Abstract
Disorders of water balance, an excess or deficit of total body water relative to body electrolyte content, are common and ascertained by plasma hypo- or hypernatremia, respectively. We performed a two-stage genome-wide association study meta-analysis on plasma sodium concentration in 45,889 individuals of European descent (stage 1 discovery) and 17,637 additional individuals of European descent (stage 2 replication), and a transethnic meta-analysis of replicated single-nucleotide polymorphisms in 79,506 individuals (63,526 individuals of European descent, 8765 individuals of Asian Indian descent, and 7215 individuals of African descent). In stage 1, we identified eight loci associated with plasma sodium concentration at P<5.0 × 10−6. Of these, rs9980 at NFAT5 replicated in stage 2 meta-analysis (P=3.1 × 10−5), with combined stages 1 and 2 genome-wide significance of P=5.6 × 10−10. Transethnic meta-analysis further supported the association at rs9980 (P=5.9 × 10−12). Additionally, rs16846053 at SLC4A10 showed nominally, but not genome-wide, significant association in combined stages 1 and 2 meta-analysis (P=6.7 × 10−8). NFAT5 encodes a ubiquitously expressed transcription factor that coordinates the intracellular response to hypertonic stress but was not previously implicated in the regulation of systemic water balance. SLC4A10 encodes a sodium bicarbonate transporter with a brain-restricted expression pattern, and variant rs16846053 affects a putative intronic NFAT5 DNA binding motif. The lead variants for NFAT5 and SLC4A10 are cis expression quantitative trait loci in tissues of the central nervous system and relevant to transcriptional regulation. Thus, genetic variation in NFAT5 and SLC4A10 expression and function in the central nervous system may affect the regulation of systemic water balance.
Keywords: hyponatremia, water-electrolyte balance, human genetics, hypernatremia
Abnormal water balance is an excess or deficit of total body water relative to electrolyte content, and it is determined by measuring the plasma sodium concentration.1 Hyponatremia (relative water excess) is the most common electrolyte abnormality among hospitalized patients2; it is highly prevalent among the acutely ill,3 patients undergoing surgery,4 and the elderly.5,6 Severe acute hyponatremia causes brain edema, seizures, and death.7 Reversible defects in cognition, coordination, and mood occur with even subtle chronic hyponatremia.8,9
Water balance is regulated by thirst and aquaporin-2–dependent water reclamation in the kidney collecting duct. Both phenomena are influenced by arginine vasopressin, the secretion of which is governed by brain regions that continually monitor the osmolality of the extracellular fluid compartment. Activation of arginine vasopressin receptor-2 (encoded by AVPR2) by circulating arginine vasopressin mediates the insertion of preformed aquaporin-2 into the luminal membrane of principal cells of the kidney collecting duct and is permissive for water reabsorption. The degree of water reabsorption, in turn, affects the concentration of plasma sodium, the principal extracellular cation and determinant of plasma osmolality or tonicity. Changes in systemic osmolality or tonicity are almost immediately transmitted to the intracellular milieu via water movement. Within cells, osmoregulation and thus, water balance are governed by the tonicity-responsive transcription factor tonicity-enhancer binding protein10, also known as the osmotic response element binding protein11 and NF of activated T cells 5 (NFAT5).12,13
Identification of molecular pathways influencing systemic water balance has implications for both understanding the pathogenesis of primary disorders of water imbalance (such as the syndrome of inappropriate antidiuresis7) and the development of novel therapies. Exceedingly rare point mutations in AQP2 (encoding aquaporin-2) and AVPR2 cause Mendelian disturbances in water balance14–19; however, little is known about the genes influencing the interindividual variability of plasma sodium concentration (i.e., water balance) in the general population. We previously showed heritability of plasma sodium concentration at the population level.20 Genome-wide association study (GWAS)–based approaches have proven to be instrumental in identifying genetic loci associating with a wide variety of diseases and physiologic traits21 and thus, novel biologic mechanisms22,23; however, the only previous GWAS of plasma sodium had limited power and did not detect any significant associations.24 Other efforts to determine the genetic architecture of water balance have been limited to candidate gene–based studies.25 We, therefore, performed a GWAS and replication on the phenotype of plasma sodium concentration in 45,889 and 17,637 individuals of European descent, respectively, with follow-up in 8765 and 7215 individuals of Asian Indian and African descent, respectively, and transethnic meta-analysis of replicated single-nucleotide polymorphisms (SNPs) in all 79,506 individuals.
Results
Study Participants
After excluding individuals with high glucose or impaired kidney function (Concise Methods, Supplemental Table 1), 63,526 individuals of European descent from 25 cohorts were included in this analysis: 45,889 individuals from 21 cohorts in stage 1 meta-analysis and 17,637 individuals from four cohorts in stage 2 meta-analysis (Table 1). Furthermore, a total of 7215 individuals from five cohorts of African descent and 8765 individuals from four Asian Indian cohorts were included in the transethnic analyses (Supplemental Table 2). Details of each cohort’s study design, genotyping methods, and quality control criteria are shown in Concise Methods and Supplemental Tables 3 and 4. Plasma sodium and glucose concentrations were within expected ranges in all cohorts (Table 1).
Table 1.
Study participant characteristics
| Study | Sample Size, n | Age, yr | Sex, % Women | Plasma Sodium, mEq/L | eGFRcrea, ml/min per 1.73 m2 | Plasma Glucose, mg/dl |
|---|---|---|---|---|---|---|
| Stage 1 discovery | ||||||
| Amish studies | 1131 | 48.1 (15.0) | 49.8 | 139.2 (2.2) | 94.1 (16.3) | 91.1 (14.0) |
| BLSA | 594 | 69.6 (15.1) | 42.9 | 141.3 (3.0) | 72.5 (17.5) | 90.1 (13.2) |
| ARIC: Europeans | 8535 | 54.1 (5.7) | 53.2 | 141.0 (2.3) | 90.0 (17.3) | 99.8 (11.0) |
| FHS | 2494 | 48.1 (15.0) | 49.8 | 139.2 (2.2) | 94.1 (16.3) | 91.1 (14.0) |
| COLAUS | 2816 | 58.3 (10.4) | 53.5 | 142.6 (1.8) | 81.1(15.4) | 107.2(21.7) |
| MrOS | 3909 | 73.8 (5.9) | 0 | 141.5 (2.6) | 77.1 (16.4) | 100.9 (12.5) |
| MICROS | 1146 | 45.2 (16.0) | 56.5 | 139.0 (1.9) | 91.2 (16.9) | 84.1 (11.7) |
| KORA F3 | 1425 | 62.2 (10.1) | 51.7 | 142.9 (4.4) | 80.2 (14.4) | 102.4 (16.1) |
| KORA F4 | 1671 | 60.7 (8.8) | 52.4 | 139.1 (2.6) | 81.6 (14.5) | 98.1 (12.6) |
| GENOA: Europeans | 1064 | 59.0 (10.2) | 43.8 | 138.2 (2.1) | 64.4(13.6) | 100.4 (14.0) |
| InCHIANTI | 1142 | 67.9 (15.6) | 55.7 | 141.8 (2.5) | 79.1 (17.9) | 90.2 (14.6) |
| LOLIPOP_EW610 | 881 | 55.8 (9.8) | 27.2 | 140.4 (2.1) | 74.4 (12.5) | 94.1 (11.9) |
| LOLIPOP_EWA | 546 | 54.1 (10.4) | 13.3 | 140.5 (2.7) | 82.9 (19.5) | 94.6 (13.6) |
| LOLIPOP_EWP | 574 | 55.4 (9.3) | 0 | 140.5 (2.5) | 81.6 (13.0) | 96.7 (13.9) |
| LURIC | 2579 | 62.2 (10.8) | 29.7 | 141.4 (2.8) | 83.4 (17.1) | 103.5 (15.3) |
| Ogliastra genetic park Talana study | 691 | 50.2 (19.1) | 58.5 | 139.1 (2.4) | 72.6 (13.6) | 90.6 (12.1) |
| Ogliastra genetic park study | 382 | 54.1 (13.5) | 0 | 137.2 (3.0) | 75.2 (13.1) | 99.6 (14.6) |
| SHIP | 3767 | 48.8 (16.1) | 51.7 | 138.8 (2.8) | 80.5 (14.1) | 96.3 (14.5) |
| SHIP-TREND | 979 | 50.0 (13.6) | 56.2 | 139.3 (2.2) | 91.8 (20.1) | 96.8 (11.3) |
| The Rotterdam study | 3415 | 69.2 (8.7) | 63 | 140.2 (3.3) | 78.0 (16.3) | 109.8 (17.9) |
| SARDINIA | 6148 | 46.1 (17.7) | 57.4 | 141.7(2.6) | 100.5 (26.12) | 88.8 (12.5) |
| Stage 2 replication | ||||||
| DIACORE | 1151 | 65.8 (8.6) | 42.3 | 139.5 (2.8) | 78.5 (23.5) | 111.3 (21.0) |
| FINCAVAS | 1761 | 60.2 (12.1) | 37.5 | 139.8 (2.6) | 88.7 (20.7) | 105.9 (15.3) |
| LifeLines Cohort Study | 12,270 | 48.8 (11.4) | 58.2 | 141.7 (1.9) | 90.3 (16.3) | 91.6 (15.7) |
| MESA | 2455 | 62.6 (10.3) | 47.4 | 147.2 (3.8) | 74.2 (14.3) | 91.1 (21.4) |
Data are given as mean (SD) or percentage.
Stage 1 Meta-Analysis of European Descent Studies
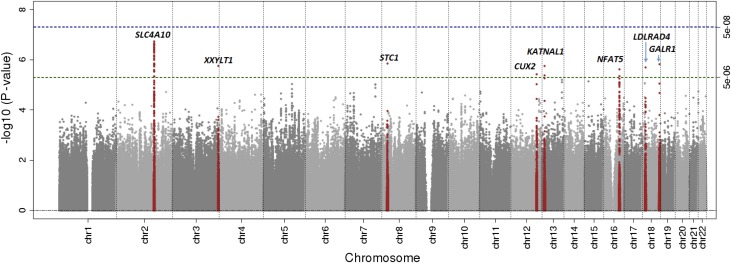
In stage 1 meta-analysis, there was little evidence of population stratification as evidenced by the quantile-quantile plot (Supplemental Figure 1) and the low genomic inflation factor λ=1.02 with study-specific inflation factors ranging from 0.9 to 1.04. Eight SNPs met the prespecified criteria for follow-up in stage 2 meta-analysis (P<5 × 10−6), with minimum P=1.9 × 10−7. These SNPs, detailed in Table 2, showed no relevant heterogeneity (I2≤30%) and good imputation quality, except for rs6565990 at the GALR1 locus (Supplemental Table 5). Figure 1 shows the −log10 P value (Manhattan) plot, and Supplemental Tables 6 and 7 list all SNPs associated with plasma sodium concentration with P<1 × 10−5.
Table 2.
Genetic association analysis results in cohorts of European descent
| SNP Identification | Chromosome | Position (Build 36) | Locus | Effect/Other Allele | Stage 1 Discovery (n=45,889) | Stage 2 Replication (n=17,637) | Stages 1 and 2 Combined (n=63,526) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect Allele Frequency | β | P Value | Effect Allele Frequency | β | One-Sided P Value | Effect Allele Frequency | β | P Value | N | |||||
| rs16846053 | 2 | 162274291 | SLC4A10,a DPP4 | G/T | 0.10 | 0.06 | 1.86 × 10−7 | 0.10 | 0.03 | 0.04 | 0.10 | 0.05 | 6.74 × 10−8 | 63,524 |
| rs753628 | 3 | 196040762 | XXYLT1, LSG1, TMEM44 | A/G | 0.37 | 0.04 | 1.75 × 10−6 | 0.38 | 0.01 | 0.24 | 0.37 | 0.04 | 2.87 × x10−6 | 60,091 |
| rs12677356 | 8 | 23696389 | STC1, SLC25A37 | T/G | 0.05 | 0.09 | 1.41 × 10−6 | 0.05 | −0.01 | 0.58 | 0.05 | 0.08 | 4.64 × x10−6 | 51,254 |
| rs10774613 | 12 | 110030548 | CUX2,a CCDC63, ATXN2 | T/C | 0.57 | 0.03 | 3.77 × 10−6 | 0.52 | 0.02 | 0.13 | 0.56 | 0.03 | 2.40 × x10−6 | 60,095 |
| rs17074418 | 13 | 29675855 | KATNAL1,a HMGB1, UBL3, SLC7A1 | T/C | 0.92 | 0.07 | 1.77 × 10−6 | 0.94 | 0.02 | 0.21 | 0.93 | 0.05 | 4.97 × x10−6 | 63,523 |
| rs9980 | 16 | 68294969 | NFAT5,a NQO1, NOB1, CYB5B | G/C | 0.14 | 0.05 | 2.40 × x10−6 | 0.15 | 0.06 | 3.05 × 10−5 | 0.14 | 0.06 | 5.55 × x10−10 | 60,108 |
| rs11662617 | 18 | 13243598 | LDLRAD4,a CEP192, RNMT | A/G | 0.48 | −0.04 | 2.02 × x10−6 | 0.48 | 0.01 | 0.72 | 0.48 | −0.02 | 2.06 × 10−4 | 54,921 |
| rs6565990 | 18 | 73350561 | GALR1, MBP | T/G | 0.49 | 0.05 | 1.51 × x10−6 | 0.51 | 0.05 | 0.06 | 0.50 | 0.05 | 2.96 × x10−7 | 39,236 |
The gene nearest the SNP is listed first. Coded allele equals effect allele. The SNPs rs12677356 on chromosome 8 and rs6565990 on chromosome 18 were not available in the LifeLines Cohort Study, and proxy SNPs with r2>0.6 could not be identified. The SNPs rs11662617 and rs6565990 (both on chromosome 18) were not available in the MESA, and proxy SNPs with r2>0.6 could not be identified. Because only directly genotyped SNP data were available from the MESA, we analyzed three proxy SNPs for stage 2 meta-analysis. For rs16846053, we used the summary statistics of rs12476631 (chromosome 2, position 162,275,182, distance =891 bp, r2=0.91, D′=1 with rs16846053). For rs9980, we analyzed rs39999 (chromosome 16, position 68,211,197, distance =83,772 bp, r2=1, D′=1 with rs9980), and for rs17074418, we analyzed rs1023104 (chromosome 13, position 29,675,301, distance =554 bp, r2=1, D′=1 with rs17074418).
The gene nearest the SNP if the SNP is located in the gene.
Figure 1.
Stage 1 genome-wide association −log10 P value (Manhattan) plot identifies candidate loci. The green dotted line indicates the P value threshold for following SNPs in stage 2 meta-analysis (P<5 × 10−6). The blue dotted line indicates the genome-wide significance threshold (P<5 × 10−8).
Stage 2 Meta-Analysis of European Descent Studies
In stage 2 meta-analysis, the minor G allele (frequency =0.15) of rs9980 in the NFAT5 locus on chromosome 16 showed a direction-consistent significant association with higher plasma sodium concentration of similar magnitude as in stage 1 meta-analysis (β=0.06, one-sided P=3.1 × 10−5; significance threshold by Bonferroni method: P<0.01). In the combined meta-analysis of stages 1 and 2 cohorts, the association of the rs9980 minor G allele (frequency =0.14) with higher plasma sodium concentration values was genome-wide significant (β=0.06 for each copy of the minor G allele, P=5.6 × 10−10) (Supplemental Figure 2A, Table 2).
The minor allele G (frequency =0.10) of rs16846053 in the SLC4A10 locus on chromosome 2 (Supplemental Figure 2B) was nominally associated with plasma sodium, and the direction of effect was consistent with stage 1 results (one-sided P=0.04). The combined stages 1 and 2 P value was lower than stage 1 results but did not reach strict genome-wide significance (P=6.7 × 10−8) (Table 2). Nonetheless, some have advocated for a more lenient genome-wide significance threshold, especially among more recently founded populations.26
The remaining SNPs selected for replication in stage 2 did not replicate (Table 2). However, although the SNP rs6565990 at the GALR1 locus showed only a near-significant P value in stage 2 replication (one-sided P=0.06), the stage 2 replication meta-analysis effect estimate was direction consistent with stage 1 meta-analysis, and the P value of the stages 1 and 2 combined meta-analysis (P=3.0 × 10−7) was lower than that of the stage 1 meta-analysis (P=1.5 × 10−6). This SNP had poor imputation quality (Supplemental Table 5) and was available in only two studies with a sample size of n=2912 in stage 2 replication, thus limiting the power of the analysis.
Transethnic Analyses
In transethnic meta-analysis combining the summary statistics of a total of 79,506 individuals of European, African, and Asian Indian ethnicity, rs9980 at NFAT5 showed direction-consistent genome-wide significant association with plasma sodium concentration across all ethnicities (β=0.06 for each copy of the minor G allele, P=5.9 × 10−12), with similar effect sizes in each studied ethnicity (Table 3). Results of our top SNPs (P<10−5) for the individual African ancestry and Asian Indian cohorts meta-analyses are shown in Supplemental Tables 8 and 9, respectively.
Table 3.
Genetic association results of transethnic meta-analysis of rs9980 at NFAT5
| Ethnic Group | Sample Size | Effect Allele | Other Allele | Effect Allele Frequency | β | P Value | Heterogeneity I2, % |
|---|---|---|---|---|---|---|---|
| European descent | 60,108 | G | C | 0.14 | 0.06 | 5.6 × 10−10 | 0 |
| Asian Indian | 8760 | G | C | 0.12 | 0.05 | 0.02 | 0 |
| African descent | 5185 | G | C | 0.03 | 0.05 | 0.40 | 0 |
| Transethnic | 74,053 | G | C | 0.14 | 0.06 | 5.9 × 10−12 | 27 |
Bioinformatic Characterization of Associated Loci
rs9980/NFAT5 Locus
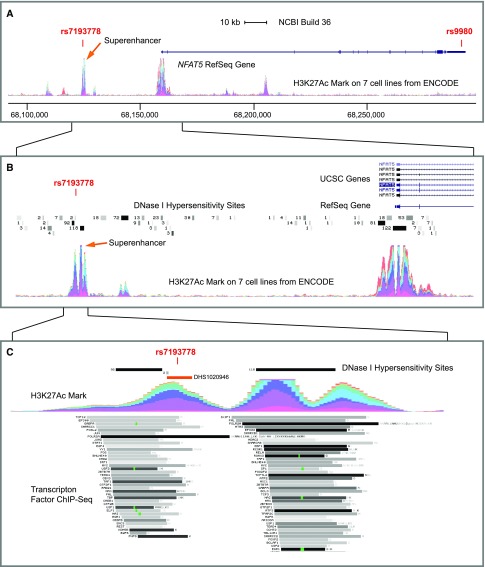
NFAT5 (Supplemental Figure 2A) encodes the ubiquitously expressed tonicity-responsive transcription factor that serves as master regulator of intracellular osmoregulation. The 3′ untranslated region of NFAT5, containing the lead variant rs9980 (Supplemental Figure 2A), confers tonicity-dependent regulation via miRNA-mediated effects on mRNA stability and protein translation.27–30 In addition, a SNP in strong linkage disequilibrium (LD) with rs9980, rs7193778 (r2=0.90, D′=1.0, within a large LD block), is significantly associated with plasma sodium concentration in stage 1 meta-analysis (minor allele C frequency =13%, β=0.05, P=4.5 × 10−6,) (Supplemental Figure 2A, Supplemental Table 6) and resides in the midst of a heavily ENCODE-annotated putative superenhancer region for NFAT5 (Figure 2, Supplemental Figure 3). A superenhancer is a group of closely spaced or even overlapping enhancers exhibiting high levels of transcription factor binding in chromatin immunoprecipitation–based approaches.31 This 3-kb region approximately 35 kb upstream of the NFAT5 transcriptional start site (TSS) is spanned by three dense peaks of chromatin modification emblematic of a transcriptional enhancer. Specifically, both H3K27Ac (Figure 2) and H3K4me1 (not shown) histone modifications are heavily enriched in the vicinity of this variant in ENCODE-tested cell lines. The putative superenhancer also encompasses two long DNaseI-hypersensitive regions present in 92 and 118 of 125 ENCODE-tested cell lines (Figure 2B). Moreover, ENCODE transcription factor ChIP-Seq experimental data showed binding of 115 of 161 ENCODE-tested transcription factors over the region (ENCODE data displayed in the UCSC Genome Browser) (Figure 2B). Because enhancers regulate spatiotemporal and tissue-specific gene expression, it is noteworthy that both rs9980 and rs7193778 are cis eQTLs for NFAT5 expression in cerebellum and temporal cortex (HaploReg v4.032 citing Zou et al.33).
Figure 2.
Sequence context of rs9980 and rs7193778 in the NFAT5 region coincides with functional genomic annotation. Depicted are ENCODE data displayed in the UCSC Genome Browser. (A) shows NFAT5 (exons connected by a blue line) and H3K27ac histone acetylation in the seven ENCODE cell lines (chromosome 16: 68087500–68307500 of human reference genome build NCBI build 36/hg18 and ENCODE histone modification track). Each color represents one of seven cell lines; peak height is the sum of activity in all cell types and proportional to the levels of enrichment of the H3K27ac histone mark across the genome as determined by ChIP-Seq assays. H3K27ac peaks coincide with the NFAT5 promoter region and the vicinity of rs7193778. SNP rs9980 resides in the 3′ untranslated region. (B) illustrates an expanded view of approximately 40 kb upstream of the NFAT5 TSS and depicts the H3K27ac triple peak of the NFAT5 superenhancer as well as the locations of ENCODE/Epigenome Roadmap experimentally confirmed DNaseI-hypersensitive sites above the H3K27ac peaks. The darkness of each site is proportional to the maximum signal strength observed in any cell line, and the adjacent numbers indicate numbers of tissues and cell lines in which hypersensitivity was detected (of a total of 125 tested tissues/cells). C illustrates the approximately 3-kb H3K27ac triple peak of the NFAT5 superenhancer region, depicting rs7193778 relative to H3K27ac marks, DNaseI-hypersensitive sites, and a partial list of the 115 ChIP-Seq–confirmed transcription factor binding sites (of 161 tested) as gray boxes. The darkness of the boxes is proportional to the maximum signal strength observed in any cell line contributing to the cluster. The small DNaseI-hypersensitive region immediately 5′ of rs7193778 (marked 2) was detected in only astrocytes of the CNS (spinal cord; HA-sp cell line) and pancreatic islets (not shown: expression of SLC4A10 is restricted to CNS and pancreatic islets). A DNaseI-sensitive region (DHS1020946) identified through the Regulatory Elements Database (http://dnase.genome.duke.edu/index.php; in the text) crosses the variant (orange bar) and maps to a cluster of motifs operative to astrocytes.
Ancillary data support functional relevance of rs7193778 to the CNS and glial/astrocytic cells in particular. In silico comparison of putative transcription factor binding sites (JASPAR 201634; http://jaspar.genereg.net) affected by rs7193778 primarily identified motifs for members of the sex-determining region Y-box (SOX) family of transcription factors (SRY and SOX2, -3, -5, -6, and -17). Of these, SOX2, -3, -5, and -6 are enriched in tissue derived from brain and/or glioma (Concise Methods). Furthermore, glial tissue (i.e., astrocytes) have been proposed as the cell type conferring central sensing of extracellular sodium concentration,35,36 in contrast to the neuronal sensing of systemic osmolality.37,38 DNaseI-hypersensitive regions (i.e., open chromatin) reported in ENCODE data further support a glial cell–specific effect for this regulatory region. Although most such regions are detected across a large number of cell types, a short DNaseI-hypersensitive region immediately upstream of rs7193778 (indicated by 2 in Figure 2C) was detected in only two tissues—the HA-sp astrocytic (glial) cell line and pancreatic islets (Figure 2C). The Regulatory Elements Database (http://dnase.genome.duke.edu/index.php) identified a DNase site (DHS1020946) physically crossing the variant (Figure 2C, orange bar) and mapping to a cluster (self-organizing map [SOM] Cluster: 977) of similar regions preferentially operative in astrocytes.39 The SOM designation refers to SOM-based clustering of DNase sites according to their profile of DNaseI hypersensitivity across diverse cell types.39
The biology of other genes in this locus (Supplemental Figure 2A) (NQO1, NOB1, WWP2, and CLEC18A) is summarized in Supplemental Table 7.
rs16846053/SLC4A10 Locus
The lead variant in SLC4A10—a gene encoding a brain-specific member of the sodium-bicarbonate transporter family—is intronic; however, the association signal spans the entire SLC4A10 gene as well as an additional 0.3 Mb 5′ of the gene (Supplemental Figure 2B). Importantly, this SNP, like rs9980 in NFAT5, is a cis eQTL for SLC4A10 expression in cerebellum and temporal cortex (Haploreg v4.032 citing Zou et al.33). Transcriptome data in the public domain (e.g., Unigene, BioGPS, and Epigenome Roadmap) support a heavily brain-enriched expression pattern for SLC4A10. Of 127 cell lines and tissues represented in the WashU Epigenome Browser implementation of the Roadmap Epigenomics Project data (http://egg2.wustl.edu/roadmap/web_portal/), SLC4A10 expression is detectable via RNA-Seq in only hippocampus, fetal brain, cultured neurospheres derived from cortex and ganglion eminence, and pancreatic islets (data not shown). Although intronic, the vicinity of rs16846053 is annotated as an active TSS or TSS flanking region in T cells and a number of other tissues. It is annotated as weakly transcribed in brain hippocampus and only in this tissue. Therefore, a novel variant of SLC4A10 may be expressed in the CNS. Moreover, the DNaseI-hypersensitive regions upstream of the gene disproportionately map to cell lines of CNS origin (e.g., SK-N-MC neuroblastoma cells and HA-h, HA-sp, and HAc astrocytic cells). Intriguingly, the lead variant at this locus affects a canonical NFAT5 DNA binding motif as determined via unbiased position-weight matrix scanning of the genomic context; moreover, the minor allele reduces the fidelity score for this predicted NFAT5 binding site (Supplemental Figure 4). An additional variant at this locus, rs16845945, maps to the SLC4A10 proximal promoter approximately 200 bp upstream of the SLC4A10 TSS. Consistent with this role, promoter-associated H3K4me3 histone modification pattern is observed in brain and pancreatic tissue (data not shown).
The known biology of additional genes in this locus (Supplemental Figure 2B) is summarized in Supplemental Table 7.
Discussion
In this GWAS meta-analysis of systemic water balance, we have identified common variants in NFAT5 associating with plasma sodium concentration in individuals of European descent, which are further supported by tentative validation in transethnic meta-analysis. Genomic functional annotation data implicate a role for genetic variation at NFAT5 and SLC4A10, the latter a locus with nominally significant association, in regulating systemic water balance through expression-level effects in glial tissue of the central nervous system. These data are the first to implicate these genes in the regulation of systemic water balance.
The NFAT5 transcription factor coordinates the response to osmotic stress at the cellular level. It transactivates genes coding for aquaporins that are permissive for water movement, and for proteins that import or synthesize osmotically protective intracellular solutes.40 NFAT5 also increases expression of heat shock proteins,41 molecular chaperones that stabilize protein conformation against the denaturing effect of increased intracellular ionic concentration.42 In addition, NFAT5 participates in the immune cell response to the varying sodium content within the skin and subcutaneous tissues.43 The novel role for genetic variation in NFAT5 in systemic osmoregulation (i.e., water balance) is likely mediated at the level of gene expression.40 The lead variant, rs9980, functions as a cis eQTL for NFAT5, such as has been observed for other disease-associated SNPs.44–47 This variant resides within the 3′ untranslated region, a known site of NFAT5 regulation by osmotic stress.27–30 NFAT5 function is also transcriptionally regulated by changes in tonicity.11,27,48 Importantly, the lead variant is in LD with variant rs7193778, which resides within a superenhancer region upstream of the TSS. This gene region exhibits remarkable enrichment for enhancer-specific histone modification, including H3K4me1 histone methylation consistent with enhancer function,49 H3K27ac histone acetylation emblematic of active (in contrast to poised) enhancers,50 and robust binding of a broad array of transcription factors (Figure 2). Our findings are consistent with the frequently observed effect of functional genetic variants on enhancer regions in disease pathogenesis51–53 and the expression of quantitative traits.54
Bioinformatic data are consistent with a potential glial/astrocytic locus of action for NFAT5 variant rs7193778 and the systemic sensing of plasma sodium concentration. Although neurons of the hypothalamus and lamina terminalis are known to have osmosensing roles,37,38 a role for non-neuronal cells in the CNS has also been proposed. Specifically, a subset of glial cells (astrocytes) senses plasma sodium concentration via the Nax channel.35,36 Interestingly, a small DNaseI-hypersensitive region immediately upstream of rs7193778 in NFAT5 is detected in astrocytes as one of only two of 125 ENCODE-tested cell lines. Moreover, the putative transcription factor binding sites affected by rs7193778 include those for SOX family members with expression highly enriched in glial tissue and glioma. Because enhancer-associated epigenomic marks are over-represented in trait-relevant tissues,55 it is plausible that rs7193778 is an eQTL for NFAT5 in glial cells and that the central sensor of plasma sodium concentration may reside in this tissue.
SLC4A10 encodes a brain-specific member of the sodium-bicarbonate transporter family, making this gene a biologically plausible participant in systemic water balance, despite lack of formal replication in stage 2 meta-analysis. The nominally significant signal at SLC4A10 thus similarly implicates CNS glial tissue in systemic osmosensing. The protein is expressed predominantly in brain, and epigenomic functional annotations disproportionately map to brain and in particular, astrocytic, cell lines. Therefore, similar to the case for NFAT5, a glial cell–specific site of action for SLC4A10 in central osmoregulation is plausible. Furthermore the lead variant at SLC4A10 affects an intronic putative NFAT5 DNA binding motif. Moreover, the variant affects the position weight matrix–defined fidelity score for the motif. Intronic enhancers have long been recognized,56 including examples in genes coding for membrane transport proteins.57–59 Notably, other members of the SLC4 family participate in volume regulation at the cellular level.60
Intriguingly, rs7193778, in LD with the lead variant at the NFAT5 locus, has previously been identified in a meta-GWAS on plasma uric acid concentration.61 Plasma uric acid level is influenced by systemic water balance and is often used to inform the diagnosis of a water-excess state, particularly in the context of the syndrome of inappropriate antidiuretic hormone.62–64
The minor allele of the NFAT5 SNP rs9980 associates with hypertonicity and thus, a reduction in either the central sensing of water loss or renal water conservation, with similar effect sizes observed in all ethnicities studied. Thus, the relative absence of this variant in African ancestry (MAF 0.03 versus 0.14 in European populations) may hint at potential selection pressure in environments where chronic or seasonal water scarcity might occur. Similarly, the minor allele of rs16846053 in SLC4A10 was under-represented in African ancestry (MAF 0.02 versus 0.10 in European populations).
Strengths of our work include the large sample size, the unbiased approach to identifying associated genetic loci by GWAS, and the bioinformatic characterization of the replicated loci. However, some limitations warrant mention. First, modest stage 2 replication and transethnic look-up sample size may have limited our ability to replicate additional loci. Second, the phenotype is based on a single measurement, potentially reducing statistical power. Third, the power for replication may have been limited by the poor imputation quality at the GALR1 locus and the limited sample size at the STC1 and LDLRAD4 loci. Fourth, although the effect direction of the association of rs9980 with sodium was consistent across all analyzed ethnicities, the association was only borderline significant in Asian Indians and was not significant in those of African descent, possibly owing to limited power. Fifth, we did not directly replicate the functionally intriguing variant at the NFAT5 locus (rs7193778), because it did not meet our a priori criteria for replication (i.e., not independent from lead signal) and is in strong LD with the rs9980 variant (D′=1 and r2=0.9). Finally, although we performed in-depth bioinformatic characterization of the identified loci, leading to important insights into potential mechanisms, the causal variant remains unknown, and we have not experimentally assessed the effects of the identified gene variants on gene function.
In summary, in this first well powered GWAS on plasma sodium concentration—the clinically measurable parameter of systemic water balance—we have identified genetic variants in NFAT5 in individuals of European descent with validation by transethnic meta-analysis. Additionally, we identified a nominally significant association with an SLC4A10 gene variant that may exert its effect through an intronic enhancer by altering its binding affinity for NFAT5. Our results and bioinformatic characterization point to a previously unknown role of genetic variation at NFAT5 and SLC4A10 in the regulation of systemic water balance via actions on gene expression within the central nervous system.
Concise Methods
Data Management
An analysis plan, detailing phenotype derivation, exclusion criteria, genome-wide association testing, and data file formatting was distributed to all participating studies. Study-specific results files were uploaded to a central server for subsequent standardized central quality control and meta-analysis.65
Phenotype Definition
Plasma sodium concentration is the principal determinant of plasma osmolality. Plasma glucose concentration also contributes to plasma osmolality, and when elevated, it will obligate water entry into the intravascular space and render the plasma sodium concentration less reflective of true plasma osmolality. Thus, individuals with plasma glucose levels >150 mg/dl at the time of plasma sodium measurement were excluded. For plasma glucose concentration <150 mg/dl, we applied the formula of Katz: transformed sodium = plasma sodium (milliequivalents per liter) + (0.016 × (glucose [in milligrams per deciliter] −100)).66 Transformed sodium was the trait used for the GWAS analysis. Because advanced CKD may impair water excretion, we excluded subjects with very low kidney function. We, thus, calculated eGFR on the basis of serum creatinine (eGFRcrea) using the four-variable Modification of Diet in Renal Disease Study equation67 and excluded those with eGFRcrea below the age-specific mean minus 2 SD. Subjects were also excluded if they were not of the predominant ethnicity in the cohort or had missing phenotypic information.
Statistical Methods
Study Design, Genotypes, and Genotype Imputation
Details of each cohort’s study design are shown in Supplemental Table 3. Details of each cohort’s genotyping methods and quality control criteria are provided in Supplemental Table 4. In stage 1, 20 studies each imputed approximately 2.5 million SNPs on NCBI build 36 with external European haplotype reference samples (HapMap release 22). One study used the HapMap release 21 reference haplotypes on NCBI build 35 (KORA F3). In stage 2, one study (the LifeLines Cohort Study) contributed association statistics on the basis of genotypes imputed with HapMap CEU release 24 haplotypes on NCBI build 36. Two studies (FINCAVAS and DIACORE) in stage 2 imputed genotypes with 1000 Genomes reference haplotypes on GRCh build 37. One study (MESA) contributed association statistics on the basis of genotyped variants annotated on NCBI build 36. We transformed the SNP information of imputed genotypes on NCBI build 35 or GRCh build 37 to NCBI build 36 to match the data with HapMap-imputed genotypes of the other studies.
Studies of individuals of African descent and Asian Indians used cosmopolitan reference haplotypes to reflect the predominant ancestry in the study: two studies of individuals of African descent used the combined CEU and YRI haplotypes from HapMap release 22 on NCBI build 36, one study of individuals of African descent used the haplotypes from the June of 2010 release of the 1000 Genomes project on NCBI build 36 (HUFS), and one study of individuals of African descent used the 1000 Genomes Phase I interim data released in June of 2011 (on GRCh build 37) and transformed SNP information to NCBI build 36. All studies of Asian Indians used the combined HapMap release 22 CEU + CHB + JPT + YRI haplotypes on NCBI build 36. Imputed genotypes were coded as the estimated number of copies of a specified allele (dosage).
GWAS
In each study, standardized residuals were obtained by applying z-score transformation on plasma sodium concentration with the covariates sex, age, the interaction of sex with age, and eGFRcrea. GWAS was then performed assuming additive genetic effects using linear regression, with the standardized residuals as the dependent variable and the SNP genotype dosage as the independent variable, including cohort-specific covariates where applicable (e.g., recruitment site and genetic principal components).
Stage 1 Meta-Analysis
A total of 21 studies of European ancestry contributed to the stage 1 meta-analysis. The summary statistics estimated from each cohort’s GWAS were combined using inverse variance weighted fixed effects meta-analysis implemented in METAL software.68 The genomic inflation factor λ69 was estimated for each study, and genomic control (GC) correction was applied if λ>1 (first GC correction). After the meta-analysis, a second GC correction on the aggregated results was applied if λaggregated>1. Between-study heterogeneity was assessed by the I2 statistic. SNPs were selected for stage 2 meta-analysis if they were available in at least 50% of all studies, they did not show an excess of heterogeneity (I2>50%), and they had a stage 1 discovery meta-analysis P ≤ 5×10−6. The SNP with the lowest P value within a window of ± 1 Mb was selected for stage 2 meta-analysis and defined as the index SNP.
Stage 2 Meta-Analysis
In stage 2, SNPs identified in stage 1 meta-analysis were followed up in four study cohorts of European ancestry using the same analysis protocol as described for stage 1 meta-analysis. If the association statistics of the lead SNP in a susceptibility locus were not available, a proxy SNP with the highest LD and D′ was used for the association analysis using the SNAP lookup tool in the HapMap release 22 dataset (https://www.broadinstitute.org/mpg/snap/ldsearch.php).70 A SNP was considered to have replicated if its effect direction was consistent with stage 1 meta-analysis, it showed a significant one-sided P value after Bonferroni correction for multiple testing (P<[0.05/number of analyzed SNPs in stage 2]), and the P value of the stages 1 and 2 combined meta-analysis was lower than the stage 1 meta-analysis P value.
Genetic Associations in Studies of Individuals of African Descent and Asian Indians
For SNPs replicated in stage 2 meta-analysis, additional validation was sought among individuals of non-European ancestry (i.e., African descent and Asian Indian cohorts) using the same protocols for GWAS and meta-analysis as described for the cohorts of European ancestry. Replication was defined by a significant one-sided P value after Bonferroni correction for multiple testing (P<0.05 divided by the number of replicated SNPs). There was little inflation in the genome-wide association meta-analysis of subjects of African and Asian ancestry (λ=1.02 in both analyses). Supplemental Tables 8 and 9 provide the summary statistics of all SNPs associated with sodium with a P<10−5 within each ethnicity.
Transethnic Meta-Analysis
We performed a transethnic meta-analysis of replicated SNPs combining summary statistics from studies of individuals of European, African, and Asian Indian descent and following the same analysis protocol as described for stage 1 meta-analysis in individuals of European descent.
Visualization of LD Blocks
To illustrate the amount of LD between highly correlated variants, we visualized LD blocks with the R package snp.plotter (https://cran.r-project.org/web/packages/snp.plotter) with the individual genotype data of the population-based KORA F4 Study (n=1814).
Functional Characterization of Replicated Loci
We evaluated SNPs at replicated loci for potential relevance to gene expression by examining for overlap with functionally annotated genomic regions from the ENCODE71 and Epigenomics Roadmap55 projects. The former was queried via the UCSC Genome Browser, whereas the latter was accessed through the WashU Epigenome Browser (http://epigenomegateway.wustl.edu/) and RoadMap Epigenome Browser (http://epigenomegateway.wustl.edu/browser/roadmap/). SNPs in LD with lead SNPs (r2>0.8)—and their corresponding functional annotations in ENCODE and the RoadMap Epigenome Project—were identified through HaploReg v4.032 citing Zou et al.33 and RegulomeDB.72 Genomic functional annotations in these public resources included the presence of DNase I-hypersensitive sites, chromatin modification (e.g., histone methylation and acetylation), transcription factor binding, RNA-Seq expression data, and algorithmically assigned chromatin functional state (e.g., active TSS, strong transcription, and enhancer region55; http://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html) in over 300 human tissues and cell lines (depending on assay platform). Putative transcription factor binding motifs were identified via JASPAR 2016 (http://jaspar.genereg.net/).34 This resource uses a predefined position weight matrix to identify a potential binding motif and assign a score reflecting the fidelity of the sequence to the canonical motif. As an alternative to the more simplistic consensus sequence, the position weight matrix reflects the frequency of occurrence of each of the four nucleotides at each position in the motif.73 In silico analysis of the NFAT5 superenhancer in the vicinity of the rs7193778 sequence context (±50 bp; obtained from dbSNP) in the JASPAR 2016 screen, with the major (t) allele, identified 21 putative transcription factor binding sites crossing the variant, of which eight corresponded to SOX family members (SOX2, -3, -5, -6, -10 [n=2], and -17 and SRY). Apart from SRY (for which there were no expressed sequence tag (EST) data presented in the UNIGENE EST Profile Viewer), all of these SOX family transcription factors were expressed in brain and 2–28 other tissues (of a total of 45 tissues tested). For SOX2, -3, -5, and -6, brain expression was relatively enriched by up to 20-fold (for SOX3). Enrichment was quantified as normalized brain EST counts/total normalized EST counts for all 45 tissues. Expression in glioma was enriched by 2.5- to 25-fold for SOX2, -3, -6, and -10 (normalized glioma EST counts/total normalized EST counts for all 26 tumor types tested). EST counts were obtained from the UNIGENE EST profile viewer (e.g., http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?UGID=155082&TAXID=9606&SEARCH=sox3).
Disclosures
None.
Supplementary Material
Acknowledgments
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the US Department of Health and Human Services.
LifeLines Cohort Study members are Behrooz Z. Alizadeh, H. Marike Boezen, Lude Franke, Pim van der Harst, Gerjan Navis, Marianne G. Rots, Harold Snieder, Morris Swertz, Bruce H.R. Wolffenbuttel, and Cisca Wijmenga (all from University of Groningen, University Medical Center Groningen).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016080892/-/DCSupplemental.
Contributor Information
Collaborators: Behrooz Z. Alizadeh, H. Marike Boezen, Lude Franke, Pim van der Harst, G. N., Marianne G. Rots, H. S., Morris Swertz, Bruce H.R. Wolffenbuttel, and Cisca Wijmenga
References
- 1.Adrogué HJ, Madias NE: Hyponatremia. N Engl J Med 342: 1581–1589, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Miller M: Hyponatremia and arginine vasopressin dysregulation: Mechanisms, clinical consequences, and management. J Am Geriatr Soc 54: 345–353, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Vachharajani TJ, Zaman F, Abreo KD: Hyponatremia in critically ill patients. J Intensive Care Med 18: 3–8, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Adrogué HJ, Madias NE: Hypernatremia. N Engl J Med 342: 1493–1499, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Kugler JP, Hustead T: Hyponatremia and hypernatremia in the elderly. Am Fam Physician 61: 3623–3630, 2000 [PubMed] [Google Scholar]
- 6.Allison SP, Lobo DN: Fluid and electrolytes in the elderly. Curr Opin Clin Nutr Metab Care 7: 27–33, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Ellison DH, Berl T: Clinical practice. The syndrome of inappropriate antidiuresis. N Engl J Med 356: 2064–2072, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Renneboog B, Musch W, Vandemergel X, Manto MU, Decaux G: Mild chronic hyponatremia is associated with falls, unsteadiness, and attention deficits. Am J Med 119: 71.e71–71.e78, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Schrier RW, Gross P, Gheorghiade M, Berl T, Verbalis JG, Czerwiec FS, Orlandi C; SALT Investigators : Tolvaptan, a selective oral vasopressin V2-receptor antagonist, for hyponatremia. N Engl J Med 355: 2099–2112, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM: Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci USA 96: 2538–2542, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko BC, Turck CW, Lee KW, Yang Y, Chung SS: Purification, identification, and characterization of an osmotic response element binding protein. Biochem Biophys Res Commun 270: 52–61, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Rodríguez C, Aramburu J, Rakeman AS, Rao A: NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA 96: 7214–7219, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stroud JC, Lopez-Rodriguez C, Rao A, Chen L: Structure of a TonEBP-DNA complex reveals DNA encircled by a transcription factor. Nat Struct Biol 9: 90–94, 2002 [DOI] [PubMed] [Google Scholar]
- 14.van Lieburg AF, Verdijk MA, Knoers VV, van Essen AJ, Proesmans W, Mallmann R, Monnens LA, van Oost BA, van Os CH, Deen PM: Patients with autosomal nephrogenic diabetes insipidus homozygous for mutations in the aquaporin 2 water-channel gene. Am J Hum Genet 55: 648–652, 1994 [PMC free article] [PubMed] [Google Scholar]
- 15.van den Ouweland AM, Dreesen JC, Verdijk M, Knoers NV, Monnens LA, Rocchi M, van Oost BA: Mutations in the vasopressin type 2 receptor gene (AVPR2) associated with nephrogenic diabetes insipidus. Nat Genet 2: 99–102, 1992 [DOI] [PubMed] [Google Scholar]
- 16.Feldman BJ, Rosenthal SM, Vargas GA, Fenwick RG, Huang EA, Matsuda-Abedini M, Lustig RH, Mathias RS, Portale AA, Miller WL, Gitelman SE: Nephrogenic syndrome of inappropriate antidiuresis. N Engl J Med 352: 1884–1890, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merendino JJ Jr., Speigel AM, Crawford JD, O’Carroll AM, Brownstein MJ, Lolait SJ: Brief report: A mutation in the vasopressin V2-receptor gene in a kindred with X-linked nephrogenic diabetes insipidus. N Engl J Med 328: 1538–1541, 1993 [DOI] [PubMed] [Google Scholar]
- 18.Pan Y, Metzenberg A, Das S, Jing B, Gitschier J: Mutations in the V2 vasopressin receptor gene are associated with X-linked nephrogenic diabetes insipidus. Nat Genet 2: 103–106, 1992 [DOI] [PubMed] [Google Scholar]
- 19.Rosenthal W, Seibold A, Antaramian A, Lonergan M, Arthus MF, Hendy GN, Birnbaumer M, Bichet DG: Molecular identification of the gene responsible for congenital nephrogenic diabetes insipidus. Nature 359: 233–235, 1992 [DOI] [PubMed] [Google Scholar]
- 20.Wilmot B, Voruganti VS, Chang YP, Fu Y, Chen Z, Taylor HA, Wilson JG, Gipson T, Shah VO, Umans JG, Flessner MF, Hitzemann R, Shuldiner AR, Comuzzie AG, McWeeney S, Zager PG, Maccluer JW, Cole SA, Cohen DM: Heritability of serum sodium concentration: Evidence for sex- and ethnic-specific effects. Physiol Genomics 44: 220–228, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H: The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42: D1001–D1006, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, et al.; ICBP Consortium, AGEN Consortium, CARDIOGRAM, CHARGe-Heart Failure Group, ECHOGen Consortium : Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7: 10023, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell’Antonio G, Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L; Swiss Kidney Project on Genes in Hypertension (SKIPOGH) team : Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med 19: 1655–1660, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer TE, Verwoert GC, Hwang SJ, Glazer NL, Smith AV, van Rooij FJ, Ehret GB, Boerwinkle E, Felix JF, Leak TS, et al.; Genetic Factors for Osteoporosis Consortium, Meta Analysis of Glucose and Insulin Related Traits Consortium : Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS Genet 6: e1001045, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian W, Fu Y, Garcia-Elias A, Fernández-Fernández JM, Vicente R, Kramer PL, Klein RF, Hitzemann R, Orwoll ES, Wilmot B, McWeeney S, Valverde MA, Cohen DM: A loss-of-function nonsynonymous polymorphism in the osmoregulatory TRPV4 gene is associated with human hyponatremia. Proc Natl Acad Sci USA 106: 14034–14039, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobota RS, Shriner D, Kodaman N, Goodloe R, Zheng W, Gao YT, Edwards TL, Amos CI, Williams SM: Addressing population-specific multiple testing burdens in genetic association studies. Ann Hum Genet 79: 136–147, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Q, Ferraris JD, Burg MB: High NaCl increases TonEBP/OREBP mRNA and protein by stabilizing its mRNA. Am J Physiol Renal Physiol 289: F803–F807, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Huang W, Liu H, Wang T, Zhang T, Kuang J, Luo Y, Chung SS, Yuan L, Yang JY: Tonicity-responsive microRNAs contribute to the maximal induction of osmoregulatory transcription factor OREBP in response to high-NaCl hypertonicity. Nucleic Acids Res 39: 475–485, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Y, Liu Y, Liu M, Wei J, Zhang Y, Hou J, Huang W, Wang T, Li X, He Y, Ding F, Yuan L, Cai J, Zheng F, Yang JY: Sfmbt2 10th intron-hosted miR-466(a/e)-3p are important epigenetic regulators of Nfat5 signaling, osmoregulation and urine concentration in mice. Biochim Biophys Acta 1839: 97–106, 2014 [DOI] [PubMed] [Google Scholar]
- 30.Li W, Kong LB, Li JT, Guo ZY, Xue Q, Yang T, Meng YL, Jin BQ, Wen WH, Yang AG: MiR-568 inhibits the activation and function of CD4+ T cells and Treg cells by targeting NFAT5. Int Immunol 26: 261–281, 2014 [DOI] [PubMed] [Google Scholar]
- 31.Pott S, Lieb JD: What are super-enhancers? Nat Genet 47: 8–12, 2015 [DOI] [PubMed] [Google Scholar]
- 32.Ward LD, Kellis M: HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 44[D1]: D877–D881, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, et al.; Alzheimer’s Disease Genetics Consortium : Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet 8: e1002707, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, Lenhard B, Sandelin A, Wasserman WW: JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44: D110–D115, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiyama TY, Watanabe E, Ono K, Inenaga K, Tamkun MM, Yoshida S, Noda M: Na(x) channel involved in CNS sodium-level sensing. Nat Neurosci 5: 511–512, 2002 [DOI] [PubMed] [Google Scholar]
- 36.Hiyama TY, Watanabe E, Okado H, Noda M: The subfornical organ is the primary locus of sodium-level sensing by Na(x) sodium channels for the control of salt-intake behavior. J Neurosci 24: 9276–9281, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Bourque CW: Osmometry in osmosensory neurons. Nat Neurosci 6: 1021–1022, 2003 [DOI] [PubMed] [Google Scholar]
- 38.Bourque CW: Central mechanisms of osmosensation and systemic osmoregulation. Nat Rev Neurosci 9: 519–531, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Sheffield NC, Thurman RE, Song L, Safi A, Stamatoyannopoulos JA, Lenhard B, Crawford GE, Furey TS: Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res 23: 777–788, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung CY, Ko BC: NFAT5 in cellular adaptation to hypertonic stress - regulations and functional significance. J Mol Signal 8: 5, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo SK, Lee SD, Kwon HM: TonEBP transcriptional activator in the cellular response to increased osmolality. Pflugers Arch 444: 579–585, 2002 [DOI] [PubMed] [Google Scholar]
- 42.Cohen DM, Wasserman JC, Gullans SR: Immediate early gene and HSP70 expression in hyperosmotic stress in MDCK cells. Am J Physiol 261: C594–C601, 1991 [DOI] [PubMed] [Google Scholar]
- 43.Machnik A, Neuhofer W, Jantsch J, Dahlmann A, Tammela T, Machura K, Park JK, Beck FX, Müller DN, Derer W, Goss J, Ziomber A, Dietsch P, Wagner H, van Rooijen N, Kurtz A, Hilgers KF, Alitalo K, Eckardt KU, Luft FC, Kerjaschki D, Titze J: Macrophages regulate salt-dependent volume and blood pressure by a vascular endothelial growth factor-C-dependent buffering mechanism. Nat Med 15: 545–552, 2009 [DOI] [PubMed] [Google Scholar]
- 44.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, Zhernakova A, Heap GA, Adány R, Aromaa A, et al. : Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 42: 295–302, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ: Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet 6: e1000888, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK: Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, Weersma RK, Hofstra RM, Buurman WA, Rensen S, Wolfs MG, Platteel M, Zhernakova A, Elbers CC, Festen EM, Trynka G, Hofker MH, Saris CG, Ophoff RA, van den Berg LH, van Heel DA, Wijmenga C, Te Meerman GJ, Franke L: Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 7: e1002197, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo SK, Dahl SC, Handler JS, Kwon HM: Bidirectional regulation of tonicity-responsive enhancer binding protein in response to changes in tonicity. Am J Physiol Renal Physiol 278: F1006–F1012, 2000 [DOI] [PubMed] [Google Scholar]
- 49.Hon GC, Hawkins RD, Ren B: Predictive chromatin signatures in the mammalian genome. Hum Mol Genet 18: R195–R201, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R: Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE: Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518: 337–343, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kioussis D, Festenstein R: Locus control regions: Overcoming heterochromatin-induced gene inactivation in mammals. Curr Opin Genet Dev 7: 614–619, 1997 [DOI] [PubMed] [Google Scholar]
- 53.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E: A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12: 1725–1735, 2003 [DOI] [PubMed] [Google Scholar]
- 54.Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, Sabo PJ, Vierstra J, Voit RA, Yuan GC, Porteus MH, Stamatoyannopoulos JA, Lettre G, Orkin SH: An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342: 253–257, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al.; Roadmap Epigenomics Consortium : Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gillies SD, Morrison SL, Oi VT, Tonegawa S: A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33: 717–728, 1983 [DOI] [PubMed] [Google Scholar]
- 57.Forrester WC, van Genderen C, Jenuwein T, Grosschedl R: Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science 265: 1221–1225, 1994 [DOI] [PubMed] [Google Scholar]
- 58.Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, Harris A: Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci USA 106: 19934–19939, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blattler A, Yao L, Witt H, Guo Y, Nicolet CM, Berman BP, Farnham PJ: Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol 15: 469, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romero MF, Chen AP, Parker MD, Boron WF: The SLC4 family of bicarbonate (HCO₃⁻) transporters. Mol Aspects Med 34: 159–182, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.et al. ; LifeLines Cohort Study, CARDIoGRAM Consortium, DIAGRAM Consortium, ICBP Consortium, MAGIC Consortium : Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 45: 145–154, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beck LH: Hypouricemia in the syndrome of inappropriate secretion of antidiuretic hormone. N Engl J Med 301: 528–530, 1979 [DOI] [PubMed] [Google Scholar]
- 63.Passamonte PM: Hypouricemia, inappropriate secretion of antidiuretic hormone, and small cell carcinoma of the lung. Arch Intern Med 144: 1569–1570, 1984 [PubMed] [Google Scholar]
- 64.Musch W, Decaux G: Utility and limitations of biochemical parameters in the evaluation of hyponatremia in the elderly. Int Urol Nephrol 32: 475–493, 2001 [DOI] [PubMed] [Google Scholar]
- 65.Fuchsberger C, Taliun D, Pramstaller PP, Pattaro C; CKDGen consortium : GWAtoolbox: An R package for fast quality control and handling of genome-wide association studies meta-analysis data. Bioinformatics 28: 444–445, 2012 [DOI] [PubMed] [Google Scholar]
- 66.Katz MA: Hyperglycemia-induced hyponatremia--calculation of expected serum sodium depression. N Engl J Med 289: 843–844, 1973 [DOI] [PubMed] [Google Scholar]
- 67.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D; Modification of Diet in Renal Disease Study Group : A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Ann Intern Med 130: 461–470, 1999 [DOI] [PubMed] [Google Scholar]
- 68.Willer CJ, Li Y, Abecasis GR: METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Devlin B, Roeder K: Genomic control for association studies. Biometrics 55: 997–1004, 1999 [DOI] [PubMed] [Google Scholar]
- 70.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI: SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24: 2938–2939, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al.; ENCODE Project Consortium, NISC Comparative Sequencing Program, Baylor College of Medicine Human Genome Sequencing Center, Washington University Genome Sequencing Center, Broad Institute, Children’s Hospital Oakland Research Institute : Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M: Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22: 1790–1797, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stormo GD, Schneider TD, Gold L, Ehrenfeucht A: Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res 10: 2997–3011, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.