Abstract
DNA charge transport (CT) chemistry provides a route to carry out oxidative DNA damage from a distance in a reaction that is sensitive to DNA mismatches and lesions. Here, DNA-mediated CT also leads to oxidation of a DNA-bound base excision repair enzyme, MutY. DNA-bound Ru(III), generated through a flash/quench technique, is found to promote oxidation of the [4Fe-4S]2+ cluster of MutY to [4Fe-4S]3+ and its decomposition product [3Fe-4S]1+. Flash/quench experiments monitored by EPR spectroscopy reveal spectra with g = 2.08, 2.06, and 2.02, characteristic of the oxidized clusters. Transient absorption spectra of poly(dGC) and [Ru(phen)2dppz]3+ (dppz = dipyridophenazine), generated in situ, show an absorption characteristic of the guanine radical that is depleted in the presence of MutY with formation instead of a long-lived species with an absorption at 405 nm; we attribute this absorption also to formation of the oxidized [4Fe-4S]3+ and [3Fe-4S]1+ clusters. In ruthenium-tethered DNA assemblies, oxidative damage to the 5′-G of a 5′-GG-3′ doublet is generated from a distance but this irreversible damage is inhibited by MutY and instead EPR experiments reveal cluster oxidation. With ruthenium-tethered assemblies containing duplex versus single-stranded regions, MutY oxidation is found to be mediated by the DNA duplex, with guanine radical as an intermediate oxidant; guanine radical formation facilitates MutY oxidation. A model is proposed for the redox activation of DNA repair proteins through DNA CT, with guanine radicals, the first product under oxidative stress, in oxidizing the DNA-bound repair proteins, providing the signal to stimulate DNA repair.
Keywords: electron transfer, iron–sulfur cluster, oxidative DNA damage
DNA-mediated charge transport (CT) from a distance to generate oxidative damage was first demonstrated in an assembly containing a tethered metallointercalator (1). In this assembly, photoinduced oxidative damage of the 5′-G of 5′-GG-3′ sites was observed; this damage pattern has since become the hallmark of DNA CT chemistry, and long-range oxidative damage has been confirmed by using a variety of pendant oxidants (2–6). Long-range oxidative DNA damage has been demonstrated over a distance of at least 200 Å (7, 8). Indeed, DNA either packaged in nucleosome core particles (9) or inside the cell nucleus (10) has been found to be susceptible to long-range oxidative damage. Chemically well defined assemblies, consisting of DNA duplexes with covalently bound oxidants, have been particularly useful in establishing the sensitivity of DNA CT to base-stacking perturbation (11–16). Recently, analogous studies probing long-range reductive chemistry on DNA has been probed both in solution (17–20) and on DNA-modified surfaces (14, 15, 21). As with oxidation chemistry, these reactions show only small variations in rate with distance but are remarkably sensitive to perturbations in the intervening base pair stack. Mechanistic descriptions for DNA CT focused first on a mixture of hopping and tunneling. A phonon-assisted polaron model has also been put forth (22). Studies as a function of temperature have shown the CT process to be gated by base pair dynamics; in fact, base pair motions are required for CT (23, 24). We have therefore described DNA CT in the context of transport among delocalized DNA domains formed and dissolved based on sequence-dependent DNA dynamics.
Given the exquisite sensitivity of DNA CT to DNA lesions and mismatches, we have recently explored a possible role for DNA CT in repair. We demonstrated that redox activity required DNA binding for MutY (25), a base excision repair (BER) enzyme from Escherichia coli that acts as a glycosylase to remove adenine from G:A and 7,8-dihydro-8-oxo-2-deoxyguanosine:A mismatches (26–33). Commonly considered a redox cofactor, [4Fe-4S]2+ clusters are ubiquitous to BER enzymes (27–37), yet redox activity in these proteins could not be detected under physiological conditions. Electrochemistry on DNA-modified electrodes showed a shift in potential for MutY to approximately +90 mV versus NHE (25), a potential characteristic of high potential iron proteins. Companion electrochemistry experiments showed furthermore that CT from the electrode surface to the [4Fe-4S] cluster requires DNA and is DNA-mediated. Electrochemical studies on DNA-modified surfaces and EPR experiments in solution testing additional BER enzymes more recently showed that this DNA-dependent redox activity of BER enzymes is general (A.K.B., unpublished work). Bound to DNA, BER enzymes containing [4Fe-4S]2+ clusters show similar redox potentials; binding to DNA shifts the [4Fe-4S]3+/2+ potential, activating the proteins toward oxidation. Based on this DNA-dependent redox activity, we have proposed a model for how BER enzymes might more quickly redistribute onto regions of the genome containing DNA lesions (25). This model depends on DNA-mediated CT among the BER enzymes and the sensitivity of DNA CT chemistry to intervening perturbations in base pair stacking, e.g., DNA mismatches and lesions.
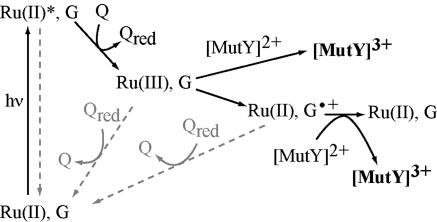
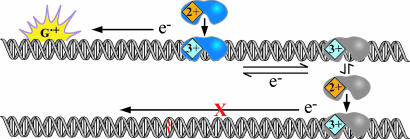
Here, we describe the redox activation of MutY by an oxidized base radical, a product of oxidative stress. We generate guanine radicals by using ruthenium flash/quench chemistry. This chemistry was first developed to probe long-range electron transfer in proteins (38). Examining DNA CT using the flash/quench technique has been particularly advantageous in that the methodology permits both spectroscopic studies to monitor formation of DNA radicals on a short time scale (16, 39–41) and biochemical analysis to determine the yield of oxidative damage occurring on a longer time scale (39–43). The flash/quench experiment for DNA typically is carried out with dipyridophenazine (dppz) complexes of Ru(II), complexes that bind avidly to DNA by intercalation (44). As illustrated in Scheme 1, the cycle is initiated by visible light, which excites the intercalated Ru(II) complex. This excited Ru(II) complex, *Ru(II), is then quenched by a nonintercalating electron acceptor, Q, such as [Ru(NH3)6]3+ or [Co(NH3)5Cl]2+, so as to form Ru(III) in situ. It is this Ru(III) species that can oxidize guanines from a distance. The oxidized guanine radical can then undergo further reaction with H2O and/or O2 to form a family of oxidative products, Gox (45). However, the lifetime of the guanine radical is relatively long (ms), and thus the guanine radical can also react with DNA-bound peptides (46) and proteins (16), or, as we demonstrate here, a BER glycosylase such as MutY.
Scheme 1.
Schematic illustration of the flash-quench technique used to generate Ru(III) in situ and subsequently to oxidize DNA-bound MutY. Back electron transfer reactions are in gray.
Experimental Procedures
Materials. All chemical reagents and starting materials were purchased from commercial sources and used as received. Phosphoramidites were purchased from Glen Research (Sterling, VA). Poly(dGC) (ε260 = 8,400 M-1·cm-1) and poly(dAT) (ε260 = 6,600 M-1·cm-1 were purchased from Amersham Pharmacia and passed through spin columns (Bio-Rad) before use. The ligands 4-butyric acid-4′-methyl-2,2′-bipyridine (bpy′) and dppz as well as [Ru(bpy′)(dppz)(phen)]Cl2 were synthesized as described (47–51).
DNA Synthesis. The oligonucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer (52, 53), purified by reverse-phase HPLC, and characterized by mass spectroscopy. The synthesis of ruthenium-modified oligonucleotides was carried out with rac-[Ru(bpy′)(dppz)(phen)]Cl2 (54). Purification of the ruthenium-modified DNA by reverse-phase HPLC yielded four isomers, which were characterized by UV-visible spectroscopy and mass spectrometry; the mixture of diastereomers was used.
Protein Preparation. MutY was used in all experiments either fused to maltose binding protein (MBP) or in a truncated form (Stop 225). Both forms are stable at concentrations much higher than the native form and thus are preferable for spectroscopic and EPR studies. Stop 225 was used in all transient absorption experiments, and MutY-MBP was used in EPR and gel electrophoresis studies. Also, C199H-MutY was expressed as an MBP fusion and used for EPR experiments. All forms of MutY were purified as reported (55).
EPR Spectroscopy. X-band EPR spectra were obtained on a Bruker EMX spectrometer equipped with a rectangular cavity working in the TE102 mode. Low-temperature measurements (10 K) were conducted with an Oxford (ES9000) continuous-flow helium cryostat (temperature range 3.6–300 K). A frequency counter built into the microwave bridge provided accurate frequency values. Solutions were prepared by adding the protein (50 μM) or protein storage buffer (20 mM NaPi/100 mM NaCl/1 mM EDTA/10% glycerol, pH 7.5) to a solution of poly(dAT) (1 mM bp), poly(dGC) (1 mM bp), or Ru-tethered duplex (25 μM) in the presence of quencher [Co(NH3)5Cl, 125 μM]. Samples were then irradiated in standard EPR quartz tubes while cooling in an unsilvered Dewar filled with liquid nitrogen; the excitation source was a focused beam from a xenon lamp (a suitable filter was used to remove light with λ <350 nm).
EPR parameters were as follows: receiver gain, 5.64 × 103; modulation amplitude, 4 G; and microwave power, 1.27 mW.
Assay of Oxidized Products. Unmetalated oligonucleotide strands were labeled at the 5′ end with 32P by using standard procedures (56). DNA duplexes were formed by mixing equal concentrations of complementary strands (30- and 42-mer) in 50 mM NaCl, 10 mM sodium phosphate, pH 7 and heating to 90°C followed by slow cooling to 20°C over 120 min. The Ru-tethered DNA strand (12-mer) was then added to the duplex, and the solution was heated to 37°C followed by slow cooling to 4°C. Samples containing 4 μM Ru-tethered DNA duplex and 80 μM quencher [Co(NH3)5Cl2+] were irradiated for 15 min at 4°C by using a He-Cd laser (≈13 mW at 442 nm). After irradiation, all samples were treated with 10% (vol/vol) piperidine at 90°C for 30 min, dried, and subjected to electrophoresis through a 20% denaturing polyacrylamide gel. The levels of damage were quantitated by using phosphorimagery (imagequant, Amersham).
Laser Spectroscopy. Time-resolved emission and transient absorption measurements used an excimer pumped dye (Coumarin 480) laser (λ = 480 nm) or a YAG-OPO laser (λexc = 470 nm) (40). Laser powers ranged from 1 to 2.5 mJ per pulse. The emission of the dppz complexes was monitored at 610 nm, and the emission intensities were obtained by integrating under the decay curve for the luminescence. MutY (20 μM) was first incubated with poly(dGC) (1 mM bp) at ambient temperature for 20 min in 5 mM sodium phosphate buffer, pH 7.5, with [Ru(NH3)6]3+ (400 μM) and [Ru(phen2dppz]2+ (20 μM).
Results
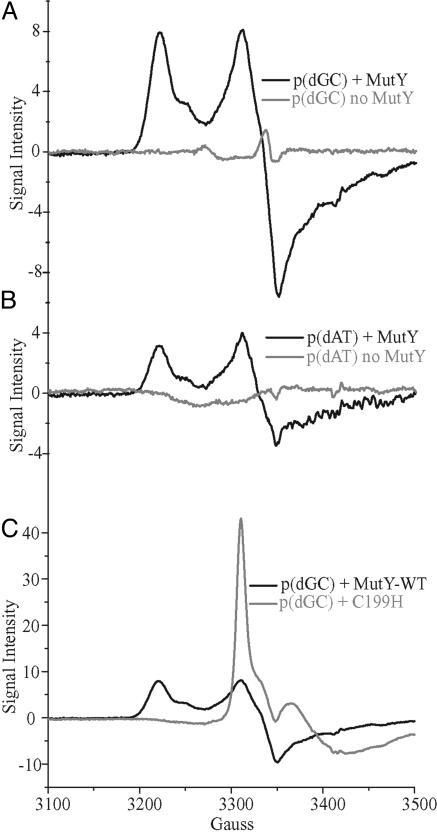
Flash-Quench Experiments with Poly(dGC) and Poly(dAT) Probed by EPR Spectroscopy. Solutions containing poly(dGC) or poly(dAT) (1 mM bp), [Ru(phen)2dppz]2+ (25 μM), and [Co(NH3)5Cl]2+ (125 μM) were irradiated in the presence or absence of MutY (50 μM). Samples were irradiated in EPR tubes while freezing in liquid nitrogen. EPR spectra were then acquired at 10 K. As shown in Fig. 1, in the absence of MutY, irradiation of poly-(dGC), [Ru(phen)2dppz]2+ and [Co(NH3)5Cl]2+ results in an EPR signal with g = 2.004; we attribute this signal, found previously, to the guanine radical (57, 58). Also as seen earlier and in contrast, with poly(dAT), this signal is not observed; [Ru(phen)2dppz]3+ has been seen to promote formation of the guanine radical but not the adenine radical cation.
Fig. 1.
EPR spectroscopy at 10 K of DNA samples after irradiation of [Ru(phen)2dppz]2+ (25 μM) with [Co(NH3)5Cl]2+ (125 μM) as quencher and poly(dGC) (1 mM bp) with and without MutY (50 μM) (A); poly(dAT) (1 mM bp) with and without MutY (50 μM) (B); and poly(dGC) (1 mM bp) with native MutY or C199H mutant (50 μM) (C).
More interesting are our observations in the presence of MutY. Irradiation results in the appearance of EPR signals with primary g values of 2.02 and 2.08 and a feature at 2.06 for both poly(dGC) and poly(dAT) (Fig. 1 A and B, respectively). The peak at g = 2.02 is characteristic of the [3Fe-4S]1+ cluster (59). Earlier studies of MutY bound to DNA and oxidized by Co(phen)33+ or MutY oxidized in the absence of DNA with ferricyanide (60) yielded the same EPR signal; the [3Fe-4S]1+ cluster can form as a decomposition product of [4Fe-4S]3+. Not seen earlier for MutY is the signal at g = 2.08 with a secondary feature at g = 2.06, and this g value is attributed to the fully intact, oxidized [4Fe-4S]3+ cluster (59, 61). In the absence of quencher, [Co(NH3)5Cl]2+, or DNA, no EPR signal is observed. Noteworthy, additionally, is that with poly(dAT) and MutY, both signals are also apparent, although at significantly lower intensity. Fluorescence experiments show that the concentration of excited Ru(II) and therefore Ru(III) is only slightly lower for poly(dAT) compared with poly(dGC). MutY therefore can be oxidized without guanine radical as an intermediate, but the formation of guanine radicals first may facilitate efficient MutY oxidation.
Also shown in Fig. 1 is the flash/quench result for poly(dGC) in the presence of the C199H mutant of MutY (60). Interestingly, this mutant yields an EPR spectrum that is characteristic only of the [3Fe-4S]1+ cluster. In addition, the signal intensity is significantly larger. In this particular mutant, the cluster is more susceptible to decomposition (60); thus it is not unexpected that this mutant only exhibits formation of the degraded cluster.
Flash-Quench Experiments with Poly(dGC) and Poly(dAT) Probed by Transient Absorption Spectroscopy. We also examined flash/quench reactions of [Ru(phen)2dppz]2+ bound to poly(dGC) with and without bound MutY on a faster time scale at ambient temperatures. Excitation of [Ru(phen)2dppz]2+ bound to poly-(dGC) by nanosecond laser pulses leads to an emission decay at 610 nm that can be fit biexponentially. This excited state is oxidatively quenched by [Ru(NH3)6]3+ in the presence (≈70% quenched) and absence (≈90% quenched) of MutY. Quenching is less efficient with bound MutY, however, likely because of restricted access of the quencher to [Ru(phen)2dppz]2+ when MutY is bound to DNA. MutY alone does not quench the excited state of [Ru(phen)2dppz]2+, indicating the absence of direct electron transfer from the protein to the [Ru(phen)2dppz]2+ excited state.
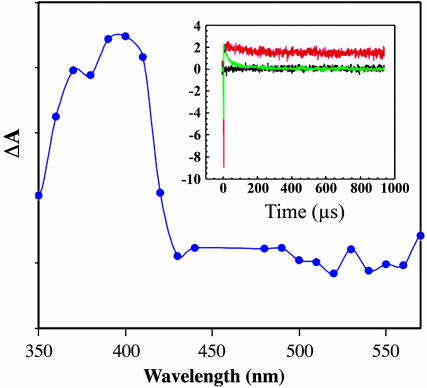
We probed these assemblies by transient absorption spectroscopy to obtain the full absorption difference spectrum with and without MutY bound to poly(dGC). At each wavelength, the transient absorption signal was fit as follows [A(t) = C0 + C1exp(-k1t)] and the coefficients for the fast phase (C1) and the slow phase (C0) were plotted against wavelength. The spectrum of the fast phase resembles the spectrum of the guanine radical in duplex DNA, with broad maxima at 390 and 510 nm (40). There appears to be less of this product in the presence of MutY, however. The spectrum of the slow phase shows evidence of the formation of a new species with an absorption maximum at ≈405 nm (Fig. 2). It is noteworthy that a [4Fe-4S]3+/2+ difference spectrum should show an absorption maximum near 405 nm (60, 62). This long-lived absorption is not observed with poly(dAT); the spectrum with poly(dAT) instead shows first a negative signal at 440 nm consistent with Ru(II) bleaching, with no long-lived signal. This long-lived signal is also not observed without inclusion of one or more of the necessary components: MutY, [Ru(phen)2dppz]2+, and [Ru(NH3)6]3+. Thus, these transient absorption data are consistent with formation first of a guanine radical upon oxidative flash/quench of [Ru(phen)2dppz]2+ bound to poly(dGC) in the presence of bound MutY, followed by a second species, likely [4Fe-4S]3+, that is very long lived.
Fig. 2.
Time-resolved transient absorption data for Ru(phen)2(dppz)2+ (20 μM) bound to poly(dGC) (1 mM bp) quenched by [Ru(NH3)6]3+ (0.4 mM) with MutY (20 μM). Shown is the absorption difference spectrum of the long-lived transient with data averaged over four experiments. (Inset) Transient absorption at 405 nm in the presence (red) and absence (green) of MutY bound to poly(dGC) or without DNA (black).
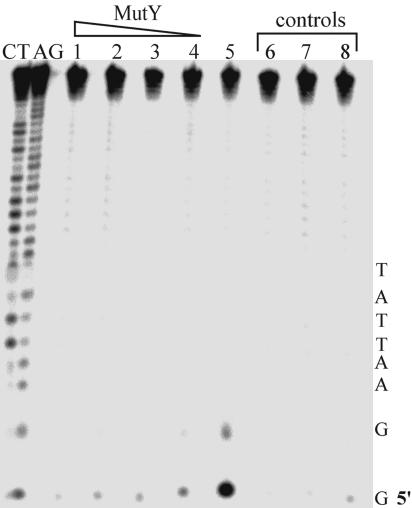
Flash-Quench Experiments with Ruthenium-Tethered Oligonucleotides. Shown in Fig. 3 are autoradiographs after denaturing PAGE of 32P-5′-end-labeled DNA duplexes covalently linked to a ruthenium intercalator, irradiated in the presence or absence of MutY. The DNA duplex was assembled from a 12-mer ruthenium-tethered strand, a 30-mer strand 32P-end-labeled containing a 5′-GG-3′ doublet, and the full 42-mer complement. In the absence of MutY, the typical 5′-G damage on the 5′-GG-3′ doublet guanine is observed; this guanine damage is expected upon oxidation from a distance through DNA-mediated CT from Ru(III) generated in situ. In the presence of 0.5–2 equivalents (2–8 μM) MutY, however, this damage is inhibited.
Fig. 3.
Autoradiogram after denaturing PAGE of 32P-5′-TTGGAATTATAATTTATAATATTAAATATT-3′ after oxidation of the ruthenium-tethered oligonucleotide duplex by flash/quench. Lanes shown are Maxam-Gilbert sequencing reactions for C + T and A + G. respectively. Lanes 1–5: Ru-DNA irradiated in the presence of cobalt quencher and 8, 6, 4, 2, or 0 μM MutY. Lane 6: Ru-DNA irradiated with 4 μM MutY but no quencher. Lane 7: Ru-DNA without MutY or quencher. Lane 8: DNA irradiated without Ru-tethered strand. Concentrations were [DNA] = 4 μM and [Co(NH3)5Cl]2+ = 200 μM. Irradiations were for 15 min. Reactions were carried out in 5 mM sodium phosphate, 50 mM NaCl, pH 7.
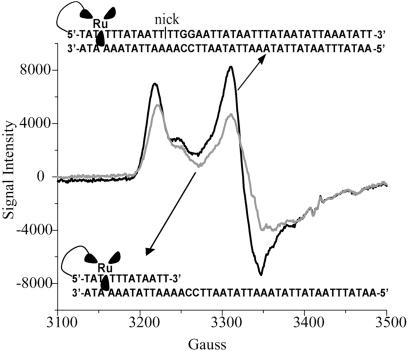
We also monitored the flash/quench reaction by EPR spectroscopy for the ruthenium-tethered oligonucleotide in the presence of MutY (Fig. 4). As with poly(dGC), here, too, at 10 K strong signals with g = 2.08, 2.06, and 2.02 are apparent, consistent with formation of the oxidized [4Fe-4S]3+ cluster as well as its decomposition product, [3Fe-4S]1+. Not apparent is any evidence of guanine radical formation in the absence of MutY; likely the lower concentration of guanine radical in this oligonucleotide assembly compared with poly(dGC) makes its detection by EPR more difficult.
Fig. 4.
EPR spectroscopy at 10 K of ruthenium-tethered DNA duplexes (25 μM, fully or partially hybridized) with MutY (50 μM) after irradiation in the presence of quencher {[Co(NH3)5Cl]2+, 125 μM}.
In addition, we examined the flash/quench reaction of the Ru-tethered duplex lacking the 30-mer strand. This assembly, composed of a short duplex region and long single-stranded segment, contains no guanines but can generate Ru(III) by flash/quench with yields comparable to that for the fully duplexed oligomer above. Yet this assembly in the presence of MutY results in an attenuated EPR signal in comparison with the fully hybridized duplex containing the 5′-GG-3′ doublet. Thus in the fully hybridized duplex, oxidation of MutY mediated by the DNA duplex must occur, and here, too, guanine radical formation appears to facilitate efficient MutY oxidation.
Discussion
DNA-Bound MutY Oxidation by the Flash/Quench Technique. Results reported here show clearly that DNA-bound Ru(III) can promote oxidation of the [4Fe-4S]2+ cluster of MutY to [4Fe-4S]3+ and its decomposition product [3Fe-4S]1+. Flash/quench experiments monitored by EPR spectroscopy reveal spectra with g values characteristic of the oxidized clusters. Earlier studies had shown a resistance to oxidation of the [4Fe-4S]2+ cluster of BER enzymes in the absence of DNA but an enhancement in oxidation in the presence of DNA (25, 63, 64). We have attributed this facility in oxidizing the DNA-bound proteins to the shift in oxidation potential associated with DNA binding.
Interestingly, these data provide a direct demonstration of the formation of [4Fe-4S]3+ in MutY. The signal with g = 2.08, 2.06 is characteristic of that seen for [4Fe-4S]3+ in high potential iron proteins (59, 61). We find some evidence for formation of the [4Fe-4S]3+ cluster in oxidation of DNA-bound uracil DNA glycosylase from Archaeoglobus fulgidus by Co(phen)33+ (unpublished data), but for Endo III and MutY from E. coli both oxidation by ferricyanide and Co(phen)33+ have produced only the oxidized but decomposed product, [3Fe-4S]1+ (60, 64).
It is useful in this context to consider our results for the C199H mutant. For this mutant, oxidative decomposition to [3Fe-4S]1+ is known to be facile, owing to the poorer coordination of the cluster by the histidine ligand (60). Our finding of a signal at g = 2.02 for C199H, characteristic of the [3Fe-4S]1+ cluster, helps us to assign the signal at g = 2.08, 2.06 for WT MutY to the one electron oxidized [4Fe-4S]3+. We suggest that the direct oxidation product is obtained by flash/quench, because this process is particularly fast. In this case, also, we use the tightly bound, well stacked DNA intercalator as oxidant rather than Co(phen)33+ or ferricyanide that do not bind deeply in the base pair stack by intercalation. Thus the direct, rapid formation of [4Fe-4S]3+ appears to be facilitated by the DNA-mediated oxidation of MutY.
The transient absorption data also provide a consistent picture. The long-lived transient, with a maximum absorption at 405 nm, is attributed primarily to formation of [4Fe-4S]3+ and possibly also [3Fe-4S]1+; both absorb more in this region than does [4Fe-4S]2+ (60–63). The shape of the spectrum has some features that resemble that of a tyrosine radical, and several tyrosine residues surround the cluster in the enzyme (36, 37), but the extinction coefficient for [4Fe-4S]3+ is expected to be significantly higher in this region, so that tyrosine radical or even guanine radical may not be distinguishable. Some tyrosine radical formation at ambient temperatures on a short time scale, or even tyrosine radical as a second intermediate, cannot be ruled out, however.
MutY Oxidation with Guanine Radical as an Intermediate. Oxidation of DNA-bound MutY does not necessitate a DNA-mediated charge transfer, but the data here illustrate that the DNA-mediated reaction can occur and guanine radical formation may facilitate MutY oxidation. The oxidation potential of guanine is -1.25 V versus NHE (65), the midpoint potential of DNA-bound MutY ([4Fe-4S]2+/3+)is + 0.1 V versus NHE (25), whereas the reduction potential of Ru(III) is 1.5 V versus NHE (40). Thus the net reaction for these charge transfer processes is thermodynamically favored. The biochemical data indicate that MutY inhibits long-range oxidative damage to guanines. The EPR data show that the flash/quench reaction promotes oxidation of the [4Fe-4S]2+ cluster in DNA-bound MutY. Taken together, these data show that MutY oxidation, the thermodynamic product, is formed at the expense of guanine radicals and accounts for the loss of irreversible oxidative DNA damage in the presence of MutY.
Cluster oxidation furthermore appears generally to occur in a DNA-mediated reaction. In the absence of DNA, no MutY oxidation occurs; DNA binding is required to shift the [4Fe-4S]3+/[4Fe-4S]2+ potential of MutY, activating it toward oxidation. Moreover, the Ru(III) oxidant must also be DNA-bound to have been generated from excited Ru(II); there is no detectable formation of Ru(II) excited state unless the complex is intercalated. Thus both MutY and the ruthenium complex must be bound to DNA. In addition, MutY oxidation was found to be greater for the full ruthenated 42-bp duplex assembly versus that lacking the 30-mer strand. Ru(III) formation is equivalent in these assemblies and the shorter duplex region along with the single-stranded tail in this assembly might be expected to facilitate direct encounters between Ru(III) and DNA-bound MutY. Yet, oxidation is greater with the longer duplex that contains a guanine site. Although some direct oxidation cannot be ruled out, oxidation mediated by a DNA duplex appears favored.
Is the cluster oxidized in competition with guanine oxidation or does guanine radical represents an intermediate in the CT process? The transient absorption spectroscopic data indicate that the guanine radical is formed on a fast time scale compared with the oxidized cluster formed in the presence of MutY. Low-temperature EPR data for polyd(GC) also indicate that [4Fe-4S]3+ and [3Fe-4S]1+ form, and the sharp organic radical signal is no longer apparent. In the case of poly(dAT), no base radical in the absence of protein has been observed; an adenine radical, if formed, would be expected to be short-lived, and the large negative bleach associated with Ru(III) makes detecting any small positive transients in this wavelength region difficult. In any case, the transient absorption data with poly(dGC) indicate quite clearly that guanine radical is formed in the presence of MutY but is depleted, and instead the [4Fe-4S]2+ cluster is oxidized.
Indeed, although a guanine radical is not required as an intermediate in MutY oxidation, its presence appears to enhance oxidation. In the absence of any guanines, both for poly(dAT) and the assembly lacking the 30-mer strand that only contains adenines and thymines, Ru(III), once generated, does oxidize DNA-bound MutY. But the yield of oxidation per Ru(III) is clearly greater with poly(dGC) than poly(dAT). Furthermore, in the assembly with the extended duplex containing a guanine site, the yield of cluster oxidation seen by EPR spectroscopy is significantly greater than in the assembly containing only a 12-mer duplex region and no guanines.
Why does the presence of intervening guanines appear to enhance the efficiency of cluster oxidation? It is reasonable to consider that guanine radical formation serves to compete with fast back electron transfer to the DNA-bound ruthenium so that there is more time for oxidation of MutY. The guanine radical lifetime in the absence of MutY is on the millisecond time scale (40). Thus a DNA-mediated oxidation of MutY can occur with or without intervening guanines, but guanine radical formation, the first DNA product under oxidizing conditions, facilitates the oxidation of DNA-bound MutY.
Implications for DNA Repair. Under conditions of oxidative stress, guanine radicals in DNA are generated and lead to the formation of 7,8-dihydro-8-oxo-2-deoxyguanosine (8-oxo-G); note that 8-oxo-G:A mismatches represent the primary substrate for MutY (37). The results presented here indicate that this signal of the need for DNA repair may in fact activate the repair machinery through protein oxidation. Fig. 5 shows our model for how DNA CT among BER enzymes may facilitate the detection of DNA lesions. The data here describe MutY oxidation, but other BER enzymes containing [4Fe-4S]2+ clusters show equivalent DNA-bound redox potentials. In our model, the BER enzyme, robust to oxidation in solution has a [4Fe-4S]2+ cluster. DNA binding shifts the cluster potential, promoting its oxidation to [4Fe-4S]3+, with DNA-mediated CT to another oxidized repair protein bound at a distal site along the duplex; reduction of this distal DNA-bound repair protein then facilitates dissociation from DNA and relocation onto another site. In this model, CT occurs effectively among the repair proteins bound along well matched, undamaged DNA and thus provides a strategy to scan the genome. However, when the protein binds to a region nearby a DNA lesion, DNA-mediated CT cannot occur, and the repair protein processively moves on a slower time scale to the site of the lesion and carries out its repair. Thus, DNA CT provides a route to redistribute the repair proteins onto regions of the genome containing DNA lesions.
Fig. 5.
Model for detection strategy for BER enzymes using DNA-mediated CT stimulated by guanine radicals. The guanine radicals, formed under oxidative stress, are reduced and hence repaired through DNA-mediated electron transfer from the BER enzyme (above). Oxidation of the repair protein then drives CT to an alternate repair protein bound at a distal site, thereby promoting the redistribution of DNA repair proteins on genomic sites. Because no DNA CT can proceed through intervening lesions, the proteins are preferentially redistributed onto sites near lesions (below). Thus guanine radicals, in oxidizing the DNA-bound repair proteins, and driving the redistribution, provide a signal to stimulate DNA repair.
Also, as illustrated in Fig. 5, guanine radicals, as effective oxidants of the repair proteins in a DNA-mediated reaction, may promote this redistribution. The guanine radicals, formed under oxidative stress, can essentially be “repaired” directly through DNA-mediated electron transfer from the repair protein. Significantly, oxidation of the repair protein through this process serves further to drive the redistribution of DNA repair proteins on genomic sites and hence preferentially onto sites near lesions. Thus guanine radicals, in oxidizing the DNA-bound repair proteins, can provide a signal to stimulate DNA repair.
DNA CT chemistry has been seen earlier to provide a route to carry out oxidative DNA damage from a distance. This chemistry also has been seen to be exquisitely sensitive to the presence of mismatches, lesions, and generally perturbations to the base pair stack, and as a result provides a sensor for mismatches and lesions in DNA. Here, we see that this chemistry may also provide a unique biological signal within the cell. Oxidative damage from a distance may itself provide a stimulus for DNA CT among DNA-bound proteins and hence for activation of DNA repair.
Acknowledgments
We thank the National Institutes of Health for their financial support of this research.
Author contributions: E.D.A.S., S.S.D., and J.K.B. designed research; E.Y., A.K.B., E.D.A.S., and E.M.B. performed research; A.L.L., V.L.O., and S.S.D. contributed new reagents/analytic tools; E.Y., A.K.B., and E.M.B. analyzed data; and E.Y., A.K.B., and J.K.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CT, charge transport; BER, base excision repair; dppz, dipyridophenazine.
References
- 1.Hall, D. B., Holmlin, R. E. & Barton, J. K. (1996) Nature 382, 731-735. [DOI] [PubMed] [Google Scholar]
- 2.Hall, D. B., Kelley, S. O. & Barton, J. K. (1998) Biochemistry 37, 15933-15940. [DOI] [PubMed] [Google Scholar]
- 3.Gasper, S. M. & Schuster, G. B. (1997) J. Am. Chem. Soc. 119, 12762-12771. [Google Scholar]
- 4.Meggers, E., Kusch, D., Spichty, M., Wille, U. & Giese, B. (1998) Angew. Chem. Int. Ed. 37, 460-462. [DOI] [PubMed] [Google Scholar]
- 5.Nakatani, K., Dohno, C. & Saito, I. (1999) J. Am. Chem. Soc. 121, 10854-10855. [Google Scholar]
- 6.Núñez, M. E., Noyes, K. T., Gianolio, D. A., McLaughlin, L. W. & Barton, J. K. (2000) Biochemistry 39, 6190-6199. [DOI] [PubMed] [Google Scholar]
- 7.Núñez, M. E., Hall, D. B. & Barton, J. K. (1999) Chem. Biol. 6, 85-97. [DOI] [PubMed] [Google Scholar]
- 8.Ly, D., Sanii, L. & Schuster, G. B. (1999) J. Am. Chem. Soc. 121, 9400-9408. [Google Scholar]
- 9.Núñez, M. E., Noyes, K. T. & Barton, J. K. (2002) Chem. Biol. 9, 403-415. [DOI] [PubMed] [Google Scholar]
- 10.Núñez, M. E., Holmquist, G. P. & Barton, J. K. (2001) Biochemistry 40, 12465-12471. [DOI] [PubMed] [Google Scholar]
- 11.Rajski, S. R., Kumar, S., Roberts, R. J. & Barton, J. K. (1999) J. Am. Chem. Soc. 121, 5615-5616. [Google Scholar]
- 12.Bhattacharya, P. K. & Barton, J. K. (2001) J. Am. Chem. Soc. 123, 8649-8656. [DOI] [PubMed] [Google Scholar]
- 13.Hall, D. B. & Barton, J. K. (1997) J. Am. Chem. Soc. 119, 5045-5046. [Google Scholar]
- 14.Boon, E. M., Ceres, D. M., Drummond, T. G., Hill, M. G. & Barton, J. K. (2000) Nat. Biotechnol. 18, 1096-1100. [DOI] [PubMed] [Google Scholar]
- 15.Boon, E. M., Salas, J. W. & Barton, J. K. (2002) Nat. Biotechnol. 20, 282-286. [DOI] [PubMed] [Google Scholar]
- 16.Wagenknecht, H.-A., Rajski, S. R., Pascaly, M., Stemp, E. D. A. & Barton, J. K. (2001) J. Am. Chem. Soc. 123, 4400-4407. [DOI] [PubMed] [Google Scholar]
- 17.Giese, B., Carl, B., Carl, T., Carell, T., Behrens, C., Hennecke, U., Schieman, O. & Feresin, E. (2004) Angew. Chem. Int. Ed. 43, 1848-1851. [DOI] [PubMed] [Google Scholar]
- 18.Breeger, S., Hennecke, U. & Carell, T. (2004) J. Am. Chem. Soc. 126, 1302-1303. [DOI] [PubMed] [Google Scholar]
- 19.Ito, T. & Rokita, S. E. (2004) J. Am. Chem. Soc. 126, 15552-15559. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, F. D., Liu, X., Miller, S. E., Hayes, R. T. & Wasielewski, M. R. (2002) J. Am. Chem. Soc. 124, 11280-11281. [DOI] [PubMed] [Google Scholar]
- 21.Kelley, S. O., Jackson, N. M., Hill, M. G. & Barton, J. K. (1999) Angew. Chem. Int. Ed. 38, 941-945. [DOI] [PubMed] [Google Scholar]
- 22.Barnett, R. N., Cleveland, C. L., Joy, A., Landman, U. & Schuster, G. B. (2001) Science 294, 567-571. [DOI] [PubMed] [Google Scholar]
- 23.O'Neill, M. A. & Barton, J. K. (2004) J. Am. Chem. Soc. 126, 11471-11483. [DOI] [PubMed] [Google Scholar]
- 24.O'Neill, M. A. & Barton, J. K. (2004) J. Am. Chem. Soc. 126, 13234-13235. [DOI] [PubMed] [Google Scholar]
- 25.Boon, E. M., Livingston, A. L., Chmiel, N. H., David, S. S. & Barton, J. K. (2003) Proc. Natl. Acad. Sci. USA 100, 12543-12547, and correction (2004) 101, 4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David, S. S. & Williams, S. D. (1998) Chem. Rev. 98, 1221-1262. [DOI] [PubMed] [Google Scholar]
- 27.Radicella, J. P., Clark, E. A. & Fox, M. S. (1998) Proc. Natl. Acad. Sci. USA 85, 9674-9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porello, S. L., Cannon, M. J. & David, S. S. (1998) Biochemistry 37, 6465-6475. [DOI] [PubMed] [Google Scholar]
- 29.Nghiem, Y., Cabrera, M., Cupples, C. G. & Miller, J. H. (1998) Proc. Natl. Acad. Sci. USA 85, 2709-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaels, M. L., Cruz, C., Grollman, A. P. & Miller, J. H. (1992) Proc. Natl. Acad. Sci. USA 89, 7022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michaels, M. L. & Miller, J. H. (1992) J. Bacteriol. 174, 6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaels, M. L., Tchou, J., Grollman, A. P. & Miller, J. H. (1992) Biochemistry 17, 10964-10968. [DOI] [PubMed] [Google Scholar]
- 33.Manuel, R. C., Czerwinski, E. W. & Lloyd, R. S. (1996) J. Biol. Chem. 271, 16218-16226. [DOI] [PubMed] [Google Scholar]
- 34.McGoldrick, J. P., Yeh, Y. C., Solomon, M., Essigmann, J. M. & Lu, A. L. (1995) Mol. Cell. Biol. 15, 989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desiraju, V., Shanabruch, W. & Lu, A. (1993) J. Bacteriol. 175, 541-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan, Y., Manuel, R. C., Arvai, A. S., Parikh, S. S., Mol, C. D., Miller, J. H., Lloyd, S. & Tainer, J. A. (1998) Nat. Struct. Biol. 5, 1058-1064. [DOI] [PubMed] [Google Scholar]
- 37.Fromme, J. C., Banerjee, A., Huang, S. J. & Verdine, G. L. (2004) Nature 427, 652-656. [DOI] [PubMed] [Google Scholar]
- 38.Chang, I.-J., Gray, H. B. & Winkler, J. R. (1991) J. Am. Chem. Soc. 113, 7056-7057. [Google Scholar]
- 39.Yoo, J., Delaney, S., Stemp, E. D. A. & Barton, J. K. (2003) J. Am. Chem. Soc. 125, 6640-6641. [DOI] [PubMed] [Google Scholar]
- 40.Stemp, E. D. A., Arkin, M. R. & Barton, J. K. (1997) J. Am. Chem. Soc. 119, 2921-2925. [Google Scholar]
- 41.Delaney, S., Yoo, J., Stemp, E. D. A. & Barton, J. K. (2004) Proc. Natl. Acad. Sci. USA 101, 10511-10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaney, S., Pascaly, M., Bhattacharya, P. K., Han, K. & Barton, J. K. (2002) Inorg. Chem. 41, 1966-1974. [DOI] [PubMed] [Google Scholar]
- 43.Arkin, M. R., Stemp, E. D. A., Coates Pulver, S. & Barton, J. K. (1997) Chem. Biol. 4, 389-400. [DOI] [PubMed] [Google Scholar]
- 44.Freidman, A. E., Chambron, J.-C., Sauvage, J.-P., Turro, N. J. & Barton, J. K. (1990) J. Am. Chem. Soc. 112, 4960-4961. [Google Scholar]
- 45.Burrows, C. J. & Muller, J. G. (1998) Chem. Rev. 98, 1109-1151. [DOI] [PubMed] [Google Scholar]
- 46.Wagenknecht, H.-A., Stemp, E. D. A. & Barton, J. K. (2000) Biochemistry 39, 5483-5491. [DOI] [PubMed] [Google Scholar]
- 47.Anderson, P. A., Deacon, G. B., Haarman, K. H., Keene, F. R., Meyer, T. J., Reitsma, D. A., Skelton, B. W., Strouse, G. F., Thomas, N. C. & Treadway, J. A. (1995) Inorg. Chem. 34, 6145-6157. [Google Scholar]
- 48.Amouyal, E., Homsi, A., Chambron, J.-C. & Sauvage, J.-P. (1990) J. Chem. Soc. Dalton Trans. 6, 1841-1846. [Google Scholar]
- 49.Della Ciena, L., Hamachi, I. & Meyer, T. J. (1989) J. Org. Chem. 54, 1731-1735. [Google Scholar]
- 50.Dupureur, C. M. & Barton, J. K. (1997) Inorg. Chem. 36, 33-43. [Google Scholar]
- 51.Strouse, G. F., Anderson, P. A., Schoonover, J. R., Meyer, T. J. & Keene, F. R. (1992) Inorg. Chem. 31, 3004-3006. [Google Scholar]
- 52.Goodchild, J. (1990) Bioconjugate Chem. 1, 165-187. [DOI] [PubMed] [Google Scholar]
- 53.Beaucage, S. L. & Caruthers, M. H. (1981) Tetrahedron Lett. 22, 1859-1862. [Google Scholar]
- 54.Holmlin, R. E., Dandliker, P. J. & Barton, J. K. (1999) Bioconjugate Chem. 10, 1122-1130. [DOI] [PubMed] [Google Scholar]
- 55.Chmiel, N. H., Golinelli, M.-P., Francis, A. W. & David, S. S. (2001) Nucleic Acids Res. 29, 553-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 57.Schiemann, O., Turro, N. J. & Barton, J. K. (2000) J. Phys. Chem. B 104, 7214-7220. [Google Scholar]
- 58.Cullis, P. M., Malone, M. E. & Merson Davies, L. A. (1996) J. Am. Chem. Soc. 118, 2775-2781. [Google Scholar]
- 59.Cowan, J. A. & Lui, S. M. (1998) Adv. Inorg. Chem. 45, 313-350. [Google Scholar]
- 60.Messick, T. E., Chmiel, N. H., Golinelli, M.-P., Langer, M. R., Joshua-Tor, L. & David, S. S. (2002) Biochemistry 41, 3931-3942. [DOI] [PubMed] [Google Scholar]
- 61.Dilg, A. W. E., Mincione, G., Achterhold, K., Iakovleva, O., Mentler, M., Luchinat, C., Bertini, I. & Parak, F. G. (1999) J. Biol. Inorg. Chem. 4, 727-741. [DOI] [PubMed] [Google Scholar]
- 62.Johnson, M. K., Duderstadt, R. E. & Dunn, E. C. (1999) Adv. Inorg. Chem. 47, 1-82. [Google Scholar]
- 63.Rogers, P. A., Eide, L., Klungland, A. & Ding, H. (2003) DNA Repair 2, 809-817. [DOI] [PubMed] [Google Scholar]
- 64.Cunningham, R. P., Asahara, H. & Bank, J. F. (1989) Biochemistry 28, 4450-4455. [DOI] [PubMed] [Google Scholar]
- 65.Steenken, S. & Jovanovic, S. V. (1997) J. Am. Chem. Soc. 119, 617-618. [Google Scholar]