Abstract
Replication of DNA requires helicase and primase activities as part of a primosome assembly. In bacteriophage T4, helicase and primase are separate polypeptides for which little structural information is available and whose mechanism of association within the primosome is not yet understood. Three-dimensional structural information is provided here by means of reconstructions from electron microscopic images. Structures have been calculated for complexes of each of these proteins with ssDNA in the presence of MgATPγS. Both the helicase (gp41) and primase (gp61) complexes are asymmetric hexagonal rings. The gp41 structure suggests two distinct forms that have been termed “open” and “closed.” The gp61 structure is clearly a six-membered ring, which may be a trimer of dimers or a traditional hexamer of monomers. This structure provides conclusive evidence for an oligomeric primase-to-ssDNA stoichiometry of 6:1.
Keywords: three-dimensional structure, DNA replication, protein hexamers, replisome
The replicative helicases and primases are members of the primosome subassembly that is a key and integral unit in the function of the replisome during DNA replication. These proteins, isolated from diverse organisms, have been the subjects of structural studies as part of a broad effort to relate their structure to their function.
The replicative helicases show common hexameric architecture. As revealed by electron micrographs, the DnaB helicase from Escherichia coli exists as an equilibrium mixture of two structural shapes: a hexamer with sixfold symmetry and a trimer of dimers with threefold symmetry (1). The hexamers are stabilized by Mg2+; the equilibrium between the two ring forms can be shifted by the addition of nucleotides, such as ATPγS, which favors a threefold symmetric ring (2).
Key crystal structures of the active helicase domain of the phage T7 gene 4 protein revealed a hexameric structure whose subunit arrangements showed some deviation from a sixfold symmetry. The variation was ascribed to structural disruption postulated to accommodate stepwise ATP binding and hydrolysis, now reflected in this “trapped” structure (3). Three-dimensional reconstruction and electron microscopy images of the intact T7 gp4 helicase/primase also revealed a hexameric structure with sixfold symmetry (4). Nuclease protection and cross-linking experiments suggest that ssDNA passes through the hole in the center of the hexameric ring. Electron microscopy examination of the SV40 large T antigen (5) identified the helicase domains also as lying within a hexameric ring in an orientation identical to that seen by means of electron microscopic reconstruction of the bacteriophage T7 gp4 helicase (6).
The crystal structure of the intact bifunctional helicase/primase from phage T7, however, revealed a heptameric ring (7). Retrospective electron microscopy showed that both hexameric and heptameric oligomeric forms were present. The heptameric ring, which contains the helicase domain, is relatively flat, but the adjoining subunits do not show true sevenfold symmetry, owing to rotational effects within the ring plane. The primase domains fan out from the helicase ring, with few self-contacts, and are connected through highly flexible linkers to the helicase domains. The expanded ring of the heptamer allows binding of dsDNA in the central channel, thereby possibly conferring a translocation activity on the heptamer (8). The flexibility of the linkage between each primase and helicase subunit allows each of the two enzymatic activities to operate nearly independently of one another yet, in the case of primase, to enjoy the increased priming activity from an oligomeric assembly versus a monomer.
In contrast, less is known about the independent helicase (gp41) and primase (gp61) proteins from bacteriophage T4. Solution studies suggested that the gp41 protein assembles into a toroidal (ring-shaped) hexameric form from monomer and dimer solution subunits upon the binding of ATP or GTP (9). Similar studies showed that the T4 gp61 is monomeric in solution. In the presence of ATP or GTP and DNA, the helicase and primase combine to form a gp41·gp61·DNA complex of presumed 6:1:1 stoichiometry (10). This complex serves an important functional role in that the polymerase activity of the primase is greatly stimulated by the helicase. Unexpectedly, we found the increase in primase activity implicated an active gp41·gp61·DNA complex of 6:6:1 stoichiometry (11).
Here, we report the results of electron microscopy studies on gp41 and gp61 of bacteriophage T4. Three-dimensional reconstructions from electron microscopic images indicate the ring-like structure and hexameric stoichiometry of each protein when complexed with DNA, as occurs in formation of the primosome.
Materials and Methods
Materials. The gp41 and gp61 proteins were purified by chitin-based affinity chromatography as described in ref. 11. The ssDNA substrate was 45 nucleotides in length and contained a GTT priming site, which is underlined here (5′-GGGTGGGAGGGAGGTTTGCAACTGATCGATGATAGTACGTCTGTG-3′).
Electron Microscopy. Equimolar ratios of protein were allowed to assemble into complexes in the presence of 45-mer ssDNA and MgATPγS. Samples were then adsorbed to thin carbon films and embedded in methylamine vanadate (Nanovan, Nanoprobes, Yaphank, NY). Transmission electron micrographs were taken with minimum dose focusing at a magnification of 63,000 using an inline energy filter. Micrographs were digitized by using a flatbed scanner to give a pixel size of 3.2 Å on the image scale.
Image Analysis. All computations were performed by using spider/web software (12). For all reconstructions, images were interactively selected and then aligned by using a reference-free algorithm. For de novo structures, hierarchical ascendant classification using principal component analysis was used to identify separate views of the particles. The gp41 structure was calculated from tilt pairs of micrographs; that is, a primary reference structure was determined by merging reconstructions calculated from classes of 55°-tilt images. Reconstruction refinement was accomplished by projecting the primary reference at 10° intervals to which 7,475 flat images were mapped. For gp61, the determination of optimal angles for merging of primary reference structures calculated from image classes was done by using an orientation search algorithm. Each of the reconstructions provided ring structures, which converged to structures with no discernable differences. These were merged into a primary reference, which was refined as described above by the mapping of 6,160 images to projections at 10° intervals.
The same primary reference structures and data sets described above for gp41 and gp61 were used to obtain symmetrized structures. The sixfold or threefold symmetry was imposed during the iterative back-projection calculations.
In all calculations of three-dimensional structures, care was taken to use the appropriate parameters and number of images in each projection class to prevent introduction of artifacts due to overrepresentation of particular frequencies or views (14). Resolution limits were determined from the 50% cutoff of the Fourier shell coefficient between reconstructions of half data sets. Thresholds were determined by using mass and partial specific volume values corresponding to a stoichiometry of six protein subunits and one DNA molecule. For presentation, structure surfaces were created by using iris explorer (Numerical Algorithms Group, Downers Grove, IL), and projections of three-dimensional reconstructions were created in web.
Results and Discussion
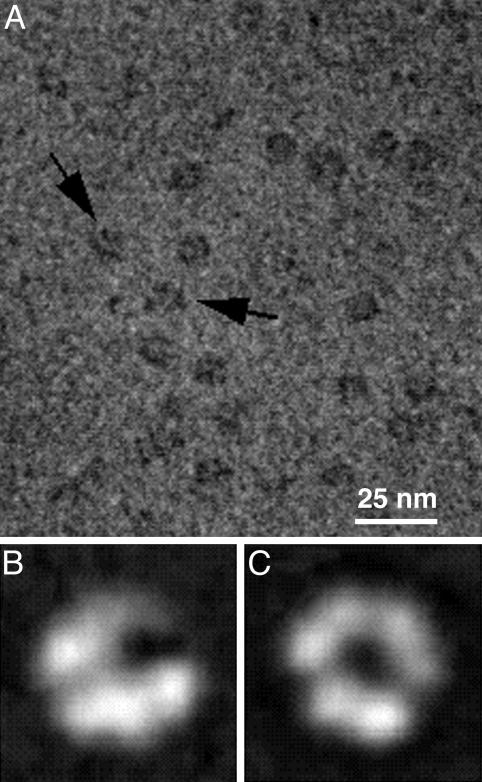
The gp41 Protein Forms a Hexameric but Nonsymmetric Ring in the Presence of ssDNA. After reaction of gp41 with ssDNA in the presence of MgATPγS, one of the major views seen in electron microscopic images of samples negatively stained with methylamine vanadate is a ring with a central hole (Fig. 1A). The ring measures ≈12 nm in diameter. Other images are much narrower but of similar diameter. These correspond to “side” views of the assembly and suggest a single-layered ring.
Fig. 1.
Images of the gp41 assembly in negatively stained electron micrographs appear as both open and closed hexameric rings. (A) A typical field of images shows complete rings and rings with a gap (arrows). (B) Averaged images of open rings. (C) Averaged images of closed rings. Each average contains ≈200 images.
When the ring-shaped views are closely examined, a particularly striking feature is observed: Some appear to have a gap (arrows in Fig. 1A), whereas others are hexagonal with regularly distributed vertices. Two-dimensional averages of gp41 images emphasize the “open” (Fig. 1B) and “closed” (Fig. 1C) appearances of the particle. The latter shows six regularly spaced densities, as is typical of electron microscopic views of hexameric protein ring assemblies. These image averages may result from different orientations of the particle; that is, the view in B appears tilted out of the plane that gives a direct view down the center of the ring as shown in the en face view in C. This interpretation is suggested by the shift of position of the central “hole” when the two averages are compared. In the en face view, the appearance of “spaces” persists, although to a much less noticeable extent. These spaces appear to varying degrees at three places around the ring, resulting from rotational averaging of the “gap” during the computational alignment process. Overall, two-dimensional views of the gp41 complex with ssDNA show that although a hexagonal ring is visible, the distribution is not symmetric, suggesting that there may actually be two conformations of the complex: one open and one closed.
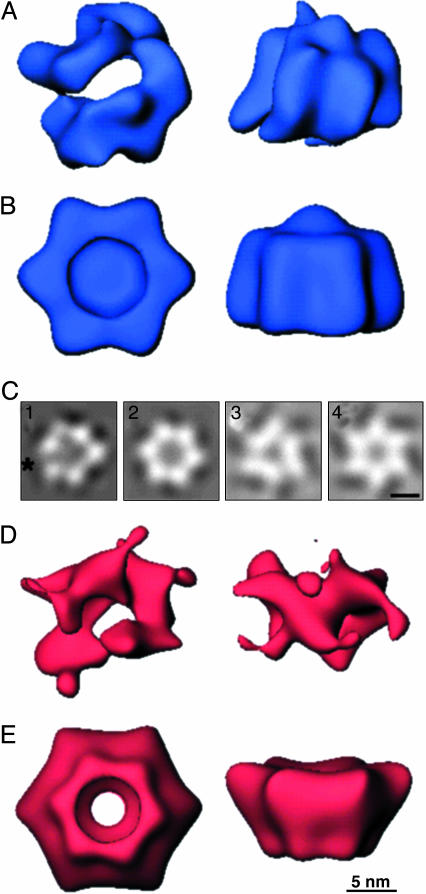
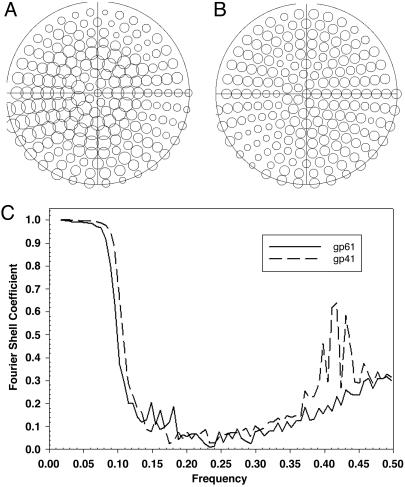
Computational reconstruction of the three-dimensional structure of the gp41 assembly (Fig. 2) also indicates an asymmetric hexagonal ring structure with a central hole. Its maximal dimensions are 12 nm in diameter and 9.5 nm in depth. The distance between the vertices around the circumference of the ring averages 7.5 nm. When no symmetry is enforced on the ring structure (Fig. 2A), there is a distinct gap, as was predicted from the two-dimensional image averages. This gap is reiterated upon projection of the three-dimensional structure from the en face view (Fig. 2C1, asterisk). That this asymmetry is not an artifact of data sampling is demonstrated by the completeness and evenness of the coverage of angular projections used for the reconstruction (Fig. 3A).
Fig. 2.
Three-dimensional structures of gp41 and gp61 complexes derived from electron microscopic images negatively stained with methylamine vanadate. Views are shown down the axis perpendicular to the plane of each ring (Left) and of a side view resulting from a 90° rotation of the en face view about its horizontal axis (Right). Each reconstruction was filtered to its resolution limit of 27 Å and is shown at a threshold corresponding to 100% of the mass of six copies of each protein and one of the 45-mer ssDNA. (A) The gp41 assembly with no rotational symmetry applied. (B) The gp41 assembly calculated with imposition of sixfold rotational symmetry. (C1–C4) En face projections corresponding to reconstruction of gp41 with no applied symmetry, with an asterisk marking the observed open interface of the hexamer (1), gp41 with sixfold rotational symmetry (2), gp61 with threefold rotational symmetry (3), and gp61 with sixfold rotational symmetry (4). (D) The gp61 assembly with no rotational symmetry applied. (E) The gp61 assembly calculated with imposition of sixfold rotational symmetry.
Fig. 3.
Demonstration of completeness of data sets and resolution limits for the reconstruction calculations. (A) Angular distribution of images used for gp41. A circle is present for each of the angular projections of the primary reference structure to which images were mapped. The diameter of the circle increases with the number of images in each projection. (B) Angular distribution of images used for gp61. (C) Plots of the Fourier shell correlation coefficients between reconstructions calculated from half data sets.
If sixfold rotational symmetry is imposed during the reconstruction of the gp41 complex, the cleft and uneven density distributions are no longer observable. The central hole now is closed. The three-dimensional structure (Fig. 2B) and its projection (Fig. 2C2) are now typical of other helicase assemblies. However, a correlation coefficient of only 0.81 between the two types of structures indicates that imposition of sixfold symmetry provides a reasonable approximation of the gp41 structure, but that it is not likely to be entirely accurate.
In a previous study of gp41 assemblies in the presence of ATP and DNA oligomers meant to mimic replication forks (9), an average of cryoelectron microscopic images also showed a hexagonal ring, but no distinct gap between vertices was apparent. This disparity could be the result of differences in imaging methods, nucleotide, and DNA used, as well as other factors. However, averaged cryoelectron microscopic images of another helicase, the E. coli rho protein, clearly show both closed and “notched” rings very similar to those presented here (15). Moreover, a model has been proposed based on both hydrodynamic and electron microscopic data (9) in which assembly of the T4 helicase around DNA is a process in which “open hexamers” are formed before “closed hexamers.” Our observation of an asymmetric ring structure with a gap is consistent with this model and with the three-dimensional reconstruction representing an average of the two states.
The gp61 Protein also Forms an Asymmetric Hexameric Ring Assembly. Reaction of gp61 with ssDNA and MgATPγS produces an assembly that appears very similar in negatively stained electron micrographs to the gp41 assembly. The only noticeable difference is that no gapped rings are observed. The most prominent view is of an apparently hexagonal ring structure with a central hole that measures ≈12 nm in diameter (data not shown). The calculated three-dimensional gp61 assembly (Fig. 2D) is of similar diameter to that of gp41, but the depth is approximately three-fourths, as expected because of the smaller mass of gp61 with respect to gp41. A central hole also is apparent. Projection from the en face view (data not shown) shows a hexagonal ring with regular density distribution at each vertex. The data set used again provides complete and even coverage of the projection angles (Fig. 3B).
Upon examination of the surface representation of the gp61 calculated when no symmetry is imposed during computation of the reconstruction (Fig. 2D), it is difficult to determine whether the gp61 structure is assembled as a trimer of dimers or a simple hexamer. Imposition of threefold or sixfold rotational symmetry during the reconstruction calculation provides structures that are highly similar in projection (Fig. 2 C3 and C4, respectively). As expected, the former has three density distributions that each combine two vertices of the hexagon and three interstitial areas of lower density. Similarly, the sixfold symmetric structure is a very even hexagonal ring. Unlike gp41, the central hole is preserved in the projection of the symmetrized structure of gp61, which is also the case when the three-dimensional reconstruction is depicted (Fig. 2E). The most prominent feature of the gp61 structure is the presence of another smaller ring protruding from one face of the particle. This smaller ring has a diameter approximately three-fourths that of the larger ring. When threefold symmetry is imposed, this ring resolves into three distinct “legs” (data not shown).
Both the threefold and sixfold symmetrized structures have a correlation with the basic structure of ≈0.6. Thus, it is likely that neither is correct. A probable explanation is that the subunits assemble in a fashion such that there is a twist in each subunit as it extends from the larger to the smaller ring. This explanation is consistent with the demonstrated domain flexibility and presence of both hexameric and heptameric primosome protein assemblies in bacteriophage T7 (7).
Conclusions
The structures obtained in this study are both of limited resolution. As determined from the Fourier shell correlation (Fig. 3C), this resolution is ≈27 Å. Undoubtedly, higher resolution studies with such methods as cryoelectron microscopy will provide more precise three-dimensional structures of both gp41 and gp61 complexes with DNA. Moreover, using the initial structure of the gp41 assembly presented here, it should now be possible to appropriately classify and separate open and closed forms and calculate the three-dimensional reconstructions of each type rather than the current hybrid structure.
Despite their limitations, the structures obtained in this study clearly demonstrate that both assemblies are fundamentally single-layer hexagonal rings. Imposition of assumed symmetries results in significant changes in overall appearance of the structures. Thus, such practices should be avoided to prevent introduction of significant artifactual structural characteristics.
Overall, these data demonstrate that both the gp41 and gp61 assemblies with DNA have a 6:1 stoichiometry. Thus, it is logical that a 6:6:1 stoichiometry exists in the active gp41·gp61·DNA complex (11). Moreover, it is likely that these rings stack concentrically in the complete primosome assembly.
Acknowledgments
This work was supported by National Institutes of Health Grants GM13306 (to S.J.B.) and GM071130 (to M.M.S.).
Author contributions: M.T.N., F.T.I., and S.J.B. designed research; M.T.N., J.A.W., M.M.S., F.T.I., and M.A.T. performed research; M.M.S. contributed new reagents/analytic tools; M.T.N. and J.A.W. analyzed data; and M.T.N. wrote the paper.
References
- 1.Bujalowski, W., Klonowska, M. M. & Jezewska, M. J. (1994) J. Biol. Chem. 269, 31350-31358. [PubMed] [Google Scholar]
- 2.Yang, S., Yu, X., VanLoock, M. S., Jezewska, M. J., Bujalowski, W. & Egelman, E. H. (2002) J. Mol. Biol. 321, 839-849. [DOI] [PubMed] [Google Scholar]
- 3.Singleton, M. R., Sawaya, M. R., Ellenberger, T. & Wigley, D. B. (2000) Cell 101, 589-600. [DOI] [PubMed] [Google Scholar]
- 4.Egelman, E. H., Yu, X., Wild, R., Hingorani, M. M. & Patel, S. S. (1995) Proc. Natl. Acad. Sci. USA 92, 3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanLoock, M. S., Alexandrov, A., Yu, X., Cozzarelli, N. R. & Egelman, E. H. (2002) Curr. Biol. 12, 472-476. [DOI] [PubMed] [Google Scholar]
- 6.VanLoock, M. S., Chen, Y. J., Yu, X., Patel, S. S. & Egelman, E. H. (2001) J. Mol. Biol. 311, 951-956. [DOI] [PubMed] [Google Scholar]
- 7.Toth, E. A., Li, Y., Sawaya, M. R., Cheng, Y. & Ellenberger, T. (2003) Mol. Cell 12, 1113-1123. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan, D. L. & O'Donnell, M. (2002) Mol. Cell 10, 647-657. [DOI] [PubMed] [Google Scholar]
- 9.Dong, F., Gogol, E. P. & von Hippel, P. H. (1995) J. Biol. Chem. 270, 7462-7473. [DOI] [PubMed] [Google Scholar]
- 10.Dong, F. & von Hippel, P. H. (1996) J. Biol. Chem. 271, 19625-19631. [DOI] [PubMed] [Google Scholar]
- 11.Valentine, A. M., Ishmael, F. T., Shier, V. K. & Benkovic, S. J. (2001) Biochemistry 40, 15074-15085. [DOI] [PubMed] [Google Scholar]
- 12.Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M. & Leith, A. (1996) J. Struct. Biol. 116, 190-199. [DOI] [PubMed] [Google Scholar]
- 13.Penczek, P. A., Zhu, J. & Frank, J. (1996) Ultramicroscopy 63, 205-218. [DOI] [PubMed] [Google Scholar]
- 14.Sorzano, C. O., Marabini, R., Boisset, N., Rietzel, E., Schroder, R., Herman, G. T. & Carazo, J. M. (2001) J. Struct. Biol. 133, 108-118. [DOI] [PubMed] [Google Scholar]
- 15.Gogol, E. P., Seifried, S. E. & von Hippel, P. H. (1991) J. Mol. Biol. 221, 1127-1138. [DOI] [PubMed] [Google Scholar]