Abstract
Unraveling cause-and-effect relationships in the nervous system is challenging because some biological processes begin stochastically, take a significant amount of time to unfold, and affect small neuronal subpopulations that can be difficult to isolate and measure. Single-cell approaches are slow, subject to user bias, and sometimes too laborious to achieve sample sizes large enough to detect important effects. Here, we describe an automated imaging and analysis system that enables us to follow the fates of individual cells and intracellular proteins over time. Observations can be quantified in a high-throughput manner with minimal user bias. We have adapted survival analysis methods to determine whether and how factors measured during longitudinal analysis predict a particular biological outcome. The ability to monitor complex processes at single-cell resolution quickly, quantitatively, and over long intervals should have wide applications for biology.
Keywords: Akt, proportional hazard analysis, survival analysis
The nervous system poses difficult experimental challenges for those seeking to understand the biochemical bases for its function in health and disease. The brain is composed of a staggering number of different cell types (1–4). Because the relevant changes often occur in a small subpopulation of neurons, the primary signals can be small and difficult to detect by standard biochemical or molecular biological assays. Moreover, changes in one subpopulation of neurons can trigger additional changes in neighboring cells or in distant but synaptically connected neuronal subpopulations. Because the different cell types are often intermixed and the sources of the signals cannot be resolved, the picture that emerges can be complex and misleading.
Elucidating pathogenic mechanisms can be especially challenging. First, the time at which a particular neuron dies from disease may be stochastic (5), which reduces the measurable signal and increases its variability. Second, the initiating pathological event may incite a myriad of homeostatic responses within the affected cell, its neighbors, and its synaptic partners. Consequently, changes associated with pathological states are necessarily abnormal but are not necessarily pathogenic. However, determining whether a change is pathogenic, an epiphenomenon, or a beneficial coping response can be a conundrum with standard approaches that rely on static “snapshots.” Third, the disease itself may limit detection. As a degenerative disease progresses, vulnerable neurons die, and fewer are available to generate detectable biochemical signals. Eventually, the predominant biochemical changes are the coping responses of the remaining cells. Finally, many pathogenic processes are slow, further reducing the signals that can be measured at any single point in time.
One strategy to overcome these limitations is to use assays that can resolve signals from single cells. However, standard single-cell assays are commonly time-consuming, subject to user bias, and often less quantitative than population-based assays. To overcome the problems, we built a system with the ability to repeatedly and accurately resolve signals within single cells over arbitrary time intervals. Our system combines the specificity of single-cell approaches with the throughput of population-based approaches and enables us to systematically and quantitatively elucidate the relationship between intermediate changes and the long-term fate of a cell.
Methods
Reagents and Plasmids. Kainic acid was from Sigma–Aldrich, ethidium homodimer (EtHD) was from Molecular Probes, and Lipofectamine2000 was from Invitrogen. We used the following plasmids: pCMV6-Myr-Akt-hemagglutinin (a gift of Thomas F. Franke, Columbia University, New York; ref. 6), pCMV4 (a gift from Warner Greene, Gladstone Institute of Virology and Immunology, San Francisco), pGW1-GFP (a gift of Don Arnold, University of Southern California, Los Angeles), pGW1-CFP and pGW1-YFP (gifts from Ricardo Dolmetsch, Stanford University, Stanford, CA), and mammalian expression plasmid encoding dsRED1 (Clontech).
Neuronal Culture. Primary cultures of cortical neurons were prepared from rat embryos (E16–18) as reported in ref. 7. Brain cortices were dissected, treated with papain and then trypsin inhibitor, and gently triturated to dissociate single neurons in Optimem/glucose medium. Cells (6 × 105) were plated in each well of a 24-well tissue culture plate; 2 h later, the plating medium was replaced by growth medium with serum. (Contact S.F. for additional details.)
Transfection. Neurons were transfected at 6–7 days in vitro as described in ref. 7. Growth medium was replaced 1 h before transfection. A Ca2+/phosphate coprecipitate of plasmid DNA (1–4 μg) was formed and added to each well of a 24-well plate. GFP was coprecipitated with AKT or empty plasmid in a 1:3 ratio. Approximately 20 min after the precipitate was added, the medium was removed, and cells were treated briefly with medium containing 2% dimethyl sulfoxide, rinsed twice, and maintained in serum-free growth medium. For some transfections, Lipofectamine2000 and DNA were mixed together, added to neurons for 5 h, and replaced with serum-free growth medium.
Robotic Microscope. The imaging system was built with a Nikon TE300 Quantex base and was equipped with Super Fluor ×40 [numerical aperture (N.A.) 1.3] and CFI Plan Achromat ×100 (N.A. 1.3) objectives, longer working distance objectives ×20 (N.A. 0.45) and ×40 (N.A. 0.6), and low-magnification ×10 (N.A. 0.3) and ×4 (N.A. 0.13) objectives. The detector was a Hamamatsu Orca II 12/14-bit digital cooled charge-coupled device camera (Middlesex, NJ). Images were collected and analyzed with metamorph (Universal Imaging, Downingtown, PA) and custom programs written with mat lab and visual c. A Uniblitz shutter (Vincent Associates, Rochester, NY) limited transmitted light illumination. The motorized stage and focus knob control (ProScan II with linear encoders) were from Prior Scientific (Rockland, MA). Illumination was from a xenon lamp (175 W) via a liquid light guide. Fluorescence excitation and emission were controlled independently with sets of optical filters (Chroma Technology, Rockingham, VT) by using two 10-position motorized filter wheels (Lambda 10–2, Sutter Instruments, Novato, CA). The system was mounted on a vibration isolation table (Technical Manufacturing, Peabody, MA). Images were stored temporarily on computer hard drives and then burned onto digital video discs for further off-line automated analysis.
Software Control of Image Acquisition and Analysis. To determine the tilt of the plate, we wrote a program to measure the relative location of the focal plane at three different locations within a single well or three different locations within a multiwell plate. The program uses the lateral distances between the three measurement points and their relative focal points to calculate the slope of the focal plane. The program that registers the position of a plate directs the stage to image the location of a fiduciary mark on the plate created by the manufacturer, focus, and then collect a reference image of the mark. We then relate every other point in the acquisition program to this fiduciary mark. When the plate is returned to the plate holder, we reposition it to restore registration of the fiduciary mark.
Threshold values were determined with the following equation:
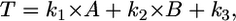 |
where T is the calculated threshold value, k1, k2, and k3 are empirically determined constants, A is the standard deviation of the array of digitized pixel values that compose an image, and B is the minimum pixel value in the array.
Results
Image Acquisition. We built an efficient wide-field epifluorescence imaging system with motorized stage and focus controls. The camera was placed at the basement port of the inverted microscope to minimize the complexity of the light path and to maximize the number of detected photons. Fixed multiband pass dichroic mirrors and 10-position excitation and emission filter wheels were used to examine fluorescent signals from selected spectra. Filter wheels were computer controlled during automated image acquisition. We also used a 14-bit charge-coupled device camera to resolve a wide range of signal intensities for a particular bandwidth of light.
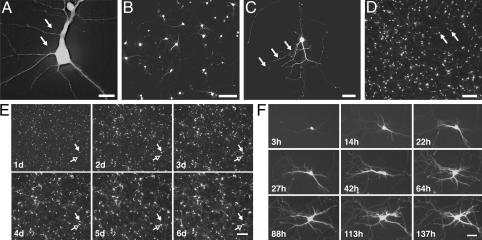
With this configuration, we generated high-resolution images of neurons grown on plastic tissue culture plates and transfected with a variety of fluorescent proteins (Fig. 1). Tissue culture plastic transmits some wavelengths of light (e.g., UV) less well than glass and can scatter more light, reducing image resolution. However, plastic has several advantages. First, neurons survive better and appear healthier when grown on plastic than on glass. Second, plastic plates are well suited to inexpensive high-throughput analysis. Finally, the direct attachment of a neuron to the plate facilitates the development of software algorithms that enable us to return to precisely the same neuron or field of neurons over arbitrary time intervals.
Fig. 1.
Highly resolved images of neurons grown on plastic tissue-culture dishes and expressing a variety of fluorescent proteins. (A) Spines (arrows) on dendrites of a cortical neuron transfected with dsRED. (×40, N.A. 0.60; scale bar, 15 μm.) (B) Cell bodies and neurites of a group of neurons containing CFP. (×10, N.A. 0.30; scale bar, 150 μm.) (C) Growth cones and neurites (arrows) on a striatal neuron expressing GFP. (×20, N.A. 0.45; scale bar, 50 μm.) (D) Cell bodies (arrows) of cortical neurons transfected with GFP. (×4, N.A. 0.13; scale bar, 300 μm.) (E) Images collected (×4) at approximately daily intervals after transfection demonstrate the ability to return to the same field of neurons. One neuron (open arrow) survives throughout the experiment; another (filled arrow) dies between days 4 and 5. (Scale bar, 300 μm.) (F) Intracellular and extracellular structures (e.g., neurites) of single neurons can be resolved and monitored over time. (×20, N.A. 0.45; scale bar, 60 μm.)
To automate image acquisition, we wrote computer programs to control the stage and focus knob motors. These programs specify the number of stepper motor movements needed to scan a single well of a multiwell plate and generate a series of nonoverlapping images with different objectives (×4, ×10, ×20, and ×40). Additional programs move the plate so that each well of a multiwell plate can be systematically scanned. For automated focusing between stage movements, we incorporated a commercially available algorithm (metamorph, Universal Imaging). It identifies the focal plane by comparing images collected at different focus knob positions. Content-based mathematical algorithms to identify the image corresponding to the focal plane among a series of adjacent images along the z-axis are described in refs. 8–10. Some of these algorithms have been implemented in high-content screening systems, as described in refs. 11–14.
Initially, automated focusing was unreliable, phototoxic, and slow, accounting for 90% of the time to execute a single cycle of imaging and lateral stage movement. This process involves image collection by the camera's charge-coupled device chip, transfer of the electronic information to the computer, analysis by the computer, and movement of the focus knob in response to the analysis. Transfer of the electronic information to the computer was the most time-consuming step. Therefore, we tested the effect of reducing spatial resolution of the image on autofocusing by varying the number of discrete elements of the charge-coupled device chip that we used. Reducing the spatial resolution by ≈95% sped up the focusing cycle 5-fold without reducing its accuracy.
Using fluorescence illumination to focus was potentially phototoxic and unreliable. For fields lacking fluorescence, the microscope could search so far from the focal plane that it never recovered. By using phase-contrast optics and a 20-ms light pulse from a low-intensity incandescent lamp, we can generate an image with sufficient spatial contrast to sharply focus the microscope whether or not the image contained fluorescence (8).
To acquire images of multiple fields of neurons within a single well of a multiwell dish, we moved the stage in a spiral pattern from the center of the well outward. However, the meniscus of the medium interfered with phase-contrast optics, degrading the image with increasing radial distance from the center of the well. Automated focusing quickly became impossible.
We engineered two related solutions that made automated focusing reliable and significantly sped up the image-acquisition routine. In one approach, we used an objective with such a low N.A. and therefore high depth of field that all objects within adjacent fields in a well of a 24-well plate remained sharply focused. This solution is especially useful for high-throughput applications (e.g., counting neurons to measure neuronal survival). The lateral spatial resolution was sufficient to resolve individual neurons, and the low magnification made it possible to image many neurons in a single field. However, this approach failed with higher N.A. objectives whose narrow depth of field could no longer keep adjacent fields in focus. Instead, we programmed the microscope to measure the tilt of the plate. We then used the measurement to introduce a compensatory vertical movement of the objective for each lateral movement of the stage, thereby keeping all adjacent fields in focus. The program uses the slope to modify the automated stage movements so that the field remains in focus, even with a high N.A. objective.
We next developed an approach to return to precisely the same neuron or field of neurons. Although the movements of the stepper motors were precise, when we removed and then replaced the plate on the holder, the contents of the microscopic fields from corresponding images were different or only partially overlapped. Attempts to fix the position of the plate within the plate holder were impractical and largely ineffective. Instead, we developed a simple program to relate the movements of our acquisition programs to a fiduciary mark on the plate. Reregistration can be performed by using automated programs described below. The strategy allows us to return to precisely the same neuron or field of neurons over almost any interval (Fig. 1 E and F).
Image Analysis. To avoid user bias, we automated the analysis by selecting “positive” pixels in an image and grouping them together as objects for further analysis. We tested whether simple features of an image could be used to accurately calculate a threshold value for a positive pixel. A relatively good estimate of a threshold value could be calculated from the minimum pixel value and the overall variance of the pixels in an image (see Fig. 5 A–E, which is published as supporting information on the PNAS web site). This program accurately selected appropriate threshold values of unrelated images and of images of the same field of neurons at different times and performed better than a commonly used commercially available algorithm on images with ≥30 objects of interest. For images with fewer objects, the latter algorithm was more reliable. This finding is important because fluorescence from green fluorescent protein (GFP) can be detected within 2–3 h after transfection, and fluorescence continues to increase for 100 h after transfection, owing to increasing steady-state GFP levels (Fig. 5 F and G). Our program compensates for the changing GFP expression levels, enabling it to identify the same objects (e.g., neurons) from images collected at different times (Fig. 5F). This compensation also makes it possible to more accurately measure the true dimensions of a neuron independently of the absolute level of GFP (Fig. 5F).
Once a cutoff was determined, contiguous pixels whose intensities exceeded this value were grouped together as distinct objects. These objects could be evaluated further with software filters, programs that measure the shape and dimensions of the object (metamorph, Universal Imaging). We constructed software filters based solely on the area and elipticity of an object, which could routinely identify and measure 93–98% of the living neurons in a microscope field while excluding >99% of fluorescent debris. If two or more positive cells are too close to each other, our threshold algorithm and the commercial one treated the contiguous positive pixels as a single object. Empiric measurement of the areas of contiguous positive pixels formed from clusters of positive neurons showed that the areas tended to fall within relatively narrow ranges, which were directly proportional to the number of neurons within the cluster. With this information, we constructed software filters to count clusters of two, three, and four neurons and thereby arrived at accurate (±1%) estimates of the number of cells within a field.
The biological response produced by a protein often depends on its concentration. Therefore, failure to capture the relationship between gene expression at the single-cell level and the biological response it produces could make it difficult to detect or characterize a relationship. The cellular heterogeneity of the brain compounds these problems because the same gene may have significantly different effects that depend on both concentration and cell type. Therefore, it would be valuable to be able to both identify transfected cells and estimate the expression level of the transfected gene within each cell over time.
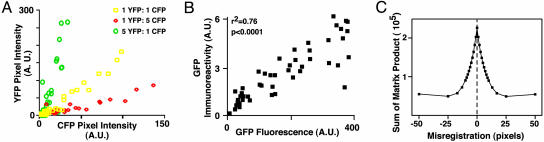
Therefore, we assessed the expression level of a marker gene as a surrogate for expression of a cotransfected gene (15). We cotransfected neurons with various amounts of yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP), measured the fluorescence of each protein in each neuron, and determined whether the expression of one gene correlated with the expression of the other. We confirmed that we could measure the fluorescence of each protein independently (see Fig. 6, which is published as supporting information on the PNAS web site). Then, we cotransfected YFP and CFP into neurons in various ratios. Although the expression of the two proteins varied significantly from cell to cell, the fluorescence of one protein was nearly always highly correlated with the fluorescence of the other (r2 = 0.99) and related directly to the ratio of transfected plasmid DNA (Fig. 2A). We then measured GFP expression by imaging GFP fluorescence and by immunocytochemistry in single neurons. The two measurements were highly correlated (16) (Fig. 2B). Thus, fluorescence of a marker gene in vivo is a good indicator of its expression level and can be used to estimate the expression of a cotransfected gene.
Fig. 2.
Boolean image analysis. (A) Neurons were cotransfected with CFP, YFP, or both. Cell-by-cell comparison of CFP and YFP fluorescence from neurons transfected with CFP and YFP in various ratios reveals that nearly all neurons are cotransfected and that the fluorescence intensities of the two transfected proteins are highly correlated (r2 = 0.99). (B) Cell-by-cell comparison of GFP expression as estimated by measuring GFP fluorescence directly and by measuring immunofluorescence against GFP. (C) A matrix algorithm to register two images. Here, two images are placed out of register in the x–y plane to various degrees and then subjected to a computer algorithm that calculates the sum of a product matrix derived from the two images. The sum is maximal when the two images are registered. A.U., arbitrary units.
In the previous experiment, we compared the fluorescence of CFP with YFP on a neuron-by-neuron basis. Accurately automating these and other (e.g., Boolean) comparisons requires that the exact pixel positions in the two images correspond. However, the automated movements of the filter wheels occasionally led to a small misregistration of different fluorescence images of the same microscope field. We developed a simple automated algorithm to correct misregistration. After each fluorescence image was collected, a 12-bit digital phase-contrast image of the same microscope field also was collected. The phase-contrast images were binarized to generate matrices in which approximately half the digits of each matrix were zeroes and half were ones. A subset of one matrix was selected and multiplied by a subset of the other matrix; the product matrix was summed. The sum is maximal when the images (and the subset matrices) are identical (i.e., when they are in perfect registration) (Fig. 2C). Thus, we can empirically sample a set of potential x–y misregistration quantities and calculate the one that produces the optimal registration. The optimal value then is used to register the portions of the original image that overlap, which can be used for direct comparisons.
Automated Longitudinal Analysis of Biological Processes. To determine whether the system can be used to longitudinally monitor neuronal survival or other long-term adaptive responses, we compared images collected periodically over several days or weeks. Occasionally, the GFP fluorescence of a neuron in one image would abruptly disappear in a subsequent image. Erb and colleagues (17) recently showed that the loss of GFP is a sensitive and specific marker of different types of death in some nonneuronal cell types. If GFP loss corresponds to the death of a neuron, it can be quantified over time as a measure of neuronal survival (17, 18). To test this possibility, we simultaneously measured the loss of GFP in transfected neurons and the loss of membrane integrity by using a membrane-impermeant nuclear dye, EtHD. EtHD staining of cell nuclei indicates death in the widely used LIVE/DEAD assay. In response to the neurotoxin kainic acid and in the presence of extracellular EtHD, the cell bodies of GFP-transfected neurons rounded and swelled, and their neurites began to retract. Eventually, the fluorescence of particular neurons disappeared abruptly, and at that moment, their nuclei stained positively with EtHD (Fig. 3A). Recently, loss of one soluble cytosolic fluorescent protein, GFP, was shown to coincide with the loss of a cotransfected soluble cytosolic fluorescent protein, red fluorescent protein (mRFP) (16). In turn, staining of neurons with the apoptosis marker annexin V immediately preceded loss of mRFP. Thus, the loss of GFP fluorescence correlates well with two widely accepted measures of cell death.
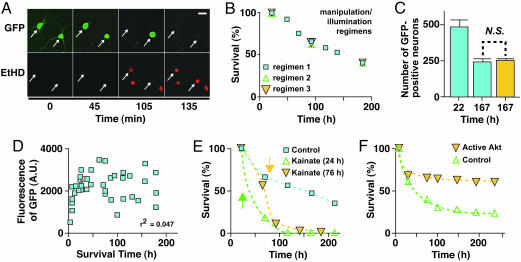
Fig. 3.
Automated imaging of neuronal survival. (A) GFP-transfected neurons (arrows) were treated with kainate in the presence of the membrane-impermeant nuclear dye EtHD. Loss of GFP fluorescence correlated with loss of membrane integrity and nuclear staining with EtHD (n = 12). (Scale bar, 50 μm.) (B) The frequency of automated imaging measurements did not detectably affect survival. Neurons in sister cultures subjected to (1) daily removal from the incubator and imaging, (2) daily removal from the incubator with imaging every third day, or (3) removal from the incubator and imaging every third day survived equally well. (C) In parallel, one culture of transfected neurons was imaged once at the end of the experiment (gold bar), and the other was imaged both 1 day after transfection and at the end of the experiment (blue bars). The survival rates were nearly identical. N.S., not significant. (D) GFP expression does not detectably affect survival. The duration of survival of each neuron was determined and plotted against the GFP expression level in that neuron before its death. GFP expression did not correlate with neuronal survival by correlation analysis (n = 2). A.U., arbitrary units. (E) Automated imaging and analysis demonstrate kainate neurotoxicity (n = 2). The survival of transfected neurons in three sister cultures was compared. In the first (blue squares), neurons were left untreated. In the second (gold triangles) and third (green triangles), kainate (10 μM) was added 24 or 76 h after transfection, respectively. Neuronal survival significantly decreased soon after kainate treatment in both treated cultures. (F) Automated imaging and analysis detects the ability of CA-Akt to promote neuronal survival (n = 3). Neurons were transfected with GFP and either an expression plasmid for CA-Akt (gold triangles) or an empty control vector (green triangles).
To monitor neuronal survival and other long-term responses, we need to know whether the periodic imaging or the GFP marker affected neuronal survival (19, 20). First, we found no significant survival difference between neurons from sister cultures that were imaged with different frequencies (Fig. 3 B and C), suggesting that our imaging methods do not perturb neuronal survival. Next, we tested whether the expression of GFP affected survival. Studies of the potential toxicity of GFP and its variants have arrived at conflicting conclusions (17–19, 21). If GFP is toxic, the level of GFP expression as measured by GFP fluorescence should correlate inversely with duration of survival. By using the robotic microscope to track GFP fluorescence and survival times of GFP-transfected neurons, we found no correlation between the survival of individual neurons and their levels of GFP expression (Fig. 3D, r2 = 0.047), suggesting no significant effect on survival.
If this approach accurately measures neuronal survival, it also should detect the effects of molecules that regulate survival. To test this possibility, we transfected neurons with GFP and periodically imaged the cells before and after mock treatment or treatment with kainic acid. As shown by automated analysis of GFP-positive neurons, neuronal survival decreased after kainate application (Fig. 3E) and increased after cotransfection of a constitutively active form of the prosurvival kinase Akt (Fig. 3F). These results suggest that the automated microscope is a sensitive and valid way to measure the effects of extracellular or intracellular molecules on neuronal survival.
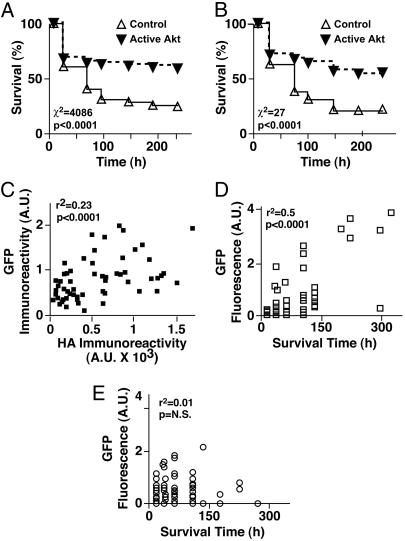
The ability to return periodically to precisely the same microscope field creates unparalleled opportunities for data analysis. Cohorts of neurons can be tracked longitudinally, and the number of neurons that die during the intervening period can be deduced by simple subtraction with an automated analysis program. By determining the survival times of particular cohorts of neurons, we lay the foundation for the application of powerful statistical techniques known as survival analysis. For the purposes of survival analysis and by convention, neurons that died during the interval were assigned a survival time equivalent to the period between transfection and their disappearance from an image. For example, we deduced survival times for cohorts of neurons transfected with empty vector or active Akt (Fig. 3F) and then used nonparametric Kaplan–Meier analysis to estimate the underlying survival function, S(t), for each condition (Fig. 4A). Analysis with the log-rank test showed that the survival functions were significantly different (χ2 = 4,086).
Fig. 4.
Applications of survival analysis. (A) Kaplan–Meier analysis of population-based Akt survival data. The number of surviving neurons transfected with CA-Akt or an empty vector was determined at different intervals from a longitudinal series of low-magnification (×4) images. The number of neurons that died during each interval was deduced by comparing identical microscope fields before and after the interval. By convention, the neurons that died during a particular interval were assigned an event (i.e., survival) time that corresponded to the end of that interval. These event times were used to construct the Kaplan–Meier cumulative survival plot shown and analyzed for statistical significance by the log-rank (Mantel–Cox) test (n = 2). (B) Kaplan-Meier analysis of single-cell-based Akt survival data. Three higher-magnification (×20) images of the same transfected neurons as in A were collected longitudinally. Individual neurons were identified in each of the three images. The approach described in A was used to determine the event time for each neuron by comparing images of those neurons collected at different time points, and a Kaplan-Meier survival plot was constructed (n = 2). (C) Neurons cotransfected with GFP and with hemagglutinin-tagged CA-Akt were immunostained, and fluorescent signals were measured and correlated cell by cell. Expression of GFP and CA-Akt are significantly correlated, suggesting that GFP fluorescence in vivo can be used as a surrogate for CA-Akt expression. (D and E) GFP fluorescence is correlated with and predicts longevity in neurons cotransfected with CA-Akt (D) but not in those cotransfected with empty vector (E). A.U., arbitrary units; N.S., not significant.
To confirm that our automated image-analysis programs accurately quantify neuronal survival, we performed longitudinal analysis manually. We randomly chose three microscope fields and examined each image of those fields over the course of the experiment. The survival time of each neuron in each field was determined and then subjected to Kaplan–Meier analysis (Fig. 4B). Although the sample size (83 control neurons and 88 neurons with Akt) was only ≈1% of that in the automated analysis, the survival curves were remarkably similar. These results support two conclusions. First, our automated analysis programs can measure neuronal survival quickly and accurately and can generate data sets whose sheer size makes the statistical analysis extremely sensitive. Second, survival analysis performed on even very small samples of neurons can detect and accurately measure biological effects.
Longitudinal Analysis of Single Cells to Identify Factors That Predict Fate. Although slower than our automated approaches, longitudinal analysis of single cells provides unique opportunities to elucidate cause-and-effect mechanisms. For example, any number of variables can be measured within an individual neuron and followed longitudinally until an important biological fate is observed. Additional statistical techniques, such as proportional-hazards regression analysis of survival data, make it possible to quantify the contribution of a particular factor to the fate of a neuron. The sensitivity and accuracy of single-cell analysis of small samples suggests that this approach is feasible and worthwhile. For survival studies, automated and single-cell analyses make it possible to identify and distinguish early changes within a neuron that predict survival or death or are unrelated to neuron fate.
Next, we determined whether expression of active Akt in a neuron predicts its longevity. We anticipated that the fluorescence of a marker protein (e.g., GFP) in vivo could be used as a surrogate for the expression level of cotransfected Akt (Fig. 2 A and B). However, because the expression plasmids that encode GFP and constitutively active Akt (CA-Akt) use different promoters, the expression of GFP and CA-Akt might not be correlated. We cotransfected neurons with GFP and a hemagglutinin-tagged version of CA-Akt, performed dual-label immunocytochemistry, and quantified the fluorescence of each tag cell by cell. GFP and CA-Akt expressions were significantly correlated (Fig. 4C). Therefore, we used longitudinal single-cell analysis to examine neurons cotransfected with GFP and empty vector or CA-Akt. GFP fluorescence from each neuron was estimated by measuring average pixel intensity values from images. GFP fluorescence and longevity were highly correlated in neurons cotransfected with CA-Akt and GFP (Fig. 4D), but not in neurons transfected with GFP and empty vector (Fig. 4E). These results suggest that the level of CA-Akt expression within individual neurons, as estimated by the level of GFP fluorescence, is the critical factor promoting survival, not GFP itself, and can predict the longevity of a neuron. That cotransfected GFP acts a surrogate for expression avoids the potential for unanticipated effects of fusing GFP to the molecule of interest and increases the system's versatility and efficiency.
Discussion
We developed a robotic microscope with several features. It can acquire images rapidly and automatically, making it suitable for high-throughput applications. Acquisition is sufficiently accurate and precise to allow fine features of a neuron or other cell types (16) to be spatially resolved and to allow the same neuron or field of neurons to be reimaged after arbitrary time intervals. The automated analysis programs can extract critical morphometric features of neurons within each microscope field, perform Boolean analyses of different fluorescence images of the same field, and follow cohorts of neurons longitudinally to determine how features of these neurons change over time. In our case, Boolean analysis refers to logical operations that determine the symbolic relationship (e.g., AND, OR, NOT) between different features of the same object as revealed by images of that object collected with different fluorescence filters (22). Such analysis can expand the amount of information derived from multichannel high-content screening. With the appropriate analysis programs, the system also should be applicable to nonfluorescent images of cells (22). Finally, our single-cell analysis methods make it possible to identify and distinguish early changes within a neuron that predict its fate, making it possible to evaluate potential cause-and-effect relationships.
Automation of image acquisition and analysis improves both the quality and the speed of our measurements. It eliminates intense fluorescence illumination during manual focusing and thereby reduces associated photobleaching and phototoxicity. As a result, low-intensity fluorophores can be observed and more images collected without perturbing neuronal health. Automated survival analysis provides an extremely rapid and sensitive way to quantify the regulatory effects of molecules on long-term biological responses. Recently, we used this system to assess the detrimental effects of disease-associated versions of the huntingtin protein (16) and found that survival of individual neurons could be predicted from the number of mutant huntingtin molecules they contained. Here, we detected the survival-promoting effect of active Akt with 10% of the number of neurons used in the original report and nevertheless achieved greater statistical significance (23). The automated survival analysis was even quicker, and the statistical significance was nearly 20-fold greater than was obtained by single-cell analysis. The speed and additional sensitivity might be especially useful for high-throughput screening. Thus, the range of the image acquisition and processing programs of the automated microscope system encompass both survival- and death-promoting genes and molecules.
Longitudinal analysis is particularly useful in neuroscience, because variability associated with static measurements can obscure important effects. Variability likely arises from multiple factors, including cell diversity, the stochastic nature of the underlying biochemical processes, and the slow pace of many long-term adaptive and maladaptive responses (20). Here, the stochastic nature of events is exploited to identify factors that contribute the most to a particular outcome. Elucidating disease mechanisms may be even more complicated than studying physiologic pathways. Disease-related changes are likely to represent a complex combination of primary pathogenic changes and helpful and harmful coping responses by the affected cell or neighboring cells. Untangling the contributions of these changes to the disease is critical to identifying potential therapeutic targets. By focusing on an outcome of interest, we can identify combinations of factors that predict a particular fate and classify those factors as pathogenic, incidental, or potentially beneficial.
Supplementary Material
Acknowledgments
We thank members of the S.F. laboratory for useful discussions, Stephen Ordway and Gary Howard for editorial assistance, and Kelley Nelson for administrative assistance. M.A. is a Ministry of Education, Science and Culture–Fulbright Fellow and is supported by the Hillblom Foundation. Primary support for this work was provided by National Institute of Neurological Disease and Stroke Grant R01 NS45491. Additional support was provided by National Institute of Aging Grant P01 AG022074, the Taube Family Foundation Program in Huntington's Disease Research, the J. David Institutes, the Hereditary Disease Foundation, and National Institute of Neurological Disease and Stroke Grant R01 NS39074.
Author contributions: M.A. and S.F. designed research; M.A. and S.F. performed research; M.A. and S.F. analyzed data; and S.F. wrote the paper.
Abbreviations: EtHD, ethidium homodimer; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein; CA-Akt, constitutively active Akt; N.A., numerical aperture.
References
- 1.Pituello, F. (1997) Curr. Biol. 7, R701-R704. [DOI] [PubMed] [Google Scholar]
- 2.Brody, T. & Odenwald, W. F. (2002) Development (Cambridge, U.K.) 129, 3763-3770. [DOI] [PubMed] [Google Scholar]
- 3.McConnell, S. K. (1995) J. Neurosci. 15, 6987-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price, J., Williams, B. P. & Götz, M. (1995) Ciba Found. Symp. 193, 71-84. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, G., Collins, R. A., Leavitt, B. R., Andrews, D. F., Hayden, M. R., Lumsden, C. J. & McInnes, R. R. (2000) Nature 406, 195-199. [DOI] [PubMed] [Google Scholar]
- 6.Franke, T. F., Yang, S.-I., Chan, T. O., Datta, K., Kazlauskas, A., Morrison, D. K., Kaplan, D. R. & Tsichlis, P. N. (1995) Cell 81, 727-736. [DOI] [PubMed] [Google Scholar]
- 7.Finkbeiner, S., Tavazoie, S. F., Maloratsky, A., Jacobs, K., Harris, K. M. & Greenberg, M. E. (1997) Neuron 19, 1031-1047. [DOI] [PubMed] [Google Scholar]
- 8.Price, J. H. & Gough, D. A. (1994) Cytometry 16, 283-297. [DOI] [PubMed] [Google Scholar]
- 9.Groen, F. C. A., Young, I. T. & Lighart, G. (1985) Cytometry 6, 81-91. [DOI] [PubMed] [Google Scholar]
- 10.Kenny, L. C. (1983) J. Microsc. 132, 97-107. [Google Scholar]
- 11.Li, Z., Yan, Y., Powers, E. A., Ying, X., Janjua, K., Garyantes, T. & Baron, B. (2003) J. Biomol. Screening 8, 489-499. [DOI] [PubMed] [Google Scholar]
- 12.Ding, G. J. F., Fischer, P. A., Boltz, R. C., Schmidt, J. A., Colaianne, J. J., Gough, A., Rubin, A. & Miller, D. K. (1998) J. Biol. Chem. 273, 28897-28905. [DOI] [PubMed] [Google Scholar]
- 13.Giuliano, K. A., Debiasio, R. L., Dunlay, R. T., Gough, A., Volosky, J. M., Zock, J., Pavlakis, G. N. & Taylor, D. L. (1997) J. Biomol. Screening 2, 249-259. [Google Scholar]
- 14.Ramm, P., Alexandrov, Y., Cholewinski, A., Cybuch, Y., Nadon, R. & Soltys, B. J. (2003) J. Biomol. Screening 8, 7-18. [DOI] [PubMed] [Google Scholar]
- 15.Hack, N. J., Billups, B., Guthrie, P. B., Rogers, J. H., Muir, E. M., Parks, T. N. & Kater, S. B. (2000) J. Neurosci. Methods 95, 177-184. [DOI] [PubMed] [Google Scholar]
- 16.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. (2004) Nature 431, 805-810. [DOI] [PubMed] [Google Scholar]
- 17.Strebel, A., Harr, T., Bachmann, F., Wernli, M. & Erb, P. (2001) Cytometry 43, 126-133. [DOI] [PubMed] [Google Scholar]
- 18.Steff, A.-M., Fortin, M., Arguin, C. & Hugo, P. (2001) Cytometry 45, 237-243. [DOI] [PubMed] [Google Scholar]
- 19.Liu, H.-S., Jan, M.-S., Chou, C.-K., Chen, P.-H. & Ke, N.-J. (1999) Biochem. Biophys. Res. Commun. 260, 712-717. [DOI] [PubMed] [Google Scholar]
- 20.Trachtenberg, J. T., Chen, B. E., Knott, G. W., Feng, G., Sanes, J. R., Welker, E. & Svoboda, K. (2002) Nature 420, 788-794. [DOI] [PubMed] [Google Scholar]
- 21.Alexander, L., Lee, H., Rosenzweig, M., Jung, J. U. & Desrosiers, R. C. (1997) BioTechniques 23, 64-66. [DOI] [PubMed] [Google Scholar]
- 22.Xu-van Opstal, W. Y., Ranger, C., Leujeune, O., Forgez, P., Boudin, H., Bisconte, J. C. & Rostene, W. (1994) Microsc. Res. Tech. 28, 440-447. [DOI] [PubMed] [Google Scholar]
- 23.Dudek, H., Datta, S. R., Franke, T. F., Birnbaum, M. J., Yao, R., Cooper, G. M., Segal, R. A., Kaplan, D. R. & Greenberg, M. E. (1997) Science 275, 661-665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.