Abstract
Residues important in the interaction between the 23-residue transmembrane (TM) domains of the integrin αIIb- and β3-subunits were identified by mutating each non-Leu residue to Leu. Leu substitutions of αIIb at G972, G976, and T981, and of β3 at I693 and G708, increased ligand binding. Substitutions with other amino acids at αIIbG972 and β3G708 could also increase ligand binding. The results are consistent with and extend the helical interface between the integrin α- and β-subunit TM domains previously defined by cysteine scanning and disulfide bond formation. We differentiated between affinity- and valency-based modes of activation by TM domain mutations. The mutant αIIb W967C forms disulfide-linked αIIb-subunits within an (αIIbβ3)2 tetramer. This tetramer behaved as an ideal model for the valency mode of regulation, because it exhibited significantly increased binding to multivalent but not monovalent ligands and basally retained the bent conformation. By contrast, the activating Leu mutants showed increased binding to the monovalent, ligand-mimetic PAC-1 Fab and increased exposure of ligand-induced binding site (LIBS) epitopes, suggesting that they partially adopt an extended conformation. Furthermore, the previously described β3G708N mutation in Chinese hamster ovary cells enhanced ligand binding affinity, not valency, and did not alter cell-surface clustering as defined by confocal microscopy. Our studies provide evidence that disrupting the integrin heterodimeric TM helix–helix interface activates ligand binding mainly by increasing the monomeric affinity for ligand, but not the receptor valency, i.e., clustering.
Integrins are noncovalently associated heterodimeric cell-adhesion molecules that transmit signals bidirectionally across the plasma membrane and regulate many biological functions including wound healing, cell differentiation, and cell migration. Integrins bind to ligands in the extracellular matrix and on cell surfaces. The “avidity,” or overall strength, of cellular adhesive interactions results from both the affinity of individual receptor–ligand bonds and the valency, or the total number of bonds formed. The affinity of integrins for ligands is conformationally regulated (1–4). On physiological cell surfaces, integrins can assume multiple conformations, each with a distinct affinity. Various studies have indicated that close apposition of the membrane-proximal regions of each subunit and the overall bent structure of the extracellular domain are hallmarks for the low-affinity state of integrins, whereas the extended conformation, with separated legs, represents the high-affinity state (5–9). On the other hand, the valency of integrins for ligands is regulated by changes in their cell-surface distribution. Although a variety of studies suggest that integrin clustering serves to enhance the propensity to form initial adhesions with multivalent substrates (10–14), others have shown that valency regulation does not precede ligand binding, but instead functions in adhesion strengthening after binding to multivalent ligands (15–23).
Although considerable progress has been made, the role of integrin transmembrane (TM) domains in integrin signaling still remains controversial. We previously used disulfide crosslinking studies to determine the helical interface between integrin α- and β-subunit TM domains and showed that restraining this specific heterodimeric helical interface with disulfide bonds maintains integrins in the low-affinity state (8). By contrast, Li et al. (11, 24, 25) have shown that in detergent micelles, integrin TM domains have the potential to form homooligomers rather than heterodimers. Based on two mutations, with most experiments on the mutation β3G708N, they hypothesized that the homomeric associations between TM segments provide a driving force for integrin cell-surface oligomerization (i.e., clustering), thereby enhancing binding to multivalent ligands (11). However, homomeric interactions were only demonstrated with model TM peptides in detergent micelles, and not with intact integrins on cells. The basis for enhanced ligand binding by the β3G708N mutation characterized in this study thus remained unclear.
Here, we use full-length integrin αIIbβ3 expressed on the surface of mammalian cells to study the effects of mutations of the TM domains. Previously we used cysteine scanning to introduce disulfides that maintained association of αIIb and β3 TM domains and showed that this prevented activation. Here we use the converse approach of Leu scanning to perturb TM domain association and screen for activating mutations. We show that substitution with Leu of residues located at the heterodimeric interface defined in the previous study activates ligand binding, suggesting that the activating effect of the TM mutations is a consequence of the disruption of the helical interface between the αIIb and β3 TM domains. We demonstrate that activation is a consequence of conformational changes in the extracellular domain and increased affinity for monomeric ligand. Contrasting results are obtained by crosslinking of αIIb TM domains to form (αIIbβ3)2 tetramers. Tetramers show no extracellular domain conformational change and increased multimeric, but not monomeric, affinity for ligand.
Materials and Methods
Plasmid Construction and Transient Transfection. Plasmids coding for full-length human αIIb and β3 were subcloned into pEF/V5-HisA or pcDNA3.1/Myc-His(+) as described in ref. 5. Mutants were made by using site-directed mutagenesis with the QuikChange kit (Stratagene), and DNA sequences were confirmed before being transfected into 293T cells by using calcium phosphate precipitation (26).
Soluble Fibrinogen, PAC-1 IgM, and PAC-1 Fab Binding Assay. Ligand-mimetic IgM PAC-1 was from Becton Dickinson, PAC-1 Fab was a generous gift from S. Shattil (27), and the activating anti-αIIb mAb PT25-2 was a generous gift from M. Handa (28). Transiently transfected 293T cells or stably transfected Chinese hamster ovary (CHO) cells in 20 mM Hepes with 150 mM NaCl (HBS) supplemented with 5.5 mM glucose and 1% BSA were incubated with fluorescein-labeled fibrinogen (30 μg/ml), PAC-1 IgM (10 μg/ml), or PAC-1 Fab (10 μg/ml) in the presence of 1 mM EDTA, 5 mM Ca2+, or 1 mM Mn2+ plus 10 μg/ml PT25-2 at room temperature for 30 min. For fibrinogen binding, cells were coincubated with Cy3 dye-conjugated anti-β3 mAb AP3 (Cy3-AP3) to a final concentration of 10 μg/ml. For PAC-1 binding, cells were washed with HBS once and suspended with HBS with 1 mM Ca2+, 10 μg/ml FITC-conjugated anti-mouse IgM, and 10 μg/ml Cy3-AP3. For PAC-1 Fab binding, cells were washed once and suspended with HBS with 1 mM Ca2+ and 10 μg/ml FITC-conjugated anti-mouse IgG. Cells were incubated at 0°C for 30 min and then subjected to flow cytometry. Binding is presented as specific mean fluorescence intensity (MFI) of FITC-conjugated anti-mouse IgM or fibrinogen expressed as a percentage of the MFI of Cy3-AP3.
LIBS Expression. The anti-LIBS mAb AP5 was from the Fifth International Leukocyte Workshop (29), LIBS1, LIBS6 (30), and PMI-1 (31) were from M. H. Ginsberg, and D3 was from L. K. Jennings (32). LIBS expression was measured as described in ref. 33. In brief, cells in HBS supplemented with 5.5 mM glucose and 1% BSA were incubated in the presence of 5 mM Ca2+ or 1 mM Mn2+ plus 100 μM GRGDSP peptide ligand for 30 min at room temperature and then with LIBS mAbs AP5, LIBS1, LIBS6, D3, and PMI-1 (10 μg/ml) at 0°C for 30 min, followed by staining with FITC-conjugated anti-mouse IgG and flow cytometry. LIBS binding is presented as MFI of FITC-conjugated anti-mouse IgG as a percentage of the MFI of Cy3-AP3.
Integrin Clustering and Confocal Microscopy. CHO cells stably expressing wild-type αIIbβ3 and mutant αIIbβ3 (11) were kindly provided by J. Bennett (University of Pennsylvania, Philadelphia). Cells in culture medium were incubated with Cy3-AP3 (10 μg/ml) at 37°C for 30 min in the absence or presence of rabbit polyclonal anti-mouse IgG (10 μg/ml) and cytochalasin-D (400 nM), followed by fixation with 3.7% formaldehyde. Confocal imagining was performed with a Bio-Rad Radiance 2000 laser-scanning confocal system coupled to an Olympus BX50WI microscope and a ×100 water immersion objective. All image processing was performed with openlab software (Improvision, Lexington, MA). For each condition, the integrin cell-surface distribution patterns of at least 200 cells, from randomly selected fields, were scored as being either “even” (i.e., exhibiting predominantly even cell-surface distribution) or “macroclustered” (i.e., exhibiting significant levels of uneven or patchy cell-surface distribution).
Results
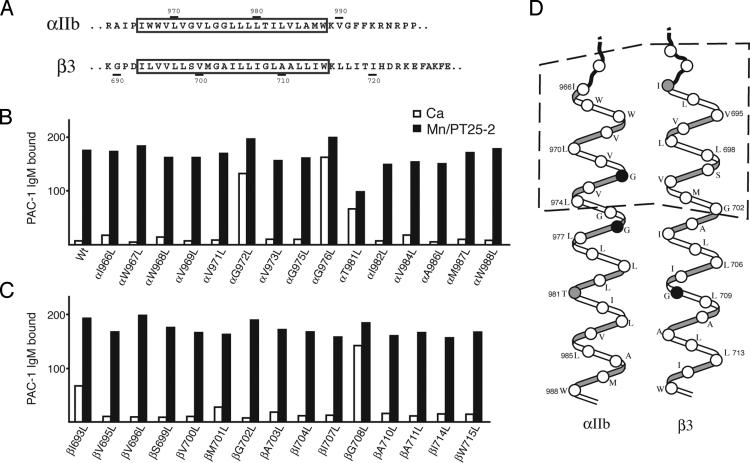
Leu Scanning Shows That Mutations of Several Residues at the Helix–Helix Interface Activate Ligand Binding. Studies on glycophorin A TM domains have suggested that Leu substitutions on average disrupt formation of TM homodimers more than substitutions with other hydrophobic residues studied, i.e., Ala, Cys, Val, Ile, and Met (34). We therefore serially mutated all non-Leu residues in the 23 residue integrin αIIb and β3 TM domains to Leu (Fig. 1A). Mutant αIIb-subunits were coexpressed with wild-type β3-subunits, mutant β3-subunits were coexpressed with wild-type αIIb-subunits, and both were screened for enhanced binding to the soluble, multivalent, ligand-mimetic PAC-1 IgM (Fig. 1 B and C). Under activating conditions (in the presence of Mn2+ and PT25-2), all mutants bound PAC-1 IgM at levels similar to wild type. In the absence of activation, i.e., in Ca2+, wild-type αIIbβ3 and most mutants exhibited essentially no PAC-1 binding. By contrast, three mutants, αIIbG972L, αIIbG976L, and β3G708L, bound PAC-1 IgM at nearly maximal levels, and two others, αIIbT981L and β3I693L, bound partially. We extended our previously generated structural model from crosslinking studies (8) to the C termini of the TM domains assuming ideal α-helices and found that all of these activating residues, with the exception of αIIbT981L, map to the αIIb–β3 interface (Fig. 1D). The inconsistency regarding αIIbT981L may suggest deviation from ideal helices associated with an interacting coiled-coil structure; however, it is noteworthy that mutant αIIbT981L was only partly activating and, in contrast to all other mutants, was expressed poorly on the cell surface relative to wild type (data not shown).
Fig. 1.
Leu scanning of the αIIbβ3 TM domains. (A) Sequences of the αIIbβ3 TM regions. The predicted TM segments are boxed. (B and C) Soluble PAC-1 IgM binding of 293T transfectants expressing integrins with Leu substitutions in the αIIb (A) or β3 (B) TM domains in the presence of 5 mM Ca2+ (open bars) or 1 mM Mn2+ plus 10 μg/ml mAb PT25-2 (filled bars). Binding was measured with two-color immunofluorescence (see Materials and Methods) and is expressed as the MFI of PAC-1 staining as a percentage of MFI of staining with AP3 mAb. Data are representative of two independent experiments. (D) Relative orientation between the αIIb and β3 TM domain α-helices as determined by disulfide scanning of residues shown between dashed lines (8). Activating residues are shown as black circles, partially activating residues are shown as gray circles, and others are shown as white circles.
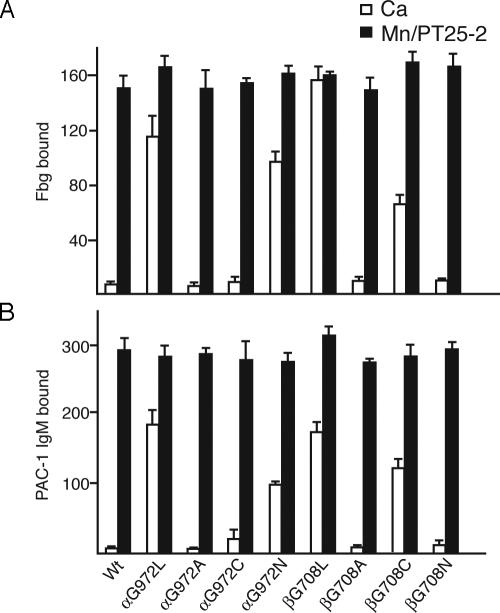
Dependence of Activation on the Specific Amino Acid Substitution. We selected two residues that were activated when mutated to Leu, αIIbG972 and β3G708, for additional mutagenesis studies. These two residues were mutated to Ala, Cys, or Asn. At residue αIIb972, the substitutions G972A and G972C were not activating, showing that in contrast to Leu, smaller hydrophobic side chains were not activating (Fig. 2 A and B). However, the more bulky, polar G972N substitution was activating, although somewhat less than the G972L substitution. At residue β3708, the G708A and G708N substitutions were not activating, whereas the G708C mutation was activating, although less so than G708L (Fig. 2).
Fig. 2.
Effects of different amino acid substitutions on αIIbβ3 activation. Fibrinogen (A) and PAC-1 IgM (B) binding to 293T transfectants was determined in the presence of 5 mM Ca2+ (open bars) or 1 mM Mn2+ plus 10 μg/ml mAb PT25-2 (filled bars). Binding was determined by immunofluorescence as described in Fig. 1.
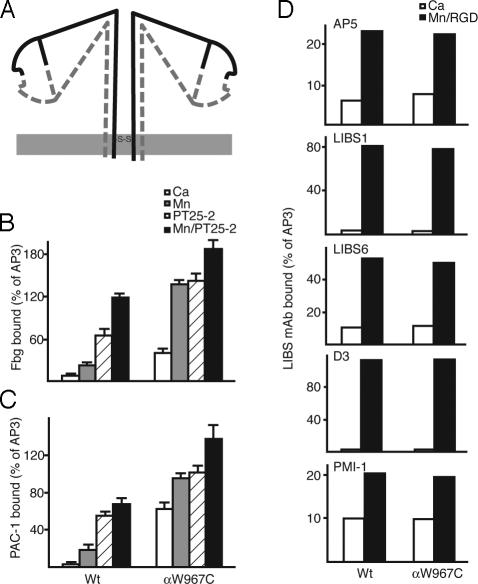
Disulfide-Bonded αIIbβ3 Tetramer Is an Ideal Model for Studying Valency Effect. We previously showed that cysteine mutant αIIbW967C spontaneously and efficiently formed a homodimeric disulfide bond when cotransfected with the β3-subunit, thereby forming a covalently linked cell-surface integrin tetramer (αIIbW967C/β3)2 (8) (Fig. 3A). In 293T transfectants, the tetramer could be recognized by all tested mAbs to constitutive αIIbβ3 epitopes, including AP3, 7E3, 10E5, HA5, and AP2 (data not shown), indicating a native fold for this disulfide-linked integrin.
Fig. 3.
Disulfide-linked αIIbβ3 tetramer provides a model for valency regulation. (A) Schematic drawing of disulfide-linked αIIbβ3 tetramer. The α- and β-subunits are continuous lines and dashed lines, respectively. The membrane is shown as a gray line. The location of the αIIb C967 disulfide bond is shown. (B and C) Soluble fibrinogen and PAC-1 IgM binding to 293T cell transfectants in the presence of Ca2+ (white bars), Mn2+ (gray bars), PT25-2 (hatched bars), or Mn/PT25-2 (black bars). Binding was determined by immunofluorescence as described in Fig. 1. (D) LIBS exposure. Binding of five different anti-LIBS mAbs is expressed as the MFI of mAb staining as a percentage of MFI of staining with AP3 mAb.
Soluble fibrinogen (Fig. 3B) and PAC-1 IgM (Fig. 3C) bound αIIbW967C/β3 more efficiently than wild-type αIIbβ3 basally in Ca2+, as well as after activation with Mn2+, PT25-2/Ca2+, or PT25-2/Mn2+. Notably, in Ca2+ the efficiency of αIIbW967C/β3 tetramer binding to PAC-1 IgM, with potentially 10 binding sites per molecule, was enhanced more than binding to fibrinogen, a divalent ligand. This result suggests that the tetrameric mutant is highly sensitive to the valency of the ligand. Indeed, when binding to the monovalent ligand PAC-1 Fab was assayed under basal conditions in Ca2+, no binding to αIIbW967C/β3 tetramer was found (Fig. 4).
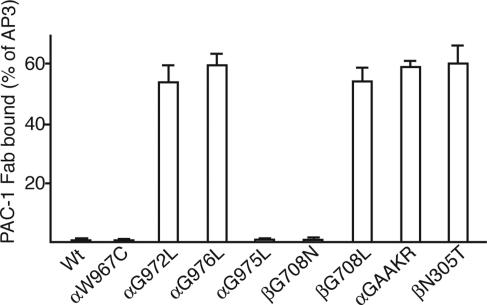
Fig. 4.
PAC-1 Fab binding activity of 293T cell transfectants. The MFI of PAC-1 Fab staining is expressed as a percentage of MFI of staining with AP3 mAb.
LIBS mAbs were used to determine the overall conformation of the αIIbW967C/β3 mutant. All of the LIBS mAbs bound poorly to the tetrameric mutant in Ca2+, similarly to wild type (Fig. 3D), suggesting that the mutant receptor is basally in the bent, resting conformation. This finding, together with the lack of effect on monomeric affinity for PAC-1 Fab, demonstrates that the observed increased binding to PAC-1 IgM and fibrinogen under basal conditions by αIIbW967C/β3 (Fig. 3 B and C) must be attributed to an effect on valency. Thus, this mutant represents an ideal model of valency-regulated ligand binding.
Monovalent Ligand-Mimetic PAC-1 Fab Binding Shows That the Activating Effect of Leu Mutations of the Integrin TM Domains Is Due to Increased Affinity. As described above, the αIIbW967C mutant did not bind to monomeric PAC-1 Fab in Ca2+ (Fig. 4). By contrast, the activating Leu mutations, αIIbG972L, αIIbG976L, and β3G708L, all bound PAC-1 Fab efficiently (Fig. 4). These mutants were activated similarly to the β3N305T mutant (Fig. 4), which introduces an N-glycosylation site into the interface between the β3 I-like and hybrid domains and stabilizes the open, high-affinity conformation of the integrin headpiece (9, 33, 35). High-affinity monomeric binding was also induced by mutation to GAAKR of the GFFKR motif at the junction between the αIIb TM and cytoplasmic domains (Fig. 4). Mutation of this motif has long been known to activate ligand binding (36) and has been shown to induce integrin TM domain separation (8) and cytoplasmic domain separation (7). Overall, the data suggest that the activating effect of the αIIbG972L, αIIbG976L, and β3G708L mutants is due to increased affinity rather than valency. As a further control, the αIIbG975L and β3G708N mutants, which did not increase multimeric affinity, also did not detectably increase monomeric affinity in 293T transfectants (Fig. 4).
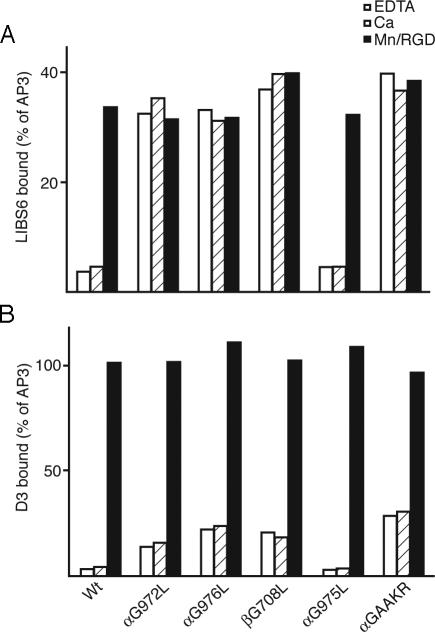
TM Domain Mutations That Increase Monomeric Affinity Partially Increase LIBS Epitope Expression. The mutant receptors were characterized for binding to two representative LIBS mAbs. Under basal conditions, the activating Leu mutants, αIIbG972L, αIIbG976L, and β3G708L, as well as the GAAKR mutant, but not the nonactivating αIIbG975L mutant, bound LIBS6 mAb to essentially maximal levels, i.e., comparably to binding in the presence of Mn2+ and RGD (Fig. 5A). The same activating Leu mutants and the GAAKR mutant, but not αIIbG975L, also elevated binding to mAb D3 under basal conditions, as shown by comparison with wild type. However, in contrast to the LIBS6 epitope, expression of the D3 epitope on all mutants could still be significantly increased by Mn2+ and RGD (Fig. 5B). The same results were obtained in EDTA as in Ca2+, excluding a contribution by ligands in the culture medium to LIBS epitope expression. By contrast to the effect of activating TM domain mutations on D3 epitope exposure, the wedge mutant β3N305T bound D3 and other anti-αIIbβ3 LIBS antibodies at maximal levels in Ca2+, i.e., comparably to binding in the presence of Mn2+ and RGD (data not shown) (33). Similarly, the β1 wedge mutant bound anti-β1 LIBS antibodies maximally (35). These results suggest that, unlike the glycan-wedge mutations that strongly stabilize the integrin in the open, extended conformation (33, 35), the activating Leu and GAAKR mutations increase affinity for ligand by shifting the equilibrium toward the extended conformation of the receptor.
Fig. 5.
Exposure of LIBS epitopes by activating TM domain mutations. The MFI of anti-LIBS mAbs LIBS6 (A) and D3 (B) is expressed as a percentage of MFI of staining with AP3 mAb.
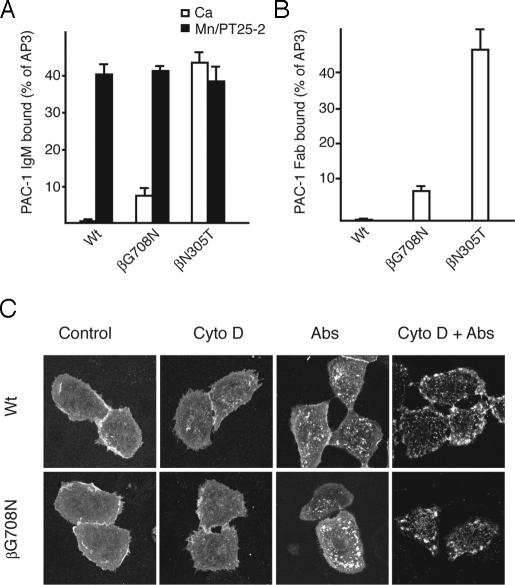
In CHO Transfectants, β3G708N Exhibits Increased Ligand Binding Activity as a Consequence of Increased Affinity Rather Than Increased Valency. Above, we showed that the β3G708N mutation has no detectable effect on ligand binding by αIIbβ3 in 293T cell transfectants. Li et al. (11) have shown that β3G708N mutation in CHO transfectants leads to increased ligand binding compared to wild type. We confirmed that in CHO αIIbβ3 transfectants, β3G708N increased binding to the multivalent ligandmimetic PAC-1 IgM, although to a level <20% of that of the wedge mutant β3N305T (Fig. 6A). The β3G708N mutant also showed increased binding to PAC-1 Fab (Fig. 6B). Binding to multimeric PAC-1 IgM and monomeric PAC-1 Fab by the β3G708N mutant was comparably increased (Fig. 6 A and B). In contrast, the tetrameric αIIbW967C mutant shows binding to multimeric PAC-1 IgM, but not to PAC-1 Fab (Fig. 4). Therefore, it is concluded that the partially activating effect of β3G708N in CHO cells is due to an effect on affinity rather than valency.
Fig. 6.
Ligand binding and clustering of selected αIIbβ3 mutants stably expressed in CHO cells. (A and B) Ligand binding. PAC-1 IgM (A) and PAC-1 Fab (B) binding of CHO cell transfectants. Binding was determined as the MFI of PAC-1 staining as a percentage of MFI of staining with AP3 mAb. (C) Confocal microscopy studies of integrin clustering on the cell surface. Cells expressing wild-type αIIbβ3 or mutant αIIbβ3G708N were incubated with Cy3-AP3 mAb in the absence (control) or presence of cytochalasin-D (Cyto D), anti-mouse IgG (Abs), or anti-mouse IgG/cytochalasin-D (Cyto D + Abs) at 37°C for 30 min, followed by fixation, and subjected to confocal microscopy.
To determine whether the β3G708N mutation induced macroclustering, i.e., areas of the cell surface with increased integrin density that are >200 nm in diameter and therefore detectable by microscopy, we conducted confocal microscopy studies of CHO-cell αIIbβ3 transfectants. Under basal conditions in Ca2+, the vast majority of the cells expressing wild-type and β3G708N αIIbβ3 demonstrated relatively evenly distributed cell-surface integrin (Fig. 6C). Only 4% and 5% of cells, respectively, showed any substantial macroclustering. We also ranked the cells by expression of αIIbβ3 and quantitated clustering in the 10% highest expressing cells in each group. These cells, which expressed about three to five more αIIbβ3 than the mean, also showed comparable macroclustering for wild type (9%) and β3G708N (12%). The presence of similar but small subpopulations of cells with macroclustering illustrates the importance of population analysis in addition to showing micrographs of individual cells. After treatment with cytochalasin-D to increase integrin cell-surface diffusivity (37), integrins remained unclustered on the vast majority of cells (Fig. 6C). Only a slight increase in the number of cells with macroclustering was seen, and the percentages were similar for wild type (13%) and β3G708N (16%). On the other hand, crosslinking cell-surface αIIbβ3 with primary and secondary antibodies promoted significant αIIbβ3 macroclustering (76% for wild type, and 73% for β3G708N) in a manner that was further enhanced by concomitant cytochalasin-D treatment (96% macroclustering for wild type, and 94% for β3G708N) (Fig. 6C). Importantly, under all conditions, no substantial difference was observed in the propensity of wild-type and β3G708N αIIbβ3 to form cell-surface macroclusters.
Discussion
This investigation has yielded insights into the role that TM domains play in integrin activation. Previously we used cysteine scanning mutagenesis to identify the α-helical interface between the N-terminal regions of the integrin α- and β-subunit TM domains and showed that disulfide bonds introduced into this interface maintained the integrin in the low-affinity state (8). Here, we use the converse approach of Leu scanning mutagenesis to perturb TM domain association. The activating mutations map to the same interface as previously defined with activation-restraining disulfides. This finding strongly suggests that activation is a consequence of disruption of the helical TM interface between the integrin α- and β-subunits. Moreover, we extended previous results by subjecting the entire TM segments to mutagenesis. With the exception of one anomalous mutation that was not well expressed and only partially activating, the results suggest that the previously defined α-helical interface extends all of the way across the membrane. We further showed that a variety of the activating mutations both increase monovalent ligand binding and induce conformational changes in the extracellular domain, consistent with our previous finding that association between the TM domains maintains the low-affinity integrin conformation.
We previously identified a mutant, αIIbW967C, that when cotransfected with wild-type β3 efficiently forms disulfide-linked integrin tetramers on the cell surface (8). We showed here that these tetramers are basally in the bent conformation and exhibit increased binding to multivalent, but not monovalent, ligands. These results are canonical for the valency (or “clustering”) mode of regulation. Thus, αIIbW967C represents an ideal model for the valency mode of activation. By contrast, Leu and several other amino acid substitutions introduced into the TM helical interface, mutation of the GFFKR motif (αIIbGAAKR), and introduction of a glycan wedge into the I-like hybrid domain interface (β3N305T) increase monomeric affinity for ligand. Thus, the αIIbW967C mutation increases valency, whereas all other activating mutations in the TM domains characterized here, including αIIbβ3G708N, act by increasing affinity, but not valency, for ligand.
Our results fail to support a major role for the TM domains in regulating cell-surface oligomerization of integrins, as has recently been suggested by others (11, 25). The mutant β3G708N has been described to stabilize homotrimerization of isolated TM domains in detergent micelles and suggested to enhance ligand binding in CHO cells by driving cell-surface integrin clustering (11). We have confirmed that this mutation partially increased binding for multimeric ligand in CHO cells; however, β3G708N binding to monomeric ligand was enhanced to the same extent. These results suggest that β3G708N activates by the affinity mode of regulation. Moreover, this mutant failed to alter the extent of cell-surface macroclustering compared with wild-type αIIbβ3 under a variety of conditions.
The two most activating mutations we found in the αIIb-subunit, G972L and G976L, are present within a GXXXG motif. GXXXG motifs appear to be generally important in association between TM helices (34), and we have previously demonstrated that this motif in αIIb is in the interface with β3 in the membrane (8). Homomeric association of truncated αIIb TM domains has been demonstrated in Escherichia coli inner membranes by reporter assays and in detergent micelles by SDS/PAGE and analytical ultracentrifugation (25). The G972L and G976L mutations decrease homomerization in these assays. We find that the same mutations activate binding of soluble monovalent ligand and expression of LIBS epitopes. The results are consistent with a similar interface involving the αIIb GXXXG motif being involved in heterodimerization with β3 and homomerization with αIIb. Furthermore, the findings that (i) G972L and G976L mutations are activating (here) and (ii) decrease αIIb homomerization (25) are compatible with activation by separation of αIIb and β3 TM domains but are incompatible with activation by αIIb homomerization.
Our results support an important role for separation of the integrin α and β TM domains in regulating the conformation and affinity for ligand of the extracellular domain. Notably, the only example in which we have been able to capture homomeric integrin TM domain interactions with disulfide bonds is in the resting, low-affinity conformation. The integrin dimer that is formed binds multivalent, but not monovalent, ligands better, demonstrating valency rather than affinity regulation. Despite a large number of α- and β-subunit TM cysteine mutations that were tested within heterodimers containing an activating α-subunit GFFKR mutation, and separation of the α- and β-subunit TM domains in these mutants, none formed disulfide-linked homomers (8). Furthermore, fluorescent resonance energy transfer between α-subunits of different integrin heterodimers, or between β-subunits of different integrin heterodimers, shows that integrin activation and separation of α and β TM domains in the plane of the membrane occurs without giving rise to homomeric α TM domain or homomeric β TM domain interactions (23). In the future, it will be interesting to learn whether micro- and macroclustering of integrins on the cell surface, which occurs only after binding to multimeric ligands (23), brings the TM domains of integrins sufficiently close together to drive formation of integrin α- or β-subunit TM domain homomers.
Acknowledgments
We thank Drs. M. H. Ginsberg, S. Shattil, M. Handa, and L. K. Jennings for generously providing antibodies; J. Bennett for providing CHO cell lines; Aideen Mulligan for laboratory management assistance; Jessica Martin for secretarial assistance; and Drs. M. H. Ginsberg and M. L. Dustin for reviewing the manuscript. This work was supported by National Institutes of Health Grant HL48675 (to T.A.S. and J.T.).
Author contributions: B.-H.L., J.T., and T.A.S. designed research; B.-H.L. and C.V.C. performed research; B.-H.L., C.V.C., J.T., and T.A.S. analyzed data; and B.-H.L., C.V.C., and T.A.S. wrote the paper.
Abbreviations: TM, transmembrane; FITC, fluorescein isothiocyanate; CHO, Chinese hamster ovary; MFI, mean fluorescence intensity; LIBS, ligand-induced binding site.
References
- 1.Hynes, R. O. (2002) Cell 110, 673-687. [DOI] [PubMed] [Google Scholar]
- 2.Liddington, R. C. & Ginsberg, M. H. (2002) J. Cell Biol. 158, 833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carman, C. V. & Springer, T. A. (2003) Curr. Opin. Cell Biol. 15, 547-556. [DOI] [PubMed] [Google Scholar]
- 4.Takagi, J. & Springer, T. A. (2002) Immunol. Rev. 186, 141-163. [DOI] [PubMed] [Google Scholar]
- 5.Takagi, J., Petre, B. M., Walz, T. & Springer, T. A. (2002) Cell 110, 599-611. [DOI] [PubMed] [Google Scholar]
- 6.Kim, M., Carman, C. V. & Springer, T. A. (2003) Science 301, 1720-1725. [DOI] [PubMed] [Google Scholar]
- 7.Vinogradova, O., Velyvis, A., Velyviene, A., Hu, B., Haas, T. A., Plow, E. F. & Qin, J. (2002) Cell 110, 587-597. [DOI] [PubMed] [Google Scholar]
- 8.Luo, B.-H., Springer, T. A. & Takagi, J. (2004) PLoS Biol. 2, 776-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao, T., Takagi, J., Wang, J.-h., Coller, B. S. & Springer, T. A. (2004) Nature 432, 59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazzoni, G. & Hemler, M. E. (1998) Trends Biochem. Sci. 23, 30-34. [DOI] [PubMed] [Google Scholar]
- 11.Li, R., Mitra, N., Gratkowski, H., Vilaire, G., Litvinov, S. V., Nagasami, C., Weisel, J. W., Lear, J. D., DeGrado, W. F. & Bennett, J. S. (2003) Science 300, 795-798. [DOI] [PubMed] [Google Scholar]
- 12.Hogg, N., Henderson, R., Leitinger, B., McDowall, A., Porter, J. & Stanley, P. (2002) Immunol. Rev. 186, 164-171. [DOI] [PubMed] [Google Scholar]
- 13.van Kooyk, Y., van Vliet, S. J. & Figdor, C. G. (1999) J. Biol. Chem. 274, 26869-26877. [DOI] [PubMed] [Google Scholar]
- 14.Constantin, G., Majeed, M., Giagulli, C., Piccib, L., Kim, J. Y., Butcher, E. C. & Laudanna, C. (2000) Immunity 13, 759-769. [DOI] [PubMed] [Google Scholar]
- 15.Plancon, S., Morel-Kopp, M. C., Schaffner-Reckinger, E., Chen, P. & Kieffer, N. (2001) Biochem. J. 357, 529-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laukaitis, C. M., Webb, D. J., Donais, K. & Horwitz, A. F. (2001) J. Cell Biol. 153, 1427-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballestrem, C., Hinz, B., Imhof, B. A. & Wehrle-Haller, B. (2001) J. Cell Biol. 155, 1319-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giannone, G., Dubin-Thaler, B. J., Dobereiner, H.-G., Kieffer, N., Bresnick, A. R. & Sheetz, M. P. (2004) Cell 116, 431-443. [DOI] [PubMed] [Google Scholar]
- 19.Loftus, J. C. & Albrecht, R. M. (1984) J. Cell Biol. 99, 822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isenberg, W. M., McEver, R. P., Phillips, D. R., Shuman, M. A. & Bainton, D. F. (1987) J. Cell Biol. 104, 1655-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliver, J. A. & Albrecht, R. M. (1987) Scanning Microsc. 1, 745-756. [PubMed] [Google Scholar]
- 22.Buensuceso, C., De Virgilio, M. & Shattil, S. J. (2003) J. Biol. Chem. 278, 15217-15224. [DOI] [PubMed] [Google Scholar]
- 23.Kim, M., Carman, C. V., Yang, W., Salas, A. & Springer, T. A. (2004) J. Cell Biol. 167, 1241-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, R., Babu, C. R., Lear, J. D., Wand, A. J., Bennett, J. S. & Degrado, W. F. (2001) Proc. Natl. Acad. Sci. USA 98, 12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, R., Gorelik, R., Nanda, V., Law, P. B., Lear, J. D., DeGrado, W. F. & Bennett, J. S. (2004) J. Biol. Chem. 279, 26666-26673. [DOI] [PubMed] [Google Scholar]
- 26.DuBridge, R. B., Tang, P., Hsia, H. C., Leong, P. M., Miller, J. H. & Calos, M. P. (1987) Mol. Cell. Biol. 7, 379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abrams, C., Deng, Y. J., Steiner, B., O'Toole, T. & Shattil, S. J. (1994) J. Biol. Chem. 269, 18781-18788. [PubMed] [Google Scholar]
- 28.Tokuhira, M., Handa, M., Kamata, T., Oda, A., Katayama, M., Tomiyama, Y., Murata, M., Kawai, Y., Watanabe, K. & Ikeda, Y. (1996) Thromb. Haemostasis 76, 1038-1046. [PubMed] [Google Scholar]
- 29.Petruzzelli, L., Huang, C. & Springer, T. A. (1995) in Leucocyte Typing V: White Cell Differentiation Antigens, eds. Schlossman, S. F., Boumsell, L., Gilks, W., Harlan, J., Kishimoto, T., Morimoto, C., Ritz, J., Shaw, S., Silverstein, R., Springer, T., et al. (Oxford Univ. Press, New York), pp. 1586-1587.
- 30.Du, X., Gu, M., Weisel, J. W., Nagaswami, C., Bennett, J. S., Bowditch, R. & Ginsberg, M. H. (1993) J. Biol. Chem. 268, 23087-23092. [PubMed] [Google Scholar]
- 31.Loftus, J. C., Plow, E. F., Frelinger, A. L. d., D'Souza, S. E., Dixon, D., Lacy, J., Sorge, J. & Ginsberg, M. H. (1987) Proc. Natl. Acad. Sci. USA 84, 7114-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jennings, L. K. & White, M. M. (1998) Am. Heart J. 135, S179-S183. [DOI] [PubMed] [Google Scholar]
- 33.Luo, B.-H., Springer, T. A. & Takagi, J. (2003) Proc. Natl. Acad. Sci. USA 100, 2403-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemmon, M. A., Flanagan, J. M., Treutlein, H. R., Zhang, J. & Engelman, D. M. (1992) Biochemistry 31, 12719-12725. [DOI] [PubMed] [Google Scholar]
- 35.Luo, B.-H., Strokovich, K., Walz, T., Springer, T. A. & Takagi, J. (2004) J. Biol. Chem. 279, 27466-27471. [DOI] [PubMed] [Google Scholar]
- 36.Hughes, P. E., Diaz-Gonzalez, F., Leong, L., Wu, C., McDonald, J. A., Shattil, S. J. & Ginsberg, M. H. (1996) J. Biol. Chem. 271, 6571-6574. [DOI] [PubMed] [Google Scholar]
- 37.Kucik, D. F., Dustin, M. L., Miller, J. M. & Brown, E. J. (1996) J. Clin. Invest. 97, 2139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]