Abstract
Although warfarin is the most widely used anticoagulant worldwide, the mechanism by which warfarin inhibits its target, human vitamin K epoxide reductase (hVKOR), remains unclear. Here we show that warfarin blocks a dynamic electron-transfer process in hVKOR. A major fraction of cellular hVKOR is at an intermediate redox state of this process containing a Cys51-Cys132 disulfide, a characteristic accommodated by a four-transmembrane-helix structure of hVKOR. Warfarin selectively inhibits this major cellular form of hVKOR, whereas disruption of the Cys51-Cys132 disulfide impairs warfarin binding and causes warfarin resistance. Relying on binding interactions identified by cysteine alkylation footprinting and mass spectrometry coupled with mutagenesis analysis, we are able to conduct structure simulations to reveal a closed warfarin-binding pocket stabilized by the Cys51-Cys132 linkage. Understanding the selective warfarin inhibition of a specific redox state of hVKOR should enable the rational design of drugs that exploit the redox chemistry and associated conformational changes in hVKOR.
Warfarin is taken by about 1% of the US and UK population to treat and prevent deep vein thrombosis, pulmonary embolism, stroke and myocardial infarction1. Warfarin, however, has a narrow therapeutic window; overdoses can cause severe haemorrhage that is often lethal. Despite extensive clinical experience, the inhibition mechanism of warfarin remains largely unknown owing to the difficulty of studying the interaction of warfarin with its target, hVKOR2,3, an enzyme residing in the endoplasmic reticulum (ER) membrane4–6.
The hVKOR enzyme catalyzes a decisive step in the vitamin K cycle that is required to sustain blood coagulation, bone metabolism7,8, and a variety of other physiological processes9,10. The cycle begins with the γ-carboxylation of selected glutamic acids in vitamin K-dependent proteins such as blood coagulation factors; this post-translational modification is required for their activation at sites of injury. The γ-carboxylase activity results in the epoxidation of vitamin K hydroquinone. The role of hVKOR is to regenerate hydroquinone by reducing vitamin K epoxide (KO) in two steps, first to quinone (K) and then to hydroquinone5,11. Each step of the reduction results in the formation of a Cys132-Cys135 disulfide bond at the active site of hVKOR.
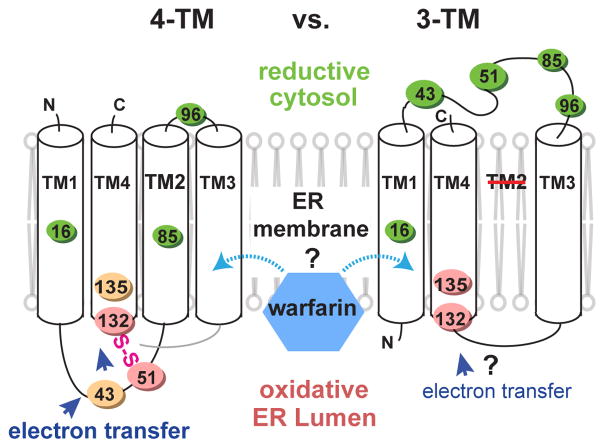
Although the initial and final states are known, the mechanism for restoring reductase activity remains unclear. If hVKOR has four transmembrane helices (TM), as in the crystal structures of its bacterial homolog12, a second pair of conserved cysteines, Cys43 and Cys51, may transfer the electrons12,13 needed to reduce the active site cysteines at the ER luminal surface14,15 (Fig. 1, left). A fundamentally different model, derived from results of several biochemical analyses14,16–18, posits that hVKOR has three TMs, and that Cys43 and Cys51 are in the cytosol with warfarin bound differently (Fig. 1, right).
Figure 1. Alternative models of hVKOR architecture to explain warfarin interaction and electron transfer.
Left, four-TM model. Cys43, Cys51, Cys132, and Cys135 are oxidized (orange and pink colors stand for oxidation levels observed in Fig. 2b) at the ER luminal side. Interactions between these four conserved cysteines permits electron transfer (blue arrows). Right, three-TM model. Cys43 and Cys51 are reduced (green) at the cytosolic side and separated from Cys132 and Cys135, and therefore cannot mediate electron transfer.
The redox cycle and associated structural mechanism of hVKOR are the basis for understanding warfarin inhibition. Warfarin was originally proposed to react with the active site cysteines19 because that tight binding would implicate irreversible inhibition20. It was later suggested, however, that warfarin binds to an oxidized and unreactive form of VKOR, given that pre-reduced VKOR is, surprisingly, less inhibited by warfarin21. In addition, warfarin-resistant mutations (WR) were identified in patients requiring higher warfarin dosage2,22,23 and in rodents resistant to warfarin-derivatized pesticides24–26; these WRs were proposed to cluster around the active site when mapped onto four-TM structures of the bacterial homolog12,27. In this homolog, the active site is surrounded by the TMs and capped by a flexible HL1-2 region (helix and loop between TM1 and TM2) that promotes electron transfer12,28. The crystal structure of hVKOR, however, has not been determined, nor has there been an effective method that can probe the electron transfer and warfarin interaction of hVKOR in vivo.
Here we develop new approaches to resolve the native architecture of hVKOR, elucidate the electron-transfer pathway, and reveal how warfarin inhibits hVKOR catalysis. Using mass spectrometric (MS) analysis to interrogate the redox status of various cysteine residues, we find that a major cellular fraction of hVKOR carries a Cys51-Cys132 disulfide. Because the active site Cys132 is located at the ER luminal surface14,15, the presence of this disulfide bond affirms the luminal location of Cys51 as in the four-TM structure (Fig. 1). Remarkably, warfarin selectively inhibits the Cys51-Cys132 form of hVKOR, stabilizing putative warfarin-binding pocket12 surrounded by known WRs2,22–26. In support of this prediction, alanine-scanning mutagenesis identifies novel WRs clustering around the pocket, with those that directly contact warfarin showing the strongest resistance. This accommodating pocket is formed during electron transfer, and the corresponding conformational state is chosen by warfarin to inhibit effectively hVKOR.
RESULTS
A majority of cellular hVKOR carries a Cys51-Cys132 disulfide characteristic of four-TM topology
The conflicting conclusions of the three-14,16–18 and four-TM29,30 models arise from topology analyses that have used modified variants of hVKOR, strongly suggesting that the native conformation can be perturbed by these modifications31,32. Here we probed the native topology of hVKOR based on the redox status of cysteines in this oxidoreductase. Because stable disulfide bonds are formed in ER, but not in the cytosol33, subcellular location of each cysteine can be revealed by its redox status. In particular, whether Cys43 and Cys51 are located in cytosol or ER is a major feature that distinguishes the three- and four-TM models (Fig. 1).
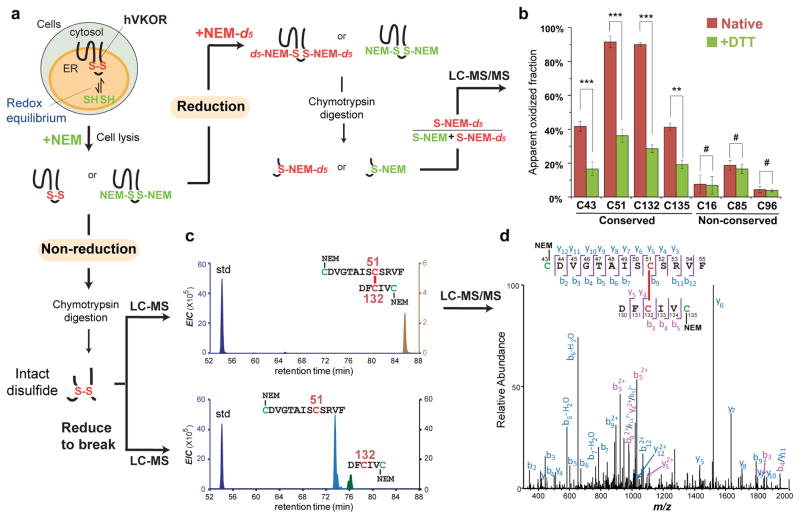
We monitored the redox status of cysteines by a MS-based in-cell footprinting method using isotope-coded N-ethylmaleimide34 (NEM; Fig. 2a and Supplementary Note). Briefly, hVKOR cysteines in reduced form were in-cell labeled by NEM, which can readily permeate the plasma and ER membranes. Subsequently, those cysteines forming disulfide bonds, which were not labeled by NEM, were reduced and then labeled by NEM-d5. Thus, relative fraction of the NEM-d5 label on a cysteine shows its oxidation level.
Figure 2. A major cellular fraction of hVKOR carries a Cys51-Cys132 disulfide bond characteristic of the four-TM topology.
(a) Scheme of MS analysis. Redox status of cysteines is quantified by differential isotope labeling before and after a reduction step (right). Intact disulfide bonds are detected under non-reducing conditions (bottom). (b) Intracellular cysteine status analyzed by quantitative MS before (red) and after DTT reduction (green; **P < 0.001, ***P < 0.0001, and #P > 0.05 by two-tailed Student’s t test). Error bars indicate s.e.m. of multiple peptides from three replicates of independent experiments (see Online Methods). MS and MS/MS spectra can be found in Supplementary Data Set 1. (c) Extracted-ion chromatograms (EIC) show a Cys51-Cys132 disulfide linked peptide (top), which separates into two individual peptides (bottom) after reduction of the disulfide. A reference peptide is used as the standard (std) for peak comparison. (d) MS/MS spectrum of the Cys51-Cys132 linked peptide (from c).
We found that ~90% of Cys51 and Cys132, and 40% of Cys43 and Cys135, are oxidized in the cellular environment (Fig. 2b). In contrast, three non-conserved35 and nonessential36,37 cysteines in hVKOR, Cys16, Cys85, and Cys96, are mostly reduced; these cysteines should not form disulfide bonds because in both topology models they are either in the cytosol or buried in the ER membrane (Fig. 1). To confirm that a high fraction of conserved cysteines is oxidized because they form disulfides (Supplementary Note), we pre-reduced the cells with dithiothreitol (DTT) to show that only the four conserved cysteines are significantly shifted to the reduced form, whereas there is no change for three non-conserved cysteines (Fig. 2b). We also showed that these disulfides are intramolecular bonds because hVKOR is monomeric5 in a non-reducing gel (Supplementary Fig. 1a). All four conserved cysteines are capable of forming disulfide bonds, which indicates that all are located in the relatively oxidizing environment of the ER lumen, as in the 4-TM model (Fig. 1, left).
Because Cys51 and Cys132 are predominantly oxidized (Fig. 2b), the species having a Cys51-Cys132 disulfide represents a major cellular form of hVKOR. Indeed, we found that Cys51 and Cys132 are disulfide-linked in peptides of various lengths, including those containing all conserved cysteines, by accurate mass measurements and MS/MS sequencing (Fig. 2c,d and Supplementary Fig. 1b,c). We confirmed these Cys51-Cys132 disulfide bonds by identifying specific individual peptides after reduction (Fig. 2c). In addition, we made a C43A C135A double mutant to eliminate interference from other possible disulfides, and we found that the Cys51-Cys132 disulfide was preserved (Supplementary Fig. 1d,e). Given that the active site Cys132 is located at the luminal surface14,15, the existence of the Cys51-Cys132 bond necessarily places Cys51 also in the ER lumen, consistent with the four-TM conformation of hVKOR (Fig. 1).
Disulfide exchange between conserved cysteines indicates electron transfer in cellular hVKOR
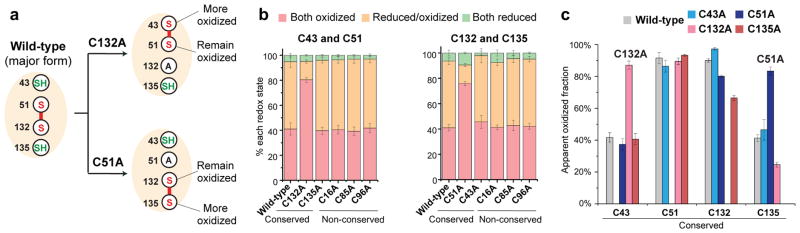
The proposed mechanism of electron transfer to maintain hVKOR activity12 requires disulfide exchange between the four conserved cysteines, with Cys51-Cys132 as an intermediate state (Supplementary Fig. 2a). Our crystallography studies of the bacterial homolog12,28 showed that mutating those conserved cysteines that are essential for electron transfer modified the disulfide bond pattern of the remaining cysteines in a predictable manner (Supplementary Fig. 2b). Using in-cell MS-based quantification, we now report similar redox changes (Fig. 3a) in hVKOR peptides containing either a cysteine pair (Fig. 3b) or a single cysteine (Fig. 3c). The most significant changes occur when the major disulfide, Cys51-Cys132, is disrupted. Mutation of C132A or C51A converts the Cys43 Cys51 or Cys132 Cys135 pair, respectively, from a reduced/oxidized mixture to mainly oxidized (Fig. 3b); this result indicates that each cysteine of this disulfide can form an alternative disulfide when its normal partner is absent. For example, the mutation C132A induces the formation of Cys43-Cys51 disulfide, and the mutation C51A induces the formation of Cys132-Cys135 disulfide (Fig. 3a). Formation of these disulfides can also be inferred from the oxidation state of peptides containing a single cysteine. The C132A mutation increases the oxidation of Cys43 while Cys51 remains highly oxidized (Fig. 3c), consistent with the formation of a Cys43-Cys51 disulfide (Fig. 3a). Similarly, the C51A mutation increases the oxidation level of Cys135 while Cys132 remains oxidized, which is consistent with the formation of a Cys132-Cys135 disulfide. These redox shifts, and a predictable pattern of shifts observed for other cysteine mutants (Supplementary Fig. 3 and Supplementary Note), support a catalytic mechanism that involves electron transfer between the four conserved cysteines in hVKOR.
Figure 3. Redox shifts induced by cysteine mutations support the electron transfer mechanism.
(a) Cysteine mutations are predicted to induce the formation of new disulfide bonds with corresponding oxidative changes. (b) Peptides containing the indicated cysteine pair are monitored. Mutation C132A (left) or C51A (right) increases the oxidation of Cys43 Cys51 or Cys132 Cys135 pairs, respectively. (c) Peptides containing a single cysteine are monitored. The mutation C132A results in predominantly oxidized Cys43 and Cys51 remains oxidized. The mutation C51A results in predominantly oxidized Cys135 and Cys132 remains oxidized (as predicted in a). Error bars, same analysis as in Fig. 2b. MS and MS/MS spectra can be found in Supplementary Data Set 1.
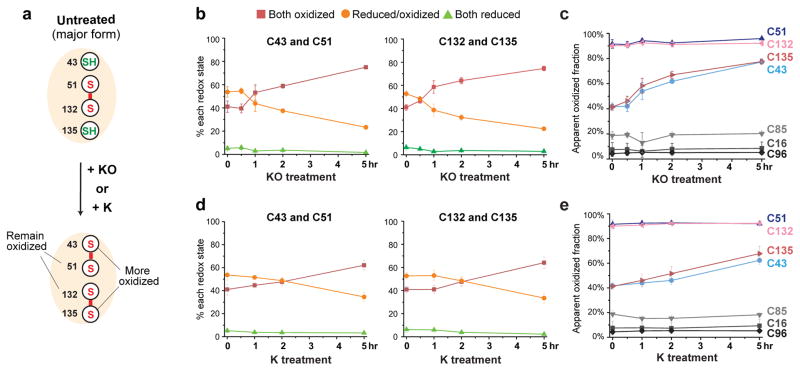
We also tracked electron transfer during hVKOR catalysis by monitoring redox-state changes induced by substrates. Treating the cells with an excess of the KO substrate shifts Cys43 Cys51 and Cys132 Cys135 pairs to a predominantly oxidized state, indicating the progressive formation of two disulfide bonds, Cys43-Cys51 and Cys132-Cys135 (Fig. 4a,b). Consistently, in peptides containing a single cysteine, the proportion of oxidized Cys43 and Cys135 is significantly increased, whereas Cys51 and Cys132 remain oxidized (Fig. 4c). A similar effect is observed with vitamin K (Fig. 4d,e), another hVKOR substrate5,11. In contrast, the three nonconserved cysteines, Cys16, Cys85, and Cys96, are unaffected (Fig. 4c,e). This oxidation pattern is expected because substrate reduction is coupled to cysteine oxidation to form the Cys132-Cys135 disulfide at the active site, followed by a subsequent disulfide exchange to generate the Cys43-Cys51 disulfide (Supplementary Fig. 2a). When these conserved cysteines are mutated, substrate-induced oxidization is blocked for cysteines downstream (Supplementary Fig. 4), because the electron-transfer pathway is hindered. In contrast, electron transfer is not affected by mutations of the three non-conserved cysteines (Supplementary Fig. 4). Taken together, the redox shifts induced by substrates indicate a dynamic process of electron transfer between conserved cysteines in hVKOR.
Figure 4. Dynamic electron transfer during hVKOR catalysis.
(a) KO or K reduction should generate two disulfide bonds, Cys43-Cys51 and Cys132-Cys135, through electron transfer (see this pathway in Supplementary Fig. 2a). (b–c) Redox changes when hVKOR catalyzes KO reduction in cells. Over time, Cys43 Cys51 and Cys132 Cys135 pairs (b) become more oxidized. In peptides containing a single cysteine (c), Cys43 and Cys135 are more oxidized, and Cys51 and Cys132 remain predominantly oxidized (as predicted in a). (d–e) Similar changes are observed when K is provided as substrate. Error bars, same analysis as in Fig. 2b. MS and MS/MS spectra can be found in Supplementary Data Set 1.
Warfarin selectively inhibits hVKOR with the Cys51-Cys132 disulfide
Establishing these redox changes during hVKOR catalysis is essential to understanding warfarin interactions, because in vitro studies suggest that warfarin preferentially inhibits oxidized VKOR21. Taking this as a starting point, we studied the effect of hVKOR redox states on warfarin binding in a cellular environment. We found that binding of warfarin and other vitamin K antagonists inhibits the alkylation of hVKOR by NEM as measured by MS quantification (Supplementary Fig. 5a and Supplementary Note) and also readily detected as a change in electrophoretic mobility (Fig. 5a). Remarkably, we found that reducing cells with DTT hinders warfarin binding to hVKOR, and the order in which DTT and warfarin are added makes a clear difference: if DTT is added first, warfarin does not bind to hVKOR, but if DTT is added after warfarin, then warfarin binding is less affected (Fig. 5b). DTT hinders warfarin binding by disrupting one or more disulfide bonds. Conversely, the presence of disulfide bond(s) in hVKOR is required for warfarin binding and inhibition, both in vitro21 and in cells.
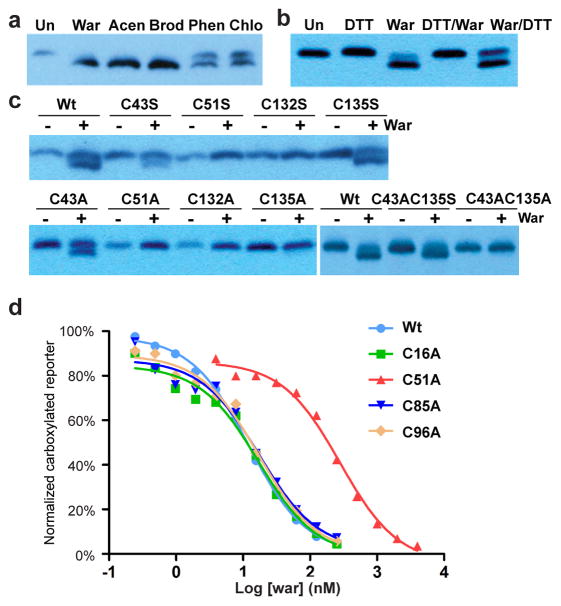
Figure 5. Warfarin selectively inhibits hVKOR with the Cys51-Cys132 disulfide.
(a) Binding of warfarin (War) and other vitamin K antagonists, acenocoumarol (Acen), brodifacoum (Brod), phenprocoumon (Phen), and chlorophacinone (Chlo), induces a gel shift at 5 μM concentration (Un: untreated cells). (b) Warfarin binding (indicated by gel shift) requires disulfide bond(s) in hVKOR. DTT reduction before (DTT/War) or after warfarin treatment (War/DTT) affects warfarin binding differently. (c) Warfarin binding is abolished by Cys51 and Cys132 mutations that disrupt the Cys51-Cys132 disulfide. In addition, C135S (top) allows warfarin binding and C135A (bottom left) does not. The same difference is observed when these Cys135 mutations are combined with C43A to force the formation of a Cys51-Cys132 disulfide bond (bottom right). (d) C51A shows warfarin resistance. Uncropped blot/gel images are shown in Supplementary Data Set 2. Source data for resistance and basal activity are available in Supplementary Data Set 3, 4.
To identify which disulfide bond is important for warfarin binding, we mutated cysteines to alanine or serine, and found that Cys51 and Cys132 mutations abolish warfarin binding (Fig. 5c and Supplementary Fig. 5b,6a). We also found that C135A, but not C135S, impairs warfarin binding (Fig. 5c), suggesting that a hydrogen bond involving Cys135 promotes warfarin binding. In fact, when a Cys51-Cys132 disulfide bond is formed, Cys135 is left unpaired with a free sulfhydryl group (see wild-type hVKOR in Fig. 3a) available for hydrogen bonding. This redox pattern required by warfarin binding is confirmed by comparing the double mutants, C43A C135A and C43A C135S, which eliminate the interference from Cys43 (Fig. 5c). Therefore, warfarin binds a specific redox state of hVKOR with a Cys51-Cys132 disulfide and a reduced Cys135 sulfhydryl, both of which are located at the active site.
Binding to this redox state is further supported by the effects of cysteine mutations on the sensitivity of hVKOR activity to inhibition by warfarin. In particular, the C51A mutant is strongly resistant to warfarin (Fig. 5d), whereas C132A and C135A are catalytically inactive (Supplementary Fig. 6b). Thus, disruption of the Cys51-Cys132 disulfide hinders warfarin inhibition. Taken together, these data show that warfarin, as a potent anticoagulant, achieves effective inhibition by binding selectively to a major cellular form of hVKOR that contains a Cys51-Cys132 disulfide.
The binding orientation of warfarin
To understand the topology of warfarin binding to the Cys51-Cys132 state of hVKOR, we compared warfarin with 4-hydroxycoumarin (4HC), which lacks the phenyl butanone side group of the warfarin structure (Fig. 6a). Although 4HC completely inhibits hVKOR at high concentration, this drug does not induce the gel shift that is characteristic of warfarin binding (Fig. 6b). Clearly, without the side group of warfarin, 4HC cannot maintain certain interactions with hVKOR that influence alkylation by NEM. The effects of 4HC and warfarin on susceptibility to NEM were quantified by MS (Fig. 6c). Compared to the untreated sample, 4HC affects the alkylation of Cys43 on HL1-2 and Cys135 at the active site. In contrast, warfarin affects two additional cysteines, Cys16 on TM1 and Cys85 on TM2. This difference shows that the additional side group of warfarin interacts with the TMs and suggests its 4HC moiety orients towards the active site and HL1-2.
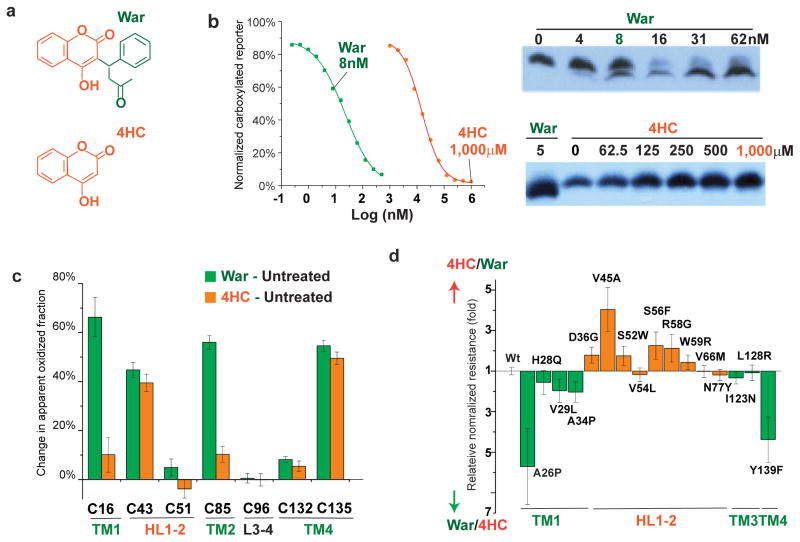
Figure 6. The orientation of warfarin binding to hVKOR.
(a) Chemical structures of warfarin (War) and 4-hydroxycoumarin (4HC), with the 4HC and the warfarin side group shown in orange and green, respectively. (b) Left, Inhibition of hVKOR activity by warfarin and 4HC. Right, 4HC does not induce the gel shift that is associated with warfarin binding. (c) Cysteine footprinting shows that the side group of warfarin interacts with the TMs of hVKOR. Each bar shows the difference in NEM alkylation at specific cysteine residues induced by adding warfarin or 4HC to cells expressing hVKOR (i.e. subtracting untreated from treated for each cysteine). Warfarin significantly changes the alkylation of four cysteines. In contrast, without the warfarin side group, 4HC has much less effect on Cys16 in TM1 and Cys85 in TM2. Error bars, same analysis as in Fig. 2b but with error propagation calculated for the subtraction. (d) Comparison of normalized resistance of hVKOR mutants to 4HC and to warfarin. WRs on TMs (green) and HL1-2 (orange) show stronger resistance to warfarin and 4HC, respectively. The calculation of relative normalized resistance and s.e.m. is described in detail in Online Methods. Uncropped blot/gel images are shown in Supplementary Data Set 2. Source data for resistance and basal activity are available in Supplementary Data Set 3, 4.
This binding orientation of warfarin is consistent with the locations of WRs, which are distributed in the TMs and in HL1-212,28. WRs in HL1-2 induce more resistance to 4HC than to warfarin, whereas WRs on TM1 and TM4 preferentially confer resistance to warfarin (Fig. 6d). This trend supports the conclusion that the 4HC and phenyl butanone side group of warfarin interact primarily with HL1-2 and the TMs of hVKOR, respectively.
Identification of novel WR mutations supports the prediction from MD simulation
To understand how warfarin interacts with VKOR in greater structural detail, we created a structural model of hVKOR that is based on a bacterial homolog (PDB code 4NV528) containing the disulfide corresponding to Cys51-Cys132 (Supplementary Fig. 2b). We initially positioned warfarin in contact with the strong WR residues and ran a 200 ns molecular dynamics (MD) simulation that reached a local equilibrium. The simulation reveals a warfarin-binding pocket that is stabilized by Cys51-Cys132 and surrounded by the naturally occurring WRs2,22–26 (Fig. 7a–c).
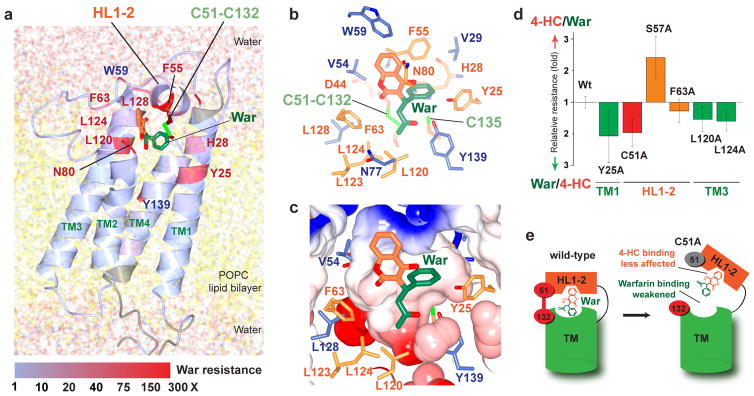
Figure 7. Identification of the warfarin binding pocket stabilized by Cys51-Cys132.
(a) The MD model of warfarin binding to hVKOR is supported by the location and relative strength of warfarin-resistant mutations. The heat representation indicates normalized resistance strengths (colored bar below) of mutations identified from alanine scanning mutagenesis (unscanned region in grey) and from naturally occurring warfarin resistant variants. The Cys51-Cys132 bond stabilizes the conformation of HL1-2. Model coordinates and validation report are available in Supplementary Data Set 5, 6. (b) In this model, warfarin closely interacts with residues that induce strong warfarin resistance when mutated (orange: WRs identified from alanine scans; blue: naturally occurring). (c) A tight warfarin-binding pocket formed by these WR residues (same view as in b). (d) The effects of novel WRs on sensitivity to warfarin and 4HC are similar to the effects of naturally occurring WRs (Fig. 6d), although C51A (red) on HL1-2 deviates from this general trend. Error bars, same analysis as in Fig. 6d. Source data for resistance and basal activity are available in the Supplementary Data Set 3, 4. (e) A cartoon explains the response of the C51A mutant. Left, in wild-type hVKOR, the putative sites that bind the 4HC (orange) and side groups (green) of warfarin are located on HL1-2 and TM region, respectively. Right, Cys51Ala (grey) breaks the Cys51-Cys132 disulfide, resulting in flexible movements that dissociate HL1-2 from the TMs. Because HL1-2 binds the 4HC group, this separation weakens warfarin binding, but has less effect on the 4HC compound. Therefore, the C51A mutant shows more resistance to warfarin than to 4HC.
Through alanine mutagenesis scans of nearly the entire protein, we identified many novel WRs (Supplementary Fig. 7a) that coincide remarkably with the proposed region of warfarin binding (Fig. 7a–c and Supplementary Video 1). The newly identified WRs, predicted by the MD model to contact warfarin directly (e.g., F55A and N80A), are also highly resistant (Fig. 7a), with mutations on HL1-2 and TMs conferring more resistance to 4HC and warfarin, respectively (Fig. 7d). Contrary to the general trend, C51A on HL1-2 imparts more resistance to warfarin, probably because loss of the Cys51-Cys132 disulfide disrupts the warfarin-binding pocket and has less effect on the binding of 4HC because 4HC binds primarily to HL1-2 (Fig. 7e).
DISCUSSION
With the novel MS-based footprinting method, we can monitor the native status of each cysteine in cellular hVKOR. The MS quantification provides direct evidence for the electron-transfer pathway. Moreover, the analysis of disulfide bonds in hVKOR effectively resolves the debate about its folding topology14,16–18,29,30. Furthermore, establishing the catalytic process and architecture of hVKOR provides the basis to understand interactions with warfarin.
The conflicting three or four-TM models derived from previous studies suggest that the native conformation of hVKOR is sensitive to perturbation, such as truncations of TM domains14 or fusions with large reporter proteins16,30. In general, caution should be taken when analyzing proteins with a flexible structure31,38,39, and this example reinforces that caution. Furthermore, measurements of enzymatic activity of modified hVKOR constructs were often not reported together with topology analyses. In contrast, here we probe the active hVKOR without any truncation or large tag. In this native state, the four conserved cysteines in hVKOR form disulfide bonds in the oxidative ER lumen instead of being reduced in cytosol33. In particular, the Cys51-Cys132 disulfide linkage to the luminal active site indicates the luminal location of a Cys51-containing loop, thereby proving the four-TM model (Fig. 1).
The electron-transfer mechanism was proposed from studies of the bacterial VKOR homolog, suggesting that the conserved Cys43 and Cys51 mediates the transfer in hVKOR12. In vitro VKOR activity can be stimulated by reduced thioredoxin40,41, which is a cytosolic protein but can function as an artificial partner42 to transfer electrons. If Cys43 or Cys51 are mutated, thioredoxin cannot maintain hVKOR activity in microsomes13. However, in a cellular assay, Cys51 mutation does not affect much of the hVKOR activity11,16 (Supplementary Fig. 6b), therefore raising the question of whether Cys43 and Cys51 mediates electron transfer in vivo.
To address this question, we tracked the cellular interaction of cysteines by their redox-state changes during catalysis, which are characteristic of the electron-transfer mechanism. With the substrates added to cells (Fig. 4), we found progressive oxidation of Cys132 Cys135 and Cys43 Cys51 pairs through catalysis at the active site and subsequent electron transfer, respectively; conversely, mutations of these conserved cysteines block the progressive oxidation (Supplementary Fig. 4). In absence of substrates, these mutations also change the redox pattern (Fig. 3 and Supplementary Fig. 3) because breaking the existing disulfides leads to the formation of new disulfides; all these disulfides are essentially electron-transfer intermediates (Supplementary Fig. 2a). The intriguing results of C51A mutation can be explained by invoking an additional pathway that also reduces the active site cysteines of hVKOR. Although the relative contribution of this alternative pathway remains unknown, our MS data have now clarified that electron transfer mediated by Cys43 and Cys51 indeed occurs in a cellular environment.
One challenge of warfarin therapy is the significant variation in the effective dose caused by genetic differences among patients, including those carrying resistant mutations. The MD model of warfarin binding pocket (Fig. 7c) is highly predictive of the location of these mutations (Fig. 7a), both for the novel WRs identified by alanine scanning (Supplementary Fig. 7a) and for previously known naturally occurring WRs. Among the strong new WRs, N80A is located on TM2 where no WR has previously been identified; F55A is on HL1-2, suggesting that this region maintains critical interactions with warfarin; Y25A (TM1), L120A, and L124A (TM3) may interact with the side group of warfarin (Fig. 7d); C51A is resistant because the Cys51-Cys132 disulfide is required for warfarin binding. The new WR residues showing strong resistance are highly conserved in eukaryotic VKOR homologs (Supplementary Fig. 7b). Therefore, these residues should form part of a conserved pocket that can bind warfarin. Taken together, the locations of these residues and their interactions with warfarin provide a unified explanation for warfarin resistance: the naturally occurring and novel WR residues are clustered at the luminal side (Fig. 7a) where they maintain the warfarin-binding pocket (Fig. 7c and Supplementary Video 1).
The selective binding of warfarin to the Cys51-Cys132 form of hVKOR explains the puzzling results in which ~ 100-fold higher concentrations of warfarin are required to inhibit the enzyme in vitro2,27,43 than in cell-based assays11,44 or estimated from clinical warfarin dosages22,27. These in vitro studies are based on an assay that initiates the hVKOR catalysis by reducing it with DTT45, which provides electrons to the active site but bypasses12 the normal electron-transfer pathway. In addition, as seen in our in-cell results (Fig. 5b), in vitro pre-reduction with DTT makes hVKOR more resistant to warfarin21. In both settings, the reduction of all disulfides by DTT disrupts the Cys51-Cys132 disulfide that is required for optimal warfarin binding. Therefore, warfarin treatment before or after DTT reduction of hVKOR in vitro gives significantly different levels of inhibition but always less than the extent of hVKOR inhibition in intact cells where a significant fraction of hVKOR exists in the oxidized Cys51-Cys132 state (Fig. 2b).
Selective inhibition of the Cys51-Cys132 state by warfarin can be explained by conformational changes in hVKOR during electron transfer. Our molecular model of hVKOR, validated by novel WRs, is based on the crystal structures of the bacterial homolog12,28, which has been determined for three different electron-transfer states. Our modeling template is the Cys51-Cys132 state, whereas previous modeling of warfarin binding27, without knowing this selective inhibition and the orientation of warfarin binding (Figs. 5, 6), used a structure of the bacterial homolog captured in a different state. Comparison of these states in the bacterial structures reveals large conformational changes in the highly flexible HL1-2 region, which contains a short amphipathic helix that forms a cap over the active site. Part of this helix undergoes a winding and unwinding motion to facilitate electron transfer to the active site. The sequence of this flexible helix is highly conserved between the bacterial homolog and hVKOR12,35, which may, therefore, use a similar structural mechanism. As an intermediate state of the electron transfer (Supplementary Fig. 2a), formation of the Cys51-Cys132 disulfide in hVKOR likely lowers the flexibility of HL1-2 and forces a longer segment of the conserved sequence to become α-helical28, thereby altering the conformation of the active site (Supplementary Fig. 2b and Supplementary Video 2). In the MD model of hVKOR, a specific conformation around the active site, stabilized by Cys51-Cys132, provides a fitting and closed pocket to bind warfarin (Fig. 7c and Supplementary Video 1). Conversely, warfarin is essentially a conformation-specific inhibitor of hVKOR at this Cys51-Cys132 state.
Finally, after more than six decades of administering warfarin, our finding should enable the design of safer anticoagulants or more warfarin antidotes that reverse a hemorrhage event caused by overdoses. Such new drugs may be designed to target specific redox and conformational states of hVKOR, with the goal to prevent warfarin side-effects and to improve treatments for thrombotic and cardiovascular diseases.
ONLINE METHODS
Cell lines
The Vkorc1 (hVKOR) and Vkorc1l1 (hVKOR-like) double-knockout cells containing a chimeric FIXgla-Protein C gene11, derived from HEK 293 cell line, was a gift from D.W. Stafford and J. Tie. The T-REx-293 cells46, derived from HEK293 and used for constructing stable cell lines, was a gift from S. Wanrooij. These cell lines have not been tested for mycoplasma.
Sample preparation for MS quantification of apparent oxidized fraction of cysteines
Stable cell lines were established in 293TRex cells to express wild-type and mutant hVKOR proteins. Cells were grown on 15 cm plates, and each MS sample required the amount of cells from approximately five plates. The cells were washed once with ice-cold phosphate-buffered saline (PBS) buffer. Subsequently, 1.8 mL labeling buffer47,48 was added to each plate; the buffer contains 20 mM NEM, 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, and a protease inhibitor cocktail (Roche). The plates were left on ice for 20 min with frequent shaking to facilitate the NEM reaction with free cysteines. The cell lysate was collected to a new tube, and 200 μL of 1 M DTT (final concentration 100 mM) was added to quench the NEM reaction. This mixture was centrifuged at 4°C for 15 min at 12,000 g. The supernatant was loaded on a G-25 desalting column to remove DTT and NEM. The wild-type or mutant hVKOR proteins, each with a flag tag and ER retention sequence, were immunopurified with anti-flag M2 resin (Sigma), precipitated by trichloroacetic acid, and applied to reducing SDS-PAGE. The protein band of hVKOR was excised and reduced in-gel with 5 mM tris-(2-carboxyethyl)phosphine (TCEP) for 30 min at 56 °C, and the cysteines not labeled by NEM were subsequently modified with 10 mM NEM-d5 isotope for 60 min at room temperature. The labeled protein was subjected to in-gel protease digestion (see below).
Sample preparation for MS detection of disulfide-bonded peptides
To maintain intact disulfide bonds, cells were treated by NEM as described above, but without DTT quenching. Extra NEM was removed from the cell lysate by a G-25 desalting column. Subsequently, the hVKOR protein was immunopurified and applied to non-reducing SDS-PAGE, and the hVKOR band was in-gel digested. After digestion, the peptide solutions were split, and one was reduced with TCEP to separate disulfide-linked peptides. The extracted ion chromatograms of the reduced and corresponding disulfide linked peptides (i.e., untreated) were compared.
In-gel digestion
A previously described protocol49 was modified to reach high sequence coverage that identified all seven cysteines of membrane-bound hVKOR. Briefly, gel pieces were digested overnight at 37°C by 0.02 μg/mL chymotrypsin (Roche) in a buffer containing 50 mM Tris-HCl pH 8.0, 10 mM CaCl2, and 0.1% RapiGest SF surfactant (Waters). The digestion reaction was terminated by treating with formic acid for 30 min at 37 °C, and the peptides released from the in-gel digestion were directly used for MS analysis.
LC-MS/MS analysis
An Eksigent 2D nanoLC system (Eksigent Technilogies, Dublin CA) and a Thermo LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA) were used for LC-MS/MS analysis of the chymotryptic peptides. The parameters were modified from those described previously50. Briefly, the peptides were loaded onto a trap column (Acclaim PepMap100, 100 μm × 2 cm, C18, 5 μm, 100 Å, Thermo Scientific Dionex) at a flow rate of 4.5 μL/min with 0.1% formic acid in water. Subsequently, the peptides were separated on a reversed-phase column (0.0075 mm × 180 mm) packed in house using Michrom Magic C18 material (5 μm particle size and 200 Å pore size). An LC gradient of solvent B (0.1% formic acid in acetonitrile) was delivered at a flow rate of 260 nL/min for 0–10% in 5 min and 10–40% in 95 min. Mass spectra (m/z range 350–2000) of the eluted peptides were acquired at high mass resolving power (50,000 for ions of m/z 400). The seven most abundant ions were selected for fragmentation by collision-induced dissociation in the linear ion trap without charge state rejection.
Criteria for selecting cysteine-containing peptides for the analysis of apparent oxidized fraction
The LC-MS/MS raw data files were converted and searched with the MASCOT search engine against a custom-built database containing wild-type and mutant hVKOR sequences. NEM-cysteine, deuterated-NEM-cysteine and oxidized methionine were selected in custom-built modification profiles. The chymotrypsin digestion generated peptide fragments of variable lengths containing the same cysteine(s), and these peptides were confirmed by the pairwise appearance of NEM- and NEM-d5-labeled species at the same LC retention time. Among the peptides identified by MASCOT, product-ion spectra were manually validated, affording six peptides for Cys16; three peptides each for Cys43, Cys51, Cys96, Cys43 Cys51 pair, and Cys132 Cys135 pair; and one peptide each for Cys85, Cys132, and Cys135. The accurate m/z of all charge states of these peptides (with NEM and NEM-d5 modifications) were consistent with the assignments as listed in Supplementary Data Set 1.
Calculation and statistics of apparent oxidized fraction and percentage of redox states
An ion chromatogram was extracted for each peptide m/z, and the peak area was measured using Xcalibur Qualbrowser. For peptides containing one cysteine, the apparent oxidized fraction was calculated by dividing the peak area of NEM-d5 modified peptide by the summed peak areas of NEM and NEM-d5 modified peptides (i.e., apparent oxidation fraction = NEM-d5/(NEM-d5 + NEM)). For peptides containing two cysteines, the percentage of each redox state (both cysteines oxidized, both reduced, or one reduced/one oxidized) was obtained by dividing the peak area of a given state by the summed peak areas of the three states.
The calculations were conducted for all the validated peptides (see above) containing the same cysteine(s). Peptides from three repeats of completely separate experiments (from sample preparation to MS analysis) were averaged together to give the final values. Therefore, the mean value and s.e.m. are from the combination of 18 peptides (6 peptides × 3 replicates) for Cys16; 9 peptides (3 peptides × 3 replicates) each for Cys43, Cys51, Cys96, Cys43 Cys51 pair, and Cys132 Cys135 pair; and 3 peptides (1 peptides × 3 replicates) each for Cys85, Cys132, and Cys135. Two-tailed Student’s t test is used, with P < 0.01 as significant P values.
Identification of disulfide-bond linked peptides
Disulfide bond-linked peptides were analyzed using the xQuest software51, as reported previously for cross-linked peptides50. The MASCOT search results for the non-reduced and TCEP-treated hVKOR samples were also compared: peptides containing free cysteines were identified only in the TCEP-treated samples, but not in the non-reduced samples. Based on the identification of peptides containing free cysteines, disulfide-bonded peptides were identified and validated by manually examining the product-ion data. An hVKOR peptide (VLGQDSILN) containing no cysteine was used as an internal standard to compare the peak intensities.
Gel-shift assay
Cells expressing wild-type hVKOR and mutants were grown in 6-well plates, and incubated with warfarin or 4HC for 24 h. The cells were washed once with ice-cold PBS, and 120 μL of labeling buffer (20 mM NEM, 50 mM Tris-HCl 7.5, 150 mM NaCl, 1% Triton X-100, and a protease inhibitor cocktail; same as above) was added to each well. The plates were left on ice with shaking for 20 min, and the cell lysates were transferred to a tube. The NEM labeling was quenched by adding 1 M DTT to a 100 mM final concentration. After centrifuging at 12,000 g 4 °C for 15 min, the supernatant was applied to SDS-PAGE under reducing conditions for subsequent western blotting using an anti-flag M2 antibody (Sigma).
Cell-based activity assay and statistics
VKOR activity assays were performed as previously described11. Briefly, hVKOR constructs were cloned into a pBudCE4.1 vector carrying a Metridia luciferase gene. The plasmids were transfected into Vkorc1 (hVKOR) and Vkorc1l1 (hVKOR-like) double-knockout cells containing a chimeric FIXgla-Protein C gene. The carboxylation level of secreted FIXgla-PC was measured by enzyme-linked immunosorbent assay (ELISA) by using the cell-culture medium, with luciferase activity serving as the control for transfection efficiency. To measure IC50s of warfarin and 4HC, hVKOR-transfected cells were treated with warfarin or 4HC at 11 different concentrations. These inhibited activities were normalized to the activity of untreated cells. The IC50s and s.e.m. were obtained from inhibition curves using Prism 5 software.
Comparison of resistance of hVKOR mutants to warfarin and 4HC
The normalized warfarin resistance (nRwar) of hVKOR mutants was obtained as follows. First, the IC50s of wild-type and mutant hVKORs to warfarin were measured by the cell-based activity assay (see above) in the same set of experiments. Next, the IC50s of the mutants were normalized to the IC50 of wild type hVKOR. The normalized 4HC resistance (nR4HC) of hVKOR mutants was obtained using the same approach. The relative normalized resistance of mutants to 4HC and to warfarin was given by the ratio of either nR4HC/nRwar if nR4HC > nRwar, or by nRwar/nR4HC if nRwar > nR4HC. All error propagations were calculated accordingly.
MD simulation
The initial homology model of hVKOR was generated with I-TASSER52 using the bacterial VKOR structure constrained with a disulfide corresponding to Cys51-Cys132 (PDB code 4NV5) as a starting point. The warfarin molecule was docked, with the 4HC group facing HL-12 and the side group facing the TMs, into the hVKOR homology model using Swissdock53. The longer HL1-2 loop in hVKOR was manually adjusted to place strong, naturally occurring WRs in contact with warfarin. The resulting hVKOR-warfarin complex was inserted into a pure POPC membrane bilayer with the CHARMM-GUI online tool54. To simulate the ER membrane, we used a surface area of 68.3 Å2 per lipid for the POPC bilayer. The MD of the hVKOR-warfarin-membrane system was conducted with the GROMACS 4.6.7 package55 and the Charmm36 force field. The force-field parameters for warfarin were generated with ParamChem based on the Charmm FF54. The system was energy minimized using a steepest descent algorithm with a 0.01 nm step size and with a 1.2 nm neighbor list cut-off distance for Coulomb and Van der Waals interactions until the maximum force on any atom in the system fell below 1000 kJ/mol/nm. The system was then equilibrated for 2.5 ns with a 0.001 fs time-step and a Berendsen thermostat to hold the system temperature at 310.15 K. A verlet cut-off scheme was employed for the neighbor list. Particle mesh Ewald was used for the Coulomb interactions. The 200 ns production run was conducted with the Parrinello-Rahman barostat and velocity-rescaling thermostat. LINCS was employed for bond restraints.
Data availability
All MS data that support the findings of this study have been deposited to the ProteomeXchange Consortium under accession code PXD003774. Supplementary data sets for figure 2b, 3b–c, 4b–e, 5d, 6b, 6c–d, 7a, and 7d, and supplementary figure 1d, 3a, 4, 5, 6b, and 7a are available with the paper online.
All other relevant data are available from the corresponding author on request.
Supplementary Material
Supplementary Figure 1. Cys51-Cys132 is an intramolecular disulfide bond.
(a) The wild-type hVKOR protein shows a monomeric band on a non-reducing SDS-PAGE gel. To prevent disulfide shuffling, the cells were treated with NEM to block reactive free cysteines. Uncropped blot/gel images are shown in Supplementary Data Set 2. (b–c) Cys51-Cys132 linked peptides of various lengths (in addition to Fig. 2c,d) have been identified by mass measurements before and after reduction of the disulfide bond (left), and by MS/MS (right). (d) Cys51-Cys132 is preserved in a C43A C135A double mutant, which eliminates alternative ways of forming disulfide bonds between the four conserved cysteines, so that Cys51-Cys132 becomes the only possible form (see inset). Both Cys51 and Cys132 are oxidized in this double mutant, and both can be reduced by DTT (as wild-type hVKOR in Fig. 2b). Error bars, same analysis as in Fig. 2b. MS and MS/MS spectra can be found in Supplementary Data Set 1. (e) A Cys51-Cys132 linked peptide is identified in this double mutant.
Supplementary Figure 2. The electron transfer pathway and associated conformational changes.
(a) Scheme of an electron transfer pathway proposed to regenerate hVKOR activity. The observed oxidation level of each conserved cysteine (Figs. 2b, 3, and 4) reflects a combination of different redox states in this pathway. State I: a pair of free cysteines (green) from a redox partner reduces (blue arrow) the Cys43-Cys51 disulfide (red) in fully oxidized hVKOR. State II to III: electron transfer (blue arrows) generates alternative disulfide bonds (red bars) and Cys51-Cys132 is formed as an intermediate state (III). State IV to V: all cysteines are reduced and hVKOR is catalytically active to reduce KO. State V to I: Reduction of KO (KO to K and K to hydroquinone KH2) generates the Cys132-Cys135 disulfide, and the Cys43-Cys51 disulfide (as in Fig. 4b–e), thereby regenerating State I. (b) Electron transfer is associated with conformational changes in the crystal structures of a bacterial VKOR homolog. Left, A C56A mutant (corresponding to C51A in hVKOR; in parentheses) generates a structure (PDB code: 3KP9) with the Cys130-Cys133 disulfide (Cys132-Cys135 in hVKOR). Right, A C50A mutant (C43A in hVKOR) generates a structure (PDB code: 4NV2 and 4NV5) with the Cys56-Cys130 disulfide (Cys51-Cys132 in hVKOR). Comparison of these two structures shows that the formation of different disulfide bonds (during electron transfer as in a) is associated with conformational changes in HL1-2 (red). In particular, the N-terminal part of the short helix alternates between extended (left) and helical (right) conformations, which directly change the VKOR active site.
Supplementary Figure 3. Cysteine mutations result in a predictable pattern of redox changes in hVKOR.
(a) Redox changes after cysteine mutations (for comparison, Fig. 3c shows only the four conserved cysteines). Error bars, same analysis as in Fig. 2b. MS and MS/MS spectra can be found in Supplementary Data Set 1. (b) Predictions (left) based on a redistribution mechanism (Supplementary Note) fit well with the observed redox changes (right) after cysteine mutations. The P values from two-tailed Student t test (right) are calculated by comparing the changes between mutants and wild-type (observed in a); the values are shown in green for cysteines that become more reduced, in red for those that become more oxidized, and in black for insignificant change s.
Supplementary Figure 4. Cysteine mutations blocks the substrate-induced oxidation through electron transfer.
The KO treatment is 5 h and these mutants are analyzed in the same way as for the wild-type hVKOR in Fig. 4. The red arrows indicate the direction of oxidation and the cross signs indicate mutations. All subsequent oxidation in the pathway are blocked by the mutation of conserved cysteines (right). In contrast, mutation of non-conserved cysteines (left) has no effect. Error bars, same analysis as in Fig. 2b. MS and MS/MS spectra can be found in Supplementary Data Set 1.
Supplementary Figure 5. MS analysis of cysteine protection induced by warfarin binding.
(a) The untreated and warfarin-treated wild-type hVKOR were alkylated with NEM and compared by quantitative MS. Warfarin inhibits the alkylation of four cysteines (**P < 0.001, ***P < 0.0001) in hVKOR. Error bars, same analysis as in Fig. 2b. (b) Cysteine mutations change the pattern of warfarin-induced protection. Wild-type hVKOR (data from a) and each cysteine mutant was monitored for the change in NEM alkylation of individual cysteines induced by warfarin binding. Each bar shows the difference (as in Fig. 6c) between apparent oxidation levels for warfarin-treated samples and untreated samples. Mutations of C51A, C132A, and C135A block warfarin-induced changes (**P < 0.001, ***P < 0.0001; compared with wild type), thereby confirming the gel shift analyses (Fig. 5c) indicating that these mutations interfere with warfarin binding. In contrast, C43A, C16A, C85A, and C96A mutations have insignificant effects. Error bars, same analysis as in Fig. 2b but with error propagation calculated for the subtraction. MS and MS/MS spectra can be found in Supplementary Data Set 1.
Supplementary Figure 6. The Cys51-Cys132 disulfide is important for warfarin binding.
(a) Cysteine mutations were tested for warfarin-induced gel shift as control experiments for Fig. 5c. Uncropped blot/gel images are shown in Supplementary Data Set 2. (b) The activity of cysteine mutants. C132A and C135A are inactive mutants because they delete active site residues. The activity of C43A is also low and does not allow reliable analysis for warfarin resistance in Fig. 5d. Error bars defined in Online Methods. Source data for basal activity are available in Supplementary Data Set 4.
Supplementary Figure 7. Confirming the MD-predicted warfarin-binding pocket by clustering of WRs.
(a) Novel WRs identified by alanine scanning mutagenesis (orange) and naturally occurring WRs (blue) together indicate that the warfarin binding region is located at the luminal portion of the hVKOR protein according to the four-TM model (bars below). These data are shown as a heat map in Fig. 7a. Error bars defined in Online Methods. Source data for resistance and basal activity are available in Supplementary Data Set 3, 4. (b) Strong WR residues are highly conserved in eukaryotic VKOR homolog proteins. The sequences of these proteins were generated from the Psi-Blast search of the human VKOR against eukaryotic proteins in a non-redundant NCBI database. Top 500 sequences from the search were used to plot the sequence logo.
Acknowledgments
We thank D.W. Stafford, J. Tie, and D. Jin from University of North Carolina for their gift of double-knockout cell lines for the activity assay and their generous help in the assay protocol; S. Wanrooij from Umeå University for the gift of T-REx-293 cell line and protocol of establishing stable cell lines; J.T., D.W.S., S. Liu, and T. Ellenberger for critical reading of the manuscript. H.Z. is supported by DOE (DE-SC0001035 to PARC). W. H. is supported by China Scholarship Council (201206235027), G.R.B. is supported by a BWF Career Award. J.E.S. is supported by NIH NHLBI (R01 HL130446 and U54 HL112303). M.L.G. and the MS measurements are supported by NIH NIGMS (P41 GM103422). W.L. is supported by NHLBI (R01 HL121718), AHA (grant-in-aid 14GRNT20310017), and an ASH Scholar Award.
Footnotes
Accession codes. MS data have been deposited to the ProteomeXchange Consortium under accession code PXD003774.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
G.S., H.Z., W.C., J.E.S., M.L.G. and W.L. conceived the studies. G.S. designed and performed experiments with contributions from W.C., W.H., H.Z., Q.L., Y.Y., J.E.S. and W.L.. W.C., G.S., H.Z. and W.L. analyzed the data. F.Z. performed MD simulation with G.R.B.’s help. W.L., G.S., M.L.G. and J.E.S. wrote the manuscript with input from the team.
COMPETING FINANCIAL INTNTERESTS
The authors declare no competing financial interests.
References
- 1.Pirmohamed M. Warfarin: almost 60 years old and still causing problems. Br J Clin Pharmacol. 2006;62:509–11. doi: 10.1111/j.1365-2125.2006.02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 3.Li T, et al. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 4.Tie JK, Stafford DW. Structure and function of vitamin K epoxide reductase. Vitam Horm. 2008;78:103–30. doi: 10.1016/S0083-6729(07)00006-4. [DOI] [PubMed] [Google Scholar]
- 5.Chu PH, Huang TY, Williams J, Stafford DW. Purified vitamin K epoxide reductase alone is sufficient for conversion of vitamin K epoxide to vitamin K and vitamin K to vitamin KH2. Proc Natl Acad Sci U S A. 2006;103:19308–13. doi: 10.1073/pnas.0609401103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hildebrandt EF, Preusch PC, Patterson JL, Suttie JW. Solubilization and characterization of vitamin K epoxide reductase from normal and warfarin-resistant rat liver microsomes. Arch Biochem Biophys. 1984;228:480–492. doi: 10.1016/0003-9861(84)90014-6. [DOI] [PubMed] [Google Scholar]
- 7.Bügel S. Vitamin K and bone health in adult humans. Vitam Horm. 2008;78:393–416. doi: 10.1016/S0083-6729(07)00016-7. [DOI] [PubMed] [Google Scholar]
- 8.Karsenty G, Ferron M. The contribution of bone to whole-organism physiology. Nature. 2012;481:314–20. doi: 10.1038/nature10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkner KL. Vitamin K-dependent carboxylation. Vitam Horm. 2008;78:131–56. doi: 10.1016/S0083-6729(07)00007-6. [DOI] [PubMed] [Google Scholar]
- 10.Oldenburg J, Marinova M, Müller-Reible C, Watzka M. The vitamin K cycle. Vitam Horm. 2008;78:35–62. doi: 10.1016/S0083-6729(07)00003-9. [DOI] [PubMed] [Google Scholar]
- 11.Tie JK, Jin DY, Tie K, Stafford DW. Evaluation of warfarin resistance using transcription activator-like effector nucleases-mediated vitamin K epoxide reductase knockout HEK293 cells. J Thromb Haemost. 2013;11:1556–1564. doi: 10.1111/jth.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, et al. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–12. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rishavy Ma, Usubalieva A, Hallgren KW, Berkner KL. Novel insight into the mechanism of the vitamin K oxidoreductase (VKOR): electron relay through Cys43 and Cys51 reduces VKOR to allow vitamin K reduction and facilitation of vitamin K-dependent protein carboxylation. J Biol Chem. 2011;286:7267–78. doi: 10.1074/jbc.M110.172213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tie JK, et al. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J Biol Chem. 2005;280:16410–16416. doi: 10.1074/jbc.M500765200. [DOI] [PubMed] [Google Scholar]
- 15.Carlisle TL, Suttie JW. Vitamin K dependent carboxylase: subcellular location of the carboxylase and enzymes involved in vitamin K metabolism in rat liver. Biochemistry. 1980;19:1161–7. doi: 10.1021/bi00547a019. [DOI] [PubMed] [Google Scholar]
- 16.Tie JK, Jin DY, Stafford DW. Human vitamin k epoxide reductase and its bacterial homologue have different membrane topologies and reaction mechanisms. J Biol Chem. 2012;287:33945–55. doi: 10.1074/jbc.M112.402941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D, Cousins E, Sandford G, Nicholas J. Human Herpesvirus 8 Viral Interleukin-6 Interacts with Splice Variant 2 of Vitamin K Epoxide Reductase Complex Subunit 1. Journal of Virology. 2012;86:1577–1588. doi: 10.1128/JVI.05782-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tie JK, Jin DY, Stafford DW. Conserved Loop Cysteines of Vitamin K Epoxide Reductase Complex Subunit 1-Like 1 (VKORC1L1) Are Involved in Its Active Site Regeneration. J Biol Chem. 2014;289:9396–9407. doi: 10.1074/jbc.M113.534446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman RB. Model Studies for a Molecular Mechanism of Action of Oral Anticoagulants. J Am Chem SOC. 1981;103:3910–3915. [Google Scholar]
- 20.Fasco MJ, Principe LM. R- and S-warfarin inhibition of vitamin K and vitamin K 2,3-epoxide reductase activities in the rat. J Biol Chem. 1982;257:4894–4901. [PubMed] [Google Scholar]
- 21.Fasco MJ, Principe LM, Walsh Wa, Friedman Pa. Warfarin inhibition of vitamin K 2,3-epoxide reductase in rat liver microsomes. Biochemistry. 1983;22:5655–60. doi: 10.1021/bi00293a031. [DOI] [PubMed] [Google Scholar]
- 22.Watzka M, et al. Thirteen novel VKORC1 mutations associated with oral anticoagulant resistance: insights into improved patient diagnosis and treatment. J Thromb Haemost. 2011;9:109–18. doi: 10.1111/j.1538-7836.2010.04095.x. [DOI] [PubMed] [Google Scholar]
- 23.Rieder MJ, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 24.Rost S, et al. Novel mutations in the VKORC1 gene of wild rats and mice--a response to 50 years of selection pressure by warfarin? BMC Genet. 2009;10:4. doi: 10.1186/1471-2156-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodroge A, Longin-Sauvageon C, Fourel I, Benoit E, Lattard V. Biochemical characterization of spontaneous mutants of rat VKORC1 involved in the resistance to antivitamin K anticoagulants. Arch Biochem Biophys. 2011;515:14–20. doi: 10.1016/j.abb.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Pelz H, et al. The genetic basis of resistance to anticoagulants in rodents. Genetics. 2005;170:1839–47. doi: 10.1534/genetics.104.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czogalla KJ, et al. Human VKORC1 mutations cause variable degrees of 4-hydroxycoumarin resistance and affect putative warfarin binding interfaces. Blood. 2013;122:2743–50. doi: 10.1182/blood-2013-05-501692. [DOI] [PubMed] [Google Scholar]
- 28.Liu S, Cheng W, Fowle Grider R, Shen G, Li W. Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer. Nat Commun. 2014;5:3110. doi: 10.1038/ncomms4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulman S, Wang B, Li W, Rapoport Ta. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci U S A. 2010;107:15027–32. doi: 10.1073/pnas.1009972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Z, et al. The membrane topology of vitamin K epoxide reductase is conserved between human isoforms and the bacterial enzyme. Biochem J. 2016;473:851–858. doi: 10.1042/BJ20151223. [DOI] [PubMed] [Google Scholar]
- 31.Lee H, Kim H. Membrane topology of transmembrane proteins: determinants and experimental tools. Biochem Biophys Res Commun. 2014;453:268–276. doi: 10.1016/j.bbrc.2014.05.111. [DOI] [PubMed] [Google Scholar]
- 32.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 33.Sevier CS, Kaiser Ca. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–47. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 34.Su D, et al. Interactions of Apurinic/apyrimidinic endonuclease with a redox inhibitor: Evidence for an alternate conformation of the enzyme. Biochemistry. 2011;50:82–92. doi: 10.1021/bi101248s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodstadt L, Ponting CP. Vitamin K epoxide reductase: homology, active site and catalytic mechanism. Trends Biochem Sci. 2004;29:2002–2005. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Jin DY, Tie JK, Stafford DW. The conversion of vitamin K epoxide to Vitamin K quinone and vitamin K quinone to vitamin K hydroquinone uses the same active site cysteines. Biochemistry. 2007;46:1–6. doi: 10.1021/bi700527j. [DOI] [PubMed] [Google Scholar]
- 37.Rost S, et al. Site-directed mutagenesis of coumarin-type anticoagulant-sensitive VKORC1: evidence that highly conserved amino acids define structural requirements for enzymatic activity and inhibition by warfarin. Thromb Haemost. 2005;94:780–6. doi: 10.1160/TH05-02-0082. [DOI] [PubMed] [Google Scholar]
- 38.Brach T, et al. Non-invasive topology analysis of membrane proteins in the secretory pathway. Plant J. 2009;57:534–541. doi: 10.1111/j.1365-313X.2008.03704.x. [DOI] [PubMed] [Google Scholar]
- 39.van Geest M, Lolkema JS. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silverman RB, Nandi DL. Reduced thioredoxin: a possible physiological cofactor for vitamin K epoxide reductase. Further support for an active site disulfide. Biochem Biophys Res Commun. 1988;155:1248–54. doi: 10.1016/s0006-291x(88)81274-9. [DOI] [PubMed] [Google Scholar]
- 41.Soute Ba, Groenen-van Dooren MM, Holmgren A, Lundström J, Vermeer C. Stimulation of the dithiol-dependent reductases in the vitamin K cycle by the thioredoxin system. Strong synergistic effects with protein disulphide-isomerase. Biochem J. 1992;281(Pt 1):255–9. doi: 10.1042/bj2810255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preusch PC. Is thioredoxin the physiological vitamin K epoxide reducing agent? FEBS Lett. 1992;305:257–9. doi: 10.1016/0014-5793(92)80681-6. [DOI] [PubMed] [Google Scholar]
- 43.Bevans CG, et al. Determination of the warfarin inhibition constant Ki for vitamin K 2,3-epoxide reductase complex subunit-1 (VKORC1) using an in vitro DTT-driven assay. Biochim Biophys Acta. 2013;1830:4202–10. doi: 10.1016/j.bbagen.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 44.Fregin A, et al. A new cell culture-based assay quantifies vitamin K 2,3-epoxide reductase complex subunit 1 function and reveals warfarin resistance phenotypes not shown by the dithiothreitol-driven VKOR assay. J Thromb Haemost. 2013;11:872–880. doi: 10.1111/jth.12185. [DOI] [PubMed] [Google Scholar]
- 45.Fasco MJ, Principe L. Vitamin K1 hydroquinone formation catalyzed by a microsomal reductase system. Biochem Biophys Res. 1980;97:1487–1492. doi: 10.1016/s0006-291x(80)80033-7. [DOI] [PubMed] [Google Scholar]
- 46.Wanrooij S, Goffart S, Pohjoismäki JLO, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci. 2009;122:4287–4295. doi: 10.1242/jcs.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Toei M, Forgac M. Analysis of the membrane topology of transmembrane segments in the C-terminal hydrophobic domain of the yeast vacuolar ATPase subunit a (Vph1p) by chemical modification. J Biol Chem. 2008;283:20696–20702. doi: 10.1074/jbc.M803258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shevchenko A, Tomas H, Havlis̆ J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes TL - 1. Nat Protoc. 2007;1 VN-re:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, et al. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science. 2013;342:1104–7. doi: 10.1126/science.1242321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walzthoeni T, et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9:901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 52.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. Journal of Computational Chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Cys51-Cys132 is an intramolecular disulfide bond.
(a) The wild-type hVKOR protein shows a monomeric band on a non-reducing SDS-PAGE gel. To prevent disulfide shuffling, the cells were treated with NEM to block reactive free cysteines. Uncropped blot/gel images are shown in Supplementary Data Set 2. (b–c) Cys51-Cys132 linked peptides of various lengths (in addition to Fig. 2c,d) have been identified by mass measurements before and after reduction of the disulfide bond (left), and by MS/MS (right). (d) Cys51-Cys132 is preserved in a C43A C135A double mutant, which eliminates alternative ways of forming disulfide bonds between the four conserved cysteines, so that Cys51-Cys132 becomes the only possible form (see inset). Both Cys51 and Cys132 are oxidized in this double mutant, and both can be reduced by DTT (as wild-type hVKOR in Fig. 2b). Error bars, same analysis as in Fig. 2b. MS and MS/MS spectra can be found in Supplementary Data Set 1. (e) A Cys51-Cys132 linked peptide is identified in this double mutant.
Supplementary Figure 2. The electron transfer pathway and associated conformational changes.
(a) Scheme of an electron transfer pathway proposed to regenerate hVKOR activity. The observed oxidation level of each conserved cysteine (Figs. 2b, 3, and 4) reflects a combination of different redox states in this pathway. State I: a pair of free cysteines (green) from a redox partner reduces (blue arrow) the Cys43-Cys51 disulfide (red) in fully oxidized hVKOR. State II to III: electron transfer (blue arrows) generates alternative disulfide bonds (red bars) and Cys51-Cys132 is formed as an intermediate state (III). State IV to V: all cysteines are reduced and hVKOR is catalytically active to reduce KO. State V to I: Reduction of KO (KO to K and K to hydroquinone KH2) generates the Cys132-Cys135 disulfide, and the Cys43-Cys51 disulfide (as in Fig. 4b–e), thereby regenerating State I. (b) Electron transfer is associated with conformational changes in the crystal structures of a bacterial VKOR homolog. Left, A C56A mutant (corresponding to C51A in hVKOR; in parentheses) generates a structure (PDB code: 3KP9) with the Cys130-Cys133 disulfide (Cys132-Cys135 in hVKOR). Right, A C50A mutant (C43A in hVKOR) generates a structure (PDB code: 4NV2 and 4NV5) with the Cys56-Cys130 disulfide (Cys51-Cys132 in hVKOR). Comparison of these two structures shows that the formation of different disulfide bonds (during electron transfer as in a) is associated with conformational changes in HL1-2 (red). In particular, the N-terminal part of the short helix alternates between extended (left) and helical (right) conformations, which directly change the VKOR active site.
Supplementary Figure 3. Cysteine mutations result in a predictable pattern of redox changes in hVKOR.
(a) Redox changes after cysteine mutations (for comparison, Fig. 3c shows only the four conserved cysteines). Error bars, same analysis as in Fig. 2b. MS and MS/MS spectra can be found in Supplementary Data Set 1. (b) Predictions (left) based on a redistribution mechanism (Supplementary Note) fit well with the observed redox changes (right) after cysteine mutations. The P values from two-tailed Student t test (right) are calculated by comparing the changes between mutants and wild-type (observed in a); the values are shown in green for cysteines that become more reduced, in red for those that become more oxidized, and in black for insignificant change s.
Supplementary Figure 4. Cysteine mutations blocks the substrate-induced oxidation through electron transfer.
The KO treatment is 5 h and these mutants are analyzed in the same way as for the wild-type hVKOR in Fig. 4. The red arrows indicate the direction of oxidation and the cross signs indicate mutations. All subsequent oxidation in the pathway are blocked by the mutation of conserved cysteines (right). In contrast, mutation of non-conserved cysteines (left) has no effect. Error bars, same analysis as in Fig. 2b. MS and MS/MS spectra can be found in Supplementary Data Set 1.
Supplementary Figure 5. MS analysis of cysteine protection induced by warfarin binding.
(a) The untreated and warfarin-treated wild-type hVKOR were alkylated with NEM and compared by quantitative MS. Warfarin inhibits the alkylation of four cysteines (**P < 0.001, ***P < 0.0001) in hVKOR. Error bars, same analysis as in Fig. 2b. (b) Cysteine mutations change the pattern of warfarin-induced protection. Wild-type hVKOR (data from a) and each cysteine mutant was monitored for the change in NEM alkylation of individual cysteines induced by warfarin binding. Each bar shows the difference (as in Fig. 6c) between apparent oxidation levels for warfarin-treated samples and untreated samples. Mutations of C51A, C132A, and C135A block warfarin-induced changes (**P < 0.001, ***P < 0.0001; compared with wild type), thereby confirming the gel shift analyses (Fig. 5c) indicating that these mutations interfere with warfarin binding. In contrast, C43A, C16A, C85A, and C96A mutations have insignificant effects. Error bars, same analysis as in Fig. 2b but with error propagation calculated for the subtraction. MS and MS/MS spectra can be found in Supplementary Data Set 1.
Supplementary Figure 6. The Cys51-Cys132 disulfide is important for warfarin binding.
(a) Cysteine mutations were tested for warfarin-induced gel shift as control experiments for Fig. 5c. Uncropped blot/gel images are shown in Supplementary Data Set 2. (b) The activity of cysteine mutants. C132A and C135A are inactive mutants because they delete active site residues. The activity of C43A is also low and does not allow reliable analysis for warfarin resistance in Fig. 5d. Error bars defined in Online Methods. Source data for basal activity are available in Supplementary Data Set 4.
Supplementary Figure 7. Confirming the MD-predicted warfarin-binding pocket by clustering of WRs.
(a) Novel WRs identified by alanine scanning mutagenesis (orange) and naturally occurring WRs (blue) together indicate that the warfarin binding region is located at the luminal portion of the hVKOR protein according to the four-TM model (bars below). These data are shown as a heat map in Fig. 7a. Error bars defined in Online Methods. Source data for resistance and basal activity are available in Supplementary Data Set 3, 4. (b) Strong WR residues are highly conserved in eukaryotic VKOR homolog proteins. The sequences of these proteins were generated from the Psi-Blast search of the human VKOR against eukaryotic proteins in a non-redundant NCBI database. Top 500 sequences from the search were used to plot the sequence logo.
Data Availability Statement
All MS data that support the findings of this study have been deposited to the ProteomeXchange Consortium under accession code PXD003774. Supplementary data sets for figure 2b, 3b–c, 4b–e, 5d, 6b, 6c–d, 7a, and 7d, and supplementary figure 1d, 3a, 4, 5, 6b, and 7a are available with the paper online.
All other relevant data are available from the corresponding author on request.