Abstract
We created a model of HIV-1 infection of conventional mice for investigation of viral replication, control, and pathogenesis. To target HIV-1 to mice, the coding region of gp120 in HIV-1/NL4-3 was replaced with that of gp80 from ecotropic murine leukemia virus, a retrovirus that infects only rodents. The resulting chimeric virus construct, EcoHIV, productively infected murine lymphocytes, but not human lymphocytes, in culture. Adult, immunocompetent mice were readily susceptible to infection by a single inoculation of EcoHIV as shown by detection of virus in splenic lymphocytes, peritoneal macrophages, and the brain. The virus produced in animals was infectious, as shown by passage in culture, and immunogenic, as shown by induction of antibodies to HIV-1 Gag and Tat. A second chimeric virus based on clade D HIV-1/NDK was also highly infectious in mice; it was detected in both spleen and brain 3 wk after tail vein inoculation, and it induced expression of infection response genes, MCP-1, STAT1, IL-1β, and complement component C3, in brain tissue as determined by quantitative real-time PCR. EcoHIV infection of mice forms a useful model of HIV-1 infection of human beings for convenient and safe investigation of HIV-1 therapy, vaccines, and potentially pathogenesis.
Keywords: animal model, AIDS, brain, vaccine
HIV-1 naturally infects human beings and can also infect a small number of nonhuman primate species (1). This restriction in host range severely limits the animal models suitable for the study of HIV-1 disease development and control. Mice are attractive candidates for HIV-1 research because of the versatility in inbred and genetically engineered strains and the extensive knowledge of the murine immune system. However, mice are not susceptible to HIV-1 and investigation of HIV-1 replication in mouse cell lines in culture was generally disappointing. Blocks at virus entry (2) and after transfection of viral DNA were shown in early studies in NIH 3T3 cells (3), a murine fibroblast cell line. Consistent with these general defects, Rev and Tat, viral proteins originally described in HIV-1, were found to function poorly in NIH 3T3 cells (4, 5). The latter defect was attributed to the inability of murine cyclin T1 to associate with Tat during the formation of a complex with TAR RNA required for efficient transcription (6). In contrast to studies in murine cell lines in culture, animals transgenic for the entire HIV-1 genome driven by the viral LTR expressed virus in skin and lymph nodes (7) or in leukocytes (8), and virus expression could be induced in the animal by physiological stimuli. These studies indicate that primary mouse cells, unlike NIH 3T3 cells, are permissive to late stages of the virus life cycle (8). Consistent with this interpretation, we and others (9, 10) have shown that bypassing restrictions in virus entry through DNA transfection or through pseudotyping permits efficient HIV-1 expression and the production of infectious progeny virus in rat cell lines or primary rat cells in culture. More recently, we have also demonstrated that murine lymphocytes, macrophages, and astrocytes produced viral RNA, protein, and infectious progeny upon infection by HIV-1/NL4-3 pseudotyped by vesicular stomatitis virus envelope glycoprotein G, and similar findings were reported when HIV-1 pseudotyped with murine leukemia virus (MLV) envelope was used (11, 12).
Because primary mouse cells are permissive to HIV-1 expression (11, 12), the principal difficulty in constructing mouse models of HIV-1 infection is inefficient virus binding and entry into murine cells. Models using mice or rats transgenic for the human receptors for HIV-1, CD4, and either CCR5 or CXCR4 were poorly susceptible to HIV-1 infection (13, 14). We adopted a different strategy based on studies of infection in culture by HIV-1 enveloped by heterologous proteins (11, 12, 15). Rather than endow mice with receptors for HIV-1, we constructed HIV-1 species with receptors for mouse cells. As described here, we converted the host species range of HIV-1 from primate to rodent by replacing the coding region of its surface envelope glycoprotein, gp120, with the envelope-coding region from ecotropic MLV that restricts the replication of the virus to rodents (16). We constructed two such chimeric viruses, EcoHIV on a backbone of clade B NL4-3 (17) and EcoNDK on a backbone of clade D NDK (18). The chimeric virus replicated in murine lymphocytes but not human lymphocytes in culture. EcoHIV and EcoNDK established systemic infection in mice after one inoculation. This experimental infection reproduced several major characteristics of HIV-1 infection of human beings, including virus targeting to lymphocytes and macrophages, induction of antiviral immune responses, neuroinvasiveness, and elevation of expression of inflammatory and antiviral factors in the brain. These findings indicate that EcoHIV and similar chimeric viruses can serve as important tools for investigation of HIV-1 disease and intervention in a versatile and convenient animal host.
Materials and Methods
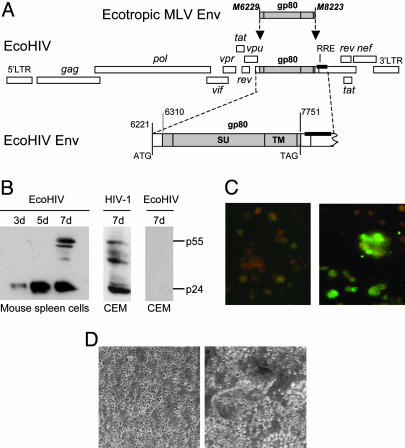
Virus Construction and Preparation. EcoHIV was constructed from pNL4-3 (17) and pNCA-WT (19), kindly provided by Stephen Goff (Columbia University College of Physicians and Surgeons, New York). The chimeric envelope flanked by a 5′ EcoRI site and a 3′ XhoI site was constructed by using PCR amplification with primers spanning the fusion of NL4-3 at nucleotide 6310 and NCA-WT at nucleotide 6229 and NCA-WT at nucleotide 8223 and NL4-3 at nucleotide 7751. This fragment was then ligated into the unique EcoRI and XhoI sites in pNL4-3, generating the chimeric virus (Fig. 1 A). The NCA-WT insert contains the complete sequence of the ecotropic envelope starting at amino acid 2. EcoNDK was similarly constructed by using a plasmid encoding NDK (18) kindly provided by I. Hirsch (Institut National de la Santé et de la Recherche Médicale, Marseille, France). Virus stocks were prepared by transfection of plasmid DNA into 293T cells (20) and titered for p24 content by using the HIV Ag kit (Coulter). Chimeric viruses were washed and resuspended in saline for injection.
Fig. 1.
Structure and replication of EcoHIV. (A) Schematic map of EcoHIV. HIV-1 env (1,405 bp) was excised and replaced by the coding region of the MLV ecotropic envelope gp80 with its stop codon in place. All HIV-1 cis-regulatory elements were preserved and expression of the entire construct was driven by the HIV-1 LTR. (B–D) Replication of EcoHIV in murine lymphocytes in culture. Mitogen-stimulated splenic lymphocytes were infected with EcoHIV. (B) Cultures were sampled over time for p24 expression by immunoblot, compared with human CEM cells infected by HIV-1 or EcoHIV. The amount of p24 detected by ELISA in EcoHIV-infected lymphocyte cultures underestimates the protein detected by Western blotting. (C and D) At 7 d after infection, cells were stained for HIV-1 antigens (C Right) and uninfected cells were stained in parallel (C Left) or cocultured with a 9-fold excess of uninfected cells and examined for syncytia after 2 d (D).
Mice, Cells, and Sample Preparation. All animal studies were conducted with the approval of the St. Luke's–Roosevelt Institutional Animal Care and Use Committee. Adult female BALB/c, 129P3, and 129X1 mice were purchased from The Jackson Laboratory. For inoculation of EcoHIV, mice were anesthetized with isoflurane, and a 0.1-ml solution of virus stock was injected into the tail vein. At killing, mice were anesthetized for bleeding and subjected to carbon dioxide asphyxiation, and macrophages were harvested by peritoneal wash, and spleens, brains, lungs, and kidneys were surgically removed. Spleen cell suspensions were prepared and cultures were established as described (11). Spleen cells were fractionated into CD4-bearing and CD4-negative pools by using Dynabeads (Dynal, Oslo), and fractionation was confirmed by flow cytometry. Splenic lymphocytes were infected in culture with 0.5 pg of p24 EcoHIV per cell and harvested later for immunoblotting or microscopy. CEM cells (transformed human T cells) infected with HIV-1 or EcoHIV served as controls. For PCR, brain, kidney, or lung tissue was weighed and then homogenized by using a disposable pestle. DNA was isolated from cells or tissues by using DNAzol (Invitrogen), precipitated with ethanol, and resuspended in water; RNA was isolated with either TRIzol (Invitrogen) or RNeasy (Qiagen).
PCR. EcoHIV DNA present in 5 × 105 lymphocytes or 1.25 mg of brain, kidney, or lung was amplified by using primers NE5 (5′-ATGATCTGTAGTGCTGCGCGT TCA ACG-3′) and EcoIII (5′-GAGCCGGGCGAAGCAGTACTGACCCCTC-3′) that span the joint between HIV-1 and MLV. Amplification was conducted as described (21), and reaction products were detected by using the 32P-labeled probe EcoP3 (5′-GGTTAACCCGCGAGGCCCCCTAATCCCC-3′). Murine β-globin was amplified in parallel to standardize DNA input. For detection of singly spliced Vif RNA, cDNA was prepared and amplified as described (21) by using as primers nucleotides 616–641 and nucleotides 5071–5091, and radiolabeled probe nucleotides 5127–5156 numbered according to NL4-3.
Quantitative Real-Time RT-PCR. For quantitative real-time RT-PCR, RNA was isolated from brain tissue by using a modified TRIzol protocol optimized to handle the high lipid component of brain homogenates (http://fgc.urmc.rochester.edu). RNA quality was assessed in the Agilent 2100 bioanalyzer. Total RNA (20 ng) was used for whole transcriptome amplification, based on Ribo-SPIAT technology from NuGEN Technologies (San Carlos, CA), to generate cDNA. The size distribution and linearity of amplification was measured before quantitative analysis. Expression of selected genes in the brain was examined by using Taqman chemistry with MGB probes and primers selected from the Applied Biosystems assay on demand program. The relative efficiency of all assays was compared to GAPDH and was within the parameters established for ΔΔCt (threshold cycle) analysis. After probe and primer optimization, all cDNAs were diluted 1:300 after amplification and used as described in ref. 22. Test transcript values were normalized to levels of GAPDH and presented as fold change vs. the levels in control brain samples.
Fluorescence Microscopy. Microscopy was conducted as described (11). Nuclei were stained with propidium iodide. Images were captured by using a Zeiss Axioplan 2 microscope with an ORCA-ER digital camera (Hamamatsu).
Immunoblotting and RIA. Immunoblotting was conducted as described (11). Solid-phase RIAs were constructed by coupling purified recombinant viral proteins (National Institutes of Health AIDS Research Reagent Repository) to Immunolon wells (Dynatech) by using 250 ng of Gag and 500 ng of Tat per well. Dilutions of mouse sera were added to wells, and Ig binding was detected by using 125I-labeled anti-mouse Ig (Amersham Pharmacia Biosciences).
Immunocytochemistry of Brain Sections. At the killing of control and EcoNDK-infected mice, a sagittal portion of the cerebral hemisphere was fixed in 10% neutral-buffered formalin, cut into an average of five coronal sections, dehydrated, and embedded in paraffin, and 6-μm sections were cut for staining and microscopy. Sections were subjected to indirect immunohistochemical staining using a biotin-avidin system as described (23) with rabbit polyclonal anti-STAT1 sc-346 (Santa Cruz Biotechnology) binding detected with diaminobenzidine as a chromogen and hematoxylin/eosin counterstaining. Negative control slides were run omitting the primary anti-STAT-1 antibody. Sections were examined in a Zeiss Photomicroscope III, and images were captured by using a Nikon DN100 digital camera.
Results
Host Range of EcoHIV in Culture. To redirect HIV-1 to infect the rodent, the ecotropic MLV gp80 gene carrying its own stop codon was inserted in-frame into the NL4-3 env gene, preserving the first 90 coding residues, deleting the subsequent 1,440, and resuming HIV-1 near the beginning of the gp41-coding region (Fig. 1A). The resulting chimeric virus, EcoHIV, contains all of the known coding and regulatory regions of the HIV-1 genome with the exception of gp120; gp41 is unlikely to be expressed because it lacks an in-frame codon for initiation of translation. We tested the biological activity of EcoHIV in culture by several approaches. Mitogen-stimulated BALB/c splenic lymphocytes were infected and harvested over 1 wk for analysis of the expression of HIV-1 p24 by Western blotting (Fig. 1B). Transformed human T cells, CEM cells, were exposed to HIV-1 or to EcoHIV as positive and negative controls, respectively. Fully processed p24 increased in amount with time after infection of mouse cells, indicating that it was newly synthesized and properly processed by HIV-1 protease. In contrast, CEM cells were not susceptible to EcoHIV. In similar studies, we infected primary mitogen-stimulated human lymphocytes or transformed mouse cells with EcoHIV and could detect no p24 production (not shown). EcoHIV-infected mouse lymphocytes were also examined for the presence of viral antigens by indirect immunofluorescence staining with AIDS patient serum or for the presence of syncytia during cocultivation with fresh splenic lymphocytes (Fig. 1 C and D). HIV-1 antigens were detected in EcoHIV-infected but not in uninfected mouse lymphocytes, at a frequency of ≈10%, similar to infection of mouse lymphocytes by pseudotyped HIV-1 (11) but less than HIV-1 infection of human lymphocytes in culture. Upon cocultivation, EcoHIV-infected cells formed large syncytia, indicating that gp80 is properly cleaved to fusion-competent proteins. These findings demonstrate that EcoHIV can productively infect primary murine lymphocytes in culture and that it acquired the host range of ecotropic MLV and is unable to infect human cells.
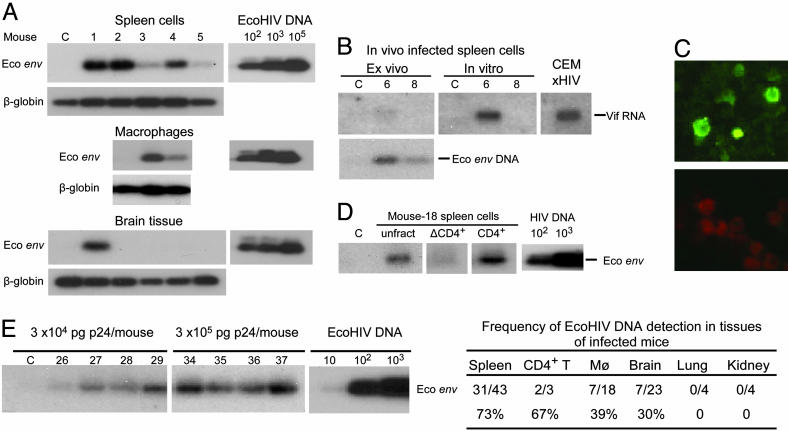
Infection of Mice by EcoHIV. Next we tested whether EcoHIV can also establish infection in vivo, in conventional mice (Fig. 2). Adult immunocompetent 129/P3 mice were inoculated by an i.v. injection of 105 pg of p24 EcoHIV. Six weeks after infection or mock-infection, mice were killed and tissues were collected for analysis (Fig. 2 A). Guided by the cell types infected by HIV-1 in human beings, we tested viral infection in lymphocytes, macrophages, and brain cells by PCR amplification for a region unique to the EcoHIV genome that spans the joint between HIV-1 and MLV. DNA from 5 × 105 spleen cells or 1.25 mg of brain tissue was run in each reaction. The entire sample of 5–10 × 105 peritoneal macrophages from each mouse was subjected to amplification. Viral DNA was detected in one or more tissues of four of the five mice injected. The peak virus burden in the spleen at 6 wk after infection, ≈1 in 1,000 cells carrying viral DNA, is similar to the range of 1 in 200 to 1 in 20,000 HIV-1 DNA-positive cells observed in resting lymphocytes in HIV-1-infected human beings (24). Unlike modest EcoHIV infection in culture, the efficiency of EcoHIV infection in vivo is comparable to that of HIV-1.
Fig. 2.
Conventional mice are susceptible to EcoHIV infection. Mice were inoculated once with EcoHIV and numbered or mock-inoculated and maintained in parallel, labeled as C. (A) Six weeks after infection, mice were killed and DNA from spleen, brain, and macrophages was subjected to PCR, amplifying a region in the chimeric envelope gene unique to EcoHIV; input was standardized by amplification of β-globin. At right is a standard curve of amplification of the indicated number of copies of plasmid DNA. (B) After infection (3 mo), mice were killed, spleens were collected, and either DNA or RNA was isolated or cells were cultured. (Upper) Amplification of singly spliced Vif mRNA in fresh cells or cells after 3-d culture. HIV-1-infected human CEM cells serve as positive controls. (Lower) Amplification of EcoHIV DNA in the same samples. (C) Spleen cells harvested 6 wk after EcoHIV infection were serially cocultured three times with uninfected cells and stained for HIV-1 antigens. (Upper) Infected cells. (Lower) Uninfected cells. (D) Spleen cells harvested 3 mo after infection were subjected to magnetic bead fractionation for CD4-bearing cells before DNA PCR; unfract, unfractionated spleen cells; ΔCD4+, CD4-negative spleen cells. (E) Mice were inoculated once with EcoHIV at the indicated doses, and spleen cells were harvested 6 wk after infection and subjected to DNA PCR. The table at the right summarizes six independent experiments testing the indicated tissues for EcoHIV DNA.
To evaluate ongoing virus replication, newly isolated or cultured spleen cells from 129/P3 mice infected for 3 mo were tested for the expression of singly spliced HIV-1 Vif RNA (Fig. 2B Upper). Spliced RNA was at the limit of detection upon isolation of cells from mouse 6, but culture with mitogen increased virus replication; spliced RNA was not detected in mouse 8; both animals carried viral DNA (Fig. 2B Lower). To evaluate the infectivity of progeny virus produced by cells infected in the animal, spleen cells were harvested 3 mo after infection and serially cocultured with uninfected spleen cells three times. The culture was then assayed for the expression of HIV-1 antigens (Fig. 2C). Brilliantly stained cells were detected at a frequency of 5–10% in cocultures of EcoHIV-infected but not uninfected cells, demonstrating that EcoHIV recovered from mice is infectious.
We also performed limited investigations of the cell and tissue tropism of EcoHIV in the mouse, testing known infected mice, and the dose–response of infection. Splenic lymphocytes were fractionated into CD4-positive and CD4-negative populations before DNA PCR for EcoHIV (Fig. 2D). CD4-positive but not CD4-negative lymphocytes carried viral DNA, and the DNA burden was higher in CD4-positive cells than in unfractionated cells. In all, two of three mice tested carried viral DNA in CD4-positive but not in CD4-negative splenic lymphocytes and none of four mice tested carried viral DNA in lungs or kidneys. The lowest dose of EcoHIV tested, 3 × 104 pg, infected three of four mice tested at 6 wk after infection, and a 10-fold higher dose infected all four mice (Fig. 2E). The table in Fig. 2 shows a summary of the results of virus detection in various tissues from six independent experiments of inoculation of 129/P3 mice with EcoHIV at doses from 3 × 104 pg to 5 × 105 pg of p24 per mouse. EcoHIV was most frequently detected in splenic lymphocytes, but in two mice virus was present in the brain but not the spleen; a total of 33 of 43 mice tested carried EcoHIV DNA in one or more tissues. In recent experiments, we found that mice carrying EcoHIV in splenic lymphocytes also carried viral DNA in peripheral blood lymphocytes, permitting longitudinal studies of virus burden (not shown). Overall, the initiation of spreading infection after a single exposure, replication competence, infectivity of progeny, and tissue distribution of EcoHIV in the mouse reproduce important features of the natural infection of human beings by HIV-1.
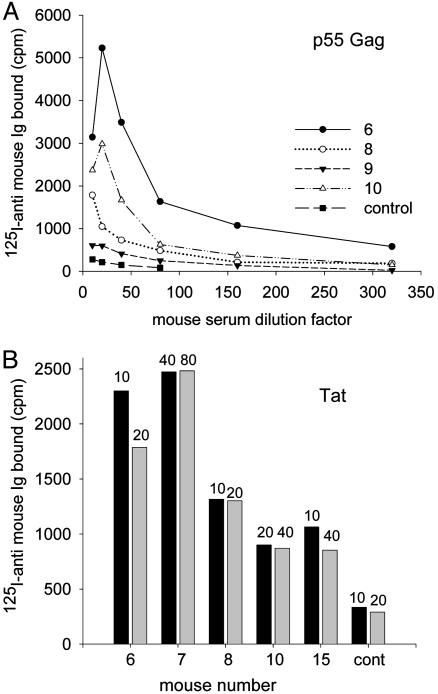
EcoHIV-Infected Mice Mount Antiviral Immune Responses. In natural virus infections, antiviral immune responses generally accompany ongoing virus replication. We therefore tested mice experimentally infected by EcoHIV for humoral immune responses to the major HIV-1 structural protein p55 and the regulatory protein Tat by RIA using mouse sera obtained at killing after 6 or 12 wk of infection (Fig. 3). All of the mice tested that carried viral DNA in the spleen also produced anti-Gag and anti-Tat antibodies (Fig. 3 and data not shown). Mouse 9 was both seronegative and negative for viral DNA (Fig. 3 and data not shown). These findings indicate that EcoHIV replication in mice has two characteristics essential for induction of immune responses: Mice are responsive to HIV-1 antigens, and viral structural and regulatory proteins are produced in sufficient quantity during EcoHIV replication in the mouse to elicit responses.
Fig. 3.
EcoHIV-infected mice produce anti-Gag and anti-Tat antibodies. RIA was conducted by using the dilutions indicated of mouse sera obtained 3 mo after infection (nos. 6–10) or 6 wk after infection (no. 15), and detecting Ig binding by using 125I-labeled anti-mouse Ig. Shown are HIV-1 Gag wells (A) and HIV-1 Tat wells (B). The dilutions of sera tested are indicated at the tops of the bars.
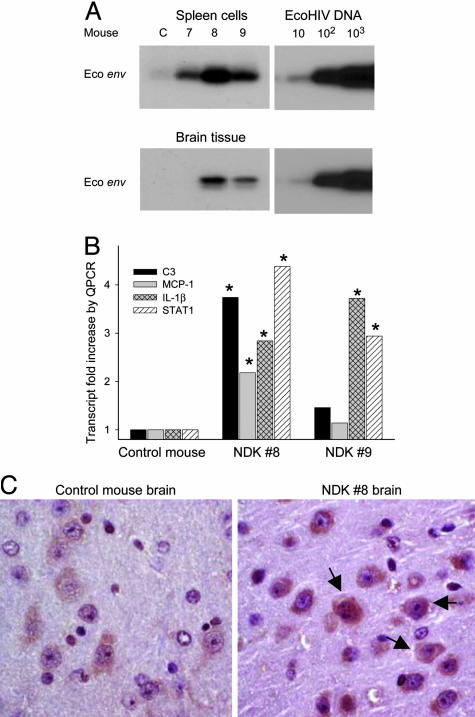
EcoNDK Is Infectious and Neuroinvasive and Alters Cellular Gene Expression in the Brain. To determine the applicability of our approach to the study of different natural HIV-1 species in mice, we constructed EcoNDK with the MLV gp80 gene inserted into NDK, a highly cytopathic clade D HIV-1 (18). EcoNDK contains the gag gene of NDK, which contributes to its high virulence in culture (25) and might enhance its activity in the mouse. Three 129X1 mice were inoculated with 105 pg of p24 EcoNDK, and after 3 wk were killed and spleen and brain were collected. Viral DNA was detected in each mouse spleen, and two of the three mice also carried viral DNA in the brain (Fig. 4A). NDK8 had ≈1 copy of viral DNA per 5,000 lymphocytes, comparable to mice infected by EcoHIV (Fig. 2) and also comparable to HIV-1-infected persons (24). It is also noteworthy that EcoNDK entered and infected mouse brain by 3 wk after inoculation, the earliest time point yet assayed for infection in the brain.
Fig. 4.
EcoNDK infects conventional mice and alters cellular gene expression in the brain. (A) PCR amplification of EcoNDK viral DNA in spleen and brain 3 wk after inoculation; standard curves of amplification of plasmid DNA are at the right. (B) Quantitative real-time RT-PCR was conducted on total cellular RNA from brain tissue of EcoNDK 8 and 9, comparing levels of transcripts with levels found in the control brain. *, Differences compared with control at P < 0.05 by t test. (C) Immunocytochemical staining for STAT-1 in mouse cortical brain sections; arrows indicate examples of more intense staining for STAT-1 in NDK 8 (Right) compared with the control brain (Left). (×360.)
Immune deficiency or neurological impairment by HIV-1 infection of human beings takes years to develop, and we have observed no overt signs of immune dysfunction or histological changes in the brain in EcoHIV-infected mice, most of which were killed 6 wk after inoculation. However, there are molecular markers of cellular abnormalities associated with simian immunodeficiency virus (SIV) infection or HIV-1 infection in the brain, some of which predict later neurological disease (26–29). To investigate subtle changes that may occur early in HIV-1 infection of the mouse, we determined the expression of complement component C3, IL-1β, IL-6, MCP-1, and STAT-1, which are among the factors that influence inflammatory or antiviral responses to HIV-1 in the brain. Quantitative real-time RT-PCR was conducted and expression was normalized to a housekeeping transcript, and the data are reported as fold increase relative to transcript levels in brain tissue of the control mouse (Fig. 4B). NDK 8, which had the highest virus burden in spleen and the brain, showed significant increases in the expression of C3, IL-1β, MCP-1, and STAT-1. Increased expression of C3 was also seen in viral DNA-positive brains from two mice infected by EcoHIV (not shown). NDK 9 had lower levels of EcoNDK in brain and spleen and showed significant increases in the expression of IL-1β and STAT-1, but not in C3 and MCP-1. IL-6 expression was similar in NDK 8, NDK 9, and the control mouse brain (not shown). Because STAT-1 in NDK 8 was the most highly induced transcript observed, we tested STAT-1 protein expression in cortical brain sections of NDK 8 vs. the control mouse (Fig. 4C). STAT-1 protein expression was increased in cytoplasm of neurons in NDK 8 relative to the control but not in other cell types. These findings indicate that the approach to construction of HIV-1 tropic to mice can be generalized to different HIV-1 backbones. Moreover, they indicate that in only a few weeks of infection, chimeric HIV-1 elicits cellular responses in mouse brain like those seen in HIV-1 or SIV infection in the brain (26–29).
Discussion
The mouse model of HIV-1 infection introduced here consists of inoculation of conventional mice with a chimeric HIV-1 that uses species-specific cellular receptors to enter mouse cells. The infection spreads to multiple organs, induces antiviral immune responses, and alters cellular gene expression in the brain. Because the ecotropic envelope that they carry does not mediate entry into human cells, EcoHIV and EcoNDK are less hazardous than are HIV-1 and SIV. These chimeric viruses will be useful for modeling in a small animal host many aspects of HIV-1 infection of human beings.
The infectivity of EcoHIV in murine lymphocytes in culture demonstrates that HIV-1 tolerates the insertion of the MLV envelope-coding region and that the essential cis-regulatory elements and all of the coding regions of HIV-1 are functional. It also indicates that the MLV envelope associates with the HIV-1 core to mediate virus entry, as has been shown in studies of a replication-competent chimera of human T lymphotrophic virus type 1 and ecotropic MLV (30) and a replication-competent chimera of SIV and amphotropic MLV (31). Because HIV-1 protease is active at the cell membrane during virion budding (32) and can cleave MLV p15 to p12 (33), fusogenic p12 is likely to be present at the infected cell surface to mediate the observed cell fusion that is not generally seen with MLV. This gain-of-function may facilitate cell-to-cell transmission of the virus in the mouse. In culture, EcoHIV infects mouse but not human lymphocytes, but only a minority of cells are infected, raising the possibility that EcoHIV targets a cellular subpopulation. Retroviruses carrying the ecotropic MLV envelope have been shown to preferentially replicate in CD4-positive murine lymphocytes in culture with particular mitogens (34). It is possible that EcoHIV has a similar cellular bias upon physiological stimulation in the animal. In the mouse, ecotropic MLV replicates in T lymphocytes with both envelope and the viral LTR contributing to tropism (35, 36) but EcoHIV replicates in macrophages and the brain, as well as in lymphocytes. Neurotropism and neuropathogenesis are common features of lentiviruses, including HIV-1, in part because of their replication in macrophages (37). The LTR present in EcoHIV does not influence HIV-1 tropism in cell culture (38), but it is possible that it affects the EcoHIV host range we observed in the animal. Further research is required to determine the basis for viral infection and expression in different tissues in EcoHIV-infected animals.
One inoculation was sufficient to establish EcoHIV infection in >75% of the mice tested, and viral DNA was detected in the major target cell types of HIV-1. EcoHIV and EcoNDK reached virus burdens in the spleen comparable to HIV-1 burdens in resting lymphocytes in human beings (24), indicating that both viruses are significantly infectious under the conditions used. The extent of infection by EcoHIV was somewhat lower at 12 wk after infection than at 3 or 6 wk (not shown). This decrease could arise from a self-limiting infection or from effective antiviral immunity. The presence of EcoHIV in multiple organs and its transmission from spleen cells in culture indicate that the progeny virus is infectious and not significantly limited in its spread. Because EcoHIV-infected mice consistently produced anti-Gag and anti-Tat antibodies, it is plausible that infected mice mount immune responses that control EcoHIV infection, at least temporarily. This host–virus balance is reminiscent of the first years of HIV-1 infection in human beings while the immune system is intact and the infection is controlled (39). However, in the absence of therapy, the balance later shifts to increased viral replication, loss of immune function, and development of disease in the immune and nervous systems. It is important to investigate the consequences of long-term infection of mice by EcoHIV and EcoNDK to determine whether a similar pathogenic shift takes place. We have investigated only two substrains of 129 mice as hosts of EcoHIV infection, and a recent study of HIV-1-infected human beings indicates that a class I MHC determinant may be pivotal in viral pathogenesis in human beings (40), recommending a broader survey of mouse strains for EcoHIV pathogenesis.
Although no overt disease was detected in mice after several weeks of infection, EcoNDK activated IL-1β, MCP-1, and STAT-1 expression in the brain. The expression of IL-1β and MCP-1 was observed in the brain during SIV-induced neurological disease (28) or in human beings suffering from HIV-1-associated dementia (27), respectively, and each protein can be induced in the brain by viral Tat itself (41, 42). Early induction of MCP-1 in the CNS has been described as a predictor of later brain disease in an SIV model of encephalitis (26). STAT-1 was induced in astrocytes, microglia, and neurons during SIV encephalitis (29), and its over-expression was detected in neurons in the brain of EcoNDK-infected mice. EcoNDK infection of mice thus mimics human or monkey infection by primate lenti-viruses in activation of cellular gene expression in the brain. These potentially pathogenic changes are particularly noteworthy, because EcoHIV and EcoNDK lack gp120 that is thought to mediate some aspects of HIV-1 pathogenesis. Further investigation of the brain cell type(s) infected by EcoNDK and producing pathogenic viral proteins, including Tat is needed, but the observed infection of peritoneal macrophages in the mouse suggests that brain microglial cells are susceptible to infection by the chimeric viruses.
In conclusion, EcoHIV infection of mice reproduces key characteristics of HIV-1 infection of human beings, including host cell range, immune responses, early neuroinvasiveness, and inflammatory and antiviral responses in the brain. Further modification of the EcoHIV construct to increase virulence may tip the balance observed during the early weeks of mouse infection from immune response to immune deficiency. However, it should be clear that the control of EcoHIV infection in the context of anti-HIV-1 immune responses is an excellent starting point for studies of induction of protective immunity by a live virus vaccine. We believe that EcoHIV provides a link between the extensive range of experimental models established in mice and the knowledge base of HIV-1 molecular genetics to investigate HIV-1 infection in a tractable animal host.
Acknowledgments
We thank Dr. Stephen Goff for MLV NCA-WT, Dr. Ivan Hirsch for HIV-1 NDK, Mr. Viraj Sanghvi for technical support on gene expression studies, Dr. Bruce Chesebro through the National Institutes of Health AIDS Research Reagent Repository for hybridoma 183-H12-5C, Dr. Nurith Kurn at NuGEN Technologies for access to the Whole Transcriptome Ribo-SPIA technology used for nucleic acid amplification, Dr. Eran Hadas for illuminating discussions, and Ms. Ilene Totillo for manuscript preparation. This study was supported by Public Health Service Grants DA 14934, DA 17618, and NS 31492 (to D.J.V.) and NS 43110 (to M.J.P.).
Author contributions: M.J.P. and D.J.V. designed research; M.J.P., W.C., G.B., N.P., M.S., J.N., P.B., L.S., A.I.B., and D.J.V. performed research; M.J.P., L.S., A.I.B., and D.J.V. analyzed data; and M.J.P. and D.J.V. wrote the paper.
Abbreviations: MLV, murine leukemia virus; SIV, simian immunodeficiency virus.
References
- 1.Gao, F., Bailes, E., Robertson, D., Chen, Y., Rodenburg, C., Michael, S. F., Cummins, L., Arthur, L. O., Peeters, M., Shaw, G., et al. (1999) Nature 397, 436-441. [DOI] [PubMed] [Google Scholar]
- 2.Maddon, P. J., Dalgleish, A. G., McDougal, J. S., Clapham, P. R., Weiss, R. A. & Axel, R. (1986) Cell 47, 333-348. [DOI] [PubMed] [Google Scholar]
- 3.Levy, J. A., Cheng-Mayer, C., Dina, D. & Luciw, P. A. (1986) Science 232, 998-1001. [DOI] [PubMed] [Google Scholar]
- 4.Zheng, Y. H., Yu, H. F. & Peterlin, B. M. (2003) Nat. Cell Biol. 5, 611-618. [DOI] [PubMed] [Google Scholar]
- 5.Winslow, B. J. & Trono, D. (1993) J. Virol. 67, 2349-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak, Y. T., Ivanov, D., Guo, J., Nee, E. & Gaynor, R. B. (1999) J. Mol. Biol. 288, 57-69. [DOI] [PubMed] [Google Scholar]
- 7.Leonard, J. M., Abramczuk, J. W., Pezen, D. S., Rutledge, R., Belcher, J. H., Hakim, F., Shearer, G., Lamperth, L., Travis, W., Fredrickson, T., et al. (1988) Science 242, 1665-1670. [DOI] [PubMed] [Google Scholar]
- 8.Browning, P. J., Wang, E. J., Pettoello-Mantovani, M., Raker, C., Yurasov, S., Goldstein, M. M., Horner, J. W., Chan, J. & Goldstein, H. (2000) AIDS Res. Hum. Retroviruses 16, 481-492. [DOI] [PubMed] [Google Scholar]
- 9.Mizrachi, Y., Sternas, L. & Volsky, D. J. (1992) Virology 186, 167-174. [DOI] [PubMed] [Google Scholar]
- 10.Keppler, O. T., Yonemoto, W., Welte, F. J., Patton, K. S., Iacovides, D., Atchison, R. E., Ngo, T., Hirschberg, D. L., Speck, R. F. & Goldsmith, M. A. (2001) J. Virol. 75, 8063-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nitkiewicz, J., Chao, W., Bentsman, G., Li, J., Kim, S.-Y., Choi, S. Y., Grunig, G., Gelbard, H., Potash, M. J. & Volsky, D. J. (2004) J. Neurovirol. 10, 400-408. [DOI] [PubMed] [Google Scholar]
- 12.Hinkula, J., Rollman, E., Lundholm, P., Benthin, R., Okuda, K. & Wahren, B. (2004) Cells Tissues Organs 177, 169-184. [DOI] [PubMed] [Google Scholar]
- 13.Sawada, S., Gowrishankar, K., Kitamura, R., Suzuki, M., Suzuki, G., Tahara, S. & Koito, A. (1998) J. Exp. Med. 187, 1439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browning, J., Horner, J. W., Pettoello-Mantovani, M., Raker, C., Yurasov, S., DePinho, R. A. & Goldstein, H. (1997) Proc. Natl. Acad. Sci. USA 94, 14637-14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page, K. A., Landau, N. R. & Littman, D. R. (1990) J. Virol. 64, 5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albritton, L. M., Tseng, L., Scadden, D. & Cunningham, J. M. (1989) Cell 57, 659-666. [DOI] [PubMed] [Google Scholar]
- 17.Adachi, A., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A. & Martin, M. A. (1986) J. Virol. 59, 284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellrodt, A., Barré-Sinoussi, F., Le Bras, P., Nugeyre, M. T., Palazzo, L., Rey, F., Brun-Vezinet, F., Rouzioux, C., Segond, P., Caquet, R., et al. (1984) Lancet i, 1383-1385. [DOI] [PubMed] [Google Scholar]
- 19.Gao, G. & Goff, S. P. (1998) J. Virol. 72, 5905-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canki, M., Thai, J. N. F., Chao, W., Ghorpade, A., Potash, M. J. & Volsky, D. J. (2001) J. Virol. 75, 7925-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury, I. H., Bentsman, G., Choe, W., Potash, M. J. & Volsky, D. J. (2002) J. Neurovirol. 8, 599-610. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S.-Y., Li, J., Bentsman, G., Brooks, A. I. & Volsky, D. J. (2004) J. Neuroimmunol. 157, 17-26. [DOI] [PubMed] [Google Scholar]
- 23.Sharer, L. R., Michaels, J., Murphey-Corb, M., Hu, F. S., Kuebler, D. J., Martin, L. N. & Baskin, G. B. (1991) J. Med. Primatol. 20, 211-217. [PubMed] [Google Scholar]
- 24.Chun, T.-W., Engel, D., Berrey, M. M., Shea, T., Corey, L. & Fauci, A. S. (1998) Proc. Natl. Acad. Sci. USA 95, 8869-8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Mareuil, J., Brichacek, B., Salaun, D., Chermann, J. C. & Hirsch, I. (1992) J. Virol. 66, 6797-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zink, M. C., Coleman, G. D., Mankowski, J. L., Adams, R. J., Tarwater, P. M., Fox, K. & Clements, J. E. (2001) J. Infect. Dis. 184, 1015-1021. [DOI] [PubMed] [Google Scholar]
- 27.Conant, K., Garzino-Demo, A., Nath, A., McArthur, J. C., Halliday, W., Power, C., Gallo, R. C. & Major, E. O. (1998) Proc. Natl. Acad. Sci. USA 95, 3117-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane, T. E., Buchmeier, M. J., Watry, D. D. & Fox, H. S. (1996) Mol. Med. 2, 27-37. [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts, E. S., Zandonatti, M. A., Watry, D. D., Madden, L. J., Henriksen, S. J., Taffe, M. A. & Fox, H. S. (2003) Am. J. Pathol. 162, 2041-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delebecque, F., Pramberger, K., Prévost, M. C., Brahic, M. & Tangy, F. (2002) J. Virol. 76, 7883-7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reiprich, S., Gundlach, B. R., Fleckenstein, B. & Uberla, K. (1997) J. Virol. 71, 3328-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan, A., Manchester, M. & Swanstrom, R. (1994) J. Virol. 68, 6782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiernan, R. & Freed, E. (1998) J. Virol. 72, 9621-9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagani, A., Riviere, I., Tan, C., Krause, A. & Sadelain, M. (1999) J. Gene Med. 1, 341-351. [DOI] [PubMed] [Google Scholar]
- 35.Rosen, C., Haseltine, W. A., Lenz, J., Ruprecht, R. & Cloyd, M. W. (1985) J. Virol. 55, 862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans, L. H. & Morrey, J. D. (1987) J. Virol. 61, 1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patrick, M. K., Johnston, J. B. & Power, C. (2002) J. Virol. 76, 7923-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pomerantz, R. J., Feinberg, M. B., Andino, R. & Baltimore, D. (1991) J. Virol. 65, 1041-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pantaleo, G. & Koup, R. A. (2004) Nat. Med. 10, 806-810. [DOI] [PubMed] [Google Scholar]
- 40.Kieplela, P., Leslie, A., Honeyborne, I., Ramduth, D., Thobakgale, C., Chetty, S., Rathnavalu, P., Moore, C., Pfafferott, K., Hilton, L., et al. (2004) Nature 432, 769-774. [DOI] [PubMed] [Google Scholar]
- 41.Philippon, V., Vellutini, C., Gambarelli, D., Harkiss, G., Arbuthnott, G., Metzger, D. & Filippi, P. (1994) Virology 205, 519-529. [DOI] [PubMed] [Google Scholar]
- 42.Pu, H., Tian, J., Flora, G., Lee, Y. W., Nath, A., Hennig, B. & Toborek, M. (2003) Mol. Cell. Neurosci. 24, 224-237. [DOI] [PubMed] [Google Scholar]