Abstract
Drosophila RacGAP50C and its homologues act as part of a complex with a kinesin-like protein (Pavarotti/Zen-4) that is essential for the formation of the central spindle and completion of cytokinesis [Mishima, M., Kaitna, S. & Glotzer, M. (2002) Dev. Cell 2, 41–54; Somers, W. G. & Saint, R. (2003) Dev. Cell 4, 29–39; Jantsch-Plunger et al. (2000) J. Cell Biol. 149, 1391–1404]. We report here that RacGAP50C corresponds to the tumbleweed (tum) gene previously identified based on its defects in dendrite development of sensory neurons [Gao, F. B., Brenman, J. E., Jan, L. Y. & Jan, Y. N. (1999) Genes Dev. 13, 2549–2561]. Using mushroom body neurogenesis and morphogenesis as a model, we show that Tumbleweed (Tum), Pavarotti, and their association are required for neuroblast proliferation. Tum with a mutation predicted to disrupt the GTPase-activating protein (GAP) activity still largely retains its activity in regulating cell division but is impaired in its activity to limit axon growth. We also provide evidence that Tum and Pavarotti regulate the subcellular localization of each other in postmitotic neurons and that cytoplasmic accumulation of both proteins disrupts axon development in a GAP-dependent manner. Taken together with previous studies of RacGAP50C in regulating cytokinesis, we propose that Tum serves as a scaffolding protein in regulating cell division but acts as a GAP to limit axon growth in postmitotic neurons.
Keywords: axon growth, axon guidance, cytokinesis, cytoskeleton, signaling
Rho GTPases act as intracellular molecular switches to regulate cytoskeletal dynamics. They are positively regulated by guanine nucleotide exchange factors and negatively regulated by Rho GTPase-activating proteins (GAPs). RhoGAPs are defined by the presence of a conserved RhoGAP domain that binds to GTP-bound Rho GTPases and subsequently speeds up their intrinsically slow GTPase activity (1). In both Drosophila and human genomes, there are severalfold more genes for RhoGAPs than for Rho GTPases they regulate (2, 3). Aside from their GAP domains, RhoGAP proteins are divergent in their overall sequences, perhaps reflecting a diversity of biological functions for Rho GTPase activity modulation. The physiological consequence of losing negative regulation of the Rho GTPases is best illustrated by the fact that mutations in two RhoGAPs are linked to two different forms of mental retardation (4, 5).
We have previously conducted a genome-wide transgenic RNAi-based screen in Drosophila to identify RhoGAPs essential for neuronal morphogenesis (6). We identified two RhoGAPs that function in the development or maintenance of normal morphology of mushroom body (MB) neurons in the Drosophila brain. The first, Drosophila p190 RhoGAP, is critical for inhibiting RhoA activity to maintain the stability of axon branches (6). The second RhoGAP identified in the RNAi screen was RacGAP50C, which is the focus of the present work.
RacGAP50C is known for its function in cytokinesis. RacGAP50C and its homologues bind directly to a mitotic kinesin-like protein, named Pavarotti (Pav) in Drosophila, to form a complex known as centralspindlin (7, 8). Based on their in vitro behavior, the RhoGAP and kinesin form a heterotetrameric complex that bundles microtubules (7), and both are necessary for the formation of midzone antiparallel microtubules critical for cytokinesis (7–9). In vitro evidence indicates that the vertebrate and worm homologues of RacGAP50C, mgcRacGAP and CYK-4, have preferential GAP activity toward Rac1 and Cdc42 (9–11), but, upon phosphorylation of the GAP domain, mgcRacGAP's activity is increased for RhoA (12). Recent work has begun to shed light on the function of the centralspindlin complex during cytokinesis (13), as well as its potential function in regulating cellular polarity (14); however, whether this RhoGAP–kinesin complex also functions in postmitotic neurons is unclear. The expression of the mammalian homologue of RacGAP50C (mgcRacGAP) in postmitotic neurons (15), the uncoordinated phenotype seen with temperature-sensitive alleles of the Caenorhabditis elegans homologue of RacGAP50C (9), and the disruption of dendritic/axonal polarity with inhibition of the mammalian homologue of Pav in vitro (16) all hint at a postmitotic function for the RhoGAP–kinesin complex in neurons. Through the use of both RNAi and loss-of-function analysis, we directly examine the function of RacGAP50C and Pav in neuronal development.
Materials and Methods
Fly Strains. UAS-GFP::PavWT, UAS-GFP::PavNLS, and pav936 were generously given to us by D. Glover (University of Cambridge, Cambridge, U.K.) (17). UAS-MYC::Tum yellow fluorescent protein was a kind gift from R. Saint (Australian National University, Canberra, Australia).
Sequencing of tum. Primers designed against the tum locus were used to PCR-amplify tum from genomic DNA from heterozygous tum adults. PCR products were sequenced and scanned for the presence of double peaks indicating a nucleotide change between WT and mutant chromosomes.
Generation of Transgenes. Bac clone BACR06M19 from the Berkeley Drosophila Genome Project was BamHI-digested to generate an ≈4.4-kb DNA fragment that includes 1.56 kb upstream of the predicted transcriptional start site of tum (331 bp upstream of the stop codon of upstream gene CG16935) and >500 bp after the 3′ UTR. The fragment was subcloned into pBluescript (pBSk-gTum) and then subcloned into the P-element transformation vector, pW8, by using EcoRI and XbaI.
pBSk-gTum was EagI- or PstI-digested to generate 5′ and 3′ end templates for generating Pav-binding (gTumΔZ4) and RhoGAP (gTumR417L) mutations. Site-directed mutagenesis using PfU Turbo DNA polymerase (Stratagene) was performed with complementary oligonucleotides and DpnI digestion of the parental template. Products were transformed in DNA adenine methylation-free bacteria, tested for loss of a ClaI site with the successful generation of the R417L mutation, and sequenced before subcloning into the pW8-gTumWT by using SacII and XbaI. Deletion of the Pav-binding domain (ΔL12-F27) was done by back-to-back PCR mutagenesis, and the deletion was identified by the ≈100-bp shortening of the 1.8-kb fragment generated with EcoRI and EagI. Clones were sequenced and subcloned into pW8-gTum with EcoRI and EagI.
UAS-MYC::TumR417L was generated by replacing the relevant fragment of UAS-MYC::TumWT with that of TumR417L generated as described above.
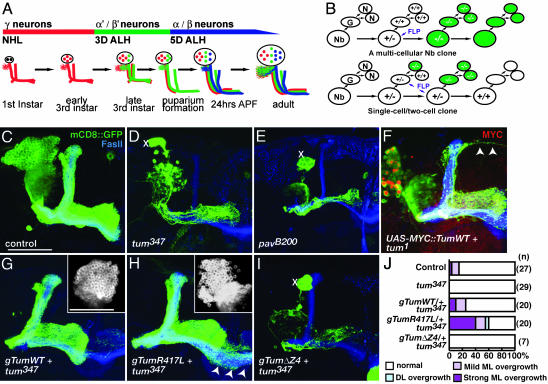
Generation of MARCM (Mosaic Analysis with a Repressible Cell Marker) Clones. pav alleles were recombined with FRT 2A, and tum alleles were recombined with FRT 42D to generate mosaic clones by using MARCM (18, 19). Flies of the following genotypes were crossed to flies carrying the appropriate FRT and tum or pav allele, or the FRT alone: hsFLP, UAS-mCD8::GFP; FRT 42D, tub-GAL80/CyO, y+; GAL4-OK107 or hsFLP, UAS-mCD8::GFP; FRT 2A, tub-GAL80/Tm3; GAL4-OK107. Progeny were heat-shocked 16–25 h after egg-laying to generate γ neuron single-cell clones and early neuroblast (Nb) clones. α/β neurons were generated by heat-shocking pupa >8 h after puparium formation.
Immunohistochemistry and Image Acquisition. Adult flies were dissected <1 week after eclosion. Immunofluorescence and image processing were performed according to the protocol described in ref. 18. Antibody conditions were as follows: anti-mCD8 (1:100; Caltag, South San Francisco, CA), fasciclin II (FasII) (1:100, 1D4; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City), GFP (1:800; Molecular Probes), MYC (9E10, 1:50; Molecular Probes). Secondary antibodies (Alex Fluor 488 goat anti-rat, Alexa Fluor 568 goat anti-mouse, and Alexa Fluor 647 goat anti-mouse; Molecular Probes) were used at 1:250. To detect FasII in addition to MYC, samples were further processed by using the Zenon Alexa Fluor kit (Molecular Probes). All Alexa Fluor 488 and Alexa Fluor 568 samples were imaged with sequential excitation.
Single-Cell Analysis of MB Axons and Dendrites. Image files were randomly coded before tracing of the axons and dendrites in neurolucida. The start of the axon terminal was defined from the end of the peduncle, and branches were categorized as extensions >1 μm. Branches were scored based on the number of terminal endings. The number of dendritic claws within the confocal z-stack and the number of dendritic branches were tabulated. Dendritic claws were defined as structures <3 μm in span that exhibit at least two filopodial-like endings that project toward each other in a claw-like configuration. Main dendritic branches were defined as processes that end with a claw or any process without a claw that is >3 μm in length that stems from the main neurite (20). Data were extracted with neuroexplorer and exported to excel (Microsoft). The values were expressed as means ± SEM, and statistical tests were performed with pairwise Student's t test.
Nb Clone Rescue Analysis. MARCM clones were generated within 16–24 h after egg-laying to generate MB Nb clones that should contain all three neuron types and represent one-quarter of all MB neurons (≈500 neurons), allowing for complete innervation of all axonal lobes (Fig. 2A). Nb clones were scored with the experimenter blind to the genotype. Nb clones that had an early failure of cytokinesis generate only γ neurons, as evident from partial innervation of the γ lobe. The degree of cytokinetic rescue was scored based on the generation of neuron types and the extent of recovery of each neuronal type. Both the appearance of enlarged cell bodies and the occurrence of axon extension beyond their normal confines were recorded for each Nb clone.
Fig. 2.
MARCM analysis of tum and pav in MB Nbs. (A) Timeline of MB neuron generation (adapted from ref. 19). NHL, newly hatched larvae; ALH, after larval hatching; APF, after puparium formation. (B) Generation of marked and homozygous mutant Nb vs. single-neuron (N) clone depends on FLP recombinase (FLP) activation in Nb or in the ganglion mother cell (G). (C) Control Nb clone generated in newly hatched larva. (D and E) tum347 and pavB200 Nb clone generated at the same stage. Xs mark presence of enlarged cell bodies, and severe decrease of cells generated from the Nb leads to the absence of all but the axons of the earliest born γ neurons. FasII highlights the axon projections of heterozygous MB neurons generated from the three other MB Nbs. (F) tum1 Nb clone expressing UAS-MYC::Tum cDNA partially rescues the cytokinetic phenotype, allowing for the generation of most but not all of the later-born neurons. Arrowheads mark axons that overextend beyond the normal dorsal lobe. (G) A transgene encompassing genomic WT tum (gTumWT) rescues tum347 Nb clone phenotypes. (Inset) Cell bodies from the same Nb and the absence of enlarged cell bodies. (H) A transgene for genomic tum with the R417L mutation also rescues proliferation defects in the tum347 Nb clone. (Inset) Cell bodies from same Nb. Arrowheads mark axons extending across the midline beyond normal ML. (I) gTum with a deletion in the Pav binding domain (ΔZ4) does not rescue Nb proliferation defect of tum347 clone. (J) Quantification of Nb clones that exhibit overextension beyond their normal lobe boundaries. (Scale bar: 50 μm.)
Gain of Function Phenotype. Samples were scored with a Nikon E600 to visualize the presence of both MYC and mCD8::GFP. Experiments without the presence of mCD8::GFP were conducted to verify GFP::Pav expression and to examine its localization. The experimenter was blind to the genotype of the samples. MB were categorized as follows. “Axons misrouted”: axons project to ectopic locations or to their normal target region via aberrant pathways; “no DL and ML”: no α′/β′ and α/β lobes; “no ML”: no β′ or β lobes; “mild morphological changes”: variations in the shape of the dorsal lobe; “normal”: presence of distinct α/β, α′/β′, and γ lobes that are fully innervated and with WT morphology.
Results
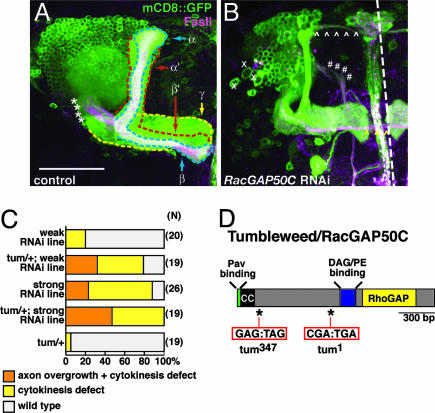
Decreased RacGAP50C Levels Lead to Defects in Nb Proliferation and Axon Guidance. We use intrinsic neurons of the Drosophila MB (hereafter referred to as MB neurons) as our model neurons to study RacGAP50C. The MB is a center for olfactory learning and memory (21) and represents an ideal structure to examine neuronal morphogenesis because its axonal projection pattern and development have been well characterized (19, 22, 23). MB neurons each have a single ventrally projecting process that extends dendrites close to the cell body in a spherical mass called the calyx. The main process continues through an axonal peduncle (asterisks in Fig. 1A) to reach its target region, where the axonal processes form distinct lobular regions segregated by neuronal type. The ≈2,500 neurons of each MB hemisphere are generated from four MB Nbs that sequentially generate three subclasses of MB neurons. The earliest born γ neurons have one major medially projecting axon branch that elaborates in the γ lobe (outlined in yellow in Fig. 1A). The second-born α′/β′ neuron innervates two distinct dorsal and medial projecting lobes (outlined in red in Fig. 1A). α/β neurons (outlined in blue in Fig. 1A) are born last and, similarly, have a major dorsal and medial projecting axon branch that can be distinguished from α′/β′ neurons by its strong expression of the cell adhesion protein FasII. The GAL4 enhancer trap, GAL4-OK107, is strongly expressed in all three MB neuron subclasses and effectively drives RNAi expression (6), along with a cellular marker, mCD8::GFP, to simultaneously knock down gene expression and visualize the gross morphology of the MB complex (Fig. 1A). In addition, with MB neurons we can examine many different aspects of neural development, including Nb proliferation, dendrite morphogenesis, and axon growth, guidance, branching, and stability, all of which are regulated by Rho GTPases (20, 24–26).
Fig. 1.
RNAi phenotypes of RacGAP50C/Tum. (A) WT MB complex labeled by GAL4-OK107 driven mCD8::GFP expression. Midline of the brain is at the right side of the image and dorsal is at the top in this and all subsequent images. The MB axons are organized in lobes outlined as: γ lobe (yellow), α′/β′ lobe (red), and α/β lobe (blue). These lobes are also distinguished by their expression of FasII (magenta). The α and α′ lobes are referred to collectively as the dorsal lobe (DL). The medial lobe (ML) includes the γ, β, and β′ lobes. Asterisks demarcate the peduncle. (Scale bar: 50 μm.) (B) Representative image illustrating phenotypes seen with RacGAP50C RNAi in the MB. Xs mark enlarged cell bodies; *caret;s follow the projection of axons overextending from the DL (white ⁁) and the ML (yellow ⁁). #s mark axons that bypass the peduncle and emerge in a medial trajectory from the dendritic region. (C) Quantitative analysis of RacGAP50C RNAi phenotypes. Heterozygous background of tum enhances MB phenotypes seen with weak and strong transgenic RacGAP50C RNAi lines. (D) Sequence analysis of two independently generated tum chromosomes revealed that they both contain nonsense mutations (tum1:R281STOP and tum347:E114STOP) in the coding region of RacGAP50C. CC, coiled-coil domain.
Knockdown of RacGAP50C in MB neurons results in reduction of cell number and axon overextension (Fig. 1B). The enlarged cells and reduction of neurons derived from the Nbs are reminiscent of cytokinesis defects seen in MB neurons that are homozygous mutant for small GTPase RhoA (25) and is consistent with previous work identifying RacGAP50C and its homologues as a critical regulator of cytokinesis (8–12). In addition to the cytokinetic phenotype, the normal axon trajectory is disrupted with RacGAP50C reduction. Dorsal projecting axon branches are normally confined within a morphologically distinct dorsal lobe that extends toward the dorsal surface of the brain (Fig. 1A). However, with reduced RacGAP50C, axons extend beyond the dorsal lobe and project toward the midline of the brain (arrowheads in Fig. 1B, and Fig. 1C). Occasionally, processes also target incorrectly and extend ventromedially from the dendritic region of the MB (#s in Fig. 1B). This phenotype suggests a previously unrecognized role for RacGAP50C activity in the postmitotic neuron: regulating neuronal projections.
RacGAP50C Corresponds to tumbleweed (tum). To confirm the RNAi phenotype of RacGAP50C and expand phenotypic analysis, we sought genetic loss-of-function mutants. Because phenotypes caused by RNAi-based knockdown of gene activity are predicted to be enhanced by losing one copy of the corresponding endogenous gene (for an example, see ref. 6), we used this strategy to look for candidate RacGAP50C alleles in the cytological position 50C region where RacGAP50C is mapped (27). We tested several candidates from our MB axon guidance screen (28), as well as candidates from a dendritic development screen (29). We found that heterozygosity of the tum mutant chromosome significantly enhanced the rough eye RNAi phenotype caused by knockdown of RacGAP50C (data not shown). We then tested the ability of candidate chromosomes to enhance the RacGAP50C RNAi phenotype in the MB and similarly found that only tum enhanced the neuronal knockdown phenotype (Fig. 1C and data not shown). These data suggests that tum is a strong candidate for the RacGAP50C gene.
To directly test this hypothesis, we sequenced the RacGAP50C region of two alleles of tum (tum1 and tum347) and found they both contain nonsense mutations (Fig. 1D). This key piece of evidence, combined with the genetic enhancement of RacGAP50C RNAi phenotype by tum mutation, the similarity of tum loss-of-function and RacGAP50C RNAi phenotypes, and the transgenic rescue of tum phenotypes by RacGAP50C genomic DNA (see below), established unequivocally that RacGAP50C is encoded by tum. According to conventions of fly genetics, we hereafter refer to RacGAP50C as Tumbleweed (Tum). Because both tum alleles are nonsense mutations before the catalytic RhoGAP domain and the predicted diacyl glycerol-binding domain (Fig. 1D), both alleles are likely null.
Tum and Its Partner in Cytokinesis, Pav, Are Required for Nb Proliferation. To examine the role of Tum and its binding partner, Pav, in the dividing Nbs and in postmitotic neurons, we generated MARCM clones in newly hatched larvae (19). MB Nb clones generated at this time produce all three neuronal types (Fig. 2 A and C). Loss of Tum or Pav causes arrest of Nb proliferation and prevents the continued generation of later-born α′/β′ and α/β neurons (Fig. 2 D and E). The presence of enlarged cells is indicative of cytokinesis defects consistent with the Tum RNAi phenotype (Fig. 1B) and the function of Tum and Pav in regulating cytokinesis (see above).
Single-cell MB neuron clones that are homozygous mutant for tum or pav are grossly normal in their axonal and dendritic projections. The number of γ neuron terminal axon branches are as follows: FRT 42D control, 12.14 ± 0.6, n = 50; FRT 42D tum347, 11.94 ± 0.47, n = 52; FRT 2A control, 14.3 ± 1.16, n = 9; and FRT 2A pavB200, 15.81 ± 0.8, n = 32. The number of dendritic claws are as follows: FRT 42D control, 11.7 ± 1.3, n = 19; FRT 42D tum347, 12.5 ± 1.3, n = 14; and FRT 2A pavB200, 11.3 ± 0.7, n = 23. The number of major dendritic branches are as follows: FRT 42D control, 5.2 ± 0.6, n = 18; FRT 42D tum347, 4.4 ± 0.5, n = 14; and FRT 2A pavB200,5 ± 0.4, n = 23. Because of the severe proliferation phenotypes, we were not able to generate Nb clones large enough to encompass later-born α′/β′ or α/β MB neurons that project to the dorsal lobes. Therefore, we cannot directly compare the axon misrouting phenotypes due to tum knockdown by RNAi (Fig. 1 B and C) and loss-of-function phenotypes. However, we did observe axon overextension defects in both Tum and Pav Nb clones expressing UAS-cDNA transgenes that partially rescue the cytokinetic phenotype (Fig. 2F and data not shown), supporting the notion that optimal Tum activity is required for correct axon projection.
RhoGAP Activity of Tum Is Not Critical for Nb Proliferation but Is Required to Limit Axon Extension. Because of the limited ability of the GAL4-dependent transgenes to rescue the Nb proliferation phenotype in MARCM clones, we generated transgenic flies by using genomic DNA encompassing the tum locus (gTum). One copy of WT gTum rescues the lethality of tum-/- and fully restores Nb proliferation as judged by increased cell number, the loss of enlarged cell bodies (Fig. 2G Inset), and the generation of late-born neurons from homozygous mutant clones (Table 1). By contrast, gTum transgenes with a deletion of the predicted Pav-binding region of Tum (8, 16) fail to rescue the proliferation phenotype associated with tum Nb clones (Fig. 2I and Table 1), supporting the necessity of Pav binding for Tum function in cytokinesis. Introduction of a point mutation (gTumR417L) of the conserved arginine residue critical for GAP activity (see below and refs. 1 and 30) abolishes the ability of gTum to rescue the lethality of tum but, surprisingly, is able to largely rescue the Nb proliferation phenotype (Fig. 2H Inset and Table 1). More than 80% of Nb clones are able to generate the latest born neuron class, and 50% of Nb fully rescue cell-proliferation defects.
Table 1. Quantification of neuroblast proliferation activity.
| Genotype | >1/2 α/β (full α′/β′ and γ), % | <1/2 α/β 1/2 α′/β′, % | <1/2 α′/β′, % | No α′/β′ >1/2 γ, % | <1/2 γ, % | n |
|---|---|---|---|---|---|---|
| Control | 85.2 | 3.7 | 3.7 | 0 | 7.4 | 27 |
| tum347 | 0 | 0 | 0 | 3.5 | 96.5 | 29 |
| gTumWT plus tum347 | 95 | 0 | 0 | 5 | 0 | 20 |
| gTumR417L plus tum347 | 45 | 30 | 10 | 10 | 5 | 20 |
| gTumΔZ4 plus tum347 | 0 | 0 | 0 | 12.5 | 87.5 | 8 |
With the rescue of Nb proliferation, it is possible to examine a larger population of neurons for their axon targeting. Restoration of later born neuron types by gTumWT or gTumR417L in tum Nb clones allowed for normal axon guidance of later born neuron types (Fig. 2 G and H). Most medial projecting axons of the MB extend toward the midline and stop. A small fraction of WT Nb clones extend their axons, mostly the α/β class, beyond the midline (Fig. 2 J). With loss of Tum's RhoGAP activity, the later born neurons are twice as likely to extend beyond their normal target region in the medial lobe compared with control Nb clones, and the degree of overextension is more severe (arrowheads in Fig. 2H and quantified in Fig. 2 J). Dorsal lobe overextension similar to the RNAi phenotypes is also occasionally seen (Fig. 2 J). Five of the 12 Nb clones with axon overextensions exhibit full rescue of Nb proliferation phenotypes as judged by the lack of enlarged cells and the full innervation of the lobes targeted by the latest born neurons, α/β lobes. This finding suggests that the axon phenotype of tum is not a secondary consequence of cytokinesis defects; rather, it reflects a role of tum in regulating axon extension in postmitotic neurons. In addition, regulation of axon extension appears to depend more on the GAP activity of Tum compared with regulation of cell proliferation.
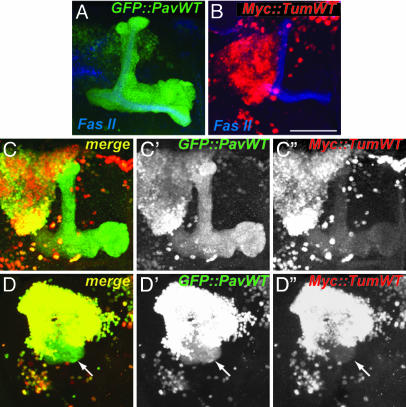
Tum and Pav Mutually Regulate Their Subcellular Localization. A role of Tum, and potentially Pav, in postmitotic neurons would predict that these proteins should be expressed there. We were not able to detect endogenous Tum or Pav in the intact CNS by using existing or our newly generated antibodies. We therefore expressed epitope-tagged transgenes of Tum and Pav, alone and in combination, to examine their subcellular localization in MB neurons. A series of GFP-tagged Pav WT (GFP::PavWT) and mutant transgenes have previously been used to study their localization in Drosophila egg chambers and live embryos (17, 31). It was found that GFP::PavWT not only rescues pav mutants but has similar localization to endogenous Pav, the ring canals (remnants of incomplete cytokinesis rings), and the oocyte nuclei (17). When expressed in MB neurons, we found that GFP::PavWT is detected in the nuclei and dendrites and is highly concentrated in the axons of the MB neurons (Fig. 3A).
Fig. 3.
Tum and Pav influence each other's localization, and their association leads to disruption of axon routing. (A) UAS-GFP::PavWT is detected in the axons as well as the nuclei of MB neurons when driven by GAL4-OK107. (B) UAS-MYC::TumWT is restricted to the nuclei of MB neurons. (C and D) Two examples of coexpression of UAS-GFP::PavWT and UAS-MYC::Tum leading to their colocalizations. C and D are merged signals for GFP (C′ and D′) and MYC (C′′ and D′′). Example in D shows that coexpression leads to axon misrouting that forms a ball-like shape close to the cell body region of the MB complex (arrow). (Scale bar: 50 μm.)
Surprisingly, UAS-MYC::TumWT is enriched in the nuclei when overexpressed in MB neurons (Fig. 3B). Expression of TumWT tagged with yellow fluorescent protein at its carboxyl terminus revealed a similar nuclear localization (unpublished observations), indicating that the nuclear localization is not an artifact of its epitope tag. MYC::TumWT localization was always predominantly nuclear in MB neurons examined in larval and pupal stages (data not shown).
Coexpression of UAS-GFP::PavWT and UAS-MYC::Tum alters the subcellular localization of both proteins (Fig. 3C). These proteins are colocalized in MB neurons, and their localization pattern is a combination of the subcellular distributions of MYC::Tum alone and GFP::PavWT alone. GFP::PavWT is more concentrated in nuclei and less concentrated in axons compared with its expression alone (compare green channels in Fig. 3 A, C′, and D′); however, MYC::Tum is now weakly detectable in axons (Fig. 3 C′′ and arrow in D′′). These observations are consistent with the notion that Tum and Pav bind to each other and regulate each other's subcellular localization in postmitotic neurons.
Accumulation of Pav/Tum in the Cytoplasm Disrupts Axon Development. Expression of UAS-GFP::PavWT alone or UAS-MYC::TumWT alone does not affect the gross MB axon projection (data not shown and Fig. 4E) as judged by coexpression with mCD8::GFP. Expression of both UAS-MYC::TumWT and UAS-GFP::PavWT leads to a high occurrence of axon misguidance (14 of 28 MBs examined). As exemplified in Fig. 3D (arrow), the majority of FasII-positive axons form a ball-like structure at their initial trajectory, and only a small subset of axons is able to extend beyond this ball-like structure into the medial lobe. In other cases, axons fail to enter their correct path at the branch point for the dorsal and medial lobe (data not shown). One possible explanation is that cytoplasmic accumulation of Tum, by means of Pav binding, disrupts axon development.
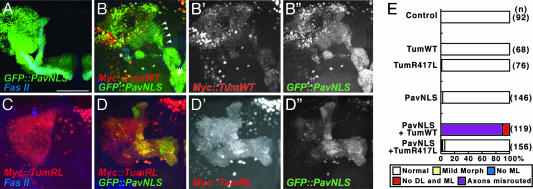
Fig. 4.
Tum/Pav coexpression phenotype is enhanced by increased cytoplasmic concentration and abolished by a mutation in the RhoGAP domain. (A) GFP::PavNLS is excluded from the nucleus and detected in the axons and dendrites of MB neurons. (B–B′′) Coexpression of UAS-GFP::PavNLS and UAS-MYC::Tum leads to their overlapping subcellular localization. Arrowheads follow misrouted axons that bypass the peduncle (*) and misguide to a region ventral to the medial lobe (#) before innervating part of the medial lobe. B is a merge of MYC (B′) and GFP (B″) signals. (C) MYC::TumR417L is restricted to the nuclei of MB neurons when expressed alone. (D–D′′) Coexpression of UAS-GFP::PavNLS and UAS-MYC::TumR417L leads to a similar overlap of their subcellular localization but does not lead to a gross change in the axonal targeting of MB neurons. D is a merge of MYC (D′) and GFP (D″) signals. (Scale bar: 50 μm.) (E) Quantification of axon misrouting with the expression of Tum and Pav transgenes. DL, dorsal lobe; ML, medial lobe. See Materials and Methods for details of analysis.
To further test this hypothesis, we made use of a Pav mutant transgene. Mutations in three of the nuclear localization signals of Pav (PavNLS) blocks the nuclear localization of Pav protein (17, 31). As predicted, the nuclear localization of GFP::PavWT, but not its dendritic or axonal localization, is disrupted in GFP::PavNLS (Fig. 4A). GFP::PavNLS does not lead to any gross morphological changes when expressed alone (Fig. 4E). As with UAS-GFP::PavWT, coexpression of GFP::PavNLS and MYC::Tum resulted in altered subcellular localization of both proteins: GFP::PavNLS is localized to the nucleus, and more MYC::Tum is “dragged” out of the nucleus (Fig. 4B, compared with Fig. 3 C and D). Interestingly, almost all MB axons were misrouted under this condition (94%; Fig. 4E). In addition to forming a cluster of axons in a ball-like form as with GFP::PavWT, axons were seen to bypass their normal path via the peduncle (asterisk in Fig. 4B) and instead project medially toward the front of the brain just below the normal medial lobe (# in Fig. 4B) and then project dorsally to innervate the medial lobe. The gain-of-function phenotype seen with coexpression of Tum and Pav suggests that they form a complex that disrupts the normal development of axons. The occurrence and severity of this axon misrouting is increased when the Pav transgene has a decreased drive to enter the nucleus, indicating that Pav may act as a transporter of Tum and that local concentrations of Tum in the axon directly influences its capacity to affect axonal development.
To test whether the GAP activity is necessary for the axon misrouting, we introduced a point mutation (TumR417L) of the conserved arginine residue critical for GAP activity (1, 30) in the UAS construct. As with the WT form, UAS-MYC::TumR417L localizes to the nuclei of MB neurons when expressed in MB neurons and does not lead to morphological phenotypes (Fig. 4 C and E). When coexpressed with UAS-GFP::PavNLS, the subcellular localization of MYC::TumR417L and GFP::PavNLS reflects a pattern similar to that seen with WT Tum coexpression, with MYC::TumR417L detected in both axons and in the nuclei of MB neurons (Fig. 4D). However, with the loss of the conserved arginine in the GAP domain, axon disruption capability of Tum is almost completely abolished (Fig. 4E), suggesting that the GAP activity of Tum mediates the gain-of-function phenotype seen with Tum and Pav coexpression.
Discussion
In this study, we show that RacGAP50C is encoded by tum, a mutation previously identified in a forward genetic screen for dendritic morphogenesis (29). By using loss-of-function mutations, we show that Tum and its binding partner, Pav, a kinesin superfamily protein, are both required for MB Nb proliferation, consistent with previously characterized functions of this protein complex in regulating cytokinesis in C. elegans, Drosophila, and mammals (7–9, 12, 13, 32, 33). Our structure-function analysis suggests that Tum's binding to Pav, but not its GAP activity, is essential for its function in cytokinesis. Recent evidence has found that the Rho-specific guanine nucleotide exchange factor, Pebble, binds directly to Tum's coiled-coil domain (8). Pebble itself is critical in cytokinesis to activate RhoA and generate the contractile ring (34). It has been proposed that Pebble's association with Tum allows for its localization to the midzone to promote the initiation of cytokinesis (8). The function of the GAP domain of Tum in promoting cytokinesis has remained enigmatic. The GAP activity of the mammalian and C. elegans homologues of Tum have little or no RhoA-specific GAP activity but preferentially regulate Rac1 and Cdc42 activity (9–11). Our finding that Tum with a mutation that disrupts the conserved arginine finger necessary for RhoGAP activity (1, 35) allows dividing Nb to generate >500 MB neurons demonstrates that the GAP activity of Tum is not essential for its function during cytokinesis. This observation supports the model that in cytokinesis, Tum acts simply as a scaffolding protein that brings Pebble to the midzone formed by the bundling of antiparallel microtubules through association of Tum with Pav.
We also provide several lines of evidence that in addition to the function of the Tum–Pav complex in cytokinesis, Tum and Pav also have a function in regulating morphogenesis of postmitotic neurons and that this function depends on the GAP activity of Tum. First, RNAi knockdown of Tum results not only in defects of Nb proliferation, but also in axon extension beyond their normal boundary. Second, introducing a tum transgene with a GAP domain mutation into tum-/- Nb clones largely restores normal proliferation but results in the overextension of its axons beyond their normal boundary. The inability to limit axon extension in the MB neuropil suggests a GAP-dependent role of Tum that is reminiscent of the neuronal function attributed to Tum through RNAi knockdown. Third, Tum and Pav regulate each other's subcellular localization, consistent with the notion that they form a complex in postmitotic neurons. Fourth, the same point mutation in the GAP domain of Tum abolishes its axon disruption activity in Tum/Pav coexpression experiments, consistent with the GAP activity being essential in regulating axon development.
Given the potential role of Tum as a GAP in regulating axon development, it is somewhat surprising that the protein should have a predominant nuclear localization when expressed in postmitotic neurons. The Pav coexpression experiments shed some light: We found that accumulation of Tum in the cytoplasm leads to severe disruption of axon development. The phenotypes are highly reminiscent of loss-of-function mutant phenotypes for the Rac GTPases, where axons collect in a ball at the base of or misroute from the cell body region (20), suggesting that cytoplasmic accumulation of Tum down-regulates Rac activity. This observation is consistent with a previous report that RacGAP50C regulates wing disk development by regulating Drosophila Rac1 (36). In contrast, loss of RhoA or Cdc42 in MB neurons does not affect axon guidance (25, 26). We speculate that in postmitotic neurons, the distribution of Tum contributes to the spatial regulation of the Rac GTPase activity essential for proper morphogenesis. Because Rac GTPases are also essential for axonal growth (20), limited and localized cytoplasmic Tum could reduce the activity of Rac GTPases and hence restrict axon growth, consistent with our finding that complete loss of Tum results in axon overgrowth. As a kinesin shown to bind to and bundle microtubules (7), Pav could act to transport Tum along microtubules to modulate Rac activity in axon growth and guidance.
Both Tum and Pav function have been implicated in dendritic morphogenesis. tum was originally identified as a mutation that leads to tangled dendritic branches in the sensory neurons of the Drosophila embryo (29). Disruption of the mammalian form of Pav, CHO1/MKLP1, in postmitotic and differentiated neurons in culture lead to the rearrangement of microtubule polarity and the subsequent loss of dendrites (16). We have not observed defects in dendritic arborization or targeting in single-cell MARCM clones that are homozygous mutant for tum or pav in either the MB neurons or the second-order olfactory projection neurons (data not shown). There are several possible explanations. First, the dendritic defects seen in tum homozygous embryos may be a secondary consequence of cytokinesis defects or due to nonautonomous effects of tum in neighboring cells. Second, the Tum/Pav pathway may be redundant with other pathways in MB or projection neurons in regulating dendritic morphogenesis. Third, perdurance of Tum/Pav proteins in single-cell clones may be sufficient to allow proper dendritic morphogenesis. Future studies using neuronal types that have more elaborate and stereotyped dendritic trees (e.g., 26) may help distinguish these possibilities.
Acknowledgments
We thank A. Maresh, P. Billuart, X. Zhao, and D. Luginbuhl for advice and help. A.Y.N.G. was a Howard Hughes Medical Institute Predoctoral Fellow. Y.-N.J. is a Howard Hughes Medical Institute Investigator. This work was supported by a grant from the National Institutes of Health (to L.L.).
Author contributions: A.Y.N.G. and L.L. designed research; A.Y.N.G. performed research; Y.-N.J. contributed new reagents/analytic tools; A.Y.N.G. and L.L. analyzed data; and A.Y.N.G. and L.L. wrote the paper.
Abbreviations: GAP, GTPase-activating protein; Pav, Pavarotti; tum, tumbleweed; Tum, Tumbleweed; Nb, neuroblast; MB, mushroom body; FasII, fasciclin II; MARCM, mosaic analysis with a repressible cell marker; gTum, genomic DNA encompassing the tum locus.
References
- 1.Scheffzek, K., Ahmadian, M. R., Kabsch, W., Wiesmuller, L., Lautwein, A., Schmitz, F. & Wittinghofer, A. (1997) Science 277, 333-338. [DOI] [PubMed] [Google Scholar]
- 2.Lander, E. S. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 3.Venter, J. C. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 4.Endris, V., Wogatzky, B., Leimer, U., Bartsch, D., Zatyka, M., Latif, F., Maher, E. R., Tariverdian, G., Kirsch, S., Karch, D. & Rappold, G. A. (2002) Proc. Natl. Acad. Sci. USA 99, 11754-11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billuart, P., Bienvenu, T., Ronce, N., des Portes, V., Vinet, M. C., Zemni, R., Roest Crollius, H., Carrie, A., Fauchereau, F., Cherry, M., et al. (1998) Nature 392, 923-926. [DOI] [PubMed] [Google Scholar]
- 6.Billuart, P., Winter, C. G., Maresh, A., Zhao, X. & Luo, L. (2001) Cell 107, 195-207. [DOI] [PubMed] [Google Scholar]
- 7.Mishima, M., Kaitna, S. & Glotzer, M. (2002) Dev. Cell 2, 41-54. [DOI] [PubMed] [Google Scholar]
- 8.Somers, W. G. & Saint, R. (2003) Dev. Cell 4, 29-39. [DOI] [PubMed] [Google Scholar]
- 9.Jantsch-Plunger, V., Gonczy, P., Romano, A., Schnabel, H., Hamill, D., Schnabel, R., Hyman, A. A. & Glotzer, M. (2000) J. Cell Biol. 149, 1391-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toure, A., Dorseuil, O., Morin, L., Timmons, P., Jegou, B., Reibel, L. & Gacon, G. (1998) J. Biol. Chem. 273, 6019-6023. [DOI] [PubMed] [Google Scholar]
- 11.Kawashima, T., Hirose, K., Satoh, T., Kaneko, A., Ikeda, Y., Kaziro, Y., Nosaka, T. & Kitamura, T. (2000) Blood 96, 2116-2124. [PubMed] [Google Scholar]
- 12.Minoshima, Y., Kawashima, T., Hirose, K., Tonozuka, Y., Kawajiri, A., Bao, Y. C., Deng, X., Tatsuka, M., Narumiya, S., May, W. S., Jr., et al. (2003) Dev. Cell 4, 549-560. [DOI] [PubMed] [Google Scholar]
- 13.Mishima, M., Pavicic, V., Gruneberg, U., Nigg, E. A. & Glotzer, M. (2004) Nature 430, 908-913. [DOI] [PubMed] [Google Scholar]
- 14.Portereiko, M. F., Saam, J. & Mango, S. E. (2004) Curr. Biol. 14, 932-941. [DOI] [PubMed] [Google Scholar]
- 15.Van de Putte, T., Zwijsen, A., Lonnoy, O., Rybin, V., Cozijnsen, M., Francis, A., Baekelandt, V., Kozak, C. A., Zerial, M. & Huylebroeck, D. (2001) Mech. Dev. 102, 33-44. [DOI] [PubMed] [Google Scholar]
- 16.Yu, W., Cook, C., Sauter, C., Kuriyama, R., Kaplan, P. L. & Baas, P. W. (2000) J. Neurosci. 20, 5782-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minestrini, G., Mathe, E. & Glover, D. M. (2002) J. Cell Sci. 115, 725-736. [DOI] [PubMed] [Google Scholar]
- 18.Lee, T. & Luo, L. (1999) Neuron 22, 451-461. [DOI] [PubMed] [Google Scholar]
- 19.Lee, T., Lee, A. & Luo, L. (1999) Development (Cambridge, U.K.) 126, 4065-4076. [DOI] [PubMed] [Google Scholar]
- 20.Ng, J., Nardine, T., Harms, M., Tzu, J., Goldstein, A., Sun, Y., Dietzl, G., Dickson, B. J. & Luo, L. (2002) Nature 416, 442-447. [DOI] [PubMed] [Google Scholar]
- 21.Heisenberg, M. (2003) Nat. Rev. Neurosci. 4, 266-275. [DOI] [PubMed] [Google Scholar]
- 22.Crittenden, J. R., Sloulakis, E. M. C., Han, K.-A., Kalderon, D. & Davis, R. L. (1998) Learn. Mem. 5, 38-51. [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, K., Awano, W., Suzuki, K., Hiromi, Y. & Yamamoto, D. (1997) Development 124, 761-771. [DOI] [PubMed] [Google Scholar]
- 24.Ng, J. & Luo, L. (2004) Neuron 44, 779-793. [DOI] [PubMed] [Google Scholar]
- 25.Lee, T., Winter, C., Marticke, S. S., Lee, A. & Luo, L. (2000) Neuron 25, 307-316. [DOI] [PubMed] [Google Scholar]
- 26.Scott, E. K., Reuter, J. E. & Luo, L. (2003) J. Neurosci. 23, 3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.FlyBase Consortium (2003) Nucleic Acids Res. 31, 172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuter, J. E., Nardine, T. M., Penton, A., Billuart, P., Scott, E. K., Usui, T., Uemura, T. & Luo, L. (2003) Development (Cambridge, U.K.) 130, 1203-1213. [DOI] [PubMed] [Google Scholar]
- 29.Gao, F. B., Brenman, J. E., Jan, L. Y. & Jan, Y. N. (1999) Genes Dev. 13, 2549-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, R., Zhang, B. & Zheng, Y. (1997) J. Biol. Chem. 272, 32830-32835. [DOI] [PubMed] [Google Scholar]
- 31.Minestrini, G., Harley, A. S. & Glover, D. M. (2003) Mol. Biol. Cell 14, 4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirose, K., Kawashima, T., Iwamoto, I., Nosaka, T. & Kitamura, T. (2001) J. Biol. Chem. 276, 5821-5828. [DOI] [PubMed] [Google Scholar]
- 33.Adams, R. R., Tavares, A. A., Salzberg, A., Bellen, H. J. & Glover, D. M. (1998) Genes Dev. 12, 1483-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prokopenko, S. N., Brumby, A., O'Keefe, L., Prior, L., He, Y., Saint, R. & Bellen, H. J. (1999) Genes Dev. 13, 2301-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vetter, I. R. & Wittinghofer, A. (2001) Science 294, 1299-1304. [DOI] [PubMed] [Google Scholar]
- 36.Sotillos, S. & Campuzano, S. (2000) Development (Cambridge, U.K.) 127, 5427-5438. [DOI] [PubMed] [Google Scholar]