Abstract
Neural stem cell (NSC) transplantation has great therapeutic potential for neurodegenerative diseases and central nervous system injuries. Successful NSC replacement therapy requires precise control over the cellular behaviors. However, the regulation of NSC fate is largely unclear, which severely restricts the potential clinical applications. To develop an effective model, we designed an assembled microfluidic system to engineer NSC niches and assessed the effects of various culture conditions on NSC fate determination. Five types of NSC microenvironments, including two-dimensional (2D) cellular monolayer culture, 2D cellular monolayer culture on the extracellular matrix (ECM), dispersed cells in the ECM, three-dimensional (3D) spheroid aggregates, and 3D spheroids cultured in the ECM, were constructed within an integrated microfluidic chip simultaneously. In addition, we evaluated the influence of static and perfusion culture on NSCs. The efficiency of this approach was evaluated comprehensively by characterization of NSC viability, self-renewal, proliferation, and differentiation into neurons, astrocytes, or oligodendrocytes. Differences in the status and fate of NSCs governed by the culture modes and micro-niches were analyzed. NSCs in the microfluidic device demonstrated good viability, the 3D culture in the ECM facilitated NSC self-renewal and proliferation, and 2D culture in the static state and spheroid culture under perfusion conditions benefited NSC differentiation. Regulation of NSC self-renewal and differentiation on this microfluidic device could provide NSC-based medicinal products and references for distinct nerve disease therapy.
I. INTRODUCTION
Neural stem cells (NSCs) originate from the central nervous system (CNS) during development and adulthood,1,2 which have the ability to self-renew through cell division and differentiate into all cell types of neural lineages including neurons, astrocytes, and oligodendrocytes.3,4 As tissue-specific stem cells, NSCs can be differentiated to replace lost or injured neurons or glial cells to reconstitute healthy neural cell populations, which has great potential as a functional stem cell therapy for CNS injuries such as ischemic stroke, spinal cord injury, and traumatic brain injury, and neurodegenerative disorders such as Parkinson's and Alzheimer's disease.5,6 NSC transplantation has already shown neuronal regeneration and functional behavior recovery in diseased animals.7 Furthermore, systematic human trials involving human NSC therapy have been conducted successfully.8 However, many obstacles still exist, which hinder the clinical application of NSCs, such as low survival rates, the lack of effective methods to control NSC fate, teratoma formation, and low tissue integration after transplantation.9
NSCs are highly sensitive to their chemical and physical microenvironments. Therefore, the importance of the NSC niche has been highlighted in previous investigations.10,11 In particular, the NSC niche integrates heterogeneity, compartmentalization, and a three-dimensional (3D) configuration. Furthermore, construction of a NSC niche requires the extracellular matrix (ECM), soluble factors, vasculature, and neighboring cellular components (neuron, glia).12,13 These niches anatomically house NSCs and functionally control their development in vivo, which in turn affect the survival, proliferation, recruitment, and fate of NSCs. More importantly, NSC niches support NSC self-renewal and, under certain circumstances, guide NSC differentiation into specific lineages by the regulation of soluble factors and intercellular/cell–ECM interactions.14–16 In practice, functional testing of NSCs in animal models has various limitations, such as low throughput, high costs, intensive labor, and experimental variations. Recently, the appearance of in vitro models incorporating NSC niche elements has facilitated studies of individual factors in NSC fate specification and possible NSC applications in regenerative medicine.17,18 Representative engineering methods used to build in vitro NSC niches include bioactive polymers functionalized with multivalent ligands or peptides,19,20 synthetically or biologically derived hydrogels,21,22 3D substrates at the microscale or nanoscale with different stiffness or topographical properties,23,24 and genetically engineered ECMs for artificial niches.25 However, the absence of physiologically realistic NSC niche models with precise spatiotemporal control and effective methods for evaluation has hindered research of NSC fates.
The intrinsic merits of microfluidic systems in stem cell research include precise control of fluidic conditions, low consumption of rare cells and reagents, high throughput screening, single cell handling, real-time analysis, perfusion and long term cultures, and high integration.26,27 Consequently, several studies on NSC niche construction using microfluidic technology have been reported, aiming to precisely and flexibly engineer physiological environments in vitro and potentially regulate the spatial and temporal progression of self-renewal or differentiation processes of NSCs. Using microfluidic methods, several individual factors regulating the NSC niche have been studied, such as cytokines, hypoxia, mechanical compression, 3D hydrogels, vessel flow, and polymer fiber scaffolds.28–33 Despite these developments, most studies have focused on the effects of a single factor on NSCs or employed a 2D culture model on the surface of microchannels in devices. Whether distinct niche locations affect NSC fates remains unclear, and the optimal culture conditions to guide NSC behaviors still need clarification. In addition, highly integrated systems with easy operation will benefit biologists and clinicians to address specific questions in stem cell biology.
Here, we developed an innovative and straightforward approach to investigate the effects of micro-niches on NSC determination using an assembled microfluidic array. In our system, we considered the dimension (2D vs. 3D) and heterogeneity of NSC culture, ECM components, and fluid shear conditions to construct various microenvironments. The advantage of our method is that all of the individual culture conditions and the corresponding effects on NSC fate could be examined with the same integrated microfluidic chip simultaneously. Under the various culture conditions, viability, self-renewal, proliferation, and differentiation behaviors of NSCs presented different phenomena. We found that NSCs in the microfluidic device demonstrated good viability, 3D culture in the ECM facilitated NSC self-renewal and proliferation, and that 2D culture in the static state and spheroid culture under perfusion conditions promoted NSC differentiation. Compared with conventional NSC culture, this study provides a novel model for investigation of the niche-induced NSC fate in vitro by screening a wide range of conditions. Additionally, NSC-based medicinal products with self-renewal or differentiation capability could be realized with our platform, which offers guidance for in vitro NSC culture to treat specific nervous system diseases.
II. MATERIALS AND METHODS
A. Design and fabrication of the microfluidic device
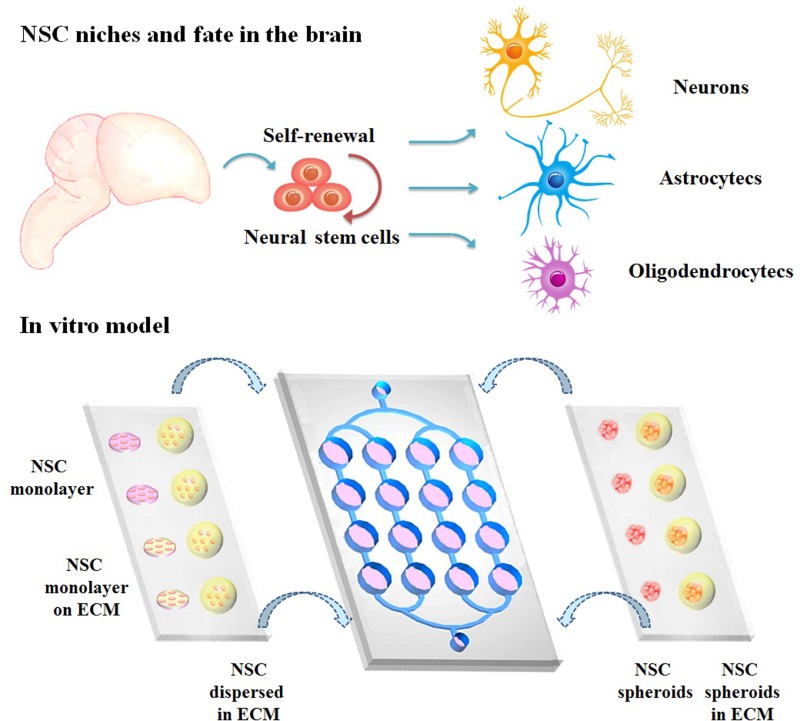
A schematic of the microfluidic device for cell culture is shown in Fig. 1. The entire microfluidic device consisted of a poly-dimethylsiloxane (PDMS, Sylgard 184, Dow Corning, USA) microfluidic chip and two PDMS-coated glass slides as the cover. The lower layer (microfluidic chip) consisted of perfusion channels and an open system with cell culture chambers as a 4 × 4 array. The array unit shared the terminal reservoir. The upper layer (glass covers) was coated with PDMS, and this hydrophobic surface was used for cell culture. The three parts were assembled together to seal the microfluidic 4 × 4 chamber array by the PDMS-PDMS surface tension and could be readily detached after the experiment for subsequent analyses by the reversible bonding.
FIG. 1.
Schematic diagram of NSC niches in the brain and in vitro model on the assembled microfluidic system. NSCs were cultured as (1) NSC monolayer, (2) NSC monolayer on the ECM, (3) NSCs dispersed in the ECM, (4) NSC spheroids, or (5) NSC spheroids in the ECM. Culture conditions were either perfusion or static.
For microfluidic chip fabrication, a positive mold patterned from SU-8 negative photoresist (3035, Micro Chem) using a standard soft lithography procedure was used to fabricate the PDMS substrate containing microchannels and chambers. After punching, plasma treatment, and sealing, the microfluidic chip was formed as depicted in Fig. 1. The chip consisted of perfusion channels with a width and height of 300 μm and a set of open chambers with a width of 5 mm, height of 2 mm, and pitch of 4 mm. The microfluidic chip was modified and sterilized to form a hydrophilic and sterile surface for smooth medium addition to the chip and maintaining cell viability. PDMS was spin-coated and solidified on the two clear glass slides. These slides were assembled to the microfluidic chip under the PDMS-PDMS bonding strength, aligning the cell culture region of the PDMS slide to the chamber region in the microfluidic chip. After sealing, only the inlet and outlet sites were open for fluidic access.
B. Isolation and culture of NSCs
NSCs were freshly isolated from the cortexes of Sprague-Dawley rat (Experimental Animal Center of Dalian Medical University, Dalian, China) fetuses at embryonic day 13 and then expanded in a cell culture dish at a density of 2 × 105 cells/ml in Dulbecco's Modified Eagle Medium/Nutrient Mixture F12 (Gibco BRL, MD, USA) supplemented with the mitogenic factors, 20 ng/ml epidermal growth factor (EGF, Gibco BRL) and basic fibroblast growth factor (bFGF, Gibco BRL) to form neurospheres at 37 °C, 5% CO2, 21% O2, and constant humidity. NSC growth medium was replaced every 48 h. For NSC differentiation, spontaneous differentiation was induced by removing EGF and bFGF.
C. Inoculation of NSCs and assembly of the microfluidic device
Five groups under different culture conditions were tested, including two 2D groups (NSC monolayer and NSC monolayer on the ECM), NSCs dispersed in the ECM, NSC spheroids, and NSC spheroids in the ECM, as shown in Fig. 1. Three days were needed for NSC inoculation. For each group, third passage neurospheres were dissociated into a single cell suspension at a seeding density of 1 × 106 cells/ml.
On the first day, a PDMS slide with an area sufficient to cover the two rows of chambers in the microfluidic chip was sterilized by ultraviolet light exposure for 1 h. The NSC suspension was manually dripped onto the PDMS slide regions, aligning the rows of chambers for NSC spheroids and NSC spheroids in the ECM in the lower microfluidic chip. Because of the hydrophobic surface, two droplet arrays with 10 μl/drop were formed on the PDMS surface. The slide was inverted and placed in a humidified chamber with 5% CO2 at 37 °C to allow the NSCs to sink and aggregate into multicellular spheroids. Then, poly-ornithine (1 mg/ml, PO, Advanced BioMatrix, USA) was dripped onto another PDMS slide with a surface area covering the two rows of chambers in the microfluidic chip, aligning the position of the chambers for the monolayer group. PO modification on the PDMS surface was achieved by overnight incubation at room temperature for the monolayer group.
On the second day, modification liquid PO was replaced with 10 μg/ml laminin (LN, Sigma, USA), followed by incubation for 4 h at 37 °C and then washing with phosphate buffered saline (PBS). A collagen hydrogel (5 mg/ml, rat tail tendon collagen type I; Shengyou Biotechnology Co., Ltd., Zhejiang, China) was used as the ECM for NSC culture. The hydrogel was diluted in a mixture of 10× PBS and distilled deionized water. The pH of the collagen solution was adjusted to 7.0 with 0.1 mol/l NaOH. For the NSC monolayer on the ECM, a collagen solution (1 mg/ml) was dripped beside the PO-LN modification array, aligning the position of the chambers for the NSC monolayer on the ECM. After solidification in a 37 °C incubator for 40 min, collagen coating was achieved. A suspension of NSCs at 10 μl/drop was manually dripped onto the modified PDMS regions for the NSC monolayer and NSC monolayer on the ECM. The seeded slide was placed in the incubator overnight for NSC attachment onto the PDMS and collagen surface.
On the third day, for the NSC monolayer, NSC monolayer on the ECM, and NSC spheroids, cell preparations were completed. For NSCs dispersed in the ECM, NSCs suspended in a collagen solution (1 mg/ml) were dripped onto the PDMS slide at 10 μl/drop, aligning the row of the chamber for NSCs dispersed in the ECM. For NSC spheroids in the ECM, after aspiration of the culture medium by pipetting, multicellular spheroids on the slide were dripped with collagen solution at 10 μl/drop. The collagen drops containing dispersed NSCs and NSC spheroids allowed them to crosslink and solidify in an incubator for 40 min.
The two PDMS slides with a 4 × 4 NSC array were assembled to the chamber region of the lower microfluidic chip by aligning the 4 × 4 NSC array towards the 4 × 4 culture chambers, and the three parts were spring clamped with stainless steel clamps. This sealed the NSC array in the microfluidic chamber for perfusion and static culture.
D. Perfusion and static culture of NSCs in the microfluidic device
After assembly of the microfluidic device, perfusion or static culture was carried out. For static culture, medium was added to the inlet using a pipette to fill the channels and culture chambers and to remove any air bubbles and immerse the NSCs in the five different conditions. Culture medium was replaced with 1.44 ml fresh medium every day. For the perfusion culture, after the procedure of static culture preparation, the culture medium was pumped into the inlet at a constant flow rate of 1 μl/min using a precision syringe pump (LSP02-2A, Baoding Longer Precision Pump Co., Ltd., Hebei, China) and polytetrafluoroethylene (PTFE) tubing (IDEX Health and Science, USA). The integrated device was placed in the humidified CO2 incubator at 37 °C for 4 days. In NSC differentiation experiments, the culture durations were 4, 7, and 14 days.
E. Cell viability assessment
The PDMS slide was detached from the lower microfluidic chip for sample collection after perfusion and static culture. The viability of NSCs under different culture conditions was evaluated by staining with Calcein-AM/PI and Hoechst 33342 (Sigma) in PBS at 1 μg/ml each. PBS washing (twice, 5 min each) was applied to the samples before and after 30 min of incubation in the staining solution. Fluorescence images were obtained using a confocal laser scanning microscope (Leica SP8, Germany). Imaging settings were constant, and each 3D specimen was divided into 30 layers for image acquisition with maintenance of the same pixel and light source intensities. Quantitative statistics were calculated by acquiring five individual stacks before z-axis projection for image analysis using Image-Pro Plus software (IPP 5.0 system, Media Cybernetics, Rockville, MD). Cell viability was evaluated by calculating the live cell percentage of the total cells. The number of cells was determined by counting cells in five randomly chosen fields.
F. Fluorescence staining and imaging
For characterization of NSC maintenance, proliferation, and differentiation, we performed immunostaining. Samples were recycled and treated with 4% paraformaldehyde (Sigma), 0.3% Triton-X100 (Sigma), and 1% bovine serum albumin in PBS and then incubated overnight with primary antibodies against nestin, β-tubulin-III, MAP2, ALDH1L1, GFAP, or SOX10 (Abcam, UK), or Ki-67 (Zhongshan Golden Bridge Biotechnology Co., Ltd, Beijing, China) at 4 °C according to the manufacturer's instructions. After rinsing with PBS, the cells were incubated with tetraethyl rhodamine isothiocyanate (TRITC)-labeled anti-mouse IgG and fluorescein isothiocyanate (FITC)-labeled anti-rabbit IgG (1:100) (Sigma) for 90 min at room temperature. A Hoechst 33258 staining solution (1:1000) (Sigma) was used to stain nuclei and indicate cell positions. Specimens of NSC spheroids, NSCs dispersed in the ECM, and NSC spheroids in the ECM were sectioned into 30 layers for image analysis. Imaging settings and quantitative statistics were the same as those used for the cell viability assessment.
G. Quantitative real-time PCR
Samples were collected from the disassembled microfluidic device for investigation of markers indicating cell lineage determination by quantitative real-time polymerase chain reaction (qRT-PCR). Briefly, total ribonucleic acid (RNA) was extracted from cultured cells. Complementary deoxyribonucleic acid (cDNA) was generated from the purified RNA using a PrimeScript™ RT reagent Kit (Takara, Japan) according to the manufacturer's instructions. qRT-PCR was performed in a Thermal Cycler Dice Real Time System by mixing 2 μl of the synthesized cDNA with SYBR® Premix Ex Taq™ II reagents (Takara, Japan) and appropriate primers according to the manufacturer's protocol. Primer sequences are summarized in Table I. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene for normalization.
TABLE I.
Primer sequences for qRT-PCR.
| Target name | Primer sequences (forward, reverse) |
|---|---|
| GAPDH | 5′-GGCACAGTCAAGGCTGAGAATG-3′ |
| 5′-ATGGTGGTGAAGACGCCAGTA-3′ | |
| MAP2 | 5′-AGCCTGCAGCTCTGCCTTTA-3′ |
| 5′-CTTCCAGTGCAGCTGTTTGTTC-3′ | |
| TUJ1 | 5′-CAGATGCTGGCCATTCAGAGTAAG-3′ |
| 5′-TGTTGCCGATGAAGGTGGAC-3′ | |
| ALDH1L1 | 5′-GTTTGCCGAGCTGACACTGAA-3′ |
| 5′-CGTGGAGCCTGTGAACCCTA-3′ | |
| GFAP | 5′-AGGCTGCTGGAGCAAGACAA-3′ |
| 5′-GCCTTAGTGGCCATTCCAGGTA-3′ | |
| MBP | 5′-ATGGTGAGATTCACCGAGGA-3′ |
| 5′-CATTGTTCTGGATCGCATCTG-3′ | |
| SOX10 | 5′-CGCACCTCCACAATGCTGA-3′ |
| 5′-GAGGTTGGTACTTGTAGTCCGGATG-3′ |
H. Statistical analysis
Quantitative data were presented as means ± standard deviation. Statistical analyses were performed using the Student's t-test. Error bars indicated the standard error of the mean relative to the control. Values of *p < 0.05 and **p < 0.01 were considered as statistically significant (α: NSC monolayer vs. NSC monolayer on the ECM both under perfusion conditions, @: NSC monolayer vs. NSC monolayer on the ECM under static conditions, π: NSCs dispersed in the ECM vs. NSC monolayer on the ECM both under perfusion conditions, *: NSCs dispersed in the ECM vs. NSC monolayer on the ECM both under static conditions, λ: NSCs dispersed in the ECM vs. NSC spheroids in the ECM both under perfusion conditions, #: NSC spheroids vs. NSC monolayer both under static conditions, &: NSC monolayer under perfusion and static conditions, %: NSC monolayer on the ECM under perfusion and static conditions, and +: NSC spheroids under perfusion and static conditions).
III. RESULTS AND DISCUSSION
A. Microfluidic array for NSC culture under multiple conditions
To establish a robust and straightforward approach for constitution of multiple NSC niches and subsequently screen the optimal culture conditions for maintenance and differentiation of NSCs, we designed and fabricated an assembled and detachable microfluidic array. The schematic of the microfluidic system for cell culture is shown in Fig. 1. The entire microfluidic device consisted of a PDMS microfluidic chip and two PDMS-coated glass slides as the cover. The lower layer (microfluidic chip) consisted of the perfusion channels with a width and height of 300 μm and an open system with cell culture chambers as a 4 × 4 array with a width of 5 mm, height of 2 mm, and pitch of 4 mm. In particular, the culture chambers were designed to be compatible with multichannel pipettes for seeding cells and changing medium, which allowed simultaneous introduction of cells or the ECM and significantly increased throughput of the cell culture and analysis. The upper layer (glass cover) was coated with PDMS, and this hydrophobic surface was used for compartmentalized cell culture under various conditions. The three parts were assembled together to seal the microfluidic 4 × 4 chamber array under the PDMS-PDMS surface tension and could be readily detached after experiments for subsequent analyses because of the reversible bonding.
In this device, five culture conditions were established, including NSCs as the monolayer, monolayer on the ECM, spheroids, dispersed cells in the ECM, and spheroids in the ECM as shown in Fig. 1. To ensure the same starting point of static or perfusion culture, the five groups of NSCs were inoculated as follows. First, for NSC spheroids and NSC spheroids in the ECM, NSC suspensions were dripped to generate two arrays on one of the PDMS slides, aligning the rows of chambers for corresponding culture groups in the lower microfluidic chip. After 2 days of sinking and aggregation, uniform NSC spheroids were generated. For monolayer NSC culture, we used PO/LN or collagen regionally modified PDMS slides. Next, NSCs suspended in the collagen solution were dripped onto the PDMS slide regions for the NSC culture dispersed in the ECM. For NSC spheroids in the ECM, after aspirating the culture medium by pipetting, multicellular spheroids on the slide were dripped with the collagen solution. The collagen drops containing dispersed NSCs and NSC spheroids were allowed to gel. Finally, the two PDMS slides with a 4 × 4 NSC array were attached to the chamber region in the lower microfluidic chip by aligning the 4 × 4 NSC array towards the 4 × 4 culture chambers. Then, perfusion or static culture was carried out for various durations.
To construct an effective niche for studying NSC fate determination, we followed several general principles. In terms of NSC self-renewal, the niche maintains NSCs in a quiescent and undifferentiated state. In contrast, for NSC differentiation, the niche must be structured so that both the type and number of differentiated cells can be modulated in response to different physiological cues. We designed the microfluidic device for flexible operation with multiple functions by taking account of the culture parameters including the dimension (2D vs. 3D) and heterogeneity of NSC culture, ECM components, and fluid shear conditions. All of these individual culture conditions as well as parallel comparisons of their effects on NSC behaviors were achieved on the same integrated microfluidic chip simultaneously in a controllable and parallel manner. This device provides a versatile model for NSC culture in vitro to facilitate screening of a wide range of conditions, which reconstructs a physiological environment such as the heterogeneity and 3D growth conditions of NSCs.
B. Viability of NSCs
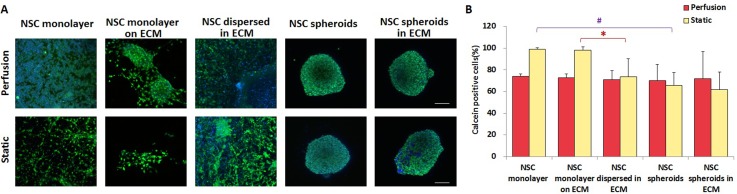
The viability of NSCs was measured by calculating the percentage of live cells stained with calcein among total cells (nuclear stained), as shown in Fig. 2(A). Under static conditions, the average live cell percentage decreased from 99.01% (NSC monolayer) and 98.06% (NSC monolayer on the ECM) to 73.56% (NSCs dispersed in the ECM), 65.84% (NSC spheroids), and 61.7% (NSC spheroids in the ECM), respectively, as illustrated in Fig. 2(B). These results imply that NSCs in spheroids or ECM-embedded culture under static conditions were prone to necrosis or apoptosis due to insufficient permeation of nutrients and oxygen. This phenomenon was particularly evident in NSC spheroids in the ECM, which had the lowest viability owing to the most complicated culture condition. In comparison, perfusion treatment partly rescued such events with almost the same percentages of live cell in the five groups, indicating that perfusion facilitates sufficient exchange of nutrients and oxygen for NSC growth regardless of either a 3D spheroid architecture or embedding in the ECM. In addition, NSC viabilities were obviously different between perfusion and static conditions in the 2D environment (NSC monolayer and NSC monolayer on the ECM), whereas the difference was not obvious in the 3D cultures. This observation suggests that the influence of shear stress on NSCs in 3D culture was less than that in 2D culture. Thus, 3D culture could effectively protect NSCs from shear force causing damage. Our microfluidic systems provided spatial and temporal control of fluid behaviors such as laminar flow and molecular transport by diffusion. It has been reported that perfusion culture can mimic the body tissue fluid flow, thereby supplying necessary nutrients and oxygen and removing waste metabolites, which stabilize physiological balances and maintain cell–cell communication and cell-ECM interactions.34,35 In addition, perfusion in 3D cell culture can significantly enhance the diffusion efficiency and confer specific shear stress to direct cell behaviors.36
FIG. 2.
Viability of NSCs cultured for 4 days in the five culture groups under perfusion and static conditions. (A) Confocal images of NSCs. Green represents live cells and blue is the nucleus. Scale bars represent 100 μm. (B) Quantitative analysis of live NSC percentages in each group, n = 5.
C. Self-renewal and proliferation of NSCs
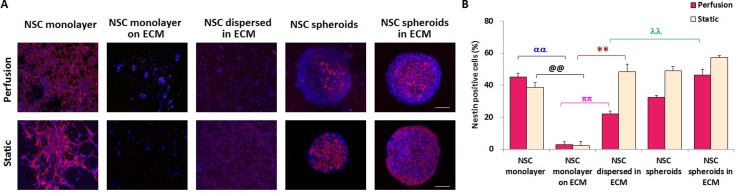
NSCs are multipotent cells that maintain their self-renewal or proliferative capacity and differentiation potential, as shown in Fig. 1. We investigated NSC self-renewal in the various niche conditions created by the microfluidic device. As shown in Fig. 3(A), immunofluorescence staining of nestin, a neuroectodermal stem cell marker, was performed in NSCs cultured for 4 days. The self-renewal of NSCs was characterized by the percentage of nestin-positive NSCs among total cells. The statistical results in Fig. 3(B) indicated that, regardless of perfusion or static conditions, NSC spheroids in the ECM exhibited the highest nestin expression with 0.02, 14.33, 1.09, and 0.44 fold higher nestin expression than in the NSC monolayer, NSC monolayer on the ECM, NSCs dispersed in the ECM, and NSC spheroids under perfusion conditions, respectively. Under the static condition, NSC spheroids in the ECM showed similarly enhanced nestin expression with 0.46, 27.50, 0.19, and 0.16 fold higher nestin expression than in the NSC monolayer, NSC monolayer on the ECM, NSCs dispersed in the ECM, and NSC spheroids, respectively. These results suggest that spheroid culture in the ECM facilitates NSC self-renewal compared with other culture conditions and that the 3D ECM microenvironment benefits the undifferentiated state of NSCs. Moreover, in 2D culture (NSC monolayer and NSC monolayer on the ECM), the numbers of nestin-positive NSCs were higher under perfusion conditions than in the static condition. Furthermore, for all of the 3D culture groups, static culture was propitious to NSC self-renewal and maintenance. Han et al.37 found that 3D collagen culture promotes maintenance of NSC self-renewal by inactivation of mTOR and over-expression of REDD1. Their study indicated that 3D ECM culture facilitates self-renewal of NSCs. These results were consistent with our findings showing the highest nestin expression in the spheroid culture in the ECM. However, Han et al.31 showed a significant reduction in nestin expression of 3D culture compared with 2D culture, which is in agreement with our results indicating higher nestin expression in the NSC monolayer than NSCs dispersed in the ECM and NSC spheroids under perfusion conditions. The proposed culture conditions for NSC self-renewal are summarized in Fig. S2 (supplementary material).
FIG. 3.
Self-renewal capability of NSCs cultured for 4 days in the five culture groups under perfusion and static conditions characterized by nestin expression. (A) Confocal images of NSCs. Red represents nestin-positive cells and blue is the nucleus. Scale bars represent 100 μm. (B) Quantitative analysis of nestin-positive NSC percentages in each group, n = 5.
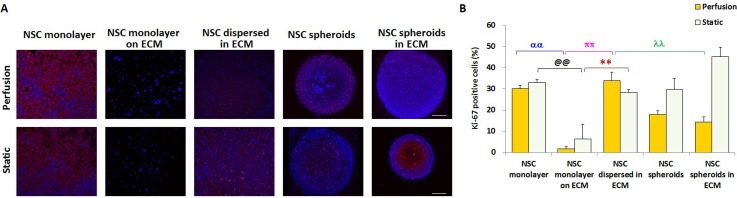
We next investigated the proliferation of NSCs. As shown in Fig. 4(A), immunofluorescence staining of Ki-67, a specific marker of proliferating cells, was performed in NSCs. The proliferative ability of NSCs was expressed by the percentage of Ki-67-positive NSCs among total cells. Regardless of perfusion or static conditions, the NSC monolayer on the ECM exhibited the lowest Ki-67 expression (Fig. 4(B)). Under perfusion conditions, NSCs dispersed in the ECM had the highest Ki-67-positive expression level, as illustrated in Fig. 4(B). Furthermore, under static conditions, the average percentages of Ki-67-positive cells in the NSC monolayer, NSCs dispersed in the ECM, and NSC spheroids were not significantly different. The most notable increase of Ki-67 expression appeared in NSC spheroids in the ECM under static conditions, which was in consistent with the variation in nestin expression. These results suggest that the synergistic effect of spheroid heterogeneity and ECM influenced the proliferation of NSCs significantly. In particular, under perfusion culture, the expression of Ki-67 in the NSC monolayer was higher than that in NSC spheroids. Moreover, compared with NSC spheroids in the ECM, NSCs dispersed in the ECM showed an increase in nestin expression. These findings suggest that individual NSCs have better proliferation under fluidic perfusion. In addition, except for NSCs dispersed in the ECM, Ki-67-positive cell ratios under perfusion conditions were lower than those under static conditions, indicating that static culture facilitates NSC proliferation.
FIG. 4.
Proliferation capability of NSCs cultured for 4 days in the five culture groups under perfusion and static conditions characterized by Ki-67 expression. (A) Confocal images of NSCs. Red represents Ki-67-positive cells and blue is the nucleus. Scale bars represent 100 μm. (B) Quantitative analysis of Ki-67-positive NSC percentages in each group, n = 5.
D. Differentiation of NSCs into neurons, astrocytes, and oligodendrocytes
Neurons, astrocytes, and oligodendrocytes all originate from NSCs. The potential for differentiation into all neural tissues via different exogenous cues from the niche makes NSCs extremely attractive for CNS regeneration therapy. Therefore, we examined the differentiation of NSCs in the niches constructed within the microfluidic array. Following the standard NSC culture protocols,3,38 static culture of NSCs was first performed to determine the optimal conditions for differentiation. To evaluate the effects of various culture conditions on NSC differentiation, we performed qRT-PCR analysis of differentiation-associated gene expression in NSC samples. The comparative relation of the five groups was set as shown in Fig. S1 (supplementary material). Briefly, the NSC monolayer and NSC monolayer on the ECM were set as two independent control groups. In addition, these two groups were also compared. Beginning with the NSC monolayer, cell fate marker expression was compared between two adjacent groups counterclockwise. Furthermore, starting from the NSC monolayer on the ECM, marker expression was compared between two adjacent groups clockwise. These two circulations were carried out simultaneously, and once marker expression of an upstream group was higher than that in the downstream group, this circulation was suspended. The set criterion of two adjacent groups was that there was only one single variable factor between the groups, thereby making them comparable. All of the comparisons for differentiation were carried out according to these criteria.
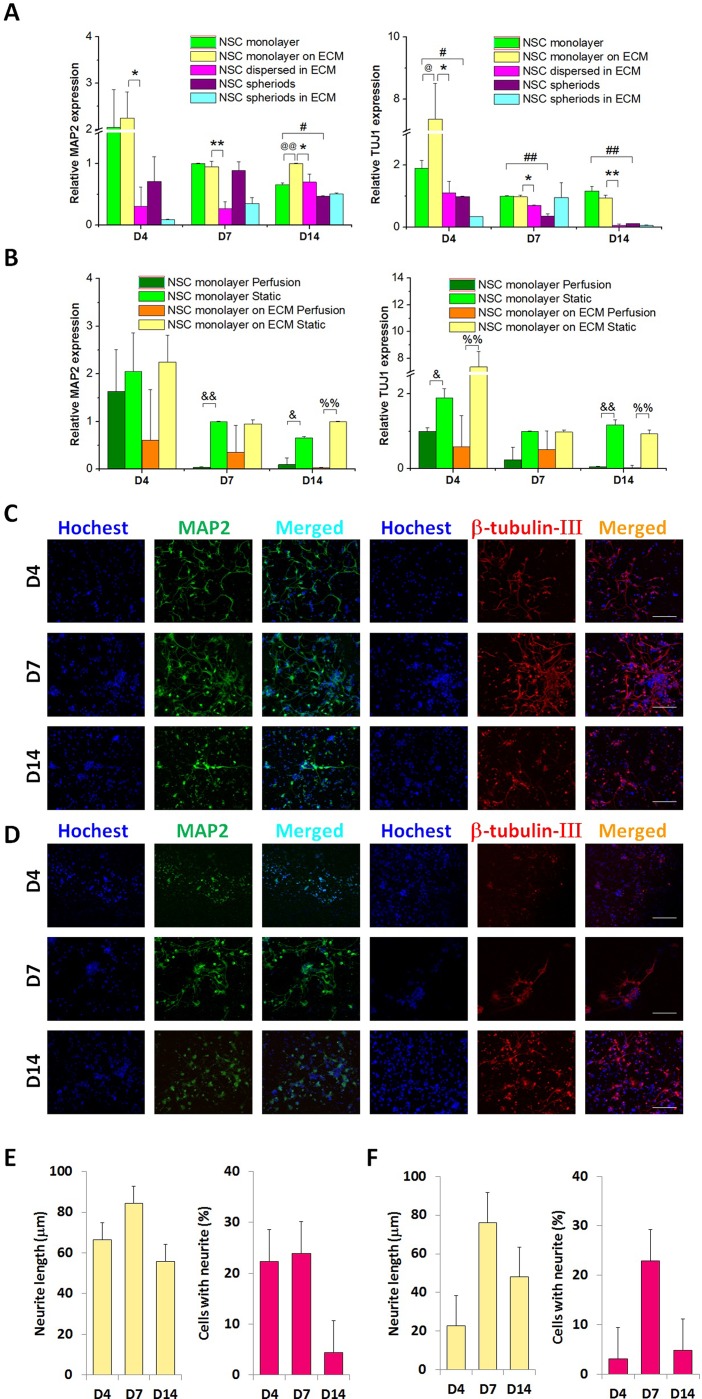
Neuronal lineage determination of NSCs was represented by MAP2 and TUJ1. Under static culture conditions, the differentiation rate of NSCs into neurons was relatively higher in the NSC monolayer and NSC monolayer on the ECM at 4, 7, and 14 days compared with that of NSC spheroids and NSCs dispersed in the ECM, in which NSCs were all cultured in 3D (Fig. 5(A)). For the NSC monolayer and NSC monolayer on the ECM, perfusion was carried out, and gene expression under perfusion was compared with that under static conditions, as shown in Fig. 5(B). We found obvious decreases in the expression of MAP2 and TUJ1 under perfusion culture, which indicated that the 2D-dependent static condition might facilitate NSC differentiation into the neuron lineage. MAP2 and β-tubulin-III, which are exclusively expressed in neurons, were used as proxies to represent the differentiation of NSCs into neurons, as shown in Figs. 5(C) and 5(D). After 4, 7, and 14 days of culture, the NSC monolayer in the static state exhibited neurite extensions with average lengths of 67, 84, and 55 μm, respectively, and a high neurite-positive percentage in total cells, as shown in Figs. 5(C) and 5(E). While in the NSC monolayer on the ECM in the static state, the average lengths of neurite extensions after 4, 7, and 14 days were 22, 76, and 48 μm, respectively. As shown in Figs. 5(C)–5(F), regardless of the neurite length or neurite-positive percentage, the NSC monolayer under static conditions displayed remarkable neuronal differentiation compared with the NSC monolayer on the ECM in the static state. Cui et al. showed inhibition of neural differentiation of NSCs in 3D collagen scaffolds compared with 2D-cultured cells,39 which is consistent with our results. In addition, Liu et al. revealed that NSCs in an adherent monolayer culture are capable of developing into neurons.40
FIG. 5.
Capability of NSCs to differentiate into neurons at 4, 7, and 14 days. (A) Quantitative analysis of MAP2 and TUJ1 mRNA expression in the five culture groups under the static condition, n = 3. (B) Quantitative analysis of MAP2 and TUJ1 mRNA expression in the NSC monolayer and NSC monolayer on the ECM in perfusion and static culture, respectively, n = 3. (C) Confocal images of the NSC monolayer in static culture. Green represents MAP2, red is β-tubulin-III, and blue is the nucleus. (D) Confocal images of the NSC monolayer on the ECM in static culture. Green represents MAP2, red is β-tubulin-III, and blue is the nucleus. Scale bars represent 100 μm. (E) Neurite length (n = 50) and the percentage of cells with neurites (n = 10) in the NSC monolayer under static conditions. (F) Neurite length (n = 50) and the percentage of cells with neurites (n = 10) in the NSC monolayer on the ECM under static conditions.
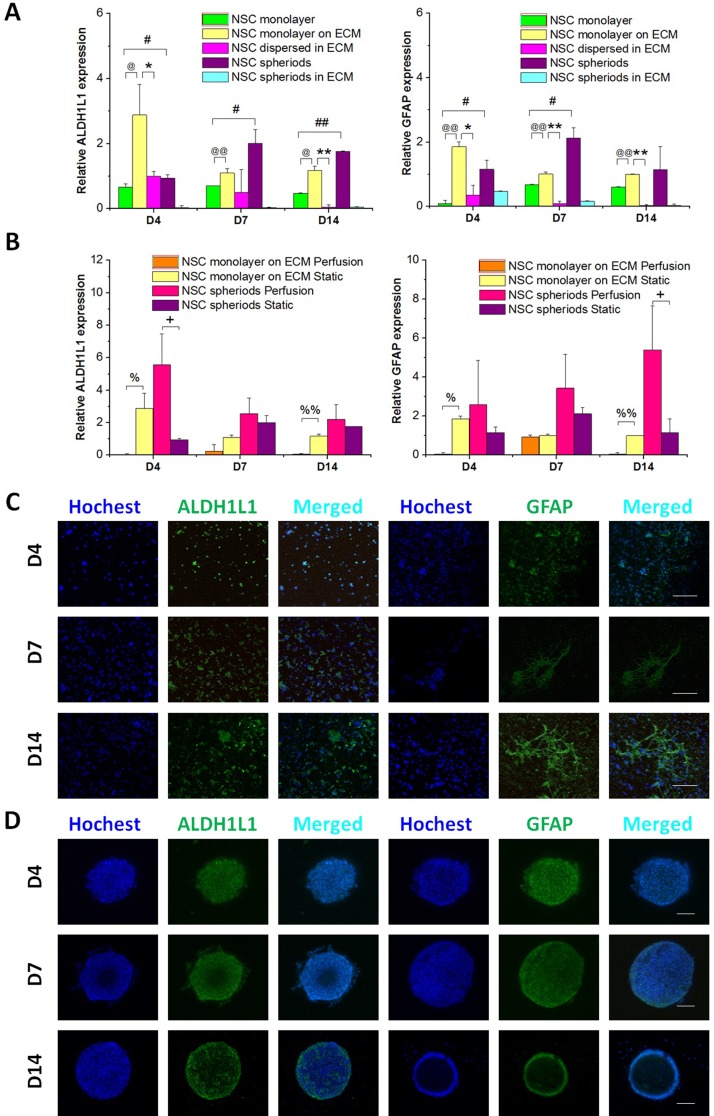
Next we examined the differentiation of astrocytes, a subtype of glial cells in the CNS. In this study, quantification of astrocytic markers ALDH1L1 and GFAP was used to characterize the differentiation of NSCs into the astrocytic lineage. As shown in Fig. 6(A), under static conditions, the NSC monolayer on the ECM expressed higher messenger RNA (mRNA) levels than NSCs dispersed in the ECM. ALDH1L1 and GFAP expressed at higher levels in NSC spheroids than in the NSC monolayer and NSC spheroids in the ECM. High expression of these markers was particularly obvious in NSC spheroids under perfusion conditions, as indicated in Fig. 6(B). However, under perfusion conditions, ALDH1L and GFAP showed nearly non-detectible expression in the NSC monolayer on the ECM. Figs. 6(C) and 6(D) show immunofluorescence staining of ALDH1L1- and GFAP-positive NSCs as a monolayer on the ECM under static culture and as spheroid aggregates under perfusion, respectively. In particular, as shown in Fig. 6(A), under static conditions at 7 and 14 days, expression of ALDH1L1 and GFAP in NSCs dispersed in the ECM and NSC spheroids in the ECM was extremely low, demonstrating an ECM-dependent inhibitive effect on differentiation of astrocytes from NSCs. Han et al.31 found a reduction of astrocytic differentiation in ECM compositions of both macro- and micro-collagen groups, which was in agreement with our results. Whilst, Zhou et al. revealed that neural progenitor cells cultured as 3D neurospheres underwent enhanced astrocyte differentiation compared with the 2D condition.41
FIG. 6.
Capability of NSCs to differentiate into astrocytes at 4, 7, and 14 days. (A) Quantitative analysis of ALDH1L1 and GFAP mRNA expression in the five culture groups under static conditions, n = 3. (B) Quantitative analysis of ALDH1L1 and GFAP mRNA expression in the NSC monolayer on the ECM and NSC spheroids in perfusion and static culture, respectively, n = 3. (C) Confocal images of the NSC monolayer on the ECM in the static state. Green represents ALDH1L1 or GFAP, and blue is the nucleus. (D) Confocal images of NSC spheroids under perfusion conditions. Green represents ALDH1L1 or GFAP, and blue is the nucleus. Scale bars represent 100 μm.
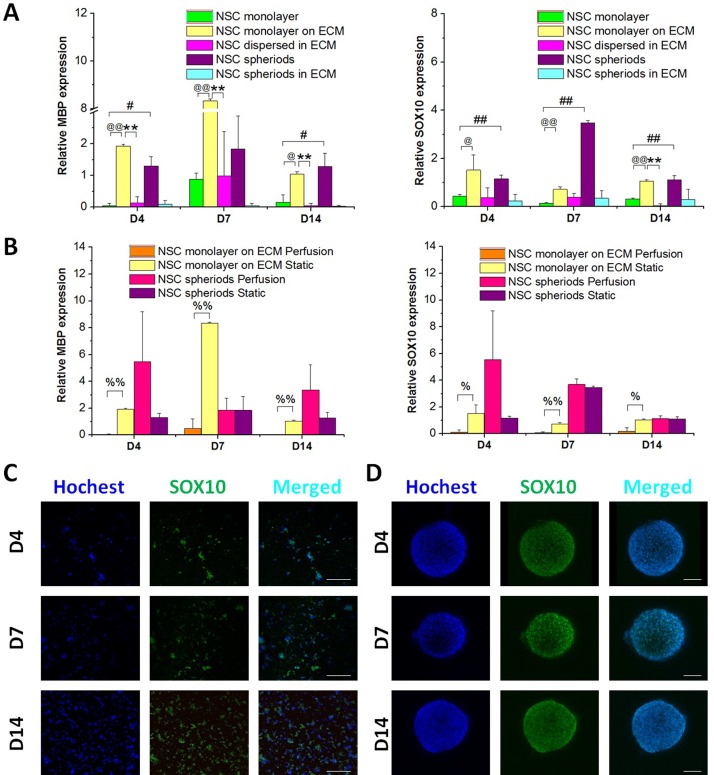
The differentiation of NSCs into oligodendrocytes was determined by MBP and SOX10 expression analysis using qRT-PCR and immunostaining at 4, 7, and 14 days. As shown in Fig. 7(A), under static conditions, notably high levels of MBP and SOX10 were observed in the NSC monolayer on ECM and NSC spheroids. However, as shown in Fig. 7(B), MBP and SOX10 were downregulated in the NSC monolayer on the ECM under perfusion, and the optimal conditions for differentiation of NSCs toward oligodendrocytes appeared to be spheroid aggregates under perfusion and a monolayer on the ECM in static culture, as indicated in Figs. 7(B)–7(D), which had the same tendency for astrocyte differentiation. These results suggest that these two culture systems facilitated glial differentiation. A summary of the proposed culture conditions for NSC differentiation is shown in Fig. S2 (supplementary material).
FIG. 7.
Capability of NSCs to differentiate into oligodendrocytes at 4, 7, and 14 days. (A) Quantitative analysis of MBP and SOX10 mRNA expression in the five culture groups under static conditions, n = 3. (B) Quantitative analysis of MBP and SOX10 mRNA expression in the NSC monolayer on the ECM and NSC spheroids in perfusion and static culture, respectively, n = 3. (C) Confocal images of the NSC monolayer on the ECM in static culture. Green represents SOX10 and blue is the nucleus. (D) Confocal images of NSC spheroids under perfusion conditions. Green represents SOX10 and blue is the nucleus. Scale bars represent 100 μm.
Neurospheres are intrinsically heterogeneous cellular entities. With the ability to aggregate and proliferate under serum-free medium conditions as well as in the presence of EGF and bFGF, the neurosphere culture system has been extensively used to isolate and expand NSCs, which could also be used as an in vitro 3D culture model for NSCs. NSCs in neurospheroids communicate with each other by directing cell–cell contact and exchanging of nutritional and signaling factors. Such a tissue-specific microenvironment enables aggregated NSCs to differentiate and develop specialized structures such as those found in brain tissue in vivo.42 The differences between the NSC monolayer and NSC spheroids and NSCs dispersed in the ECM and NSC spheroids in the ECM suggest a role of cell–cell interactions in NSC behaviors.
ECMs create a 3D microenvironment in which NSCs are instructed to form micro-tissues by guiding cell behaviors through environmental cell–matrix and cell–cell communications.9 In this micro-scale system, factor distribution is generated by accumulated factors secreted by NSCs embedded in the ECM, which is similar to real tissues. Moreover, our microfluidic system is general and suitable for other ECMs that are critical components in the regulation of NSC fate. Various 3D formats can be incorporated flexibly. Comparisons between the NSC monolayer and NSC monolayer on the ECM, and NSC spheroids and NSC spheroids in the ECM were used to illustrate the effects the ECM on NSC fates.
The influence of continuous flow shear stress on NSC fate was determined by comparing cellular behaviors in static and perfusion cultures. Wang et al. showed that shear stress generated in a microfluidic channel does not enhance differentiation of monolayered NSCs compared with 2D culture,43 which is in agreement with our results demonstrating that 2D culture in the static state facilitated differentiation towards neurons, astrocytes, and oligodendrocytes.
IV. CONCLUSIONS
In summary, we successfully established a microfluidic array to construct NSC niches in vitro in a controllable format by integrating micro-scale engineering techniques. This approach enables synchronous analysis of NSC fate under various culture conditions. The findings indicate that 3D culture in ECM facilitated NSC self-renewal and proliferation, and 2D culture under static state benefited NSC differentiation into the neuron lineage, whilst, monolayer culture on the ECM under static state and spheroid culture under perfusion are in favor of NSC differentiation into the glial cells. Differences of NSC fate induced by the cell organization and cell-cell interactions in this system may be an inspiration for NSC differentiation.42,44,45 Systematic study of NSC culture conditions will further our understanding of the mechanisms of NSC fate and facilitate the design of new platforms that encourage potential development of NSC-based nervous system therapies. Follow-up studies are underway to further optimize differentiation toward a homogeneous population of a specific cell type, and broader applications of this system may be realized in tumor research and organ-on-chip fields.
V. SUPPLEMENTARY MATERIAL
See supplementary material for Figure S1: Diagram of the control group setting and comparison process for NSC differentiation in the five culture conditions. Figure S2: The proposed culture conditions for NSC self-renewal and differentiation.
ACKNOWLEDGMENTS
This work was funded by the National Natural Science Foundation of China (Grant Nos. 81271412 and 81471308), International S&T Cooperation Project of the Ministry of S&T of China (Grant No. 2010DFR30850), and the Scientific Research Foundation for Returned Overseas Chinese Scholars. We greatly appreciate Professor Kun Zhang of the University of California San Diego and Professor Bing Song of Cardiff University, for their assistance in writing the manuscript.
The authors declare no financial or commercial conflicts of interest.
References
- 1. Gage F. H., Science 287(5457), 1433 (2000). 10.1126/science.287.5457.1433 [DOI] [PubMed] [Google Scholar]
- 2. Wen S., Li H., and Liu J., Cell Adhes. Migr. 3(1), 107 (2009). 10.4161/cam.3.1.7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chojnacki A. and Weiss S., Nat. Protoc. 3(6), 935 (2008). 10.1038/nprot.2008.55 [DOI] [PubMed] [Google Scholar]
- 4. Temple S., Nature 414(6859), 112 (2001). 10.1038/35102174 [DOI] [PubMed] [Google Scholar]
- 5. Martino G. and Pluchino S., Nat. Rev. Neurosci. 7(5), 395 (2006). 10.1038/nrn1908 [DOI] [PubMed] [Google Scholar]
- 6. Lindvall O. and Kokaia Z., Nature 441(7097), 1094 (2006). 10.1038/nature04960 [DOI] [PubMed] [Google Scholar]
- 7. Uchida N., Chen K., Dohse M., Hansen K. D., Dean J., Buser J. R., Riddle A., Beardsley D. J., Wan Y., Gong X., Nguyen T., Cummings B. J., Anderson A. J., Tamaki S. J., Tsukamoto A., Weissman I. L., Matsumoto S. G., Sherman L. S., Kroenke C. D., and Back S. A., Sci. Transl. Med. 4(155), 155ra136 (2012). 10.1126/scitranslmed.3004371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Teixeira A. I., Duckworth J. K., and Hermanson O., Cell Res. 17(1), 56 (2007). 10.1038/sj.cr.7310141 [DOI] [PubMed] [Google Scholar]
- 9. Li X., Katsanevakis E., Liu X., Zhang N., and Wen X., Prog. Polym. Sci. 37(8), 1105 (2012). 10.1016/j.progpolymsci.2012.02.004 [DOI] [Google Scholar]
- 10. Regalado-Santiago C., Juarez-Aguilar E., Olivares-Hernandez J. D., and Tamariz E., Stem Cells Int. 2016, 1513285. 10.1155/2016/1513285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nern C. and Momma S., Stem Cell Rev. 2(3), 233 (2006). 10.1007/s12015-006-0051-6 [DOI] [PubMed] [Google Scholar]
- 12. Conover J. C. and Notti R. Q., Cell Tissue Res. 331(1), 211 (2008). 10.1007/s00441-007-0503-6 [DOI] [PubMed] [Google Scholar]
- 13. Taupin P., Stem Cell Rev. 2(3), 213 (2006). 10.1007/s12015-006-0049-0 [DOI] [PubMed] [Google Scholar]
- 14. Lin R. and Iacovitti L., Brain Res. 1628(Pt B), 327 (2015). 10.1016/j.brainres.2015.04.029 [DOI] [PubMed] [Google Scholar]
- 15. Garzon-Muvdi T. and Quinones-Hinojosa A., ILAR J. 51(1), 3 (2009). 10.1093/ilar.51.1.3 [DOI] [PubMed] [Google Scholar]
- 16. Booth B. W., Mack D. L., Androutsellis-Theotokis A., McKay R. D., Boulanger C. A., and Smith G. H., Proc. Natl. Acad. Sci. U. S. A. 105(39), 14891 (2008). 10.1073/pnas.0803214105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu C. Y., Westerlund U., Svensson M., Moe M. C., Varghese M., Berg-Johnsen J., Apuzzo M. L., Tirrell D. A., and Langmoen I. A., J. Hematother. Stem Cell Res. 12(6), 689 (2003). 10.1089/15258160360732713 [DOI] [PubMed] [Google Scholar]
- 18. Yao S., Liu X., Wang X., Merolli A., Chen X., and Cui F., Prog. Nat. Sci.: Mater. Int. 23(2), 103 (2013). 10.1016/j.pnsc.2013.02.009 [DOI] [Google Scholar]
- 19. Conway A., Vazin T., Spelke D. P., Rode N. A., Healy K. E., Kane R. S., and Schaffer D. V., Nat. Nanotechnol. 8(11), 831 (2013). 10.1038/nnano.2013.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dai X., Huang Y. C., Leichner J., Nair M., Lin W. C., and Li C. Z., Biomed. Mater. 10(6), 065013 (2015). 10.1088/1748-6041/10/6/065013 [DOI] [PubMed] [Google Scholar]
- 21. Hsieh F. Y., Lin H. H., and Hsu S. H., Biomaterials 71, 48 (2015). 10.1016/j.biomaterials.2015.08.028 [DOI] [PubMed] [Google Scholar]
- 22. Wang Y., Wei Y. T., Zu Z. H., Ju R. K., Guo M. Y., Wang X. M., Xu Q. Y., and Cui F. Z., Pharm. Res. 28(6), 1406 (2011). 10.1007/s11095-011-0452-3 [DOI] [PubMed] [Google Scholar]
- 23. Li N., Zhang Q., Gao S., Song Q., Huang R., Wang L., Liu L., Dai J., Tang M., and Cheng G., Sci. Rep. 3, 1604 (2013). 10.1038/srep01604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leipzig N. D. and Shoichet M. S., Biomaterials 30(36), 6867 (2009). 10.1016/j.biomaterials.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 25. Liu C. Y., Apuzzo M. L., and Tirrell D. A., Neurosurgery 52(5), 1154 (2003). 10.1227/01.NEU.0000057747.93237.8F [DOI] [PubMed] [Google Scholar]
- 26. Zhang Q. and Austin R. H., BioNanoScience 2(4), 277 (2012). 10.1007/s12668-012-0051-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu H. W., Lin C. C., and Lee G. B., Biomicrofluidics 5(1), 13401 (2011). 10.1063/1.3528299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chung B. G., Flanagan L. A., Rhee S. W., Schwartz P. H., Lee A. P., Monuki E. S., and Jeon N. L., Lab Chip 5(4), 401 (2005). 10.1039/b417651k [DOI] [PubMed] [Google Scholar]
- 29. Yang K., Han S., Shin Y., Ko E., Kim J., Park K. I., Chung S., and Cho S. W., Biomaterials 34(28), 6607 (2013). 10.1016/j.biomaterials.2013.05.067 [DOI] [PubMed] [Google Scholar]
- 30. Esfandiari L., Paff M., and Tang W. C., Nanomed.: Nanotechnol., Biol., Med. 8(4), 415 (2012). 10.1016/j.nano.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 31. Han S., Yang K., Shin Y., Lee J. S., Kamm R. D., Chung S., and Cho S. W., Lab Chip 12(13), 2305 (2012). 10.1039/c2lc21285d [DOI] [PubMed] [Google Scholar]
- 32. Shin Y., Yang K., Han S., Park H. J., Seok Heo Y., Cho S. W., and Chung S., Adv. Healthcare Mater. 3(9), 1457 (2014). 10.1002/adhm.201300569 [DOI] [PubMed] [Google Scholar]
- 33. Wallin P., Zanden C., Carlberg B., Hellstrom Erkenstam N., Liu J., and Gold J., Biomicrofluidics 6(2), 24131 (2012). 10.1063/1.4729747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pampaloni F., Reynaud E. G., and Stelzer E. H. K., Nat. Rev. Mol. Cell Biol. 8(10), 839 (2007). 10.1038/nrm2236 [DOI] [PubMed] [Google Scholar]
- 35. Yamada K. M. and Cukierman E., Cell 130(4), 601 (2007). 10.1016/j.cell.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 36. Li Z. and Cui Z., Biotechnol. Adv. 32(2), 243 (2014). 10.1016/j.biotechadv.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 37. Han J., Xiao Z., Chen L., Chen B., Li X., Han S., Zhao Y., and Dai J., Biomaterials 34(8), 1921 (2013). 10.1016/j.biomaterials.2012.11.063 [DOI] [PubMed] [Google Scholar]
- 38. Casarosa S., Bozzi Y., and Conti L., Mol. Cell. Ther. 2, 31 (2014). 10.1186/2052-8426-2-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cui Y., Xiao Z., Chen T., Wei J., Chen L., Liu L., Chen B., Wang X., Li X., and Dai J., Stem Cells Dev. 23(4), 393 (2014). 10.1089/scd.2013.0342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu F., Xuan A., Chen Y., Zhang J., Xu L., Yan Q., and Long D., Mol. Med. Rep. 10(4), 1739 (2014). 10.3892/mmr.2014.2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou S., Szczesna K., Ochalek A., Kobolák J., Varga E., Nemes C., Chandrasekaran A., Rasmussen M., Cirera S., Hyttel P., Dinnyés A., Freude K. K., and Avci H. X., Stem Cells Int. 2016, 4937689. 10.1155/2016/4937689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Limongi T., Cesca F., Gentile F., Marotta R., Ruffilli R., Barberis A., Dal Maschio M., Petrini E. M., Santoriello S., Benfenati F., and Di Fabrizio E., Small 9(3), 402 (2013). 10.1002/smll.201201377 [DOI] [PubMed] [Google Scholar]
- 43. Wang B., Jedlicka S., and Cheng X., Plos One 9(10), e109815 (2014). 10.1371/journal.pone.0109815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Solozobova V., Wyvekens N., and Pruszak J., Stem Cell Rev. Rep. 8(3), 813 (2012). 10.1007/s12015-012-9381-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen S., Bremer A. W., Scheideler O. J., Na Y. S., Todhunter M. E., Hsiao S., Bomdica P. R., Maharbiz M. M., Gartner Z. J., and Schaffer D. V., Nat. Commun. 7, 10309 (2016). 10.1038/ncomms10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for Figure S1: Diagram of the control group setting and comparison process for NSC differentiation in the five culture conditions. Figure S2: The proposed culture conditions for NSC self-renewal and differentiation.