Abstract
Heterogenous nuclear ribonucleoprotein (hnRNP) A1 is an alternative splicing factor that is mainly nuclear, although it shuttles rapidly between nuclear and cytoplasmic compartments. Cells stressed by osmotic shock (OSM) activate the mitogen-activated protein kinase kinase3/6-p38 signaling pathway, which in turn results in accumulation of hnRNP A1 in the cytoplasm. This effect modulates alternative splicing regulation in vivo and correlates with increased hnRNP A1 phosphorylation. We have characterized the molecular mechanism involved in the cytoplasmic accumulation of hnRNP A1 in NIH 3T3 cells subjected to OSM. This treatment results in serine-specific phosphorylation within a C-terminal peptide, dubbed the “F-peptide,” which is adjacent to the M9 motif that mediates bidirectional transport of hnRNP A1. Analysis of mutants in which the F-peptide serines were replaced by aspartic acids or alanines showed that F-peptide phosphorylation is required for the subcellular redistribution of hnRNP A1 in cells subjected to OSM. Furthermore, F-peptide phosphorylation modulates the interaction of hnRNP A1 with transportin Trn1. Our findings suggest that the phosphorylation of F-peptide by cell-signaling pathways regulates the rate of hnRNP A1 nuclear import.
Keywords: shuttling, alternative splicing, stress signaling, transportin, p38 kinase
Heterogenous nuclear ribonucleoprotein (hnRNP) plays an important role in all of the steps of mRNA metabolism (1, 2). The human hnRNP family consists of at least 24 members, which are among the most abundant nuclear proteins (3, 4). hnRNP A1, a member of the hnRNP A/B subfamily, has been studied extensively and participates in the regulation of transcription, splicing, and mRNA export.
hnRNP A1 binds RNA through two RNA recognition motif modules at its N terminus (amino acids 1–196). The C-terminal domain (amino acids 197–320) comprises several RGG repeats, which also contribute to RNA binding. The C terminus also includes a 38-aa sequence, the M9 motif (amino acids 268–305), that is involved in hnRNP A1 nuclear import and export (5–8). Although at steady state hnRNP A1 is predominantly nuclear, it shuttles rapidly between the nucleus and cytoplasm (9). The shuttling of hnRNP A1 is subject to regulation and is thought to play a role in cell proliferation, survival, and differentiation of normal and transformed cells (10). The shuttling of hnRNP A1 also is required for normal myelopoiesis and BCR/ABL leuke-mogenesis (10).
The molecular mechanism that regulates the nuclear export of hnRNP A1 is unknown. In contrast, the mechanism of hnRNP A1 nuclear import is better understood. Two transport receptors of the karyopherin-β family, Trn1 and Trn2b, interact directly with the M9 sequence and mediate hnRNP A1 import (11–13). hnRNP A1 transport requires an intact M9 motif, and a single amino acid change, G274A, abolishes both import and export of the protein (14).
The subcellular distribution of hnRNP A1 is the result of steady-state control of its transport between the nucleus and the cytoplasm. However, it is not clear whether the nuclear accumulation of hnRNP A1 is caused by its nuclear retention or rather is a consequence of a faster rate of nuclear import, compared with its export to the cytoplasm. The former hypothesis is supported by experiments in early mouse embryos showing that nascent transcripts are required to localize hnRNP A1 in the nucleus (15). Moreover, inhibition of RNA polymerase II with actinomycin D results in accumulation of hnRNP A1 in the cytoplasm (9). On the other hand, studies of hnRNP A1 shuttling by a heterokaryon assay showed that its nuclear import is faster than its export (9).
The localization of hnRNPs also is regulated by posttranslational modifications. In budding yeast, the arginine methyl transferase Hmt1p methylates Npl3p and Hrp1p (two hnRNP proteins) and is essential for their export from the nucleus (16). In mammals, hnRNP A1 undergoes several modifications, e.g., methylation, sumoylation, ubiquitination, and phosphorylation (17–21). In vivo, four arginine residues (R193, R205, R217, and R224) within the RGG repeats of hnRNP A1 are methylated. This modification is thought to be constitutive and stoichiometric in HeLa cells and to influence the RNA-binding properties of the RGG repeats (17, 22). A single hnRNP A1 peptide (comprising S199) was characterized as an in vitro substrate of PKC-ζ and PKA kinases (20, 23). Overexpression of either kinase increases the cytoplasmic level of hnRNP A1 (20, 23). Endogenous hnRNP A1 is only very weakly phosphorylated in cells grown under normal conditions (21). However, we showed previously that osmotic shock (OSM) treatment of cells induces cytoplasmic accumulation of hnRNP A1, concomitant with an increase in its phosphorylation (21). We therefore proposed that specific phosphorylation might play a role in the regulation of the hnRNP A1 nucleocytoplasmic distribution.
Here, we show that mammalian cells subjected to OSM exhibit an increase in serine-specific phosphorylation of hnRNP A1. We mapped the site(s) of phosphorylation to the C-terminal domain, between amino acids 301 and 318, a segment dubbed the “F-peptide.” Mutation of the serines in the F-peptide to aspartic acids, mimicking a hyperphosphorylated state, resulted in accumulation of hnRNP A1 in the cytoplasm, even without OSM. In contrast, mutation of all of the F-peptide serines to alanines abolished OSM-induced phosphorylation of hnRNP A1 and resulted in its nuclear retention. Furthermore, we found that OSM weakens the interaction between hnRNP A1 and Trn1. In vitro and in vivo assays of transportin interactions with hnRNP A1 mutant proteins suggest that hnRNP A1/Trn1 binding is negatively regulated by OSM-induced phosphorylation. Thus, the cytoplasmic accumulation of phosphorylated hnRNP A1 in cells stressed by OSM probably results from its impaired nuclear import.
Materials and Methods
Plasmids. The mammalian expression plasmid pCGT-A1 carries a T7-tagged version of hnRNP A1 (24). The F1 and F2 mutants were generated by PCR and inserted into the pCGT plasmid (XbaI–BamHI) by using the following reverse primers: F1 (5′-ccatcgggatccTTAAAATCTTCTGCCgtcGCCATAatcgtcatcgtcatcgtcACCGCCATAGCCACCTTGGTTTCGTGG-3′) and F2 (5′-ccatcggaattcTTAAAATCTTCTGCCagcGCCATAtgcagcggcagctgcggcACCGCCATAGCCACCTTGGTTTCGTGG-3′). Plasmids expressing GST fusions to hnRNP A1, or to the F1 and F2 mutants, were generated by PCR and subcloning into the pGex-5 × 1 vector (Amersham Pharmacia). The hnRNP A1 cDNA was inserted into the BamHI site, and the F1 and F2 mutants were inserted between the BamHI and EcoRI sites. All constructs were verified by sequencing.
[32P]-Labeling, Phosphoamino Acid Analysis, and Immunoprecipitation. For metabolic labeling, 500 μCi/ml (1 Ci = 37 GBq) 32Pi was used per 60-mm dish of proliferating NIH 3T3 cells. The cells were preincubated in phosphate-depleted medium for 1 h before addition of 32Pi. OSM involved addition of 600 mM sorbitol for 3 h. Cell lysates were made with RIPA buffer (0.15 M NaCl/0.05 M Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) containing 10 μg/ml leupeptin and aprotinin, 1 mM PMSF, 0.25 mM orthovanadate, 20 mM β-glycerophosphate, and 10 mM sodium fluoride. Endogenous proteins were immunoprecipitated with mAbs 4B10 (hnRNP A1), MC3 (U2AF65), or 7–53 [protein phosphatase 2Cγ (PP2Cγ)] (46). The 4B10 and MC3 antibodies were kindly provided by G. Dreyfuss (University of Pennsylvania School of Medicine, Philadelphia) and J. Valcárcel (Centre for Genomic Regulation, Barcelona), respectively (25, 26). The immunoprecipitated proteins were separated by SDS/PAGE and transferred onto a polyvinylidene difluoride membrane. Detection of hnRNP A1 by Western blotting was with mAb A1/55, which recognizes both hnRNP A1 and its isoform A1B (L. Manche and A.R.K., unpublished data). Phosphoamino acid analysis was done by using the method of Kamps (http://pingu.salk.edu/~sefton/Hyper_protocols/Paa.html).
Cell Fractionation. Nuclear and cytoplasmic extracts were prepared from four 60-mm dishes of NIH 3T3 cells at ≈60% confluence. The cells were collected by scraping in PBS plus 1 mM EDTA and spun at 250 × g for 5 min at 4°C. The packed cells were resuspended in six volumes of buffer A (10 mM Hepes, pH 7.9/50 mM NaCl/0.5 M sucrose/0.1 mM EDTA/0.5% Triton X-100/1 mM DTT/17.5 mM β-glycerophosphate/100 mM sodium fluoride/10 μg/ml leupeptin/4 μg/ml aprotinin) and incubated on ice for 5 min. The cells were centrifuged at 230 × g for 10 min, and the supernatant (cytoplasmic extract) was cleared by centrifugation at 15,300 × g for 15 min. The pellet (nuclei) was washed with six volumes of buffer B (10 mM Hepes/10 mM KCl/0.1 mM EDTA/0.1 mM EGTA/1mMDTT plus protease inhibitors) and spun in a swinging-bucket centrifuge at 160 × g for 5 min. The nuclei were resuspended in four volumes of buffer C (10 mM Hepes/500 mM NaCl/0.1 mM EDTA/0.1 mM EGTA/0.1% Nonidet P-40/1 mM DTT plus protease inhibitors), and the nuclear proteins were extracted by vortexing at 4°C for 15 min. The nuclear extract was clarified by centrifugation at 15,300 × g for 10 min. The cytoplasmic fraction and nuclear extract were adjusted to 10% (vol/vol) glycerol and frozen at -80°C.
MALDI-TOF MS. hnRNP A1 was immunoprecipitated with a mix of mAbs 4B10 and 9H10 (25) and analyzed by SDS/PAGE. The gel was stained with GelCode Blue Stain Reagent (Pierce), and the band corresponding to hnRNP A1 was excised for in-gel trypsin digestion. The digests were concentrated, and the resulting peptides were mixed with α-cyano-4-hydroxycinnamic acid and analyzed by MALDI-TOF MS (Voyager-DE RP, Applied Biosystems). The spectral data were analyzed by using the profound search engine (27).
Purification of GST Recombinant Proteins and Pull-Down and Coimmunoprecipitation Assays. GST fusion proteins were expressed in Escherichia coli BL21 and purified on glutathione-Sepharose beads (Amersham Pharmacia). Each purified protein was dialyzed against buffer D [20 mM Hepes-KOH, pH 8.0/100 mM KCl/0.2 mM EDTA/20% (vol/vol) glycerol/0.5 mM PMSF/1 mM DTT)]. [35S]methionine-labeled Trn1 was synthesized by using the plasmid pET28b-Trn1 (kindly provided by Rui-Ming Xu, Cold Spring Harbor Laboratory) and a coupled reticulocyte lysate system (TNT T7, Promega). For protein-binding assays, 2 μg of GST recombinant protein was incubated for 15 min at 4°C with 15 μl of glutathione-Sepharose in 500 μl of binding buffer [20 mM PBS, pH 7.4/1% (vol/vol) Triton/1 mM PMSF]. The beads were washed twice with binding buffer and once with buffer E [20 mM Hepes, pH 7.9/250 mM KCl/0.05% (vol/vol) Nonidet P-40/100 μg/ml BSA/1 mM PMSF]. The beads were incubated with 4 μl of [35S]methionine-labeled Trn1 in 250 μl of buffer E for 1 h at 4°C, washed three times for 15 min in buffer E, and boiled in SDS/PAGE sample buffer. The bound proteins were separated by SDS/PAGE, and the labeled Trn1 was detected by using autoradiography and quantitated by using a phosphorimager (BAS2000, Fuji).
Immunofluorescence Microscopy. Immunofluorescence microscopy was carried out as described in ref. 21. HeLa cells were transfected by electroporation with plasmids expressing T7-tagged proteins and cultured on glass coverslips. At 24–36 h posttransfection, cells were either left untreated or exposed to 600 mM sorbitol for 2.5 h, fixed with 4% p-formaldehyde in PBS for 30 min, and permeabilized with 0.2% Triton X-100 for 5 min. The T7-tagged proteins were detected as described in ref. 24.
Immunoprecipitation and Immunoblotting. To analyze the interaction between Trn1 and hnRNP A1, cytoplasmic fractions from control or stressed HeLa cells were prepared as described in ref. 33. Cells were resuspended in RSB100 (10 mM Tris·HCl, pH 7.4/2.5 mM MgCl2/100 mM NaCl) containing 35 μg/ml digitonin, 10 mM β-glycerophosphate, 10 mM NaF, and a protease inhibitor mixture (Roche). The suspension was incubated on ice for 5 min and disrupted by repeated passage through 25-gauge needles. Centrifugation at 4,000 × g for 15 min yielded a cytoplasmic supernatant fraction. Endogenous or T7-tagged hnRNP A1 and mutants were immunoprecipitated from this fraction by incubation for 15 min at 4°C with mAb 4B10 linked to protein A-Sepharose beads. After extensive washing with RSB100, the bound fraction was eluted by boiling in SDS/PAGE sample buffer. Equivalent amounts of immunoprecipitated hnRNP A1 proteins were separated by SDS/PAGE, transferred to a polyvinylidene difluoride membrane, and probed with the following antibodies: 4B10 (1:10,000) and D45 (1:1,000) from BD Transduction Laboratories, anti-T7 tag (1:1,000) from Novagen, or anti-phosphoserine (1:100) from Abcam (Cambridge, U.K.).
Results
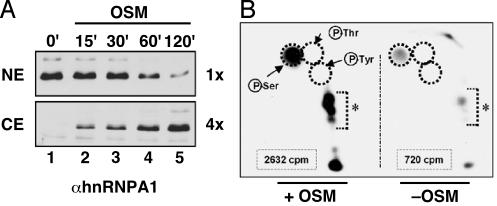
OSM Induces Serine-Specific Phosphorylation of hnRNP A1. We previously demonstrated by using immunofluorescence microscopy that NIH 3T3 cells subjected to OSM in 600 mM sorbitol undergo cytoplasmic mislocalization of endogenous hnRNP A1 (21). To address what fraction of the endogenous hnRNP A1 relocalizes in response to OSM stress, we performed cell fractionation at different time points after OSM treatment and analyzed the distribution of hnRNP A1 in nuclear and cytoplasmic extracts (Fig. 1A). Western blot analysis showed a shift of hnRNP A1 from the nuclear to the cytoplasmic fractions, in agreement with our immunofluorescence results described in ref. 21. Furthermore, >90% of hnRNP A1 was detected in the nucleus without OSM treatment (Fig. 1 A, lane 1), whereas >50% accumulated in the cytoplasm after 120 min of OSM treatment (Fig. 1 A, lane 5). Therefore, this stress stimulus has a major effect on the endogenous hnRNP A1 pool.
Fig. 1.
Subcellular distribution and specific phosphorylation of hnRNP A1 in cells subjected to OSM. (A) Nuclear (NE) and cytoplasmic (CE) extracts were prepared from NIH 3T3 cells treated with 600 mM sorbitol for the indicated times. The distribution of endogenous hnRNP A1 was detected by Western blotting with mAb A1/55 by using a 4-fold excess of CE over NE total protein. (B) Phosphoamino acid analysis of metabolically labeled hnRNP A1 by 2D electrophoresis after immunoprecipitation from cells treated with OSM or untreated. Equal amounts of hnRNP A1 (estimated by Ponceau red staining) were excised, quantitated by Cerenkov counting (indicated for each condition), and processed for phosphoamino acid analysis. The positions of phosphoserine (P-Ser), phosphotyrosine (P-Tyr), or phosphothreonine (P-Thr) standards are shown as dotted circles. *, Partially digested peptides.
The cytoplasmic accumulation of hnRNP A1 in OSM-treated cells correlates with an increase in its phosphorylation (21). We have now determined the specificity of this phosphorylation. We compared hnRNP A1 phosphorylation with that of two other nuclear mRNA-processing factors, PP2C-γ and U2AF65, which can be immunoprecipitated from cell lysates (26, 28, 46). Phosphorylation of hnRNP A1 was greatly enhanced in OSM-treated cells, whereas PP2C-γ and U2AF65 were unaffected (data not shown). Both hnRNP A1 and U2AF65 are shuttling proteins (9, 28), whereas PP2C-γ is nuclear (46). Therefore, OSM-induced phosphorylation does not target all shuttling or nuclear splicing factors.
To identify which residues undergo OSM-induced phosphorylation, we performed phosphoamino acid analysis of [32P]-labeled hnRNP A1 immunopurified from treated or untreated cells. Our 2D chromatography of equal amounts of hydrolyzed protein showed that hnRNP A1 is phosphorylated on serine under both conditions (Fig. 1B). OSM treatment resulted in a strong increase in the level of serine phosphorylation, and no radioactivity was detected corresponding to either phosphothreonine or phosphotyrosine.
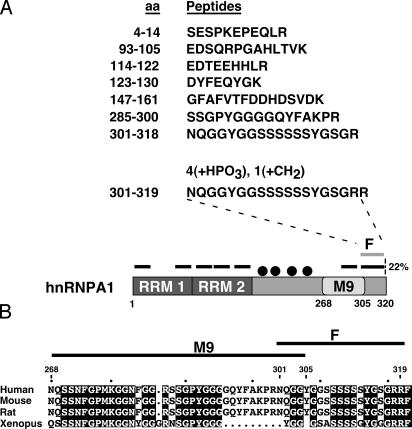
Identification of hnRNP A1 Phosphopeptide (F-Peptide) in OSM-Treated Cells. To map the OSM-specific phosphorylation site(s), we used MALDI-TOF and compared the spectra of hnRNP A1 purified from untreated vs. OSM-treated cells to detect posttranslationally modified peptides. Theoretically, trypsin digestion of hnRNP A1 can produce 39 peptides, 14 of which have sizes that can be detected by MALDI-TOF. In practice, however, we recovered only seven peptides after exhaustive trypsin digestion of immunopurified hnRNP A1, accounting for 22% of the hnRNP A1 sequence (Fig. 2A). Among the missing peptides, four cannot be generated by trypsin digestion because of hnRNP A1 arginine methylation in HeLa cells at positions R193, R205, R217, and R224 (17, 29, 30). Methylated arginines are not cleaved by trypsin, resulting in larger peptides that are not detected by MALDI-TOF. In cells treated by OSM, we found an additional peptide corresponding to an extreme C-terminal fragment of hnRNP A1 (amino acids 301–319, F-peptide) that, according to its mass, underwent posttranslational modification with covalent addition of four phosphates and one methyl group. Moreover, we also detected a shorter, unmodified version of the F-peptide (amino acids 301–318) in OSM samples, suggesting that at least two forms of hnRNP A1, with and without modification of the F-peptide, are present in OSM-treated cells. The F-peptide comprises six consecutive serines (S308–S313), an additional serine (S316), and two consecutive arginines (R318 and R319). We conclude that four of the seven serines and almost certainly one of the two arginines are modified by phosphorylation and methylation, respectively. The MALDI-TOF analysis does not allow us to determine the precise four serine residues that undergo phosphorylation, and it is possible that there is heterogeneity in terms of which of the six consecutive serines undergo modification. On the other hand, given that trypsin cleaved after R319 to generate the modified F-peptide, we conclude that R318 is the likely site of arginine methylation. Alignment of the C-terminal protein sequences from several hnRNP A1 orthologs (Fig. 2B) shows that both the M9 and F-peptide sequences are highly conserved in vertebrates.
Fig. 2.
hnRNP A1 peptides identified by MALDI-TOF. (A) The identified peptides are listed with their coordinates in the 320-aa hnRNP A1 sequence. The positions of the seven peptides also are shown as black bars over a diagram of the hnRNP A1 protein, and the modified F-peptide is shown as a gray bar. The four black dots represent the methylated arginines R193, R205, R217, and R224. (B) Comparative alignment of the M9 sequence (268–305) and the F-peptide (301–319). The C termini of several vertebrate hnRNP A1 orthologs were aligned by using clustalw (www.ebi.ac.uk/clustalw). Residues identical in all four species are highlighted.
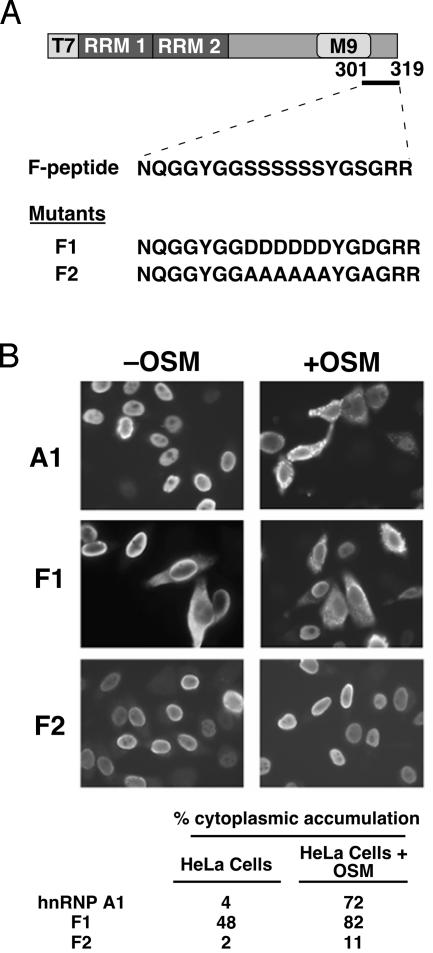
Serines Within F-Peptide Are Involved in Localization and Phosphorylation of hnRNP A1. To determine whether there is a functional link between OSM-induced cytoplasmic accumulation of hnRNP A1 and phosphorylation of serines in the F-peptide, we generated two mutant versions of hnRNP A1 in which all of the serines in the F-peptide were replaced either by aspartic acids (mimicking phosphorylation) or alanines (preventing phosphorylation) (Fig. 3A). HeLa cells were transfected with T7-tagged versions of wild-type or mutant hnRNP A1 cDNAs, followed by immunofluorescence detection of the tagged proteins to determine their subcellular localization (Fig. 3B). T7-tagged wild-type hnRNP A1, as a control, exhibited the same subcellular distribution in OSM cells as observed for endogenous hnRNP A1 in ref. 21; OSM resulted in cytoplasmic accumulation of T7-hnRNP A1 in 72% of transfected cells. The mutant mimicking constitutive phosphorylation of the F-peptide (F1 mutant) localized in the cytoplasm, even without OSM treatment. However, this effect was only observed in 48% of the transfected cells and increased after OSM treatment (to 82%), which may reflect competition with endogenous hnRNP A1 for binding to cytoplasmic transportin. In contrast, mutation of the serines to alanines (F2 mutant) strongly inhibited the OSM effect on hnRNP A1 localization, because only 11% of the transfected cells accumulated the F2 mutant protein in the cytoplasm.
Fig. 3.
Localization of transfected wild-type or mutant hnRNP A1. (A) The domain structure of T7-tagged hnRNP A1 and F1/F2 mutants is shown schematically. The mutations of serines (S308–S313 and S316) to aspartic acids (F1) or alanines (F2) in the F-peptide are shown. (B) Immunofluorescence micrographs showing the localization of T7-hnRNP A1, T7-F1, and T7-F2 in HeLa cells, with or without OSM treatment for 2.5 h. The tabulated data indicate the percentage of transfected cells in which the tagged proteins accumulated in the cytoplasm. Approximately 500 cells were counted for each condition.
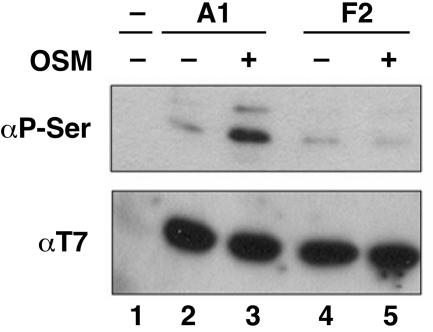
Our data showed that phosphorylation of the F-peptide was sufficient for the cytoplasmic accumulation of hnRNP A1 even in the absence of stress stimuli, although the effect was seen in a reduced percentage of transfected cells (48% vs. 82%; see below). We suggest two possible explanations for this effect not being quantitative. First, MS did not provide full-sequence coverage of hnRNP A1, so there might be additional OSM-induced phosphorylation site(s) and partial functional redundancy between these sites. Second, other events besides phosphorylation also might modulate hnRNP A1 localization. To address the first possibility, we immunoprecipitated hnRNP A1 and the F2 mutant version from OSM-treated and untreated cells and measured their phosphoserine level (Fig. 4). The T7-tagged hnRNP A1 behaved in the same manner as endogenous hnRNP A1 (compare Figs. 1B and 4, lanes 2 and 3) and showed increased phosphorylation in cells subjected to OSM. In contrast, the F2 mutant remained hypophosphorylated even after OSM treatment (Fig. 4, lanes 4 and 5). Thus, the F-peptide is the main site of hnRNP A1 phosphorylation during OSM. We conclude that other mechanisms besides hnRNP A1 phosphorylation also might contribute to the extent of OSM-induced relocalization of the protein, which will require further investigation.
Fig. 4.
Mutation of the F-peptide's seven serines to alanines abolishes the increase of hnRNP A1 phosphorylation in cells subjected to OSM. HeLa cells were transfected with T7-hnRNP A1 or T7-F2 plasmids and treated with OSM for 3 h. The transiently expressed proteins were immunoprecipitated from total protein lysates by using a T7-tag mAb and separated by SDS/PAGE. The levels of serine phosphorylation and immunoprecipitated proteins were determined by Western blotting with anti-phosphoserine and anti-T7 antibodies, respectively.
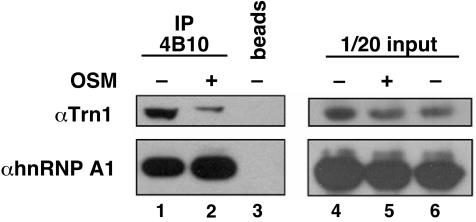
Hyperphosphorylation of F-Peptide Reduces the Interaction of hnRNP A1 with Transportin. Next, we investigated the molecular mechanism by which hnRNP A1 accumulates in the cytoplasm of stressed cells. hnRNP A1 transport depends on the M9 sequence, which serves both as a nuclear import and export signal, and mediates the interaction of hnRNP A1 with transportin Trn1 (11, 12). To test whether OSM affects this interaction, endogenous hnRNP A1 was immunoprecipitated from treated or untreated cells, and the presence of Trn1 in the pellet was detected by Western blotting (Fig. 5). The amount of coimmunoprecipitated Trn1 was greatly reduced in OSM-treated cells (Fig. 5A, lanes 1 and 2). This effect was not caused by OSM-induced Trn1 degradation, because equal amounts of Trn1 were present in cell lysates before immunoprecipitation (Fig. 5A, lanes 4 and 5). We conclude that hnRNP A1 has reduced affinity for Trn1 in OSM-treated cells.
Fig. 5.
Decreased association of hnRNP A1 with Trn1 in OSM-stressed cells. hnRNP A1 was immunoprecipitated from cytoplasmic extracts with mAb 4B10. The immunoprecipitated proteins were separated by SDS/PAGE, and Trn1 and hnRNP A1 were detected by Western blotting with D45 and 4B10 antibodies, respectively. In the panels on the right, 1/20th of the amount of each protein extract used for immunoprecipitation was analyzed as a control for protein expression level.
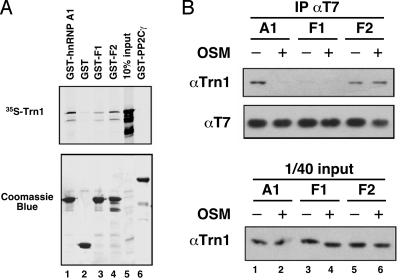
Because OSM changed the extent of F-peptide phosphorylation, we tested whether this effect, in turn, influences the interaction between hnRNP A1 and Trn1. To this end, we performed in vitro binding assays between wild-type or mutant hnRNP A1 fused to GST and in vitro-translated [35S]-Trn1. Trn1 interacted specifically with GST-hnRNP A1 (Fig. 6A, lane 1) but not with GST alone (Fig. 6A, lane 2) or GST-PP2Cγ (Fig. 6A, lane 6), which has a classical bipartite nuclear localization signal and no homology with the M9 sequence. The GST-F1 mutant mimicking phosphorylation of the F-peptide reduced the binding to [35S]-Trn1 (Fig. 6A, lane 3), whereas the affinity of GST-F2 for Trn1 was similar to that of GST-hnRNP A1 (Fig. 6A, lanes 1 and 4).
Fig. 6.
Hyperphosphorylation of F-peptide reduced the interaction of hnRNP A1 with Trn1. (A) The indicated GST recombinant proteins were bound to gluthatione-Sepharose and assayed for binding to in vitro-translated [35S]-Trn1; The proteins were analyzed by SDS/PAGE (Coomassie blue; Lower), and [35S]-Trn1 was detected by using autoradiography ([35S]-Trn1; Upper). (B) T7 plasmids expressing hnRNP A1 or F1/F2 mutants were transfected into HeLa cells, and the transiently expressed proteins were immunoprecipitated from total protein lysates with a T7-tag mAb. The T7-tagged proteins and the coimmunoprecipitated endogenous Trn1 were detected by Western blotting with T7 and D45 antibodies, respectively. A fraction of the total protein lysate (1/40th of input) also was analyzed to determine the level of endogenous Trn1 expressed under each condition.
We further assayed the coimmunoprecipitation of Trn1 with transiently expressed versions of hnRNP A1, F1, and F2 from cytoplasmic lysates. T7-tagged hnRNP A1 behaved as endogenous hnRNP A1, and its interaction with Trn1 decreased in cells treated with OSM (compare Fig. 6B, lanes 1 and 2, with Fig. 5, lanes 1 and 2). As expected, the T7-F1 mutant had lower affinity for Trn1 than did wild-type hnRNP A1, and OSM did not affect its interaction with Trn1 (Fig. 6B, lanes 3 and 4). On the other hand, the interaction of the F2 mutant with Trn1 in untreated cells was comparable to that of wild-type T7-hnRNP A1, but it did not change after OSM (Fig. 6B, lanes 5 and 6). The GST pull-down and coimmunoprecipitation experiments gave qualitatively consistent results, although the effect of the F1 mutation was stronger by the second assay; this quantitative difference may reflect the different nature of the assays, although the lack of methylation of hnRNP A1 expressed in bacteria also might play a role. Taken together, these results demonstrate that although the F-peptide is distinct from the M9 sequence, its hyperphosphorylation regulates hnRNP A1 binding to Trn1.
Discussion
The regulation of gene expression by cell-signaling pathways has largely focused on transcription factors. However, some factors involved in mRNA processing, such as hnRNP K and hnRNP A1, also are subject to regulation by cell-signaling pathways (21, 31). The polypyrimidine tract-binding protein/hnRNP I is a substrate for phosphorylation by PKA, and the phosphorylation state likewise controls the nuclear/cytoplasmic distribution of the protein (32). Similarly, changes in calcium concentration leading to hyperphosphorylation of Tra2β1 result in its relocalization to the cytoplasm (33). We reported in ref. 21 that cells stressed by OSM activate the mitogen-activated protein kinase kinase (MKK)3/6-p38 signaling pathway, resulting in an altered subcellular distribution of hnRNP A1. Here, we have characterized the molecular mechanism responsible for this redistribution. We found that OSM induces phosphorylation of hnRNP A1 within a stretch of serine residues in a tryptic peptide (F-peptide), resulting in a decreased interaction of hnRNP A1 with its import receptor, transportin. We propose that hyperphosphorylation of the F-peptide reduces the rate of hnRNP A1 nuclear import.
The shuttling activity of hnRNP A1 requires the M9 sequence (7, 14), which mediates its interaction with Trn1 and Trn2b (11–13, 34). Transportins also are import receptors for other proteins that lack homology to M9 (34–37). Thus, it is thought that transportin recognizes each import substrate through secondary and/or tertiary structural features, rather than primary sequence (34). Our results suggest that phosphorylation of the F-peptide affects the binding of M9 to Trn1 and decreases hnRNP A1 nuclear import. The M9 sequence alone does not have the same affinity and specificity for transportin as full-length hnRNP A1 (13). Taken together, these results suggest that the F-peptide may affect structural features of the M9/Trn interaction. Because the F-peptide and the M9 motif are juxtaposed near the C terminus of hnRNP A1 (Fig. 2B), we propose that the extent of F-peptide phosphorylation regulates the accessibility of M9. For example, the interaction surface of M9 may be masked by phosphorylated F-peptide, or a conformational change in M9 may reduce its affinity for Trn1. At present, the structure of the C-terminal domain of hnRNP A1 remains unknown.
hnRNP A1 regulation by phosphorylation has not been extensively studied in vivo, because it is difficult to detect the low steady-state level of phosphorylated protein or protein that is phosphorylated only under certain conditions. The characterization of F-peptide as an in vivo phosphorylation site in hnRNP A1 is a step toward the understanding of hnRNP A1 regulation by OSM-activated signaling pathways. The next step will be the identification of the kinase(s) responsible for this modification. We have already reported that p38 does not directly phosphorylate hnRNP A1 (21). Other kinases known to phosphorylate hnRNP A1 in vitro or in vivo, such as PKA, casein kinase II, and PKCζ (20, 23, 38), are not part of the MKK3/6-p38 signaling cascade. However, PKCζ and/or PKA can induce cytoplasmic localization of hnRNP A1, so cells treated with OSM may activate these kinases to regulate the hnRNP A1 interaction with transportin. As a part of the process of identifying the relevant hnRNP A1 kinase(s), it is of interest to determine whether the F-peptide is phosphorylated in the nucleus or in the cytoplasm. We have compared the phosphorylation of hnRNP A1 in cytoplasmic and nuclear extracts prepared from cells subjected to OSM and observed increased phosphorylation in the cytoplasmic extracts (data not shown). However, the specificity of this phosphorylation and the link between the kinase(s) and the MKK3/6-p38 signaling pathway will require further study.
In addition to F-peptide phosphorylation, we also have found a new site of arginine methylation in hnRNP A1, R318 (Fig. 2 A). This methylation event was detected only in hnRNP A1 isolated from cells treated by OSM, suggesting that it is an OSM-specific posttranslational modification. It is currently unknown whether or not methylation and phosphorylation of F-peptide are coregulated. In Saccharomyces cerevisiae, methylation of hnRNP proteins was proposed to regulate their nuclear export (16). An attractive hypothesis is that OSM-specific methylation enhances the nuclear export of hnRNP A1, whereas phosphorylation impairs its nuclear import. This possibility might explain our observation that the F1 and F2 mutations did not affect hnRNP A1 localization in 100% of the cells. However, by using MS, we did not find peaks corresponding to F-peptide modified only by either methylation (R318) or serine phosphorylation, suggesting that if these modification states do exist, they may be highly transitory. On the other hand, we cannot exclude the possibility that the longer transit time of hnRNP A1 in the cytoplasm of OSM-treated cells may indirectly increase its methylation, because an arginine methylase specific for hnRNP A1 is present in the cytoplasm (39). The first model seems more attractive, but further study will be required to distinguish between these two possibilities.
hnRNP A1 is involved in pre-mRNA splicing regulation as an antagonist of SR proteins, and hnRNP A1 regulation by the MKK3/6-p38 signaling pathway affects the control of alternative splice-site selection in vivo (21). However, it is not known whether OSM also affects the activity of SR proteins. Although we have shown that two members of this protein family (SF2/ASF and SC35) remain in the nucleus in cells treated by OSM (21), it is possible that OSM nevertheless affects their phosphorylation and shuttling activity. The interaction between SR proteins and import receptors is modulated by phosphorylation (40–42). In addition, at least two SR proteins (SF2/ASF and 9G8) are exported to the cytoplasm in a hypophosphorylated state (43, 44). Thus, an increase of SR protein phosphorylation may inhibit their nuclear export and/or increase their nuclear import rate. Finally, SF2/ASF was recently shown to be involved in translation regulation (45). By analogy, it will be interesting to determine whether hnRNP A1 in cells subjected to OSM has a distinct activity when bound to cytoplasmic mRNA.
Acknowledgments
We thank Svetlana Dokudovskaya for helpful comments on the manuscript. This work was supported by National Cancer Institute Grant CA13106 (E.A. and A.R.K.) and the Medical Research Council (S.G. and J.F.C.).
Abbreviations: OSM, osmotic shock; hnRNP, heterogenous nuclear ribonucleoprotein; MKK, mitogen-activated protein kinase kinase; PP, protein phosphatase.
References
- 1.Dreyfuss, G., Kim, V. N. & Kataoka, N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195-205. [DOI] [PubMed] [Google Scholar]
- 2.Krecic, A. M. & Swanson, M. S. (1999) Curr. Opin. Cell Biol. 11, 363-371. [DOI] [PubMed] [Google Scholar]
- 3.Piñol-Roma, S., Choi, Y. D., Matunis, M. J. & Dreyfuss, G. (1988) Genes Dev. 2, 215-227. [DOI] [PubMed] [Google Scholar]
- 4.Dreyfuss, G., Matunis, M. J., Piñol-Roma, S. & Burd, C. G. (1993) Annu. Rev. Biochem. 62, 289-321. [DOI] [PubMed] [Google Scholar]
- 5.Michael, W. M., Siomi, H., Choi, M., Piñol-Roma, S., Nakielny, S., Liu, Q. & Dreyfuss, G. (1995) Cold Spring Harb. Symp. Quant. Biol. 60, 663-668. [DOI] [PubMed] [Google Scholar]
- 6.Siomi, H. & Dreyfuss, G. (1995) J. Cell Biol. 129, 551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izaurralde, E., Jarmolowski, A., Beisel, C., Mattaj, I. W., Dreyfuss, G. & Fischer, U. (1997) J. Cell Biol. 137, 27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weighardt, F., Biamonti, G. & Riva, S. (1995) J. Cell Sci. 108, 545-555. [DOI] [PubMed] [Google Scholar]
- 9.Piñol-Roma, S. & Dreyfuss, G. (1992) Nature 355, 730-732. [DOI] [PubMed] [Google Scholar]
- 10.Iervolino, A., Santilli, G., Trotta, R., Guerzoni, C., Cesi, V., Bergamaschi, A., Gambacorti-Passerini, C., Calabretta, B. & Perrotti, D. (2002) Mol. Cell. Biol. 22, 2255-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollard, V. W., Michael, W. M., Nakielny, S., Siomi, M. C., Wang, F. & Dreyfuss, G. (1996) Cell 86, 985-994. [DOI] [PubMed] [Google Scholar]
- 12.Fridell, R. A., Truant, R., Thorne, L., Benson, R. E. & Cullen, B. R. (1997) J. Cell Sci. 110, 1325-1331. [DOI] [PubMed] [Google Scholar]
- 13.Rebane, A., Aab, A. & Steitz, J. A. (2004) RNA 10, 590-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michael, W. M., Choi, M. & Dreyfuss, G. (1995) Cell 83, 415-422. [DOI] [PubMed] [Google Scholar]
- 15.Vautier, D., Chesne, P., Cunha, C., Calado, A., Renard, J. P. & Carmo-Fonseca, M. (2001) J. Cell Sci. 114, 1521-1531. [DOI] [PubMed] [Google Scholar]
- 16.Shen, E. C., Henry, M. F., Weiss, V. H., Valentini, S. R., Silver, P. A. & Lee, M. S. (1998) Genes Dev. 12, 679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, S., Merrill, B. M., Rajpurohit, R., Kumar, A., Stone, K. L., Papov, V. V., Schneiders, J. M., Szer, W., Wilson, S. H., Paik, W. K., et al. (1997) Biochemistry 36, 5185-5192. [DOI] [PubMed] [Google Scholar]
- 18.Li, T., Evdokimov, E., Shen, R. F., Chao, C. C., Tekle, E., Wang, T., Stadtman, E. R., Yang, D. C. & Chock, P. B. (2004) Proc. Natl. Acad. Sci. USA 101, 8551-8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perrotti, D., Iervolino, A., Cesi, V., Cirinni, M., Lombardini, S., Grassilli, E., Bonatti, S., Claudio, P. P. & Calabretta, B. (2000) Mol. Cell. Biol. 20, 6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Municio, M. M., Lozano, J., Sanchez, P., Moscat, J. & Diaz-Meco, M. T. (1995) J. Biol. Chem. 270, 15884-15891. [DOI] [PubMed] [Google Scholar]
- 21.van der Houven van Oordt, W., Diaz-Meco, M. T., Lozano, J., Krainer, A. R., Moscat, J. & Cáceres, J. F. (2000) J. Cell Biol. 149, 307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams, K. R., Stone, K. L., Lopresti, M. B., Merrill, B. M. & Planck, S. R. (1985) Proc. Natl. Acad. Sci. USA 82, 5666-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobianchi, F., Calvio, C., Stoppini, M., Buvoli, M. & Riva, S. (1993) Nucleic Acids Res. 21, 949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cáceres, J. F., Misteli, T., Screaton, G. R., Spector, D. L. & Krainer, A. R. (1997) J. Cell Biol. 138, 225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreyfuss, G., Choi, Y. D. & Adam, S. A. (1984) Mol. Cell. Biol. 4, 1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gama-Carvalho, M., Krauss, R. D., Chiang, L. J., Valcárcel, J., Green, M. R. & Carmo-Fonseca, M. (1997) J. Cell Biol. 137, 975-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, W. Z. & Chait, B. T. (2000) Anal. Chem. 72, 2482-2489. [DOI] [PubMed] [Google Scholar]
- 28.Gama-Carvalho, M., Carvalho, M. P., Kehlenbach, A., Valcárcel, J. & Carmo-Fonseca, M. (2001) J. Biol. Chem. 276, 13104-13112. [DOI] [PubMed] [Google Scholar]
- 29.Merrill, B. M., Lopresti, M. B., Stone, K. L. & Williams, K. R. (1987) Int. J. Pept. Protein Res. 29, 21-39. [DOI] [PubMed] [Google Scholar]
- 30.Baldwin, G. S. & Carnegie, P. R. (1971) Science 171, 579-581. [DOI] [PubMed] [Google Scholar]
- 31.Habelhah, H., Shah, K., Huang, L., Ostareck-Lederer, A., Burlingame, A. L., Shokat, K. M., Hentze, M. W. & Ronai, Z. (2001) Nat. Cell Biol. 3, 325-330. [DOI] [PubMed] [Google Scholar]
- 32.Xie, J., Lee, J. A., Kress, T. L., Mowry, K. L. & Black, D. L. (2003) Proc. Natl. Acad. Sci. USA 100, 8776-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daoud, R., Mies, G., Smialowska, A., Olah, L., Hossmann, K. A. & Stamm, S. (2002) J. Neurosci. 22, 5889-5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siomi, M. C., Eder, P. S., Kataoka, N., Wan, L., Liu, Q. & Dreyfuss, G. (1997) J. Cell Biol. 138, 1181-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truant, R., Kang, Y. & Cullen, B. R. (1999) J. Biol. Chem. 274, 32167-32171. [DOI] [PubMed] [Google Scholar]
- 36.Bachi, A., Braun, I. C., Rodrigues, J. P., Pante, N., Ribbeck, K., von Kobbe, C., Kutay, U., Wilm, M., Görlich, D., Carmo-Fonseca, M., et al. (2000) RNA 6, 136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jäkel, S. & Görlich, D. (1998) EMBO J. 17, 4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao, Z. H., Metherall, J. & Virshup, D. M. (2000) Biochem. Biophys. Res. Commun. 268, 562-566. [DOI] [PubMed] [Google Scholar]
- 39.Liu, Q. & Dreyfuss, G. (1995) Mol. Cell. Biol. 15, 2800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun, C. Y., Velázquez-Dones, A. L., Lyman, S. K. & Fu, X. D. (2003) J. Biol. Chem. 278, 18050-18055. [DOI] [PubMed] [Google Scholar]
- 41.Allemand, E., Dokudovskaya, S., Bordonne, R. & Tazi, J. (2002) Mol. Biol. Cell 13, 2436-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai, M. C., Lin, R. I. & Tarn, W. Y. (2001) Proc. Natl. Acad. Sci. USA 98, 10154-10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang, Y., Yario, T. A. & Steitz, J. A. (2004) Proc. Natl. Acad. Sci. USA 101, 9666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai, M. C. & Tarn, W. Y. (2004) J. Biol. Chem. 279, 31745-31749. [DOI] [PubMed] [Google Scholar]
- 45.Sanford, J. R., Gray, N. K., Beckmann, K. & Cáceres, J. F. (2004) Genes Dev. 18, 755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray, M. V., Kobayashi, R. & Krainer A. R. (1999) Genes Dev. 13, 87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]