Summary
Susceptibility to obesity is linked to genes regulating neurotransmission, pancreatic β-cell function, and energy homeostasis. Genome-wide association studies identified an association between body mass index and two loci near Cell Adhesion Molecule1 (CADM1) and Cell Adhesion Molecule2 (CADM2), genes encoding membrane proteins mediating synaptic assembly. We show these respective risk variants associate with increased CADM1 and CADM2 expression in the hypothalamus of human subjects. Expression of both genes is elevated in obese mice and induction of Cadm1 in excitatory neurons facilitated weight gain while exacerbating energy expenditure. Loss of Cadm1 protected mice from obesity and tract-tracing analysis revealed Cadm1-positive innervation of POMC neurons via afferent projections originating from beyond the arcuate nucleus. Reducing Cadm1 expression in the hypothalamus and hippocampus promoted a negative energy balance and weight loss. These data identify roles for Cadm1-mediated neuronal input in weight regulation and provide insight into the central pathways contributing to human obesity.
Obesity is one of the leading global health threats with a projected burden on public health that appears interminable1. Numerous reports support a crucial role for genetic susceptibility to the risk for obesity and related metabolic disorders including type 2 diabetes2,3,4. Recent studies have now identified ~100 susceptibility loci, which associate with body mass index and map near genes known to function in the central nervous system5,6,7. Among the genes identified in these studies are CADM1 and CADM2 (also known as SynCAM1 and SynCAM2, respectively)7, which encode two immunoglobulin domain-containing, adhesion proteins that mediate synaptic assembly in the central nervous system8,9,10. In this study, we illustrate how increased expression of both Cadm1 and Cadm2 in regions of the brain facilitates weight gain and how targeting their expression can provide new insight into the mechanisms leading to human obesity.
SNPs associate with increased CADM genes
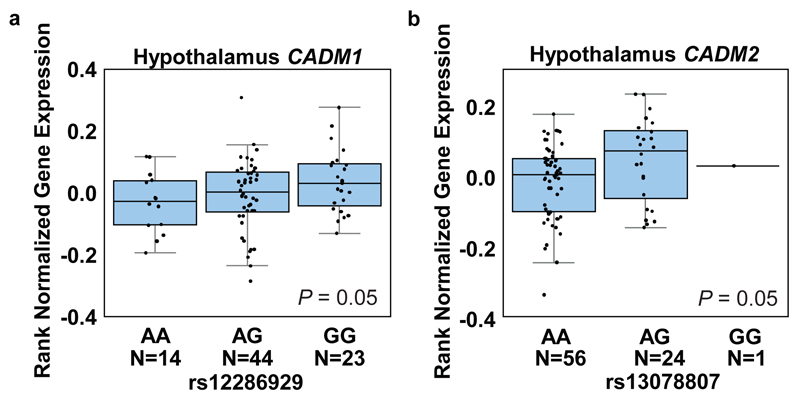
Recent genome-wide association studies (GWAS) identified two single nucleotide polymorphisms (SNPs) (rs12286929) and (rs13078807) in proximity to CADM1 and CADM2, respectively, which associate with increased body mass index (BMI) in human subjects6,7. The SNP rs12286929 is located in an intergenic region 17 kb downstream of the 3’ end of CADM1 and the SNP rs13078807 is located in the second intron of CADM2. We analyzed expression quantitative trait locus (eQTL) data from 10 distinct regions of the brain available from the GTEx consortium and observed that both risk alleles (the alleles associated with increased BMI) are associated with increased mRNA expression of their proximal genes in the hypothalamus and cerebellum of human subjects: CADM1 (hypothalamus (Allelic effect: 10% s.d., P=0.05) and cerebellum (Allelic effect: 17% s.d., P=0.02)) and CADM2 (hypothalamus (Allelic effect: 19% s.d., P=0.05) and cerebellum (Allelic effect: 30% s.d., P=0.004)) (Fig. 1a,b and Supplementary Fig. 1a,b)11. Cadm1 and Cadm2 have been shown to interact via their extracellular domains (Supplementary Fig. 2a)9,12 and thus might act synergistically. We therefore evaluated how altered expression of these genes contributes to the regulation of body weight using rodent models.
Figure 1.
BMI risk SNPs associate with increased CADM1 and CADM2 expression in the hypothalamus of human subjects. Boxplots show the 25% and 75% quantiles of normalized mRNA expression levels (y-axis), solid horizontal lines indicate the median, and whiskers indicate the 10% and 90% quantiles. (a) Elevated expression of CADM1 associates with risk allele (G) of rs12286929 in hypothalamus. (b) Genotype dependent expression levels of CADM2 for the SNP rs13078807. The risk allele (G) is associated with higher expression levels in human hypothalamus. Statistical analyses are described in the Methods and Supplementary Table 3.
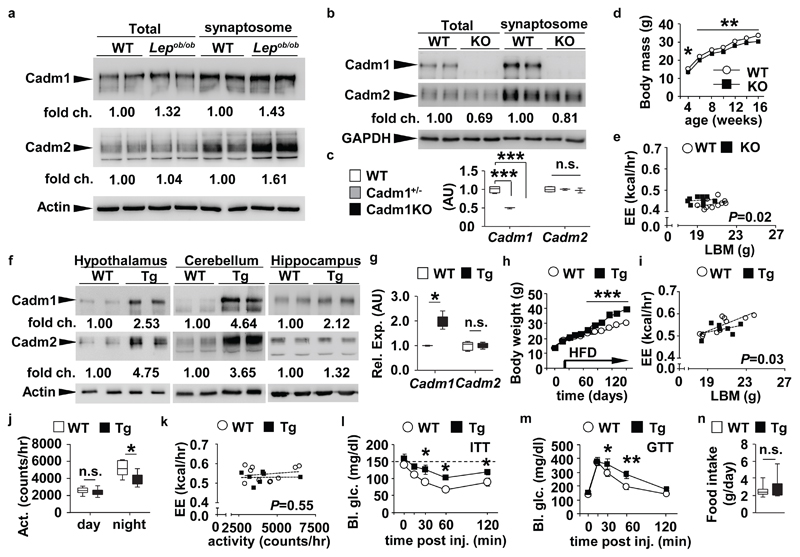
Consistent with our eQTL analysis, we observed increased Cadm1 and Cadm2 protein levels in isolated synaptosomes from hypothalamus, cerebellum, and hippocampus of obese and insulin-resistant Lepob/ob mice compared to lean littermate controls (Fig. 2a and Supplementary Fig. 2b,c and 10,11). Since administration of a low-carbohydrate, ketogenic diet has been shown to improve insulin sensitivity and random blood glucose levels in Lepob/ob animals13, we tested its impact on neuronal Cadm1 expression. Feeding 16-week-old Lepob/ob animals this diet for 60 days reduced the expression of Cadm1 in the hippocampus (hpc) and cerebellar regions, whereas the diet elicited no change in wild-type (WT) mice (Supplementary Fig. 2d-g and 10-14).
Figure 2.
Induction of Cadm1 in excitatory neurons increases body weight. (a) Western blot analysis of Cadm1 and Cadm2 in total and synaptosome-enriched lysates from hypothalamus of 4-week-old Lepob/ob mice and littermate controls. (b) Western blot analysis of Cadm1 and Cadm2 in total and synaptosome-enriched lysates from hippocampus of 3-week-old Cadm1KO and control littermates. (c) qRT-PCR analysis of Cadm1 and Cadm2 expression in hippocampus of 5-week old Cadm1KO mice, Cadm1−/+ (heterozygous) and control littermates (WT) (n=3-4). (d) Body weight curves of Cadm1KO mice (n=10) and littermate controls (n=10). (e) Energy expenditure of individual animals plotted against lean body mass (LBM) in 12-week old Cadm1KO (n=9) and littermate controls (n=10). (f) Western blot analysis of Cadm1 and Cadm2 in total lysates from hypothalamus, cerebellum, and hippocampus of 14-week-old Tg-Cadm1 mice and control littermates. (g) Quantification of Cadm1 and Cadm2 expression from cultured primary hippocampal neurons from Tg-Cadm1 mice (n=5) and wild-type littermate controls (n=5) after 5 days of treatment with doxycycline. (h) Body weight curves of Tg-Cadm1 mice (n=5) and littermate controls (n=8) upon doxycycline treatment at 4 weeks of age. High fat diet feeding was initiated on day 25 of doxycycline treatment. (i) Energy expenditure of individual animals plotted against lean body mass (LBM) in 16-week old Tg-Cadm1 mice (n=7) and littermate controls (n=10). (j) Quantification of locomotor activity measured in 16-week old Tg-Cadm1 mice (n=7) and controls (n=8). (k) Energy expenditure of individual animals plotted against locomotor activity in 16-week old Tg-Cadm1 mice (n=7) and littermate controls (n=10). (l) Blood glucose measurements during an insulin tolerance test (ITT) on 20-week old Tg-Cadm1 mice (n=5) and control littermates (n=6). (m) Glucose measurements during a glucose tolerance test (GTT) on 18-week old Tg-Cadm1 mice (n=5) and littermates (n=7). (n) Quantification of food intake in 16-week old Tg-Cadm1 mice (n=7) and littermates (n=8). All results are presented as mean ± s.e.m. *P<0.05, ** P<0.01, *** P<0.001. Boxplots show median, lower and upper quartiles (box), and minimum and maximum (whiskers). Statistical analyses are described in the Methods and Supplementary Table 3.
These results show that increased neuronal expression of Cadm1 and Cadm2 in the human and mouse brain associates with elevated body weight and indicate that these two genes may play a significant role in maintaining energy balance. The reduction of Cadm1 expression in multiple brain regions of Lepob/ob mice on a ketogenic diet also underscores the highly-regulated nature of Cadm1 and suggests it may contribute to adaptive or homeostatic plasticity within neuronal networks in accordance to nutrient state.
Cadm1 regulates energy homeostasis
To understand the endogenous function of Cadm1, we first characterized a global loss-of-function mouse model (Cadm1KO) and observed a reduction in body weight compared to littermate controls starting by post-natal day 3 (Fig. 2b-d and Supplementary Fig. 3a). A concomitant decrease in Cadm2 expression independent of changes on its mRNA level indicates Cadm1 may contribute to the stability of Cadm2 and its function (Fig. 2c), consistent with their physical association. Cadm1KO mice had a lower proportion of body fat compared to controls but indistinguishable random glucose and insulin levels (Supplementary Tables 1 and 2). Body length was slightly lower in KO mice (9.1±0.1 WT vs. 8.5±0.1 cm Cadm1KO mice, P-value<0.05 at age 12 weeks) in addition to the body weight to length ratio suggesting the decrease in body mass was due to both reduced growth rate and adipose mass (Supplementary Fig. 3b and Supplementary Table 2). We performed insulin, glucose, and pyruvate tolerance tests (ITT, GTT, and PTT, respectively) and observed lower glucose levels in Cadm1KO mice compared to controls suggesting systemic insulin sensitivity was improved (Supplementary Fig. 3c-e). We next tested sensitivity to leptin, another potent regulator of energy homeostasis. Intraperitoneal administration of leptin resulted in a greater decrease of both body mass and food intake in Cadm1KO mice (Supplementary Fig. 3f,g). Moreover, consistent with improved sensitivity to leptin, basal plasma leptin levels were significantly lower and plasma adiponectin levels were elevated in mutant animals compared to controls (Supplementary Table 1).
As insulin and leptin signaling within the brain both influence body mass and energy homeostasis14,15,16, we next addressed whether energy expenditure rates were altered in Cadm1KO animals. O2 consumption, CO2 production, energy expenditure and locomotor activity were all higher in Cadm1KO mice, while food intake, core body temperature and nutrient partitioning (based on respiratory exchange rate (RER)) were normal (Supplementary Fig. 3h-k). Evaluation of energy expenditure using analysis of covariance (ANCOVA), as plotted in relation to lean body mass, confirmed the increased rates in Cadm1KO animals (Fig. 2e)17. When energy expenditure was plotted in relation to locomotor activity, this comparison confirmed Cadm1KO animals maintain increased energy expenditure and this elevated rate is directly correlated to increased activity (Supplementary Fig. 3l,m). Similar to Cadm1KO mice, global deletion of Cadm2 in mice (Cadm2KO) also resulted in lower body weight, and increased insulin sensitivity, glucose tolerance, and energy expenditure without any change in food intake; further suggesting Cadm1 and Cadm2 may interact to coordinately contribute to body weight regulation (Supplementary Fig. 3n-y and 14 and Supplementary Table 2).
Pro-opiomelanocortin (POMC)-expressing neurons are among the most studied cells in the brain due to their regulatory role in glucose utilization and energy expenditure18,19. In wild-type C57BL/6 mice, we observed ~12% of POMC-expressing neurons were positive for Cadm1 immunostaining (Supplementary Fig. 4a,b). Meanwhile no changes in the expression of the hypothalamic neuropeptides Pomc, Npy, and Agrp were observed in Cadm1KO mice in contrast to Lepob/ob controls (Supplementary Fig. 4c). As several studies have now reported on Cadm1 function in neuronal signaling and plasticity20,21, we sought to determine whether the POMC neurons within the arcuate nucleus (ARC) were functionally altered in Cadm1KO mice and we first analyzed miniature excitatory and inhibitory post-synaptic currents (EPSC and IPSC, respectively). Recordings of IPSCs in POMC neurons from Cadm1KO mice in comparison to littermate controls, showed a significant reduction in IPSC frequency without any effect on mean amplitude, while no difference was observed in EPSCs derived from their glutamatergic inputs (Supplementary Fig. 4d,e and 11). We then quantified the number of synapses in contact with these neurons. Loss of Cadm1 resulted in a significant increase in the number of asymmetric excitatory synapses (2.3±0.5 in WT and 7.1±0.9 synapses in Cadm1KO, P-value<0.001) at POMC neurons (Supplementary Fig. 4f-h), distinct from the reported very mild reduction of excitatory synapse number in hippocampal CA1 neurons20. The increase in excitatory contacts at POMC neurons in Cadm1KO mice and the absence of any change in mEPSCs frequency in these neurons may indicate that many of the observed excess connections are ‘silent’ and thereby yield an increased capacity for synaptic plasticity22. Homeostatic changes in synaptic plasticity and/or the imbalance of excitation and inhibition in the brain20,23, may conceivably underlie the effects on energy homeostasis in Cadm1KO mice, a hypothesis further tested below.
Induction of Cadm1 promotes weight gain
To date, the contribution of Cadm1 and Cadm2 in both glutamatergic and GABAergic populations to energy homeostasis has not been directly addressed (Supplementary Fig. 5a-e). Previous studies have shown the role of Cadm1 in synaptic plasticity and the balance of network excitability is derived from its expression in excitatory neurons20,21. Based on these results, we directly tested whether increasing Cadm1 expression in a subset of glutamatergic neurons contributes to energy homeostasis by generating a mouse line bearing a transgene encoding Cadm1 under the control of seven tandem doxycycline-responsive elements (Tg-Cadm1) (Fig. 2f and Supplementary Fig. 5f). These mice were crossed to also carry alleles expressing vesicular glutamate transporter 2 (Slc17a6)-Cre (where the recombinase is under control of the endogenous Slc17a6 promoter) and lox-rtTA (where the reverse tetracycline transactivator is expressed in the presence of Cre recombinase) and treatment of doxycycline via drinking water resulted in over-expression of Cadm1 in many brain regions including a ~2-3-fold increase in the hypothalamus (Fig. 2f,g and Supplementary Fig. 12). Tg-Cadm1 mice exhibited increased body weight and adipose mass ~30 days after initiation of high-fat-diet feeding compared to controls (Fig. 2h and Supplementary Table 2). The induction of Cadm1 expression in Vglut2-expressing excitatory neurons reduced energy expenditure and locomotor activity and attenuated insulin sensitivity and glucose tolerance further suggesting the higher Cadm1 levels observed in Lepob/ob mice contributed to the obesity, insulin resistance, and hyperglycemia (Fig. 2i-m and Supplementary Fig. 5f,g). Increasing Cadm1 levels promoted Cadm2 expression as well as their co-localization, without any effect on Cadm2 mRNA levels, providing further evidence that the stability of Cadm2 is dependent upon Cadm1 expression (Fig. 2g and Supplementary Fig. 5h-j). As observed in Cadm1KO and Cadm2KO mice, the effect on body weight in Tg-Cadm1 mice was associated with an alteration in energy expenditure and not in food intake (Fig. 2n).
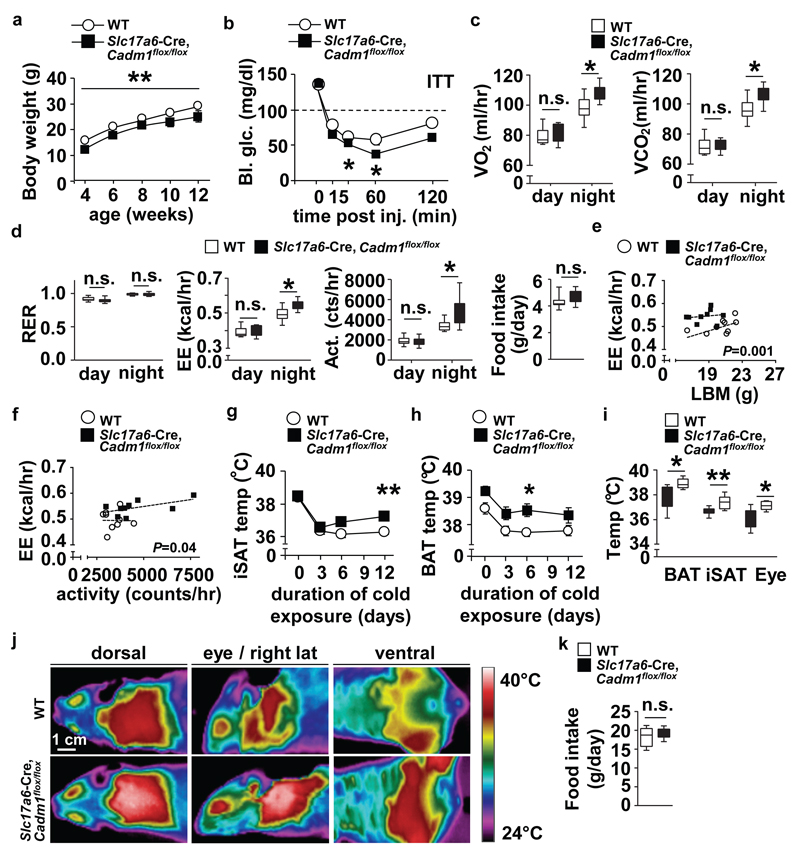
To further distinguish the function of Cadm1 in specific neuronal populations, we next eliminated its expression in either Vglut2-expressing excitatory or Vgat-expressing inhibitory neurons (by crossing Slc17a6-Cre and vesicular gamma-aminobutyric acid (GABA) transporter (Slc32a1)-Cre mouse lines with a Cadm1 conditional allele, Slc17a6-Cre, Cadm1fl/fl and Slc32a1-Cre, Cadm1flox/flox, respectively) (Supplementary Fig. 6a-d and 14)19. Similar to Cadm1KO animals, Slc17a6-Cre, Cadm1flox/flox mice recapitulated the effects on body mass, insulin and pyruvate challenge, energy expenditure, and activity (Fig. 3a-f, Supplementary Fig. 6e,f, and Supplementary Table 2). Infrared image analysis showed Slc17a6-Cre, Cadm1flox/flox mice exhibited elevated body temperatures in the eye, inguinal subcutaneous adipose tissue (iSAT) and interscapular brown adipose tissue (BAT) regions after exposure to cold without affecting food intake (Fig. 3g-k). In comparison to Slc17a6-Cre, Cadm1flox/flox mice, loss of Cadm1 in Slc32a1-Cre+ neurons resulted only in reduced insulin sensitivity compared to controls (Supplementary Fig. 6g-l). No changes were observed in body weight, glucose tolerance, energy expenditure, or food intake indicating that the effect of altered Cadm1 expression on body weight and energy homeostasis appears to be derived from its expression in Vglut2-expressing excitatory neurons.
Figure 3.
Loss of Cadm1 in excitatory neurons impacts body weight, energy homeostasis and thermogenesis. (a) Body weight curves of Slc17a6-Cre, Cadm1flox/flox mice (n=12) and littermate controls (n=14) from age 4-12 weeks. (b) Glucose measurements during an ITT on 12-week old Slc17a6-Cre, Cadm1flox/flox mice (n=5) and control littermates (n=6). (c, d) Quantification of O2 consumption, CO2 production, RER, energy expenditure, locomotor activity, and food intake in 12-week old Slc17a6-Cre, Cadm1flox/flox mice (n=9) and littermate controls (n=10). (e) Energy expenditure of individual animals plotted against lean body mass from 12-week old Slc17a6-Cre, Cadm1flox/flox mice (n=9) and littermate controls (n=10). (f) Energy expenditure of individual animals plotted against locomotor activity in 12-week old Slc17a6-Cre, Cadm1flox/flox mice (n=9) and littermate controls (n=10). (g) Inguinal subcutaneous adipose tissue (iSAT) temperature measurements from 12-week old Slc17a6-Cre, Cadm1flox/flox mice (n=5) and control littermates (n=8) during cold challenge. (h) Brown adipose tissue (BAT) temperature measurements from 12-week old Slc17a6-Cre, Cadm1flox/flox mice (n=5) and control littermates after (n=8) during cold challenge. (i) Temperature measurements of BAT, iSAT and eye regions from 12-week old Slc17a6-Cre, Cadm1flox/flox mice (n=5) and control littermates after (n=8) after one week exposure to cold. (j) Representative near-infrared analysis of 12-week old Slc17a6-Cre, Cadm1flox/flox mice and control littermates after one week cold exposure. (k) Quantification of daily food intake of Slc17a6-Cre, Cadm1flox/flox mice (n=5) and control littermates after (n=8) after cold exposure. All results are presented as mean ± s.e.m. *P<0.05 and **P<0.01. Boxplots show median, lower and upper quartiles (box), and minimum and maximum (whiskers). Statistical analyses are described in the Methods and Supplementary Table 3.
Reduction of Cadm1 protects from obesity
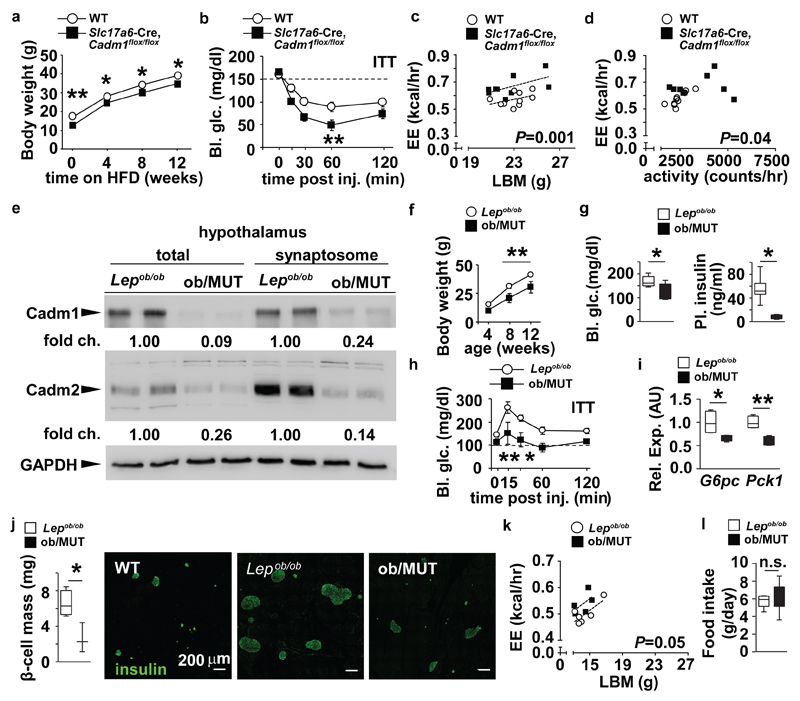
Underscoring the functional importance of Cadm1 in excitatory neurons, Slc17a6-Cre, Cadm1flox/flox mice were protected from diet-induced obesity and insulin resistance after challenging these mice with a high-fat regimen (Fig. 4a-d). To accurately assess the impact of Cadm1 on systemic insulin sensitivity and tissue-specific glucose turnover rates in response to this diet, we performed hyperinsulinemic-euglycemic clamp studies on Slc17a6-Cre, Cadm1flox/flox animals after 8 weeks of feeding. Basal blood glucose and insulin levels did not differ with Cadm1 status; however, after clamping glucose levels at 120 mg/dl, the glucose infusion rate was significantly elevated in Slc17a6-Cre, Cadm1flox/floxl animals confirming improved overall insulin sensitivity (Supplementary Fig. 6m,n). While basal hepatic glucose output was unchanged, clamped hepatic output was significantly decreased (Supplementary Fig. 6o). In addition, consistent with improved hepatic insulin sensitivity, no differences were observed in hepatic glucose uptake, rate of glycolysis or glycogen synthesis or insulin levels at the end of the clamp between Slc17a6-Cre, Cadm1flox/flox and WT animals (Supplementary Fig. 6p). The ability to reverse obesity and improve glucose homeostasis after targeting neuronal Cadm1 expression was further shown after we crossed Slc17a6-Cre, Cadm1flox/flox mice onto the Lepob/ob background (ob/MUT) and observed lower body mass, circulating random glucose and plasma insulin levels compared to Lepob/ob littermates (Fig. 4e-g, Supplementary Fig. 12 and Supplementary Table 2). Results from an ITT, and energy expenditure and liver gene expression analysis also showed ob/MUT mice were more insulin sensitive and the increased β-cell mass symptomatic of an insulin-resistant state was significantly reversed in comparison to Lepob/ob controls (Fig. 4h-k). While Lepob/ob mice exhibit several metabolic alterations beyond weight gain and hyperglycemia, these results further illustrate loss of Cadm1 in excitatory neurons protected mice from both diet and genetically-induced obesity and insulin resistance by improving energy expenditure, glucose homeostasis, and insulin sensitivity independent of any change in food intake (Fig. 4l). Importantly, these observations establish Cadm1 as a potential target in the brain for therapeutic intervention in alleviating the complications associated with diabetes and obesity.
Figure 4.
Loss of neuronal Cadm1 protects from diet and genetically-induced obesity and insulin resistance. (a) Body weight curves of Slc17a6-Cre, Cadm1flox/flox mice (n=10) and littermate controls (n=6) on high fat diet (HFD). (b) Glucose measurements during an ITT from 16-week old Slc17a6-Cre, Cadm1flox/flox mice (n=7) and control littermates (n=10) after 10 weeks on HFD. (c) Energy expenditure from individual animals plotted against lean body mass in 16-week old Slc17a6-Cre, Cadm1flox/flox mice (n=9) and littermate controls (n=10) after 10 weeks on HFD. (d) Energy expenditure from individual animals plotted against locomotor activity in 16-week old Slc17a6-Cre, Cadm1flox/flox mice (n=9) and littermate controls (n=10) after 10 weeks on HFD. (e) Western blot analysis of Cadm1 and Cadm2 in total and synaptosome-enriched lysates from hypothalamus of 12-week-old Slc17a6-Cre, Cadm1flox/flox crossed onto the Lepob/ob background (ob/MUT) mice and Lepob/ob littermates. (f) Body weight curves of ob/MUT mice (n=5) and Lepob/ob littermate controls (n=9) from 4-12 weeks of age. (g) Random glucose and plasma insulin measurements in ob/MUT mice (n=4) and Lepob/ob littermate controls (n=8). (h) Glucose measurements during an ITT on 12-week old ob/MUT mice (n=5) and control Lepob/ob littermates (n=9). (i) qRT-PCR analysis of G6pc and Pck1 expression in liver of 12-week old ob/MUT mice (n=4) and control Lepob/ob littermates (n=4). (j) Quantification of pancreatic β-cell mass (βCM) in ob/MUT mice (n=3) and littermate Lepob/ob controls (n=4) from 12 weeks of age. Immunohistochemistry of representative pancreatic sections from wild-type (WT), Lepob/ob and ob/MUT mice after detection of insulin (green). (k) Energy expenditure from individual animals plotted against lean body mass in 12-week old ob/MUT mice (n=5) and Lepob/ob littermates (n=6). (l) Quantification of food intake measured in 12-week old ob/MUT mice (n=5) and littermate Lepob/ob controls. (n=6). All results are presented as mean ± s.e.m. *P<0.05 and **P<0.01. Boxplots show median, lower and upper quartiles (box), and minimum and maximum (whiskers). Statistical analyses are described in the Methods and Supplementary Table 3.
Cadm1 functions in multiple neuronal circuits
Cadm1 is present throughout the brain and has been shown to regulate synapse formation, morphology, plasticity, and network excitability20,24,21. The broad expression pattern of Slc17a6-Cre suggests Cadm1 may contribute to energy homeostasis via multiple brain regions. To elucidate the circuits that contribute to Cadm1 function in systemic energy homeostasis we analyzed the effects of Cadm1 loss in Vglut2+ excitatory neurons on hippocampal synapse number and function. In spite of its described role in hippocampal synapse formation during the post-natal period, synapse numbers in this region were unchanged in adult Slc17a6-Cre, Cadm1flox/flox mice compared to littermate controls (Supplementary Fig. 7a,b)20. Post-synaptic density length and postsynaptic density protein 95 (PSD-95) expression were decreased in the hippocampus of Slc17a6-Cre, Cadm1flox/flox animals suggesting that Cadm1 function may vary with age, cellular context, or metabolic state (Supplementary Fig. 7c,d and 15). Electrophysiologic analysis revealed increased basal synaptic transmission supporting resent observations on Cadm1 in synaptic transmission by balancing excitatory and inhibitory transmission (Supplementary Fig. 7e)21. In addition, we observed increased long-term depression (LTD) and long-term potentiation (LTP) supporting previous observations on the role of Cadm1 in synaptic plasticity (Supplementary Fig. 7f, g)20 and on the promotion of hippocampal plasticity by locomotor activity in rodents25,26. These observations suggest that Cadm1 regulates energy and glucose homeostasis by modulating excitatory synapse function in the hippocampus.
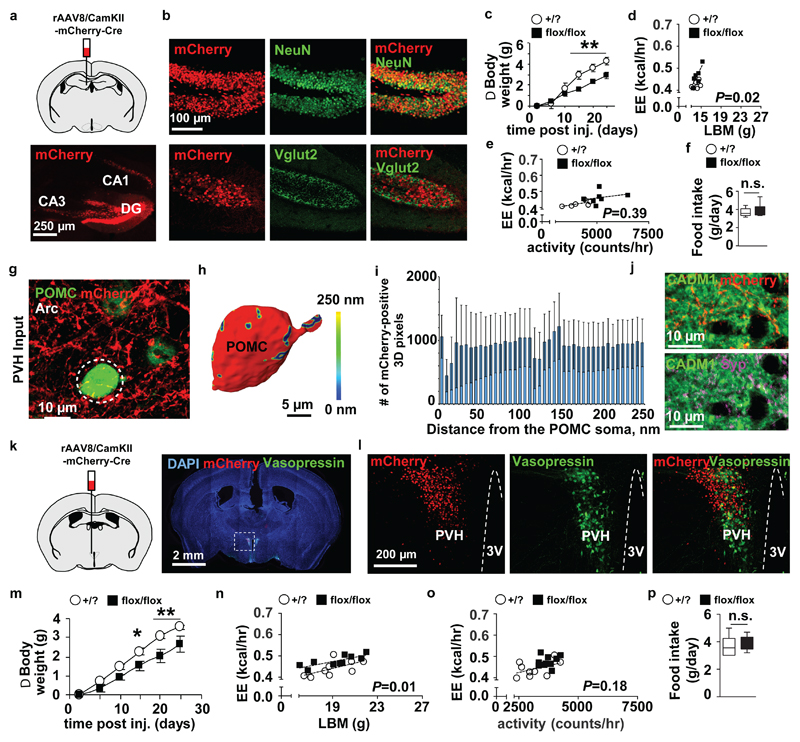
To directly address the role of Cadm1 in the regulation of body weight and energy homeostasis in hippocampal excitatory neurons, we specifically eliminated its expression in this brain region via stereotactic injection of a Cre recombinase-expressing adeno-associated virus (rAAV8/CamKII-mCherry-Cre) (Fig. 5a,b). The AAV co-expressed mCherry which allowed visualization of Cre expression in the dentate gyrus and to a lesser extent in the CA3 region of the hippocampus. Approximately 15 days after delivery of the AAV, floxed Cadm1 mice (flox/flox) mice exhibited lower body weight compared to controls (Fig.5c and Supplementary Fig. 8a and 15). Similar to previous Cadm1 loss-of-function models, energy expenditure rate was observed elevated after targeting Cadm1 in the hippocampus and confirmed with ANCOVA after plotting against lean body mass and locomotor activity (Fig. 5d,e and Supplementary Fig. 8b). These data directly demonstrate a crucial role for Cadm1 expressed in excitatory hippocampal neurons in the regulation of body weight and energy expenditure that is independent of an effect on feeding behavior (Fig. 5f).
Figure 5.
Targeting Cadm1 expression in the hippocampus and hypothalamus reduces body weight. (a) Representative coronal brain section of Cadm1 floxed mice after stereotaxic rAAV8-CaMKIIa-mCherry-Cre (AAV-Cre) injection in the dentate gyrus (DG) and CA3 regions to target Cadm1 expression in hippocampal neurons. (b) Co-localization of mCherry with NeuN and Vglut2 after delivery of AAV- CaMKIIa-Cre to the hippocampus. (c) Body weight curves of floxed Cadm1 mice (flox/flox) (n=6) and control littermates (+/?) (n=6) after delivery of AAV-Cre to the hippocampus. (d) Energy expenditure from individual animals plotted against lean body mass in 9-week old floxed Cadm1 mice (flox/flox) (n=7) and control littermates (+/?) (n=6) after delivery of AAV-Cre to the hippocampus. (e) Energy expenditure from individual animals plotted against activity measured in 9-week old floxed Cadm1 mice (flox/flox) (n=7) and control littermates (+/?) (n=6) after delivery of AAV-Cre to the hippocampus. (f) Quantification of food intake measured in floxed Cadm1 mice (flox/flox) (n=7) and control littermates (+/?) (n=6) after delivery of AAV-Cre to the hippocampus. (g) Stereotactic delivery of a mCherry-expressing AAV to paraventricular nucleus (PVH) (red) reveals PVH efferent projections to eGFP-positive POMC neurons (green) within the Arc region of the hypothalamus. Dotted white circle outlines the cell body of an eGFP-POMC-positive cell, used for 3D reconstruction. (h) Surface rendering of Amira 3D reconstruction of an eGFP-POMC-positive cell body receiving afferent input from the PVH. Cell is represented in red, while the synaptic input is color-coded, with the cold to warm colors spreading from 0 to 250nm distance between axonal varicosities and the soma (see color-coded horizontal bar for the distance definition). (i) Histograms show the number of anterograde AAV-mCherry-labelled PVH axonal varicosities found within the distance of 250nm from the POMC-positive cell body. (j) Double immunostaining of Cadm1 and mCherry or synaptophysin (Syp) indicates that PVH regions provide Cadm1-positive efferent projections to the ARC region of hypothalamus. POMC-eGFP transgenic mice received an AAV-mCherry injection to the PVH, and were subsequently immunostained for Cadm1 (green) and mCherry (red) or synaptophysin (magenta). (k) Representative coronal brain section of Cadm1 floxed mice after stereotaxic injection of AAV8-CaMKIIa-mCherry-Cre to hypothalamic neurons in the PVH, and ventromedial hypothalamus (VMH) regions to drive Cre expression to excitatory neurons in these regions. Section is showing mCherry expression and staining for vasopressin (green) and nuclei counterstained with DAPI (blue). (l) Confocal imaging of mCherry (red) and vasopressin (green). (m) Body weight curves of floxed Cadm1 mice (flox/flox) (n=9) and control littermates (+/?) (n=11) after delivery of AAV-Cre to the hypothalamus. (n) Quantification of energy expenditure (EE) adjusted to lean body mass (LBM) in 10-week old floxed Cadm1 mice (flox/flox) (n=10) and control littermates (+/?) (n=9) after delivery of AAV-Cre to the hypothalamus. (o) Quantification of energy expenditure (EE) adjusted to locomotor activity in 10-week old floxed Cadm1 mice (flox/flox) (n=10) and control littermates (+/?) (n=9) after delivery of AAV-Cre to the hypothalamus. (p) Quantification of food intake measured in floxed Cadm1 mice (flox/flox) (n=10) and control littermates (+/?) (n=9) after delivery of AAV-Cre to the hypothalamus. All results are presented as mean ± s.e.m. *P<0.05 and **P<0.01. Boxplots show median, lower and upper quartiles (box), and minimum and maximum (whiskers). Statistical analyses are described in the Methods and Supplementary Table 3.
We executed a similar strategy in the hypothalamus to delineate the contribution of Cadm1 to the regulation of body weight in this brain region and we first performed tracing experiments to identify the origins of axonal projections to POMC neurons in the ARC (Supplementary Fig. 8c). Stereotactic delivery of a mCherry-expressing AAV to the paraventricular nucleus of the hypothalamus (PVH) region identified axonal inputs to POMC neurons expressing Cadm1 (Fig. 5g-j and Supplementary Fig. 10). We then directly eliminated hypothalamic Cadm1 expression by injection of the AAV-Cre into the PVH and the ventromedial nucleus of the hypothalamus (VMH) of floxed Cadm1 mice (flox/flox) (Fig. 5k,l and Supplementary Fig.8d). Similar to Cadm1 deletion in the hippocampus, lower body weight and increased energy expenditure was observed ~15 days post-injection in floxed Cadm1 mice compared to controls (Fig. 5m-o and Supplementary Fig. 8e,f and 15). Neither insulin sensitivity nor food intake was altered indicating these neuronal regions may be distinct with respect to Cadm1 function in energy homeostasis (Fig. 5p and Supplementary Fig. 8g). Despite the fact that our tracing analysis did not identify a significant number of hippocampal inputs to POMC neurons in the arcuate nucleus, we did observe hippocampal inputs to the PVH in the hypothalamus (Supplementary Fig.8h). Tracing analysis also identified axonal tracts originating from the hypothalamus to the dentate gyrus of the hippocampus suggesting the existence of a closed-loop circuit between these regions expressing Cadm1 being involved in energy homeostasis (Supplementary Fig. 8i). Additionally, Cadm1-positive afferent inputs originating from the habenular nuclei, a key site regulating voluntary locomotor activity and motivational behavior, were observed on POMC-neurons (Supplementary Fig. 8j-m)27. Given the fact that the medial habenula is functionally connected to the hippocampus via strong efferent projections from septal nuclei28, habenular input to POMC-neurons provides an important bridge between the hippocampus and hypothalamus, indicating that Cadm1 function in energy homeostasis extends to circuits beyond the ARC.
To determine the contribution of Cadm1 in energy homeostasis in more distinct hypothalamic populations, we conditionally deleted its expression using established Cre-expressing mouse lines. The specific deletion of Cadm1 using transgenic Pomc, Agrp, and Sim1-Cre lines (which express Cre recombinase under control of the respective promoters in neurons of the ARC or PVH) all resulted in no significant changes in metabolic parameters including insulin sensitivity and in the case of POMC neurons, may reflect the low abundance of Cadm1 in these specific cell types29,30,31 (Supplementary Fig. 9a-c). Cadm1 was deleted in Lepr-Cre+ cells (Lepr-Cre, Cadm1flox/flox) and in spite of broader expression beyond the hypothalamus, the mutant mice in the steady state did not exhibit any phenotypic changes (Supplementary Fig. 9d-g)32. However, crossing this line into the Lepob/ob background (ob/Lepr-Cre, Cadm1flox/flox) resulted in increased random-fed and fasted blood glucose levels as well as increased random plasma insulin levels (Supplementary Fig. 9h,i). Ob/Lepr-Cre, Cadm1flox/flox mice consistently showed higher glucose levels in all tolerance tests indicating an exacerbation of insulin resistance compared to Lepob/ob controls (Supplementary Fig. 9j-l). Meanwhile, body composition, pancreatic β-cell mass, energy expenditure rate, locomotor activity, and food intake were all indistinguishable between ob/Lepr-Cre, Cadm1flox/flox and Lepob/ob littermates (Supplementary Fig. 9m-o and Supplementary Table 2). These results demonstrate that the role of Cadm1 in Slc32a1-Cre+ and Lepr-Cre+ cells is distinct from its function in Slc17a6-Cre+ cells with respect to the maintenance of insulin sensitivity, energy expenditure, and glucose homeostasis and the function of Cadm1 is dependent on a diverse set of neurons distributed throughout the brain beyond the traditional neuronal circuits controlling energy expenditure and body weight within the hypothalamus.
The precise mechanisms causing obesity remain unknown. It is clear that neuronal pathways regulating energy homeostasis and signaling are key contributors and genome-wide association studies have been successful in identifying genes essential to understanding fundamental aspects of our metabolism33,34. Here we illustrate how increased expression of Cadm1 in the brain is relevant to weight gain and that reducing its expression in both the hippocampus and hypothalamus can trigger a negative energy balance. The expression of both Cadm1 and Cadm2 in several neuronal circuits may indicate the maintenance of our body weight is intertwined with motor control and coordination, motivational and learning behavior, spatial navigation, cognition and memory. This is further supported by several studies now showing Cadm1 contributes to synaptic plasticity and the excitatory and inhibitory balance of specific networks20,21. Spatially modulated cells (i.e. place cells, border cells and grid cells) in hippocampal-entorhinal circuits contribute to the formation of environment-specific memory maps during spatial orientation and may constitute key circuits in the regulation of energy homeostasis35,36. Future investigations will determine how the brain is capable of integrating these neuronal circuits together with our physiology and whether targeting Cadm1 in specific regions is a viable therapeutic strategy for reversing obesity in humans37.
Methods
Animals
Mice were housed in groups of 3-5 animals and maintained on a 12-hour light/dark cycle with ad libitum access to regular chow food, high fat diet (containing 60% kcal fat, cat. no. E15741-347, ssniff Spezialdiäten GmbH), or ketogenic diet (cat. No. E15149-30, ssniff Spezialdiäten GmbH) in accordance with the Landesamt für Gesundheit und Soziales (LAGeSo). All experimental procedures were approved under protocols G 0357/10, G 0204/14, O 0405/09, and T 0436/08. Cadm1KO mice38 were characterized after backcrossing for four generations to C57BL/6 and then crossed to Lepob/ob and POMC-eGFP expressing mice (Jackson Labs). Cadm2KO mice were generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (www.komp.org). Slc17a6-Cre, Slc32a1-Cre, Lepr-Cre, Agrp-Cre, Sim1-Cre, and Pomc-Cre expressing mice were purchased (Jackson Labs) and directly crossed to Cadm1-floxed mice that were previously crossed to C57BL/6 (Jackson Labs) for four generations. To generate Tg-Cadm1 mice, full-length Cadm1 cDNA flanked by seven repeats of the doxycycline-responsive element was inserted into pTRE-Tight vector (Clontech) prior to microinjection. Positive founders were crossed to Slc17a6-Cre and ROSA26-rtTA mice (Jackson Labs). All experiments implemented Cre-expressing mice as controls to account for potential metabolic abnormalities 39. Results were consistent in both genders however data from female mice is not shown.
Analytic Procedures
Quantification of metabolic parameters
Guinea pig anti-insulin (Dako A0564, 1:500), and rabbit anti-glucagon (Millipore AB932, 1:500) antibodies for immunofluorescence were used on paraffin-embedded pancreata fixed in 4% paraformaldehyde (PFA). Plasma glucose levels were obtained with the One Touch glucometer (Bayer). Plasma and total pancreatic insulin measurements were measured by radioimmunoassay or ELISA as described (Millipore, Crystal Chem)40. Plasma leptin was measured by ELISA (Peprotech) and adiponectin by radioimmunoassay (Millipore).
Antibodies for western blot analysis
The following primary antibodies were used for western blotting at 1:1000 dilution: Cadm1 (MBL CM004-3), Cadm2 (Sigma SAB4501053), β-Actin (Sigma A1978), GAPDH (Abcam ab8245), γ-Tubulin (Sigma T6557), PSD95 (NeuroMab UCDavis 075 028). Variance in the banding patterns of Cadm proteins in different panels may result from variation in acrylamide percentage or levels of protein glycosylation. Image densitometry of 16-bit TIF images for all western blots was performed using ImageJ.
Primary cell cultures
Primary neurons were derived from hippocampi of mice at age post-natal day 2 and dissected in cold HBSS (Invitrogen), followed by digestion with papain. After centrifugation, tissue pellet was resuspended in Neurobasal (Invitrogen) supplemented with B27 (Invitrogen) and Glutamax (Invitrogen) for plating. Hippocampal cells were cultured in 24-well dishes with 12 mm coverslips (5000 cells per coverslip) coated with poly-D-lysine and laminin and immunohistochemistry was performed as described previously 41. After post-fixation in 4% PFA, sections were blocked with 10% BSA/PBS, then incubated with primary antibody overnight at 4°C, followed by the fluorochrome-conjugated secondary antibodies for 1 hour. Fluorescence was imaged under a confocal microscope (Zeiss LSM700). Digtal images were analyzed with Fiji/ImageJ.
Mouse Phenotyping
All phenotyping analysis was performed in a ‘blinded’ manner; genotypes were unknown to the investigator during the experimentation and age of animals is stated in figure legends. All genotypes were present during all experiments and randomization was implemented to the extent that all animals were identified by number prior to analysis.
Body composition and energy expenditure analysis
Body composition analysis was measured using Minispec Model LF90 II (6.5 mHz) (Bruker Instruments). VO2, VCO2, food intake and locomotor activity were measured using the PhenoMaster System (TSE, Germany) and energy expenditure was analyzed as previously described using ANCOVA 17. Animals were placed into individual cages with weight sensors quantifying ad libitum access to food. VO2 and VCO2 level were measured for one minute in a 9 min interval for 4 consecutive days and locomotor activity was measured continuously by breaks of light beams. The first 24 hours of measuring time was excluded from the analysis to allow for acclimation to the new cage environment. Measurement of energy expenditure was normalized to lean body mass as previously described42.
Tolerance tests
Glucose and pyruvate tolerance tests were performed following an overnight fast (16 hours) and injected intraperitoneally with either glucose (2g/kg body weight) or pyruvate (2g/kg body weight in saline) as described 40. Insulin tolerance tests were performed after same day fast (6 hours) by injecting insulin (Sigma) intraperitoneally (0.75 U/kg body weight). Murine leptin (Peprotech) (0.75 μg/g body weight) was injected intraperitoneally twice daily (09:00 and 19:00) for 3 days. Body weight and food intake were measured daily at 08:30.
In vivo clamp studies
Clamp studies were performed as described 43. Plasma [3H] glucose was measured by scintillation counting of ZnSO4/Ba(OH)2 deproteinized serum, dried to remove 3H2O. Calculations were obtained as follows: Basal and insulin-stimulated whole body glucose turnover rates = ([3-3H] glucose infusion (in dpm)) / (plasma glucose specific activity (dpm per mg)) at the end of the basal period and during the final 30 min of the clamp, respectively. Hepatic glucose production = (rate of total glucose appearance) – (glucose infusion rate). Plasma concentration of 3H2O = 3H counts [wet] – [dry]. Whole body glycolysis was defined by the rate of increase in plasma 3H2O concentration by linear regression of measurements between 90-140 minutes. Whole body glycogen synthesis = (whole body glucose uptake) - (whole body glycolysis). Tissue 14C-2-deoxyglucose-6-phosphate content was measured following sample homogenizing, and the supernatant’s 14C-2-deoxyglucose-6-phosphate was separated from 2-deoxyglucose by ion-exchange column.
In vivo cold exposure analysis
Cold challenge was performed as described previously 44. Briefly, 12-week-old mice were kept at 6°C for 14 consecutive days in a light and humidity-controlled climate chamber (TSE Systems) and fed chow diet ad libitum.
Tracing analysis and AAV injections
Stereotaxic injections were performed on 5-8-week old mice and all data incorporated into the analysis was obtained from animals that were examined histologically for mCherry expression in the appropriate nuclei. For POMC-eGFP/Slc17a6-Cre mice, rAAV2/EF1a-DIO-hChR2(H134R)-mCherry (UNC Vector Core, titer 6.6x1012 vg/ml) was unilaterally injected into the dentate gyrus (DG) of the hippocampus, and paraventricular nucleus of the hypothalamus (PVH) respectively (n=4); rAAV5/EF1a-DIO-hChR2(H134R)-mCherry (UNC Vector Core, titer 5x1012 vg/ml) was unilaterally injected into the habenular nuclei (Hb) (n=4). Briefly, the mouse was anesthetized with ketamine (120 mg/kg), Xylazine (10 mg/kg) and an i.c. injection of analgesic Methamizol (200 mg/kg). Next, an incision on the head skin was performed and animals were placed in a stereotactic frame (Angle Two, Leica) while maintained at a temperature of 37.5 °C with a heating pad during the whole operation. A small craniotomy was performed over the DG with coordinates: 1.8 mm posterior, 1.3 mm lateral to Bregma; Hb: 1.6 mm posterior, 0.36 lateral to Bregma; PVH: 0.7 mm posterior, 0.24 lateral to Bregma. A glass injection pipette with a ~10-20 μm diameter tip containing the AAV solution was then inserted to the following depths: 1.8 mm (for DG), 2.7 mm (for Hb), and 4.8 mm (for PVH). Using an oil piston (MO-10; Narishige) connected to this injection glass pipette 0.1 to 0.4 μl of virus was injected at a rate of 50 to 100 nl per minute. The injection pipette stayed in place for about 10 min to allow the pressure to equilibrate after the injection and then removed. Post-injection, the incision site was stitched with suture thread and the mice recovered on a heating pad at 37.5 °C with access to water with Methamizol (0.5 mg/ml) for 24 hours. Floxed Cadm1 (flox/flox) mice and littermate controls were stereotaxically injected with rAAV8/CamKII-mCherry-Cre resulting in expression of the mCherry-Cre fusion protein (UNC Vector Core). Mice were anesthetized with Ketamine HCl/Xylazine HCl (50/5 mg/kg, intraperitoneally) and restrained in a Kopf stereotaxic frame (David Kopf Instruments). Small holes with the size of the injection needle were drilled into the skull, and injections were done bilaterally with 1µl of vector per brain region. The rAAV8/CamKII-mCherry-Cre was administered (0.5 μL for the PVH and 1 µl for the dentate gyrus, titer: 4.7×1012 virus genome/ml) into the dentate gyrus (coordinates: −1.7 mm from bregma, ±1.5 mm lateral, −2.2 mm from the surface of the skull) and PVH (coordinates: −0.82 mm from bregma, ±0.5 mm lateral, −5.0 mm from the surface of the skull). Animals/samples were excluded if proper injection of the AAVs could not be validated by reporter expression and immunohistochemistry.
Immunohistochemistry and image analysis
Mice were transcardially perfused with 50 ml saline solution (0.85% NaCl, 0.025% KCl, 0.02% NaHCO3, pH 6.9, 0.01% heparin, body temperature) followed by 50 ml cold (7-15ºC) freshly depolymerized 4% PFA in 0.1 M PBS, pH 7.4. Brains were post-fixed overnight in the same fixative and placed in a mixture of 20% glycerol and 2% dimethyl sulfoxide in 0.4 M PBS for 24 h for cryoprotection. Frozen horizontal or coronal sections (40 µm) were collected in 0.1 M PBS. The following antibodies were used for immunohistochemistry: Cadm1 (MBL CM004-3, 1:500), Cadm2 (Sysy 243203, 1:500), POMC (Phoenix H-029-30, 1:500), Synaptophysin (Sysy 100011, 1:500), Vasopression (Millipore AB1565, 1:1000), NeuN (Abcam ab177487, 1:500; Millipore MAB377, 1:100), Map2 (Sysy 188004, 1:1000), Vgat (Sysy 131004, 1:500), mCherry (Abcam ab125096, 1:500), GFP (Thermo Fisher A11120, 1:500) and VGLUT2 (Sysy 135402, 1:500) were used. Sections were washed 3x in PBS (3x15 min each) followed by washing several times in PBS containing 0.3 % Triton-X-100 (9x20 min each). Sections were pre-incubated with PBS containing 5% NGS and 0.3 % Triton X-100 for 1 h, and subsequently incubated with primary antibody at 4°C for 48 h. Then, sections were washed 9 times for 20 min each in 0.3 % Triton-X-100 in 0.1 M PBS, and incubated with Alexa-conjugated secondary antibodies (1:400) for 12 h using standard techniques. Finally, sections were mounted on gelatin-coated glass slides and cover-slipped in Immu-Mount. Immunofluorescent images were acquired using a Leica SP5 confocal microscope, equipped with a 488 nm argon laser, a 561 nm DPSS laser and a 633 red Helium Neon laser. To detect fluorescence emission sections were scanned at a resolution of 1024 x 1024 pixels with 8-bit sampling in sequential scanning frame-by-frame mode. Cadm1 expression levels were quantified in ImageJ by measuring the mean gray value of fluorescence intensity within the ROI using 20x Plan-Apochromat objective. For high-resolution 3D analysis, samples were scanned using Plan-Apochromat 63x/1.32 Oil DIC objective at a resolution of 1024 x 1024 pixels with 8-bit sampling in sequential scanning frame-by-frame mode. Single optical sections were acquired using identical acquisition settings, with the pinhole of 1 Airy Unit. Stacks of 8-29 optical sections yielded voxel dimensions between 100 and 400 nm for the X, Y and Z planes. 3D reconstructions were generated with Amira Software (FEI Visualization Sciences). First, the cell body of the POMC-positive neurons was reconstructed using Amira surface editor. Synaptic input was defined by the reconstruction of AAV-mCherry positive varicosities in the same image using Isosurface module. The likely synaptic contact was defined by color-coding the surface of AAV-mCherry positive varicosities found within 250 nm from POMC-positive cell body. Subsequently, the surface of “250nm-distant” AAV-mCherry positive varicosities was mapped onto POMC-positive cell body using “map distance” tool.
Analysis of Cadm1 and Vglut2 fluorescent profiles
Overlap of fluorescent intensity profiles of Cadm1 and Vglut2 was measured using the line scan function of ImageJ on an x–y plane of a single optical section. Subsequently line profiles were analyzed using custom-developed Python script. In detail, the percentage of overlap between Cadm1 and Vglut2 was analyzed according to the following algorithm: Firstly, all fluorescent values were normalized by scaling them from 0-1000 and the penalty values were calculated by taking the absolute value of the difference between the lead curve (Vglut2) and the follow curve (Cadm1) The “Events” of fluorescent puncta were defined by the presence of high y-values of the lead curve. To account for the overlap of two curves in these areas the penalty values were multiplied with the square of the lead y-value, giving the term: Next, a measurement for the differences between the two curves was generated by taking the sum of all penalty values (sum1): The penalty was restricted to the interval 0-100% by dividing the actual penalty value by the maximum possible penalty (sum2):
Electron microscopy analysis
8-week old mice were anesthetized and transcardially perfused with 4% formaldehyde and 2.5% glutaraldehyde in PBS. Brains were isolated and post-fixed in the same solution overnight at 4°C after rinsing in PBS, and then embedded into 5% agar and sliced coronally (200 μm sections). Vibratome slices containing hippocampi were post-fixed in 1% OsO4 and 1.5% potassium hexacyanoferrat, followed by dehydration in a methanol gradient and propylene oxide, and flat embedded in epoxy resin. After polymerization the CA1 pyramidal cell layer and adjacent stratum radiatum were trimmed and ultrathin sectioned. Sections were collected on coated slotted grids and analyzed using a Zeiss 900 transmission electron microscope. Synaptic density was determined by the dissector method 45. Neuropil images with an area of ≈190 µm2 from adjacent sections with defined thickness were aligned, a dissector grid was superimposed onto the images and the appearance of the postsynaptic density was used as a “counting cap”. At least 20 dissectors were analyzed per animal and density of synaptic profiles was estimated in 2-D by counting number of profiles in 160 neuropil fields.
Quantification of excitatory and inhibitory synapses was performed as previously described 46. Mice were perfused using Somogyi-Takagi fixative; the brain was removed and a tissue block containing the hypothalamus was sectioned. EGFP (derived from the POMC-eGFP transgene) was stained using an anti-GFP antibody for signal amplification (Molecular Probes A-11120, 1:500) and ABC Elite Kit (Vector Labs). Sections were subsequently osmicated, dehydrated and ultrathin sections obtained using a microtome. Images were acquired using a Tecnai 12 Biotwin (FEI Company) electron microscope. Non-symmetric (excitatory) and symmetric (inhibitory) synapse numbers on POMC neurons were quantified in a double-blinded fashion using the dissector technique. Synapses were only quantified if a clear identification of pre- and postsynaptic membranes was possible and synaptic vesicles in the presynaptic bouton were visible. Synapses without a clear non-symmetric or symmetric specialization were excluded from the analysis.
Electrophysiological analysis
For EPSCs and IPSCs, the entire brain from 3-week old animals was removed and immediately submerged in ice cold, carbogen-saturated (95% O2 / 5% CO2) aCSF (in mM: 2.5KCl, 126 NaCl, 1.3 MgCl2, 2.0 CaCl2, 1.2 KH2PO4, 21.4 NaHCO3, 10 glucose). Coronal sections (300 μm) were cut with a Leica VT1000S Vibratome and then incubated in oxygenated aCSF at room temperature for at least 1 hr before recording. Whole-cell voltage-clamp recordings from POMC-eGFP neurons (identified with the POMC-eGFP transgene)47 were performed using a Multiclamp 700B amplifier (Molecular Devices) and filtered with an eight-pole Bessel filter at 6 kHz. Both voltage and current signals were sampled at 50 kHz using an Axon 1440A interface device (Molecular Devices) and the data were acquired using Clampex 10.2 software (Molecular Devices). Electrodes were pulled using a micropipette puller (Sutter Instruments) from thick-walled borosilicate glass GC150F capillaries (Harvard Apparatus) to a resistance of ~3 MΩ. Series resistance compensation ranged from 50 to 70% and recordings were discarded if the series resistance exceeded 10MΩ. Electrodes were filled with an internal solution of composition (in mM) 140 CsCl, 4 NaCl, 0.5 CaCl2, 10 HEPES, 5 EGTA, 2 Mg-ATP, QX-314-Br 3, pH 7.3 with CsOH, and osmolarity of 290mOsM. Cells were visualized using an infrared differential interference contrast (DIC) optics equipped with equipped ORCA-R2 camera (Hamamatsu) mounted on a Slicescope (Scientifica) with a 40X water-immersion objective. Neurons were voltage clamped at −70 mV. In order to record IPSCs, the aCSF composition included 3 mM kynurenic acid (Sigma) to block excitatory glutamatergic activity. EPSCs were recorded in presence of Picrotoxin (Tocris) 100 μM in order to block both GABA and Gly evoked currents. Both experimental conditions included TTX 0.5 μM (Tocris) to isolate miniature events. Events were excluded from the analysis if their amplitude was less than 3 standard deviations of the baseline noise and if there were overlapping events within 50 ms (for IPSCs) or 10ms (for EPSCs). All miniature events were automatically detected with Clampfit 10, followed by manual inspection of single events. For frequency distributions a minimum of 200 events were analyzed.
For MPP-DG recordings, 6-8-week old WT and Cadm1Slc17a6 mice were used for acute hippocampal field electrophysiology. Mice were decapitated after cervical dislocation and the brain extracted into ice cold dissection ‘cutting’ solution containing (in mM): 2.5 KCl, 1.25 NaH2PO4, 24 NaHCO3, 1.5 MgSO4, 2 CaCl2, 25 glucose, 250 sucrose. 350 µm sagittal brain slices prepared from both hemispheres were transferred to a resting chamber containing solution in which 250 mM sucrose was replaced with 120 mM NaCl (pH = 7.35-7.4) and incubated to recover before recordings. Recordings performed at room temperature (22-24°C) in the submerged chamber, supplied with continuously bubbled solution with an exchange of 3-5 ml per min. Stimulating (1-1.5 MΩ) and recording (1.5-2.5 MΩ) electrodes filled with solution were placed in inner part of molecular layer of DG and medial perforant path (MPP) to DG field excitatory postsynaptic potentials (fEPSPs) were recorded. Basal stimulation of 0.2 ms electrical pulses was delivered at every 30 sec for at least 10 minutes to monitor stabile baseline recordings. Only the responses which show paired pulse depression were considered as MPP and were studies further to evaluate synaptic transmission and plasticity. The stimulus response curves made as a measure of basal synaptic transmission in which stimulation intensity was increased from 10 to 100 µA, reaching maximal responses at around 60-70 µA for both genotypes. For LTP and LTD recordings, baseline stimulation was set to elicit 50 % of maximal fEPSP (30-40 µA) and GABAergic antagonist picrotoxin (100 μM) was applied to recording solution at least 30 minutes before the LTP and LTD induction. Stimulation intensity was doubled during LTP, LTD induction using stimulation intensities at which maximal fEPSPs were detected (60-70 µA). 5 x HFS were delivered every 30 seconds each HFS containing 100 pulses at 100Hz for LTP induction and LFS containing 900 pulses at 1Hz stimulation was used for LTD induction. LTP and LTD was calculated as % increase of fEPSP slope 50-60 min after application of induction protocol as compared to the initial -10-0 min of baseline. The data were recorded at a sampling rate of 10 kHz, low-pass filtered at 3 kHz and analyzed using PatchMaster software and EPC9 amplifier (Heka Elektronics).
eQTL analysis
We have downloaded currently unpublished eQTL data from the GTEx consortium analysis version 6, including results from ten distinct brain regions48. We specifically queried the data for association tests that were performed for genes CADM1 and CADM2 against SNPs rs12286929 and rs13078807. Adjustment for multiple hypothesis testing using the method of Benjamini and Hochberg showed that all associations were significant (FDR <= 15%)49. Boxplots were obtained through the GTEx portal.
Statistical analysis
All results are expressed as mean ± s.e.m. Comparisons between data sets with two groups were evaluated using an unpaired Student’s t-test. One-way and two-way repeated-measures ANOVA analysis has been performed using GraphPad Prism Software Version 6.07 for comparisons of three or more groups. Post hoc statistics were performed using Sidak’s multiple comparison test. A P-value of less than or equal to 0.05 was considered statistically significant. The presented data met the assumptions of the statistical tests used. Normality and equal variances were tested using GraphPad Prism software. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications 17, 29–31, 40, 43.
Supplementary Material
Acknowledgements
This work was funded by the Helmholtz Gemeinschaft, the Helmholtz Metabolic Dysfunction Consortium, European Research Council (ERC-2010-StG-260744 to M.N.P. and ERC-2013-StG-336607 to M.T.), the Swiss National Science Foundation Professorship (PP00P3_144886 to M.T.), the Deutsche Forschungsgemeinschaft (NeuroCure/Exc-257 and SFB958/A01 to V.H.), and the Kay Kendall Leukemia Foundation (KKLF Fellowship to L.vdW). AAV reagents were provided by the UNC Vector Core facility and used with permission by K. Deisseroth (Stanford University). The authors would like to thank Dr. M. Gruhn and the Biozentrum Imaging Facility, University of Cologne, for access to Amira Software, N. Zampieri, A. Plested, T. Breiderhoff, D. Matthäus, I. Park, C. Teng, T. Klüssendorf, H. Wessels, T. Willnow, and M. Gotthardt for helpful discussions and assistance in the conduct of this work.
Footnotes
Data Availability.
All primary data supporting the findings of this study are available on reasonable request.
Author Contributions
T.R. and M.N.P. conceived this study. T.R., X.Y., N.K., M.K., K.S., L.F., V.T., D.P., G.K., S.B., L.V., K.S., C.Y., S.C.S., S.G.T., A.S.C., M.M., J.S., A.H., L.v.d.W., A.L.B., T.N., J.F.P., T.L.H., M.H.T., M.H., M.T., V.H., and M.N.P. designed and performed the experiments with help from all authors. M.H.T., V.H., and M.N.P. wrote the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Finkelstein EA, et al. The lifetime medical cost burden of overweight and obesity: implications for obesity prevention. Obes Silver Spring Md. 2008;16:1843–1848. doi: 10.1038/oby.2008.290. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep. 2009;9:164–171. doi: 10.1007/s11892-009-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant SFA, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 4.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 5.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biederer T, et al. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 9.Fogel AI, et al. SynCAMs organize synapses through heterophilic adhesion. J Neurosci Off J Soc Neurosci. 2007;27:12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogel AI, et al. N-Glycosylation at the SynCAM (Synaptic Cell Adhesion Molecule) Immunoglobulin Interface Modulates Synaptic Adhesion. J Biol Chem. 2010;285:34864–34874. doi: 10.1074/jbc.M110.120865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biederer T. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics. 2006;87:139–150. doi: 10.1016/j.ygeno.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Badman MK, Kennedy AR, Adams AC, Pissios P, Maratos-Flier E. A very low carbohydrate ketogenic diet improves glucose tolerance in ob/ob mice independently of weight loss. Am J Physiol - Endocrinol Metab. 2009;297:E1197–E1204. doi: 10.1152/ajpendo.00357.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias CF, et al. Leptin differentially regulates NPY and POMC neurons projecting to the lateral hypothalamic area. Neuron. 1999;23:775–786. doi: 10.1016/s0896-6273(01)80035-0. [DOI] [PubMed] [Google Scholar]
- 15.Brüning JC, et al. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289:2122–2125. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 16.Ahima RS, Bjorbaek C, Osei S, Flier JS. Regulation of neuronal and glial proteins by leptin: implications for brain development. Endocrinology. 1999;140:2755–2762. doi: 10.1210/endo.140.6.6774. [DOI] [PubMed] [Google Scholar]
- 17.Tschöp MH, et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2012;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huo L, et al. Leptin-Dependent Control of Glucose Balance and Locomotor Activity by POMC Neurons. Cell Metab. 2009;9:537–547. doi: 10.1016/j.cmet.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vong L, et al. Leptin Action on GABAergic Neurons Prevents Obesity and Reduces Inhibitory Tone to POMC Neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbins EM, et al. SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron. 2010;68:894–906. doi: 10.1016/j.neuron.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park KA, et al. Excitatory Synaptic Drive and Feedforward Inhibition in the Hippocampal CA3 Circuit Are Regulated by SynCAM 1. J Neurosci Off J Soc Neurosci. 2016;36:7464–7475. doi: 10.1523/JNEUROSCI.0189-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerchner GA, Nicoll RA. Silent synapses and the emergence of a postsynaptic mechanism for LTP. Nat Rev Neurosci. 2008;9:813. doi: 10.1038/nrn2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turrigiano G. Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu Rev Neurosci. 2011;34:89–103. doi: 10.1146/annurev-neuro-060909-153238. [DOI] [PubMed] [Google Scholar]
- 24.Perez de Arce K, et al. Topographic Mapping of the Synaptic Cleft into Adhesive Nanodomains. Neuron. 2015;88:1165–1172. doi: 10.1016/j.neuron.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- 26.van Praag H, Christie BR, Sejnowski TJ, Gage FH. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc Natl Acad Sci U S A. 1999;96:13427–13431. doi: 10.1073/pnas.96.23.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu Y-WA, et al. Role of the dorsal medial habenula in the regulation of voluntary activity, motor function, hedonic state, and primary reinforcement. J Neurosci Off J Soc Neurosci. 2014;34:11366–11384. doi: 10.1523/JNEUROSCI.1861-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianco IH, Wilson SW. The habenular nuclei: a conserved asymmetric relay station in the vertebrate brain. Philos Trans R Soc B Biol Sci. 2009;364:1005–1020. doi: 10.1098/rstb.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balthasar N, et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Balthasar N, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 31.Hill JW, et al. Direct insulin and leptin action on pro-opiomelanocortin neurons is required for normal glucose homeostasis and fertility. Cell Metab. 2010;11:286–297. doi: 10.1016/j.cmet.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeFalco J, et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- 33.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loos RJF, Yeo GSH. The bigger picture of FTO – the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10:51–61. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moser M-B, Rowland DC, Moser EI. Place Cells, Grid Cells, and Memory. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a021808. a021808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartley T, Lever C, Burgess N, O’Keefe J. Space in the brain: how the hippocampal formation supports spatial cognition. Phil Trans R Soc B. 2014;369 doi: 10.1098/rstb.2012.0510. 20120510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeltser LM, Seeley RJ, Tschöp MH. Synaptic plasticity in neuronal circuits regulating energy balance. Nat Neurosci. 2012;15:1336–1342. doi: 10.1038/nn.3219. [DOI] [PubMed] [Google Scholar]
- 38.van der Weyden L, et al. Loss of TSLC1 causes male infertility due to a defect at the spermatid stage of spermatogenesis. Mol Cell Biol. 2006;26:3595–3609. doi: 10.1128/MCB.26.9.3595-3609.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harno E, Cottrell EC, White A. Metabolic pitfalls of CNS Cre-based technology. Cell Metab. 2013;18:21–28. doi: 10.1016/j.cmet.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Tattikota SG, et al. Argonaute2 Mediates Compensatory Expansion of the Pancreatic β Cell. Cell Metab. 2014;19:122–134. doi: 10.1016/j.cmet.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glynn MW, McAllister AK. Immunocytochemistry and quantification of protein colocalization in cultured neurons. Nat Protoc. 2006;1:1287–1296. doi: 10.1038/nprot.2006.220. [DOI] [PubMed] [Google Scholar]
- 42.Butler AA, Kozak LP. A Recurring Problem With the Analysis of Energy Expenditure in Genetic Models Expressing Lean and Obese Phenotypes. Diabetes. 2010;59:323–329. doi: 10.2337/db09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birkenfeld AL, et al. Deletion of the mammalian INDY homolog mimics aspects of dietary restriction and protects against adiposity and insulin resistance in mice. Cell Metab. 2011;14:184–195. doi: 10.1016/j.cmet.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevalier C, et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell. 2015;163:1360–1374. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Jeffrey M, et al. Synapse loss associated with abnormal PrP precedes neuronal degeneration in the scrapie-infected murine hippocampus. Neuropathol Appl Neurobiol. 2000;26:41–54. doi: 10.1046/j.1365-2990.2000.00216.x. [DOI] [PubMed] [Google Scholar]
- 46.Pinto S, et al. Rapid Rewiring of Arcuate Nucleus Feeding Circuits by Leptin. Science. 2004;304:110–115. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- 47.Cowley MA, et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411:480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 48.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.