. doi: 10.1093/hmg/8.7.1337. [DOI] [PubMed] [Google Scholar]
- 9.Satoh Y, Nakadate H, Nakagawachi T, Higashimoto K, Joh K, Masaki Z, et al. Genetic and epigenetic alterations on the short arm of chromosome 11 are involved in a majority of sporadic Wilms’ tumours. Br J Cancer. 2006;95:541–7. doi: 10.1038/sj.bjc.6603302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan E, Li CM, Yamashiro DJ, Kandel J, Thaker H, Murty VV, et al. Genomic profiling maps loss of heterozygosity and defines the timing and stage dependence of epigenetic and genetic events in Wilms’ tumors. Mol Cancer Res. 2005;3:493–502. doi: 10.1158/1541-7786.MCR-05-0082. [DOI] [PubMed] [Google Scholar]
- 11.Mummert SK, Lobanenkov VA, Feinberg AP. Association of chromosome arm 16q loss with loss of imprinting of insulin-like growth factor-II in Wilms tumor. Genes Chromosomes Cancer. 2005;43:155–61. doi: 10.1002/gcc.20176. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe N, Nakadate H, Haruta M, Sugawara W, Sasaki F, Tsunematsu Y, et al. Association of 11q loss, trisomy 12, and possible 16q loss with loss of imprinting of insulin-like growth factor-II in Wilms tumor. Genes Chromosomes Cancer. 2006;45:592–601. doi: 10.1002/gcc.20321. [DOI] [PubMed] [Google Scholar]
- 13.Alderborn A, Kristofferson A, Hammerling U. Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Res. 2000;10:1249–58. doi: 10.1101/gr.10.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat Genet. 1994;7:433–9. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 16.Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, et al. Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat Genet. 1994;7:440–7. doi: 10.1038/ng0794-440. [DOI] [PubMed] [Google Scholar]
- 17.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 18.DeBaun MR, Niemitz EL, McNeil DE, Brandenburg SA, Lee MP, Feinberg AP. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am J Hum Genet. 2002;70:604–11. doi: 10.1086/338934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uejima H, Lee MP, Cui H, Feinberg AP. Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat Genet. 2000;25:375–6. doi: 10.1038/78040. [DOI] [PubMed] [Google Scholar]
- 20.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattem 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 21.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]