Abstract
Serum response factor (SRF) was found to be involved in the phenotypic transition and fibrosis of the peritoneal membrane during treatment with peritoneal dialysis (PD), but the exact mechanism remains unclear. SRF regulates microRNAs (miRNAs) that contain the SRF-binding consensus (CArG) element in the promoter region. Therefore, we investigated whether the miR-199a/214 gene cluster, which contains a CArG element in its promoter, is directly regulated by SRF. High-glucose (HG) treatment significantly unregulated the expression of the miR-199a-5p/214–3p gene cluster in human peritoneal mesothelial cells (HPMCs). By chromatin immunoprecipitation and reporter assays, we found that SRF binds to the miR-199a-5p/214–3p gene cluster promoter after HG stimulation. In vitro, in HPMCs, silencing of miR-199a-5p or miR-214–3p inhibited the HG-induced phenotypic transition and cell migration but enhanced cell adhesion, whereas ectopic expression of mimic oligonucleotides had the opposite effects. Both miR-199a-5p and miR-214–3p targeted claudin-2 and E-cadherin mRNAs. In a PD rat model, treatment with an SRF inhibitor silenced miR-199a-5p and miR-214–3p and alleviated HG-PD fluid–induced damage and fibrosis. Overall, this study reveals a novel SRF–miR-199a/miR-214–E-cadherin/claudin-2 axis that mediates damage and fibrosis in PD.
Keywords: peritoneal dialysis, fibrosis, claudins, miR-199a/214 gene cluster
Peritoneal fibrosis is a final common pathway in all progressive forms, which disturbs long-term peritoneal dialysis (PD) by causing morphologic and functional changes in the peritoneal membrane (PM) that is damaged by high concentrations of glucose from PD fluids (PDFs).1 Previous studies have shown that the PM does not undergo any apparent changes during the first 2 years of PD with the exception of a phenotypic transition, such as the epithelial-to-mesenchymal transition (EMT) of human peritoneal mesothelial cells (HPMCs). It was suggested that high-glucose (HG) may induce the peritoneal mesothelial transition into a characteristic myofibroblastic phenotype by modulating complex gene expression, ultimately leading to cytoskeletal organization, cell adhesion, and ECM production. Our previous work revealed that serum response factor (SRF) is involved in the phenotypic transition and fibrosis of the PM; however, the underlying mode of action of SRF in HPMCs after high-glucose damage remains unclear.2
SRF is not only a master switch for the expression of contractile and cytoskeletal genes but is also a regulator of numerous microRNAs (miRNAs) which contain the CArG element in the promoter region.3 According to current literature, a large number of miRNAs might have been linked to peritoneal fibrosis, suggesting that they may be involved in peritoneal fibrosis.4 MiR-199a is located on chromosome 1 and is embedded in the antisense strand of intron 14 of Dynamin 3 (DNM3). DNM3OS, which stands for “DNM3 opposite strand,” gives rise to miR-199a and another miRNA, miR-214. Sometimes the two miRNAs, which contain a common CArG element in the promoter, are referred to as members of the miR-199a/214 gene cluster5,6 and may be directly regulated by SRF. Some data also suggest that miR-199a and miR-214 sculpt developmental processes by mediating the effects on cell-cycle proteins.7–9 To date, the exact mechanism in peritoneal fibrosis remains completely unknown. In this study, we further examine the roles of SRF and the miR-199a/214 cluster in peritoneal fibrosis during PD treatment.
Results
SRF Directly Regulates Transcription of the miR-199a/214 Gene Cluster in HG-HPMCs
We found that all of the possible SRF target genes contain one or more highly conserved CArG elements, which are commonly found in the promoters of miRNA genes.10 On the basis of further prediction, we selected the miR-199a/214 gene cluster from the serum response element (SRE)-containing miRNAs to further evaluate their roles in peritoneal fibrosis.
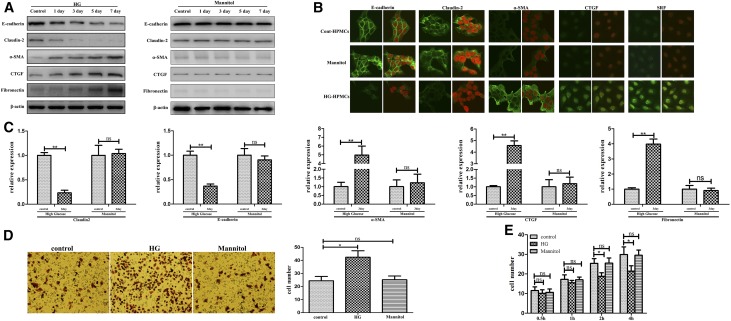
We found that the total and active protein levels of the transcription factor SRF were increased after HG stimulation in both a time- and dose-dependent manner (Figure 1A, Supplemental Figure 1). Mannitol-treated HPMCs were used as control. Quantitative RT-PCR (real-time PCR) showed that SRF, miR-199a-5p, and miR-214–3p expression levels were significantly elevated in a time-dependent manner in HG-induced HPMCs, compared with control cells (Figure 1, B–D). The other matched oligonucleotides, miR-199a-3p and miR-214–5p, were also significantly overexpressed in HG-induced HPMCs (Supplemental Figure 2).
Figure 1.
The miR-199a/214 cluster is regulated by SRF in HG-induced HPMCs. (A) The expression of SRF in HG-induced HPMCs (HG-HPMCs) was evaluated by western blot analysis. Levels of total SRF and phosphorylated SRF (p-SRF) were assessed in HG-HPMCs induced for 1, 3, 5, and 7 days and compared with control HPMCs. Mannitol-treated HPMCs were followed as osmotic pressure control timely. β-actin was used as a loading control. (B–D) Real-time PCR showing the expression levels of SRF, miR-199a-5p, and miR-214–3p in HG-HPMCs at 1, 3, 5, and 7 days, compared with control HPMCs. P values were determined using paired t test. Mannitol-treated HPMCs were followed as osmotic pressure control. (E and F) ChIP analysis showing the association between miR-199a/214 and SRF expression in HG-HPMCs compared with control HPMCs. Endogenous SRF protein in HG-HPMCs was immunoprecipitated using an anti-SRF antibody and subjected to RT-PCR analysis with miR-199a/214 primers. (G) The conserved upstream region of the miR-199a/214 gene was cloned into the pGL3 luciferase expression plasmid and transfected into control HPMCs or HG-HPMCs. SRF potently activated the luciferase reporter linked to the miR-199a/214 upstream region with the wild-type CArG box, whereas the mutant CArG box abolished responsiveness to SRF. There were no differences in luciferase expression in wild-type versus mutant or deleted CArG box constructs between the HG-HPMCs and control HPMCs. Detail data are as follows that: pGL3-basic: control HPMCs 0.169±0.017, HG-HPMCs 0.419±0.026; pGL3-del-miR-199a: control HPMCs 0.491±0.038, HG-HPMCs 0.465±0.075; pGL3-mut-miR-199a: control HPMCs 0.467±0.027, HG-HPMCs 0.413±0.042; pGL3-miR-199a: control HPMCs 1.417±0.058, HG-HPMCs 4.365±0.273. *P<0.05; **P<0.01. luc, luciferase; SRE, serum response element.
It was found that there is a predicted SRE site (CArG element) from -1880 to -1869 in the promoter of the miR-199a/214 gene cluster on chromosome 1 that may bind to SRF protein (Figure 1E, Supplemental Table 1). ChIP analysis was used to assess the binding of SRF to CArG elements upstream of the miR-199a/214 genes (Figure 1F). We predicted that SRF binds to a conserved CArG element in the promoter regions of miR-199a/214 at -332 sites. We next designed real-time PCR amplicons near the transcription start sites of miR-199a/214 transcripts to assay it. A 50-fold enrichment was used as a threshold for positive SRF binding in all studies. Signals above this threshold were obtained near the transcription start sites for each of the two pri-miRNAs assayed, which strongly suggests that SRF associates with the promoter. Consistent with these findings, the binding signals were greatly increased in the HG-HPMC cell lines compared with the control HPMCs.
Luciferase activity assays were performed to further determine whether miR-199a/214 expression patterns are directly regulated by SRF. We first constructed wild-type, mutated, and deleted miR-199a/214 luciferase reporter vectors (Figure 1G), which were then cotransfected into low- and high-SRF–expressing HG-stimulated HPMCs. SRF was also cotransfected with the miR-199a/214 constructs into HPMCs. Luciferase activity was increased in HG-stimulated HPMCs when the wild-type 5′ UTRs of miR-199a/214 were present. Furthermore, luciferase activity was found to be stronger in HG-stimulated HPMCs compared with control HPMCs. But the transcriptional activation of miR-199a/214 was abolished by mutation or deletion of the CArG box. Thus, it is indicated that SRF is able to bind to the promoter regions and induce the expression of genes within the miR-199a/214 gene cluster.
Expression of the miR-199a/214 Cluster is Upregulated in HG-Induced HPMCs, Resulting in Phenotypic Transition and Enhanced Migration but Reduced Adhesion In Vitro
To further explore the significance of the miR-199a/214 cluster in peritoneal fibrosis, we studied the phenotypic change of HG-HPMCs, including migration, adhesion, and phenotypic transition, which may contribute to peritoneal fibrosis after HG damage. Mannitol-treated HPMCs were used as control (Figure 2). We determined the expression levels of the epithelial-to-epithelial adhesion protein marker E-cadherin, the cell-to-matrix protein marker claudin-2, the mesenchymal marker α-SMA, and the fibrosis markers CTGF and FN, by immunofluorescence, western blotting, and real-time PCR (Figure 2, A–C). Concordant with our previous work, HPMCs demonstrated a loss of E-cadherin and claudin-2 expression but increased expression of α-SMA, CTGF, and FN at both the protein and mRNA levels in response to HG stimulation in a time-dependent manner. Transwell assays showed that, compared with control HPMCs, HG-stimulated HPMCs exhibited a remarkable increase in migration and deformation potential (Figure 2D). Interestingly, HG-stimulation could also significantly suppress the adhesion of HG-stimulated HPMCs in adhesion assay, which might lead to lost intercellular junctions of these HPMCs which were in preparation to transform into spindle fibroblastic-shaped cells (Figure 2F). Altogether, these findings indicate that HG stimulation plays an important role in promoting migration and loss of adhesion as well as in the occurrence of a phenotypic transition of HPMCs, which may ultimately contribute to peritoneal fibrosis.
Figure 2.
MiR-199a/214 cluster was significantly upregulated in HG-induced HPMCs in vitro. (A) Expression of E-cadherin, claudin-2, α-SMA, CTGF, and FN in HG-HPMCs was evaluated at 1, 3, 5, and 7 days by western blot analysis, and compared with control HPMCs. β-actin was used as a loading control. Mannitol-treated HPMCs were followed as osmotic pressure control. (B) Subcellular localization of SRF, E-cadherin, and claudin-2 in the HG-HPMCs treated with HG for 3 days and comparison with the control by immunohistochemical analysis. HPMCs were cultured with HG cell culture medium containing 10% serum, with or without 50 mmol/L D-glucose, fixed with 4% PFA, permeabilized, and blocked with 5% FBS for 1 hour at 20°C. Mannitol-treated HPMCs were followed as osmotic pressure control. Endogenous SRF, E-cadherin, claudin-2, α-SMA, and CTGF were detected using anti-rat monoclonal antibody and FITC-conjugated (green) secondary antibody, and then the nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; red, the right slide is the merged image for each group). Original magnification, ×400. (C) Real-time PCR showing the expression levels of E-cadherin, claudin-2, α-SMA, FN, CTGF, and collagen I in HG-HPMCs at 3 days compared with control HPMCs. Mannitol-treated HPMCs were followed as osmotic pressure control. (D) Cell migration assays. Representative fields of invasive cells on the membrane. Average numbers of migrated cells in three independent experiments. Original magnification, ×200. Bar graphs represent the average numbers of cells on the underside of the membrane ±SEM. P values were determined by paired t tests. (E) Cell adhesion assays. Bar graphs represent the average numbers of adhesive cells on the Matrigel after 0.5, 1, 2, and 4 hours ±SEM. P values were determined by t tests. *P<0.05; **P<0.01. ns, not significant.
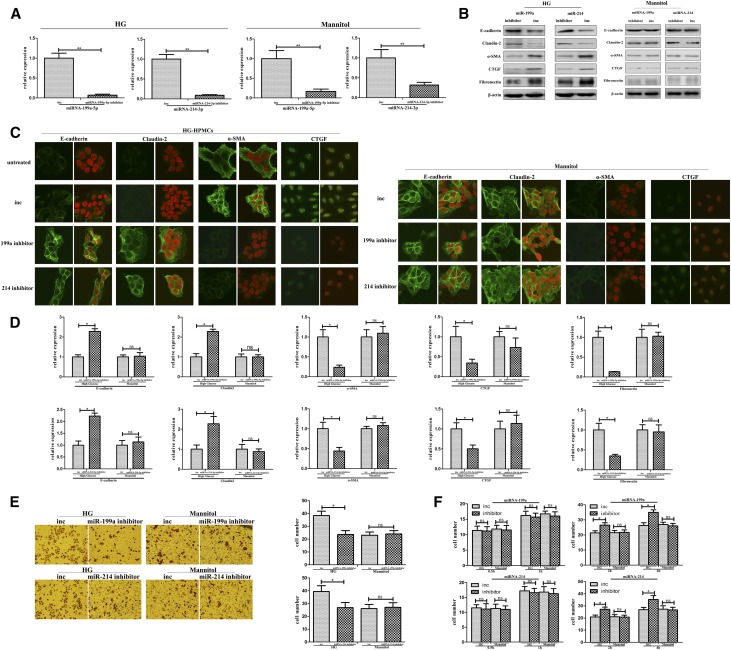
Silencing miR-199a/214 Decreased HG-Induced Phenotypic Transition and Migration but Enhanced Adhesion of HPMCs In Vitro
To obtain further insights into the biologic relevance of the miR-199a/214 gene cluster, we analyzed its effect on phenotypic transition, fibrosis, migration, and adhesion in immortal HPMCs after translation with an miR-199a or miR-214 inhibitor (Figure 3, Supplemental Figure 3). Mannitol-treated HPMCs were used as control. Immunohistochemistry showed that E-cadherin and claudin-2 protein levels were significantly increased, whereas α-SMA, CTGF, and fibronectin protein levels were significantly decreased after transfection with an inhibitor of miR-199a-5p or miR-214–3p in HG-induced HPMCs. Western blotting and real-time PCR showed similar results (Figure 3, A–D). These data show that the downregulation of miR-199a-5p or miR-214–3p can inhibit the phenotypic transition and fibrosis of HG-induced HPMCs. Transwell assay illustrated that cell migration was decreased by the miR-199a-5p or miR-214–3p inhibitor in HG-induced HPMCs compared with control (Figure 3F). Results of the adhesion assay showed that cell adhesion was strongly increased in HG-induced HPMCs after transfection with an miR-199a-5p or miR-214–3p inhibitor compared with the control HG-induced HPMCs (Figure 3F). Mannitol-treated HPMCs had no change here. Altogether, these data show that inhibition of miR-199a-5p or miR-214–3p decreases the phenotypic transition and fibrosis, inhibits the cell migration, and enhances the adhesion ability of HG-induced HPMCs. However, the other matched oligonucleotides, miR-199a-3p and miR-214–5p, play a minor role in the HG damage process of peritoneal fibrosis (Supplemental Figure 3).
Figure 3.
MiR-199a-5p and miR-214-3p inhibitors could block phenotypic transition and migration but increase adhesion of HG-HPMCs in vitro. (A) Real-time PCR shows the level of miR-199a-5p or miR-214–3p in HG-HPMCs after transfection with the miR-199a-5p or miR-214–3p inhibitor or the control oligonucleotide (inc). Mannitol-treated HPMCs were followed as osmotic pressure control. (B) Expression of E-cadherin, claudin-2, α-SMA, CTGF, and fibronectin in HG-HPMCs transfected with the miR-199a-5p or miR-214–3p inhibitor was evaluated by western blot analysis and compared with the control oligonucleotide. β-actin was used as a loading control. (C) Immunohistochemical analysis of E-cadherin and claudin-2 expression in HG-HPMCs transfected with the miR-199a-5p or miR-214–3p inhibitor compared with the control oligonucleotides. Original magnification, ×400. (D) Real-time PCR showing the mRNA levels of E-cadherin, claudin-2, α-SMA, CTGF, and fibronectin transfected with the miR-199a-5p or miR-214–3p inhibitor compared with the control oligonucleotide. (E) Cell migration assays were performed using HG-HPMCs transfected with the miR-199a-5p inhibitor, miR-214–3p inhibitor, or control oligonucleotides. Mannitol-treated HPMCs were followed as osmotic pressure control. Representative fields of migrated HG-HPMCs on the membrane. Original magnification, ×200. Average numbers of migrated cells in three independent experiments. P values were determined using t tests. (F) Cell adhesion assays. Bar graphs represent the average adhesive numbers of cells on the Matrigel after 0.5, 1, 2, and 4 hours ±SEM. P values were determined using t tests. *P<0.05; **P<0.01.
Ectopic Expression of the miR-199a/214 Gene Cluster Enhances the Phenotypic Transition and Migration but Decreases the Adhesion of HPMCs In Vitro
Using mimic oligonucleotides, we upregulated the expression of miR-199a-5p, miR-199a-3p, miR-214–3p, and miR-214–5p to study the role of the miR-199/214 gene cluster in the phenotypic transition of HPMCs, peritoneal fibrosis, cell migration, and adhesion (Figure 4).
Figure 4.
Phenotypic transition and migration of HPMCs are promoted but cell adhesion is lost by the miR-199a/214 mimic transfection in HPMCs in vitro. (A) Real-time PCR shows the level of miR-199a-5p or miR-214–3p in HPMCs after transfection with miR-199a-5p (up) or miR-214–3p (down) mimic or the control oligonucleotide (mnc). (B) Immunohistochemical analysis of E-cadherin, claudin-2, α-SMA, and CTGF expression in HPMCs transfected with the miR-199a-5p or miR-214–3p mimic compared with the control oligonucleotide. Original magnification, ×400. (C) Expression of E-cadherin, claudin-2, α-SMA, CTGF, and FN in HPMCs transfected with miR-199a-5p or miR-214–3p mimic was evaluated by western blot analysis compared with the control oligonucleotide. β-actin was used as a loading control. (D) Real-time PCR showing the mRNA levels of E-cadherin, claudin-2, α-SMA, CTGF, and fibronectin transfected with miR-199a-5p (up) or miR-214–3p (down) mimic compared with the control oligonucleotide. (E) Cell migration assays were performed using HPMCs transfected with miR-199a-5p mimic, miR-214–3p mimic, or control oligonucleotide. Representative fields of migrated HPMC cells on the membrane. Original magnification, ×200. Average numbers of migrated cells in three independent experiments. P values were determined using the paired t test. (F) Cell adhesion assays. Bar graphs represent the average adhesive numbers of cells on the Matrigel after 0.5, 1, 2, and 4 hours ±SEM. P values were determined using paired t tests. *P<0.05; **P<0.01.
Immunofluorescence and western blot analysis showed that the upregulation of miR-199a-5p or miR-214–3p in immortal HPMCs led to significant downregulation of E-cadherin and claudin-2, but upregulation of α-SMA, CTGF, and FN in HPMCs (Figure 4, A–D). The results of the transwell and adhesion assays showed that when miR-199a-5p or miR-214–3p was upregulated by mimic oligonucleotides, cell migration was significantly enhanced but cell-cell and cell-matrix adhesion were alleviated (Figure 4, E and F). Therefore, miR-199a-5p or miR-214–3p promotes phenotypic transition, fibrosis, and cell migration of HPMCs but alleviates cell adhesion by downregulating E-cadherin and claudin-2, but upregulating α-SMA, CTGF, and FN. However, miR-199a-3p and miR-214–5p play minor roles in the process of peritoneal fibrosis when HPMCs are transfected with miR-199a-3p or miR-214–5p mimic oligonucleotides.
The miR-199a-5p/214–3p Gene Cluster Positively Correlates with SRF but Negatively Correlates with E-Cadherin and Claudin-2 in Primary HPMCs from Patients Receiving Continuous Ambulatory PD Ex Vitro
We next investigated the expression levels of the miR-199a-5p/miR-214–3p gene cluster, E-cadherin, claudin-2, and α-SMA by real-time PCR in cast-off HPMCs ex vivo. We collected HPMCs ex vivo from 28 patients receiving PD (G1–G4) and omentum-derived HPMCs from five patients with ESRD who underwent a catheter operation for further PD therapy (G0) as controls.
The results showed that the expression of SRF and miR-199a/214 gene cluster was significantly elevated in HPMCs from G2, G3, and G4 patients receiving continuous ambulatory peritoneal dialysis (CAPD) compared with G0 patients (Figure 5, A–C). However, we observed decreased expression of E-cadherin and claudin-2 in G2, G3, and G4 patients who underwent >1 year of treatment compared with G0 and G1 patients who underwent short-term CAPD, consistent with previous results (Figure 5, D and E). Thus, similar to SRF, the miR-199a-5p/214–3p gene cluster may be associated with PM injury by HG during long-term PD. MiR-199a-5p and miR-214–3p positively correlated with SRF (Figure 5, F and G) but negatively correlated with E-cadherin and claudin-2 (Figure 5, H and I). Additionally, miR-199a-5p and miR-214–3p positively correlated with each other (Figure 5J) but negatively correlated with both E-cadherin and claudin-2 (Figure 5, K–N). E-cadherin and claudin-2 also positively correlated with each other (Figure 5O). Thus, SRF and the miR-199a-5p/214–3p gene cluster may be associated with the phenotypic transition and cell-cell adhesion of HPMCs after the injury of PMs by exposure to HG during long-term PD.
Figure 5.
The miR-199a/214 gene cluster and E-cadherin and claudin-2 were inversely expressed in HPMCs ex vivo. (A–E) Real-time PCR showing the expression of SRF, miR-199a-5p, miR-214–3p, claudin-2, and E-cadherin in effluent-derived HPMCs from 28 patients receiving PD from different groups (G1–G4) that underwent PD treatment, compared with omentum-derived HPMCs from five patients with ESRD who underwent catheter operation for further PD therapy as a control (G0). (F–O) The correlation analysis of SRF and miR-199a-5p, SRF and miR-214–3p, SRF and claudin-2, SRF and E-cadherin, miR-199a-5p and miR-214–3p, miR-199a-5p and claudin-2, miR-199a-5p and E-cadherin, miR-214–3p and claudin-2, miR-214–3p and E-cadherin, and claudin-2 and E-cadherin in effluent-derived HPMCs from G0 to G4. R Sq Line ar, R-squared line-adjusted rate.
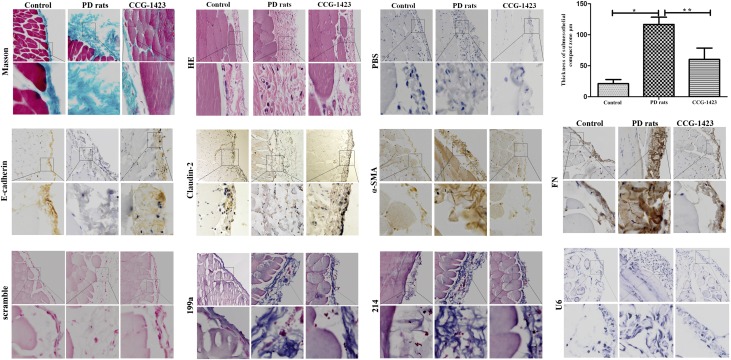
Expression of the miR-199a-5p/214–3p Gene Cluster Is Attenuated in PD Rats Treated with the SRF Inhibitor CCG-1423
We next established an animal model of PD in which rats were injected with HG-PDF (4.25% glucose dialysis solution; Baxter, Chicago, IL) or normal saline solution over a period of 6 weeks (n=8 rats in each group) (Figure 6). To examine the effect of SRF inhibition in PD, we performed PD in rats treated with the SRF inhibitor, CCG-1423, for 6 weeks. A loss of the mesothelial cell monolayer with a thicker PM was observed in the parietal peritoneum from the untreated PD group compared with the saline control group, whereas CCG-1423 significantly alleviated peritoneal thickness in the treated group as observed by H&E staining. Masson staining and overexpression of α-SMA, FN, and CTGF revealed that HG-PDF induced injury of HPMCs and PM fibrosis in the PD animal model, which was significantly inhibited by the administration of CCG-1423. Analysis of the miR-199a-5p/214–3p gene cluster by in situ hybridization and SRF, E-cadherin, claudin-2, α-SMA, and FN by immunohistochemistry indicated that HG-PDF upregulated SRF, the miR-199a-5p/214–3p cluster, α-SMA, and FN, and downregulated E-cadherin and claudin-2 in the PM of our PD animal model. In addition, CCG-1423 significantly attenuated the HG-PDF–induced expression of SRF, miR-199a-5p/214–3p cluster, α-SMA, and FN, but upregulated the expression of E-cadherin and claudin-2, and ultimately alleviated the thickness of the PM.
Figure 6.
MiR-199a/214 is decreased expression in PD rats by SRF inhibitor CCG-1423 in vivo. As measured by Masson staining, saline exposure resulted in little change in PM in the control group. Exposure to 4.25% PDF resulted in an increase in the thickness of the PM. CCG-1423 significantly reduced HG-PD. Histogram shows the thickness of Masson staining in the submesothelial compact zone (mean±SEM, n=8). Staining of the fibrosis marker FN in peritoneal samples by immunohistochemistry reveals that PDF exposure induces FN whereas CCG-1423 treatment significantly ameliorated it. Expression of miR-199a-5p and miR-214–3p by in situ hybridization in PD rats, or PD rats treated by SRF inhibitor CCG-1423, compared with the control. U6 was tested as positive control and scramble probe was used as negative control. Expression of α-SMA, E-cadherin, claudin-2, and FN protein by immunohistochemistry in PD rats and treatment of PD rats with the SRF inhibitor CCG-1423 compared with the control. Original magnifications, ×100 (top panels) and ×400 (bottom panels). Histogram shows the thickness for hematoxylin and eosin staining in the submesothelial compact zone in PD rats (116.63±11.69) or PD rats treated with CCG-1423 (60.25±18.36) compared with control (21.29±6.57). *P<0.05 versus control rats; **P<0.05 versus PD rats injected with control. HE, hematoxylin-eosin; PBS, phosphate buffer saline.
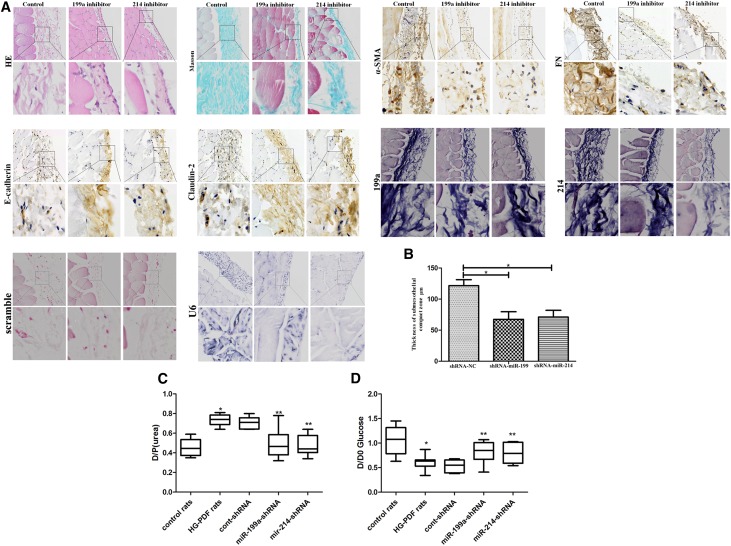
The miR-199a/214 shRNA Lentivirus Prevents Peritoneal Fibrosis in PD Rats In Vivo
We further investigated treatment with an miR-199a-5p or miR-214–3p shRNA lentivirus compared with a control vector in the PD animal model (n=8 rats in each group) (Figure 7, Supplemental Figure 5). Immunohistochemistry analysis of miR-199a-5p, miR-214–3p, E-cadherin, claudin-2, α-SMA, and FN indicated that injection of the miR-199a-5p or miR-214–3p shRNA lentivirus in PD rats caused downregulation of miR-199a-5p, miR-214–3p, α-SMA, and FN, but upregulation of E-cadherin and claudin-2 in the PM (Figure 7A). A thinner PM of PD rats, 67.38±8.44 or 71.0±9.41 mm, was observed in the parietal peritoneum after miR-199a-5p or miR-214–3p shRNA lentivirus treatment compared with the control group, in which the PM was 121.5±5.57 mm (Figure 7B).
Figure 7.
MiR-199a-5p or miR-214-3p shRNA lentiviruses blocked phenotypic transition and fibrosis in PD rats in vivo. (A) H&E and Masson staining, in situ hybridization showing the expression of miR-199a-5p and miR-214–3p, and immunohistochemistry showing the expression of α-SMA, E-cadherin, claudin-2, and FN proteins in HG-PDF rats treated with miR-199a-5p or miR-214–3p shRNA lentivirus compared with control lentivirus in vivo. U6 was tested as positive control and scramble probe was used as negative control. Original magnifications, ×100 (top panels) and ×400 (bottom panels). (B) The histogram shows the thickness of H&E staining in the submesothelial compact zone in miR-199a-5p shRNA (67.38±8.44) or miR-214–3p shRNA PD rats (71.00±9.41) compared with control lentivirus HG-PDF rats (121.50±5.57). Data are given as means±SEM (n=8). *P<0.05. (C and D) Peritoneal equilibration test of the quotient of dialysate and plasma concentration of urea nitrogen. Dialysate-to-plasma ratio of urea (D/P urea), and the initial dialysate–to–end dialysate ratio of glucose (D/D0 glucose), were used to evaluate the transport status of a PM in control rats, HG-PDF rats, and HG-PDF rats injected with cont, miR-199a-5p-shRNA, or miR-214–3p shRNA lentivirus. *P<0.05 versus control rats; **P<0.05 versus HG-PDF rats injected with control lentivirus. The detail data of D/P (urea) were: control rats 0.458±0.081, HG-PDF rats 0.735±0.054, cont-shRNA 0.708±0.057, miR-199a-shRNA 0.494±0.136, miR-214-shRNA 0.468±0.099; D/D0 glucose: control rats 1.055±0.270, HG-PDF rats 0.609±0.140, cont-shRNA 0.53±0.117, miR-199a-shRNA 0.825±0.207, miR-214-shRNA 0.795±0.195. cont, control; HE, hematoxylin-eosin.
We further analyzed the correlation between miR-199a and miR-214 expression ex vivo in HPMCs using a peritoneal equilibration test. At 240 minutes, the dialysate-to-plasma ratio of urea (D/P urea) and the initial dialysate–to–end dialysate ratio of glucose (D/D0 glucose) were measured (Figure 7, C and D). This showed a significant increase in the 240-minutes D/P urea in HG-PDF rats compared with control rats; however, there was a decrease in miR-199a-shRNA or miR-214-shRNA compared with cont-shRNA rats. Our data also showed a decrease in D/D0 glucose in HG-PDF rats compared with control rats, and an increase in D/D0 glucose in miR-199a-shRNA or miR-214-shRNA compared with cont-shRNA rats.
CDH1/CLDN2 Are the Targets of the miR-199a/214 Gene Cluster in HG-Stimulated HPMCs
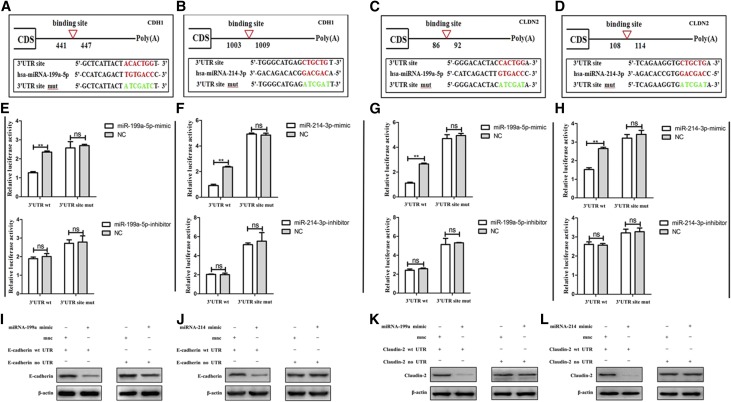
We next analyzed the expression of potential downstream target genes of miR-199a-5p and miR-214 using three prediction algorithms. We focused on phenotypic transition-related genes and found that CDH1 and CLDN2, which encode the E-cadherin and claudin-2 proteins, are involved in cell-cell or cell-matrix adhesion and may be the target genes of miR-199a-5p or miR-214, respectively (Figure 8). To determine whether CDH1 or CLDN2 is regulated by miR-199a-5p or miR-214 via direct binding to the CDH1 or CLDN2 3′ UTR, we predicted the binding site of CDH1 or CLDN2 to the miR-199a/214 cluster separately and constructed wild-type and mutant fragments of the CDH1 or CLDN2 mRNA 3′ UTR and cloned them into the region immediately downstream of the luciferase reporter gene (Figure 8, A–D). Next, miR-199a-5p or miR-214 mimic oligonucleotides were cotransfected with the different luciferase 3′ UTR constructs into HG-stimulated HPMCs (Figure 8, E–H). We found that both miR-199a-5p and miR-214 decreased the luciferase activity of the wild-type CDH1 or CLDN2 3′ UTRs, respectively. However, the luciferase activity of the mutant CDH1 or CLDN2 3′ UTRs remained constant. Therefore, we showed that the inactivation of E-cadherin or claudin-2 by miR-199a-5p or miR-214 depended on the CDH1 or CLDN2 3′ UTR.
Figure 8.
MiR-199a-5p or miR-214–3p directly targets CDH1 and CLDN2 in HPMCs. (A–D) Predicted duplex formation between the human CDH1 and CLDN2 3′ UTRs and the miR-199a-5p binding site within the wild-type (wt) CDH1 and CLDN2 3′ UTRs and mutant (mut) CDH1 and CLDN2 3′ UTRs. (E–H) Luciferase activity of wt and mut CDH1(E and F) and CLDN2 3′ UTR reporter genes in HPMCs transfected with the miR-199a-5p and miR-214–3p mimics (top) and inhibitors (bottom) compared with empty mimic or inhibitor oligonucleotides. (I–L) E-cadherin and claudin-2 expression in HPMCs transfected with E-cadherin and claudin-2 plasmids containing the wild-type 3′UTR or lacking the 3′UTR, along with miR-199a-5p and miR-214–3p, compared with the control oligonucleotide (mnc) was detected by western blotting. *P<0.05; **P<0.01. CDS, coding sequence; NC, nonspecific control; ns, not significant; UTR, untranslated regions; −, negative; +, positive.
We further transfected HG-stimulated HPMCs with E-cadherin or claudin-2 plasmids containing the wild-type 3′UTR or lacking the 3′ UTR (Figure 8, I–L). When miR-199a or miR-214 was cotransfected into HG-stimulated HPMCs, E-cadherin or claudin-2 expression was markedly reduced in cells transfected with the wild-type 3′-UTR, but not in those transfected with the construct lacking the 3′UTR.
Discussion
Increasing evidence indicates that cell migration, loss of adhesion, and the phenotypic transition, which was originally thought to be limited to tumor development and metastasis, also occur in other diseases, including kidney, liver, lung, and peritoneal fibrosis.11–14 Recently, HPMC phenotypic transition in the peritoneum and effluents of patients undergoing PD was determined to be associated with the recurrent use of HG and was linked to a decline in peritoneal function due to peritoneal fibrosis.1,15–17 The HG-induced phenotypic transition of HPMCs is characterized by a loss of epithelial markers, such as E-cadherin and claudins, and the acquisition of mesenchymal transition markers, such as α-SMA, vimentin, Snail, FN, and CTGF.18–20 So HG in the PD effluent could induce cell morphology change, promote cell migration and phenotypic transition, and lead to a loss of adhesion of HPMCs.20–22
According to previous work, CArG-containing SRF target genes are believed to contribute to vascular and visceral smooth muscle cell diseases and tumor metastasis.3,23–25 Surprisingly, some studies found that one-third of 500 or more miRNA genes in mammalian genomes contain at least one CArG box in their promoter regions, which means that hundreds and thousands of CArG-like boxes of miRNAs are potential SRF targets.3,10,26 This suggests that the regulation of miRNAs by SRF represents a common theme in which SRF functions as a sensor of cellular signals to control cellular processes.10,27,28 For example, SRF directly regulates the expression of miR-1–1/133a-2 and miR-1–2/133a-1 clusters in developing muscle cells, which contain three CArG elements.23 Some miRNAs were also mentioned in peritoneal fibrosis. For example, miR-30a, which also contains three CArG elements, negatively regulates the TGF-β1–induced epithelial-mesenchymal transition and peritoneal fibrosis by targeting Snail1.16
According to the Sanger miRBase sequences, pri-miR-199a is transcribed as the following two mature sequences: miR-199a-3p and miR-199a-5p. Pri-miR-214 is also transcribed as the following two mature sequences: miR-214–3p and miR-214–5p.29,30 The two miRNAs sometimes are referred to as members of the miR199a/214 cluster.6,7,30–32 Here, we show that the activation of SRF promotes the transcription of the CArG box–containing miR-199a/214 gene cluster in HPMC in vitro and ex vivo. Only miR-199a-5p and miR-214–3p, but not miR-199a-3p and miR-214–3p, led to increased phenotypic transition, fibrosis, and migration as well as decreased cell adhesion by downregulation of E-cadherin and claudin-2 (Figure 9). Therefore, tissue-specific expression of the SRF-regulated miR-199a/214 gene cluster may yield a detailed regulatory pathway in peritoneal fibrosis that could form the rationale for developing drugs that counteract the progressive deterioration of the PM that occurs in patients receiving CAPD.
Figure 9.
The schematic summary of the SRF-miR-199a/214-CDH1/CLDN2 axis in HG-HPMCs during peritoneal fibrosis. Upon exposure to HG from PDF, HPMCs undergo profound phenotypic changes, including the gain of mesenchymal characteristics and transition to a fibroblast-like morphology. Active SRF is translocated from the cytoplasm into the nuclei of HPMCs, where it binds to the CArG element and promotes the expression of the miR-199a/214 gene cluster. Increased miR-199a-5p and miR-214–3p, but not miR-199a-3p and miR-214–5p, can bind to CDH1 and CLDN2 mRNA, resulting in the loss of epithelial adherens junction protein, E-cadherin, and the tight junction protein, claudin-2. Thereafter, HPMCs with increased mobility may migrate into the submesothelial zone through a disrupted PM and contribute to the increased deposition of extracellular matrix, ultimately leading to PM fibrosis.
Concise Methods
Ethics Statement
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of Xijing Hospital (Xi’an, Shaanxi, China). Written informed consent was obtained from each patient. The Ethics Committee for Animal Experiments of the Fourth Military Medical University (Xi’an, Shaanxi, China) approved all animal work (permit number 20120023), and the experimental protocols strictly complied with the institutional guidelines and the criteria outlined in the Guide for the Care and Use of Laboratory Animals.
HPMC Isolation and Cell Culture
The immortal human HPMC line (ATCC; Rockville, MD), which is an untransformed HPMC line that yields data similar to the results obtained using primary cells, was cultured in Earle M199 medium.2,22 Phenotypic transition was induced in HPMCs with HG (0.5 ng/ml) (R&D Systems, Minneapolis, MN), as previously described.2,22
Primary human mesothelial cells obtained from the effluents of patients undergoing CAPD in our PD center in the Department of Nephrology, Xijing Hospital, were collected and cultured using a previously described method.2,22 This study was conducted in accordance with international guidelines for the use of human tissues. The inclusion criteria were as follows: age<65 years, duration of CAPD longer than 1 month, no peritonitis in the last 6 months, the use of 1.5% glucose dialysis solution, and no history of abdominal surgery or long-term use of an immunosuppressant (Table 1). Patients receiving CAPD were divided into five groups (G for group) according to the duration of dialysis. G0 was made up of omentum-derived primary HPMCs from five patients with ESRD who underwent catheter operation for further PD therapy, which served as a control; G1 included patients who had received 1–6 months of dialysis; G2 included patients who had received 7–24 months of dialysis; G3 included patients who had received 25–48 months of dialysis; and G4 included patients who had received >48 months of dialysis (Supplemental Table 2).
Table 1.
The morphology of primary HPMCs from patients receiving CAPD
| Morphology of Primary HPMCsa | CAPD Times (mo) | |||
|---|---|---|---|---|
| 1–6 (n=8) | 6–24 (n=7) | 24–48 (n=7) | >48 (n=6) | |
| Cobblestone | 7 | |||
| Mixed | 1 | 5 | 3 | |
| Fibroblast-like | 2 | 4 | 6 | |
P<0.001.
Lentivirus Infection and Oligonucleotide Transfection
MiR-199a-5p and miR-214–3p were purchased from GeneChem (Shanghai, China). Constructs containing the miR-199a-5p and miR-214–3p mimic and inhibitor sequences, including 100-base upstream and downstream flanking sequences, were cloned into the pGCSIL-GFP vector. Target cells (1 × 105) were infected with 1 × 107 lentivirus transducing units in the presence of 10 mg/ml polybrene. An empty lentiviral vector was used as a negative control. The miR-199a-5p inhibitor (miR20000231–1-5), miR-214–3p inhibitor (miR200004564–1-5), and negative control were designed and synthesized by RiboBio (Guangzhou, China). Target cells were transfected with the miR-199a-5p and miR-214–3p inhibitor and the negative control using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Cells were collected 48 hours after transfection.
Human embryonic kidney 293 T cells were transfected with the two expression vectors, PLVX-IRES-mCherry and PLVX-shRNA2, and the two packaging plasmids, PMD and psPAX2, at 4, 1, and 3 mg of DNA per 150-mm plate. After 6 hours, fresh DMEM was added to the cells. Forty-eight hours later, supernatants from four plates were pooled, filtered, and stored at 4°C for short-term storage. Lentivirus titers were determined by transfecting human embryonic kidney 293 T cells with a dilution series of the viral suspension, and lentivirus samples with a titer of 4 × 108 transfection U/ml were stored at 80°C2,22.
RNA Isolation, miRNA Quantification, and In Situ Hybridization
For the quantitative analysis of miRNAs, two-step TaqMan real-time PCR analysis was performed using primers and probes (MS00007602: hsa-miR-199a-3p; MS00006741: hsa-miR-199a-5p; MIMAT0000271: miR-214–5p; and MIMAT0004564: miR-214–3p) obtained from Qiagen (Germany)33–35 (Supplemental Table 1). For in situ hybridization, scramble control probe (50-GTGTAACACGTCTATACGCCCA-30) was purchased from Exiqon (Vedbaek, Denmark). MiR-199a-5p and miR-214–3p were performed in paraformaldehyde-fixed, paraffin-embedded peritoneal sections as described previously.5,10
Peritoneal Exposure Model in Rats and Immunofluorescence Analysis of the Peritoneum
A chronic infusion model of nonuremic animal PD and the SRF inhibitor CCG1423 were used. The parietal peritoneum of the abdominal wall was fixed and embedded as previously described.2,22
An additional 24 HG-PDF male Sprague-Dawley rats were divided into three groups that were separately treated, with an empty lentivirus vector (control-shRNA), a lentivirus vector containing miR-199a-5p shRNA (miR-199a-shRNA), or a lentivirus vector containing miR-214–3p shRNA (miR-214-shRNA), intraperitoneally via the peritoneal access port at a dose of 4 × 108 transfection units every 7 days, three times during the HDF-PD injection experiment.
Western Blot Analysis
Ten to fifty milligrams of total protein extracts from fresh tissue or cultured cells were loaded onto SDS-PAGE gels for western blotting. Total protein for western blot and IP was extracted using cell lysis buffer (Beyotime, Jiangsu, China). The antibodies used were as follows: anti-SRF (Santa Cruz Biotechnology) diluted 1:80, anti–p-SRF (p-SRF) (Cell Signaling Technology) diluted 1:500, anti–claudin-2 (Santa Cruz Biotechnology) diluted 1:100, anti–E-cadherin (Santa Cruz Biotechnology) diluted 1:50, anti-CTGF (Santa Cruz Biotechnology) diluted 1:80, anti-FN (Santa Cruz Biotechnology) diluted 1:80, and anti–β-actin (Sigma) diluted 1:4000. Antibody staining was performed as previously described.2,22
In Vitro Migration and Adhesion Assays
Cell migration and adhesion assay were assessed as previously described.10 For migration assays, infected or transfected cells were harvested and resuspended in serum-free medium, and 1 × 105 cells were placed into Boyden chambers (Corning, Tewksbury, MA) with an 8.0-μm pore membrane. The chambers were then inserted into the wells of a 24-well plate and incubated for 24 hours in medium with 10% FBS before examination. The cells remaining on the upper surface of the membrane were removed, and the cells adhering to the lower surface were fixed, stained in a dye solution containing 0.05% crystal violet, and counted under a microscope (Olympus Corp., Tokyo, Japan) to determine their relative numbers. For cell adhesion, the ability of HPMC cells to adhere to Matrigel (50 μg/ml; BD Biosciences) was assessed in 24-well plates as previously described. The plate surface was coated with 0.2 ml of Matrigel and incubated for 2 hours, and then the supernatant was removed. A 0.5-ml suspension of cells (1 × 105/ml) was transferred into the covered wells. After 0.5, 1, 2, and 4 hours of incubation at 37°C, the adhesive cells were stained with crystal violet and evaluated at 200× magnification in ten random fields of each well according to the manufacturer’s instructions (Life Technologies).
ChIP and Quantitative PCR
ChIP was carried out as previously described using an anti-SRF antibody (Santa Cruz Biotechnology).10 All signals were normalized to the input chromatin signals. Total cellular RNA was extracted from whole cells using the RNeasy Kit (Qiagen). Real-time PCR was performed using an ABI 7900 Sequence Detection System and the SYBR Green PCR Core Reagent Kit (Life Technologies).
Reporter Vector Construction and Luciferase Assay
To create a dual-luciferase reporter plasmid, the promoter fragment of miR199a/214 containing putative binding sites for SRF was PCR-amplified from the genomic DNA of normal HPMCs. The PCR products were gel-purified (Dongsheng Biotech, China), digested (New England Bio), and ligased (Takara, Japan) into the digested pRL-SV40 vector (Promega) between the XhoI and HindIIII sites, defined as pRL-miR199. Luciferase assays were performed as previously described.2,10,22 Briefly, HPMCs and HG-induced HPMCs were transfected using OPTI-MEM reagent (Invitrogen) according to the manufacturer’s instructions (Roche). The transfections were performed in duplicate and repeated three times. Forty-eight hours after transfection, the cell extraction was assayed for luciferase activity assay using a Luciferase Assay Kit (Promega). The ratio of Renilla to firefly luciferase was measured with the Dual-Luciferase Reporter Assay System (Promega). The primer sequences used to clone the miR-199a/214 segments are provided (Figure 1).
To create a dual-luciferase reporter plasmid, the 3′-UTR fragment of CDH1 or CLDN2 containing putative binding sites for miR-199a/214 was PCR-amplified from the genomic DNA of normal HPMCs. The PCR products were gel-purified (Dongsheng Biotech, China), digested (New England Bio), and ligased (Takara, Japan) into the digested psiCheck-2 plasmid (Promega) between the XhoI and NotI sites, defined as psi-CHECKTM-2-CDH1 or psi-CHECKTM-2-CLDN2. CDH1– or CLDN2–3′-UTR–targeted site mutations were generated using the KOD-plus mutagenesis kit (Toyobo, Japan), according to the manufacturer’s protocol. The constructs of mutCDH1 and mutCLDN2 were established using the same protocol of psi-CHECKTM-2-CDH1 or psi-CHECKTM-2-CLDN2. Then, HG-induced HPMCs were cultured in 96-well plates and cotransfected with 1 µl miR-199a-5p or miR-214–3p (20 µM), 0.5 µg of wild-type (wt) or mutant (mut) CDH1 or CLDN2 luciferase reporter vector, and 1 μl Lipofectamine 2000 (Invitrogen). Forty-eight hours after cotransfection, luciferase activity was detected by the Dual-Glo Luciferase Reporter Assay kit (Promega) (Figure 8). The transfections were performed in duplicate and were repeated three times.10
Statistical Analyses
The bands from western blotting and real-time PCR were quantified using Quantity One software (Bio-Rad, Hercules, CA). The numeric data are presented as the mean±SEM and were generated using SPSS 12.0 software (Chicago, IL). Differences between the means were assessed using paired t test, ANOVA, or the chi-squared test. P<0.05 was considered statistically significant.
Disclosures
None.
Supplementary Material
Acknowledgments
L.H. conceived and designed the experiments. M.C., T.S., S.F., H.L., and X.Z. performed the experiments. L.H. and H.W. analyzed the data. N.F., W.L., J.D., G.T., C.H., G.X., Q.Q., and S.S. contributed reagents/materials/analysis tools. L.H. wrote the paper.
This study was supported in part by grants from the National Scientific Foundation of China (81270849, 81500581, XJZT14M03, and 81270768) and Beijing Municipal Administration of Hospitals’ Youth Programme (QML 20160701).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016060663/-/DCSupplemental.
References
- 1.Krediet RT, Struijk DG: Peritoneal changes in patients on long-term peritoneal dialysis. Nat Rev Nephrol 9: 419–429, 2013 [DOI] [PubMed] [Google Scholar]
- 2.He L, Lou W, Ji L, Liang W, Zhou M, Xu G, Zhao L, Huang C, Li R, Wang H, Chen X, Sun S: Serum response factor accelerates the high glucose-induced epithelial-to-mesenchymal transition (EMT) via snail signaling in human peritoneal mesothelial cells. PLoS One 9: e108593, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niu Z, Li A, Zhang SX, Schwartz RJ: Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol 19: 618–627, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morishita Y, Yoshizawa H, Watanabe M, Imai R, Imai T, Hirahara I, Akimoto T, Ookawara S, Muto S, Nagata D: MicroRNA expression profiling in peritoneal fibrosis. Transl Res 169: 47–66, 2015 [DOI] [PubMed] [Google Scholar]
- 5.Williams KC, Renthal NE, Gerard RD, Mendelson CR: The microRNA (miR)-199a/214 cluster mediates opposing effects of progesterone and estrogen on uterine contractility during pregnancy and labor. Mol Endocrinol 26: 1857–1867, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen BF, Suen YK, Gu S, Li L, Chan WY: A miR-199a/miR-214 self-regulatory network via PSMD10, TP53 and DNMT1 in testicular germ cell tumor. Sci Rep 4: 6413, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin G, Chen R, Alvero AB, Fu HH, Holmberg J, Glackin C, Rutherford T, Mor G: TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene 29: 3545–3553, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T: Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn 237: 3738–3748, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB: Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res 37: 123–128, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, He L, Li T, Lu Y, Miao Y, Liang S, Guo H, Bai M, Xie H, Luo G, Zhou L, Shen G, Guo C, Bai F, Sun S, Wu K, Nie Y, Fan D: SRF expedites metastasis and modulates the epithelial to mesenchymal transition by regulating miR-199a-5p expression in human gastric cancer. Cell Death Differ 21: 1900–1913, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiery JP, Sleeman JP: Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7: 131–142, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Kalluri R, Neilson EG: Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 112: 1776–1784, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG: Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest 110: 341–350, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Dong Z, Liu H, Zhu J, Liu F, Chen G: Transition of mesothelial cell to fibroblast in peritoneal dialysis: EMT, stem cell or bystander? Perit Dial Int 35: 14–25, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strippoli R, Loureiro J, Moreno V, Benedicto I, Lozano ML, Barreiro O, Pellinen T, Minguet S, Foronda M, Osteso MT, Calvo E, Vázquez J, Cabrera ML, Del Pozo MA: Caveolin-1 deficiency induces a MEK-ERK1/2-Snail-1-dependent epithelial-mesenchymal transition and fibrosis during peritoneal dialysis. EMBO Mol Med 7: 102–123, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Yang M, Lan H, Yu X: miR-30a negatively regulates TGF-β1-induced epithelial-mesenchymal transition and peritoneal fibrosis by targeting Snai1. Am J Pathol 183: 808–819, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Lei P, Jiang Z, Zhu H, Li X, Su N, Yu X: Poly(ADP-ribose) polymerase-1 in high glucose-induced epithelial-mesenchymal transition during peritoneal fibrosis. Int J Mol Med 29: 472–478, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Yu MA, Shin KS, Kim JH, Kim YI, Chung SS, Park SH, Kim YL, Kang DH: HGF and BMP-7 ameliorate high glucose-induced epithelial-to-mesenchymal transition of peritoneal mesothelium. J Am Soc Nephrol 20: 567–581, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Peso G, Jiménez-Heffernan JA, Bajo MA, Aroeira LS, Aguilera A, Fernández-Perpén A, Cirugeda A, Castro MJ, de Gracia R, Sánchez-Villanueva R, Sánchez-Tomero JA, López-Cabrera M, Selgas R: Epithelial-to-mesenchymal transition of mesothelial cells is an early event during peritoneal dialysis and is associated with high peritoneal transport. Kidney Int Suppl 4: S26–S33, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Aroeira LS, Aguilera A, Sánchez-Tomero JA, Bajo MA, del Peso G, Jiménez-Heffernan JA, Selgas R, López-Cabrera M: Epithelial to mesenchymal transition and peritoneal membrane failure in peritoneal dialysis patients: Pathologic significance and potential therapeutic interventions. J Am Soc Nephrol 18: 2004–2013, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Lee YC, Hung SY, Liou HH, Lin TM, Tsai CH, Lin SH, Tsai YS, Chang MY, Wang HH, Ho LC, Chen YT, Wu CF, Chen HC, Chen HP, Liu KW, Chen CI, She KM, Wang HK, Lin CW, Chiou YY: Vitamin D can ameliorate chlorhexidine gluconate-induced peritoneal fibrosis and functional deterioration through the inhibition of epithelial-to-mesenchymal transition of mesothelial cells. Biomed Res Int 2015: 595030, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Che M, Hu J, Li S, Jia Z, Lou W, Li C, Yang J, Sun S, Wang H, Chen X: Twist contributes to proliferation and epithelial-to-mesenchymal transition-induced fibrosis by regulating YB-1 in human peritoneal mesothelial cells. Am J Pathol 185: 2181–2193, 2015 [DOI] [PubMed] [Google Scholar]
- 23.Park C, Hennig GW, Sanders KM, Cho JH, Hatton WJ, Redelman D, Park JK, Ward SM, Miano JM, Yan W, Ro S: Serum response factor-dependent microRNAs regulate gastrointestinal smooth muscle cell phenotypes. Gastroenterology 141: 164–175, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie C, Huang H, Sun X, Guo Y, Hamblin M, Ritchie RP, Garcia-Barrio MT, Zhang J, Chen YE: MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev 20: 205–210, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T, Yang SJ, Hwang D, Song J, Kim M, Kyum Kim S, Kang K, Ahn J, Lee D, Kim MY, Kim S, Seung Koo J, Seok Koh S, Kim SY, Lim DS: A basal-like breast cancer-specific role for SRF-IL6 in YAP-induced cancer stemness. Nat Commun 6: 10186, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L, Hu N, Du X, Wang W, Chen H, Li W, Wei S, Zhuang H, Li X, Li C: Upregulation of miR-483-3p contributes to endothelial progenitor cells dysfunction in deep vein thrombosis patients via SRF. J Transl Med 14: 23, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN: MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev 23: 2166–2178, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA: MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer 9: 293–302, 2009 [DOI] [PubMed] [Google Scholar]
- 29.Hu Y, Liu J, Jiang B, Chen J, Fu Z, Bai F, Jiang J, Tang Z: MiR-199a-5p loss up-regulated DDR1 aggravated colorectal cancer by activating epithelial-to-mesenchymal transition related signaling. Dig Dis Sci 59: 2163–2172, 2014 [DOI] [PubMed] [Google Scholar]
- 30.Mussnich P, Rosa R, Bianco R, Fusco A, D’Angelo D: MiR-199a-5p and miR-375 affect colon cancer cell sensitivity to cetuximab by targeting PHLPP1. Expert Opin Ther Targets 19: 1017–1026, 2015 [DOI] [PubMed] [Google Scholar]
- 31.Baumgarten A, Bang C, Tschirner A, Engelmann A, Adams V, von Haehling S, Doehner W, Pregla R, Anker MS, Blecharz K, Meyer R, Hetzer R, Anker SD, Thum T, Springer J: TWIST1 regulates the activity of ubiquitin proteasome system via the miR-199/214 cluster in human end-stage dilated cardiomyopathy. Int J Cardiol 168: 1447–1452, 2013 [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Martin JF: Macro advances in microRNAs and myocardial regeneration. Curr Opin Cardiol 29: 207–213, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoy AM, Lundie RJ, Ivens A, Quintana JF, Nausch N, Forster T, Jones F, Kabatereine NB, Dunne DW, Mutapi F, Macdonald AS, Buck AH: Parasite-derived microRNAs in host serum as novel biomarkers of helminth infection. PLoS Negl Trop Dis 8: e2701, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang S, He L, Zhao X, Miao Y, Gu Y, Guo C, Xue Z, Dou W, Hu F, Wu K, Nie Y, Fan D: MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS One 6: e18409, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Dou W, He L, Liang S, Tie J, Liu C, Li T, Lu Y, Mo P, Shi Y, Wu K, Nie Y, Fan D: MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene 32: 1363–1372, 2013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.