Abstract
Tumor development is influenced by stromal cells in aspects including invasion, growth, angiogenesis, and metastasis. Activated fibroblasts are one group of stromal cells involved in cancer metastasis, and one source of activated fibroblasts is endothelial to mesenchymal transformation (EndMT). EndMT begins when the endothelial cells delaminate from the cell monolayer, lose cell-cell contacts, lose endothelial markers such as vascular endothelial-cadherin (VE-cadherin), gain mesenchymal markers like alpha-smooth muscle actin (α-SMA), and acquire mesenchymal cell-like properties. A three-dimensional (3D) culture microfluidic device was developed for investigating the role of steady low shear stress (1 dyne/cm2) and altered extracellular matrix (ECM) composition and stiffness on EndMT. Shear stresses resulting from fluid flow within tumor tissue are relevant to both cancer metastasis and treatment effectiveness. Low and oscillatory shear stress rates have been shown to enhance the invasion of metastatic cancer cells through specific changes in actin and tubulin remodeling. The 3D ECM within the device was composed of type I collagen and glycosaminoglycans (GAGs), hyaluronic acid and chondroitin sulfate. An increase in collagen and GAGs has been observed in the solid tumor microenvironment and has been correlated with poor prognosis in many different cancer types. In this study, it was found that ECM composition and low shear stress upregulated EndMT, including upregulation of mesenchymal-like markers (α-SMA and Snail) and downregulated endothelial marker protein and gene expression (VE-cadherin). Furthermore, this novel model was utilized to investigate the role of EndMT in breast cancer cell proliferation and migration. Cancer cell spheroids were embedded within the 3D ECM of the microfluidic device. The results using this device show for the first time that the breast cancer spheroid size is dependent on shear stress and that the cancer cell migration rate, distance, and proliferation are induced by EndMT-derived activated fibroblasts. This model can be used to explore new therapeutics in a tumor microenvironment.
I. INTRODUCTION
Cancer is a complex disease with many unknown mechanisms of growth and progression. By the year 2020, cancer is predicted to affect 17 to 18 million people worldwide.1,2 A 2010 study estimated the treatment cost based on incidence and survival in the United States to be approximately 125 billion dollars. The national cost of cancer care was projected to be 158 billion dollars in 2020, modeled from the 2010 data.3 Most common clinical cancer therapies rely on radiation, chemotherapy, and surgical resection. Currently, various novel therapeutic strategies are under investigation for targeting cancer-associated biomarkers. Advances in therapeutic strategies such as drug development utilizing animal models and standard in vitro static cultures are limited due to incomplete knowledge of the disease progression. Drugs that target cellular pathways fail to address the mechanical and chemical signals that play a critical role in the initiation and progression of the disease. Additionally, these therapeutics are not always effective because the treatment is not sufficiently personalized. Thus, a strong incentive exists for better understanding of the pathophysiological conditions that lead to aggressive early stage cancer progression.4,5
Traditionally, cancer research has focused on altered epithelial cell molecular mechanisms. However, recent work has revealed that the effects of biochemical and biomechanical factors on the interactions between cancer cells and neighboring cells and tissues are also critical to the development and progression of the disease.6 Components of the tumor stroma include fibroblasts, endothelial and immune cells, the extracellular matrix (ECM), networks of blood vessels, and soluble factors, and these are linked to cancer progression, angiogenesis, invasion, and metastasis. However, the mechanisms by which these biochemical and biomechanical factors affect the tumor stroma are still poorly understood. One common cell type involved in cancer progression and in many other pathologies such as atherosclerosis, wound healing, and cardiac fibrosis is activated fibroblasts. One source of activated fibroblasts is endothelial to mesenchymal transformation (EndMT). EndMT can generate up to 40% of cancer associated fibroblasts (CAF), which promotes tumor growth.7 CAF adopt a myofibroblastic phenotype, produce a reactive ECM that is significantly different from normal ECM, and secrete a variety of biochemical and biomechanical factors promoting tumor migration to other tissues and tumor angiogenesis. An in vitro model that can recapitulate the biomechanical and biochemical interplay between tumor cells, the endothelium, and the surrounding ECM will have a significant impact on our understanding of EndMT and tumor-endothelial cell (EC) interactions. Earlier in vitro models recreated biomechanical and biochemical factors influencing tumor-vessel interactions;8,9 here, we investigate the addition of tissue properties on EndMT and cancer cell proliferation.
Fluid-induced shear stresses resulting from fluid flow in the proximity of tumor tissue are relevant to both cancer metastasis and treatment effectiveness. Low and oscillatory fluid-induced shear stress rates have been shown to enhance the invasion of metastatic cancer cells through specific changes in actin and tubulin remodeling. Additionally, fluid-induced shear stress and altered tissue properties within the tumor stroma microenvironment are relevant to the formation of activated fibroblasts.10,11 Altered ECM compositions also include the presence and production of glycosaminoglycans (GAGs), which include hyaluronic acid (HA) and chondroitin sulfate (CS). GAGs have been observed in the solid tumor microenvironment and have been correlated with poor prognosis in many different cancer types.12,13 Dysregulated ECM deposition of collagen and GAGs by myofibroblasts plays a role in fibrosis and tumor progression.7,14
Previous work has shown that flow-induced shear stress can significantly impact breast cancer spheroid morphology and migration. Changes in the morphology, cell cycle, and genetic and protein profiles of biomarkers associated with metastatic progression are observed as a result of hydrodynamic forces.15–18 In this study, we used a microfluidic system to examine the effects of flow on endothelial cell behavior and cancer cell migration rate and distance. We previously found that low shear stress and transforming growth factor beta 1 (TGF-β1) induced EndMT, while high shear stress and TGF-β1 inhibited EndMT in a three-dimensional (3D) culture microfluidic device.19 This previously developed microfluidic device with an incorporated 3D culture chamber allows for recapitulation of both steady, low fluid-induced shear stress and ECM of varying composition and stiffness. Here, we investigate the interplay between low shear stress and tissue properties involved in cancer cell-mesenchymally transformed cell interaction. The conditions for which EndMT quantification was found to be the highest were included in order to study the effect on the tumor-mesenchymally transformed endothelial cell (EC) migration rate and distance and on cancer cell proliferation. The 3D culture microfluidic device allows for direct control over important biomechanical and biochemical factors influencing tumor-endothelium interactions in the investigation of metastasis and can model some of the complex cell-cell and cell-ECM interactions in a tumor environment.
II. MATERIALS AND METHODS
A. Cell culture and cancer spheroid formation
Human umbilical vein endothelial cells (HUVEC, Lonza®, Allendale, NJ, USA) were cultured at 37 °C and 5% CO2 in endothelial cell basal medium-2 (EBM®-2) supplemented with the EGM™-2 reagents (Lonza, Basal, Switzerland). Cells between passages 2 and 9 were used for all experiments. Cell cultures were maintained in a humidified incubator at 37 °C and 5% CO2 and grown to 80%–90% confluence before passage. Cells were passaged every 4–5 days using 0.025% trypsin-EDTA for cell detachment and trypsin neutralization solution (TNS) to neutralize trypsin (Lonza). Cells were then pelleted by centrifugation at 220 relative centrifugal force (RCF) for 5 min, and a cell count was performed using a hemocytometer with Trypan-blue exclusion. For routine culture, HUVEC were seeded at a density of 10 000 cells/cm2. For tumor-endothelial cell interaction experiments, to distinguish between the cell co-culture, HUVEC were stained with CellTrace™ CSFE green 488 (5 μM 1:1000 v/v in phosphate-buffered saline (PBS), ThermoFisher Scientific, Waltham, MA), incubated for 20 min at 37 °C and 5% CO2 protected from light. Next, five times the original staining volume of culture medium was added and the cells were incubated for 5 min to remove any free dye remaining in the solution. Lastly, the cells were pelleted by centrifugation and suspended in fresh pre-warmed Dulbecco's Modified Eagle's Medium (DMEM). For static experiments, HUVEC were seeded at 2 × 105 cells per cm2 onto the 3D collagen gel in 4-well tissue culture plates (1.9 cm2 growth area; Thermo Scientific Nunc). For shear experiments, HUVEC were seeded at 5 × 105 cells per cm2 onto the 3D collagen gel by injection into the microfluidic device inlet in a 30 μl cell suspension. HUVEC were allowed to attach onto the 3D collagen gel for 4 h in the incubator at 37 °C and 5% CO2. For tumor-endothelial models, HUVEC were seeded onto breast cancer cell (BCC) spheroid embedded-3D collagen hydrogels for 48 h under static and shear conditions.
For the tumor-endothelial cell interaction study, MDA-MB-231 human breast adenocarcinoma cells (BCCs) were cultured in 1× Dulbecco's Modified Eagle's Medium (DMEM) with 4.5 g/l of D-glucose, L-Glutamine, and phenol red (ThermoFisher Scientific) supplemented with 10% fetal bovine serum (FBS; ThermoFisher Scientific) at physiological conditions (humidified 37 °C and 5% CO2). Cells were passed using 0.25% trypsin-EDTA (ThermoFisher Scientific) for cell detachment, followed by neutralization with 1× DMEM. Cells were then pelleted by centrifugation at 125 relative centrifugal force (RCF) for 9 min, and a cell count was performed using a hemocytometer with Trypan-blue exclusion (Life Technologies, Carlsbad, CA, USA). For breast cancer spheroid formation (cell aggregates), the hanging drop method was used. First, MDA-MD-231 cells were stained with CellTrace Far Red 633 as per manufacturer's protocol (1 μM diluted 1:1000 v/v in PBS; ThermoFisher Scientific). Then, they were incubated for 20 min at 37 °C and 5% CO2 while protected from light. Next, five times the original staining volume of culture medium was added to the cells and they were incubated for 5 min to remove any free dye remaining in the solution. Lastly, the cells were pelleted by centrifugation and suspended in fresh pre-warmed 1× DMEM for seeding. The cells were seeded at 1 × 104 cells per μl onto the top of an inverted Petri dish filled with 20 ml of PBS (shown in supplementary material Fig. 1(a)]. Using a micropipette, 20 μl total volume (cell + DMEM medium) were added per drop. The top of the Petri dish was then inverted and the cells formed aggregates in the culture dish gradually over 48 h at 37 °C and 5% CO2 prior to being embedded in the 3D collagen hydrogels for static and shear experiments. After 48 h of cell aggregation in the hanging fluid drop, tumor spheroids were formed with an average horizontal dimension of 0.56 ± 0.38 mm and a vertical dimension of 0.54 ± 0.30 mm. The average circularity of the spheroids was measured using ImageJ 1.48v (National Institutes of Health, USA)20 to be 0.952, where 1 is a perfect circle [shown in Fig. 1(b) of the supplementary material]. To embed the spheroids in the 3D collagen hydrogels, the BCC spheroids were observed under a Leica L2 dissecting microscope (Wetzlar, Germany) and individually transferred in 15 μl of surrounding medium using a 2–20 μl micropipette inserted into the pre-gel solution. Two spheroids were added to every 100 μl of the pre-gel solution. For the static experiments, 6 spheroids were mixed with 300 μl of the pre-gel solution. For the shear experiments, 4 spheroids were mixed with 250 μl of the pre-gel solution. The spheroids-pre-gel mixture was incubated for 1 h in a humidified chamber at 37 °C and 5% CO2 until the gel was cross-linked, and then, HUVEC were seeded onto the spheroid embedded gels (the experimental setup is shown in Fig. 2 of the supplementary material).
B. Fabrication of 3D hydrogels
To study the role of altered ECM composition and stiffness in EndMT in HUVEC, 3D collagen gel constructs composed of 1.5 mg/ml, 2.0 mg/ml, or 2.2 mg/ml collagen-only or collagen +1 mg/ml, 10 mg/ml, or 20 mg/ml GAGs were used. The stiffness of the collagen-only ECM was matched to the collagen + GAG gels as described previously.21 Supplementary material Fig. 3 illustrates the types of ECM tissue properties that were studied within the microfluidic device chamber.
1. Collagen-only hydrogels
The concentration of collagen-only gels was varied to achieve 1.5, 2.0, or 2.2 mg/ml collagen composition. Collagen-only gels were formulated based on the collagen concentration by mixing ice-cold Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen™, Carlsbad, CA, USA) at concentrations of 3× for a low collagen concentration of 1.5 mg/ml in solution or 10× for a high collagen concentration of 2.0 or 2.2 mg/ml, 10% FBS (Gemini Bio-Products GemCell™ triple 0.1 μm sterile-filtered 500 ml, West Sacramento, CA, USA), sterile 18 MΩ water, 0.1 M NaOH, and rat tail collagen type I (BD4 Biosciences, Franklin Lakes, NJ, USA).
2. Collagen + GAG hydrogels
The component concentration of collagen + GAG gels was varied to obtain 1.5 mg/ml collagen + 1, 10, or 20 mg/ml GAGs. The two groups of GAGs that were utilized to alter the ECM composition were Chondroitin Sulfate A Sodium Salt from Bovine trachea (CS; Sigma-Aldrich, St. Louis, MO) and Hyaluronic Acid sodium salt from Streptococcus equi (HA; Sigma-Aldrich, St. Louis, MO).
3. Hydrogel mechanical measurement
Dahal et al. measured the collagen gel mechanical stiffness to provide a comparison between native and pathological tissue stiffness.21 From the hydrogel stiffness tests, they determined the elastic modulus (Young's modulus) of collagen-only hydrogels and collagen + GAG hydrogels. A rheometer (AR 1000, TA Instruments, New Castle, DE) was utilized to measure various collagen-only concentrations (1.5, 2.0, and 2.2 mg/ml collagen) and 1.5 mg/ml collagen +GAGs (1, 10, and 20 mg/ml HA or CS) elastic modulus. From the measurements, the Young's modulus value of the lowest collagen-only concentration hydrogel was found to be statistically the same as the lowest collagen + GAG hydrogel. Likewise, the stiffness increased in the same manner as the collagen concentration of collagen-only hydrogels and the collagen + GAG concentration increased. The tests resulted in a correlation between stiffness and concentration in the collagen-only and collagen + GAG cases. The measured Young's modulus values were used to formulate collagen-only samples with the same stiffness as sample controls for collagen + GAG, thus making it possible to determine if stiffness or GAG content induced EndMT. Table I shows elastic modulus measurements. For the lowest concentration (1.5 mg/ml collagen-only control), elastic modulus was determined to be 2.45 ± 0.45 kPa and for the highest concentration of 2.2 mg/ml collagen-only control to be 37.28 ± 5.12 kPa.21 Depending on location, composition, and type of tissue, in vivo conditions exhibit a gradient of stiffnesses (e.g., elastic modulus of brain tissue ranges from 0.1 to 1 kPa and muscles ranges from 8 to 17 kPa).22,23 The stiffness (elastic modulus) of blood vessels ranges from 0.2 to 4.0 kPa.24,25 The 1.5 mg/ml collagen-only gel stiffness of 2.45 ± 0.45 kPa falls within this range. Breast tumor tissue has been shown in transgenic mice to have a stiffer microenvironment (4.049 ± 0.938 kPa) compared to the healthy breast tissue (0.167 ± 0.031 kPa).26 In humans, the mean stiffness of breast tumors with a size <8 mm was found to be 58.7 ± 36 kPa.27 The 2.2 mg/ml collagen-only gel stiffness of 37.28 ± 5.12 kPa falls within the range of breast cancer masses found in vivo of transgenic mice and humans.
TABLE I.
3D collagen-only stiffness controls and collagen + GAG hydrogels and their corresponding Young's modulus.21
| Sample | Young's modulus (kPa) |
|---|---|
| 1.5 mg/ml collagen-only control | 2.45 ± 0.45 |
| 1.5 mg/ml collagen + 1 mg/ml CS | 4.99 ± 1.57 |
| 1.5 mg/ml collagen + 1 mg/ml HA | 4.48 ± 0.97 |
| 2 mg/ml collagen-only control | 7.44 ± 0.86 |
| 1.5 mg/ml collagen + 10 mg/ml CS | 13.08 ± 1.55 |
| 1.5 mg/ml collagen + 10 mg/ml HA | 13.04 ± 1.69 |
| 2.2 mg/ml collagen-only control | 37.28 ± 5.12 |
| 1.5 mg/ml collagen + 20 mg/ml CS | 21.57 ± 1.34 |
| 1.5 mg/ml collagen + 20 mg/ml HA | 34.01 ± 1.88 |
4. Extracellular matrix composition in the static and fluidic experiments
For the static experiments used to study the role of the altered ECM composition in EndMT, static culture well-plates were filled with 300 μl of collagen-only pre-gel solution of various collagen concentrations (1.5, 2.0, or 2.2 mg/ml collagen). After 1 h of incubation at 37 °C and 5% CO2, the collagen solution crosslinked to form a hydrogel. After 1 h of incubation at 37 °C and 5% CO2, the collagen solution crosslinked to form a hydrogel. After ECs were seeded onto collagen-only controls or collagen + GAG gels in 300 μl culture medium, the wells were incubated at 37 °C and 5% CO2 for 4 h. For the 3D static collagen gel HUVEC-only experiments, collagen gels were plated into 4-well tissue culture plates (1.9 cm2 growth area; Thermo Scientific Nunc™).
For flow-induced steady shear stress experiments, a previously described microfluidic device with an incorporated 3D ECM chamber was utilized.19 After device fabrication, the finished device was autoclaved before collagen-only or collagen + GAGs pre-gel solution was injected into the 3D chamber and then incubated at 37 °C and 5% CO2 for 1 h. A monolayer of HUVEC was seeded on the top of the 3D gels. For the tumor-endothelial cell interaction models, BCC spheroids were embedded in the 3D collagen gels. Experiments were conducted for 48 h under static conditions in the well plates or under shear conditions in the microfluidic device.
C. Real Time-quantitative Polymerase Chain Reaction (RT-qPCR)
After 48 h, gels were removed from 4-well plates under static conditions or from the device under shear conditions. HUVEC RNA was extracted and purified from each collagen gel using the RNeasy® Mini Kit (Qiagen, Valencia, CA). cDNA was generated using the iScript™ cDNA Synthesis Kit (BioRad, Hercules, CA). Real Time-quantitative Polymerase Chain Reaction (RT-qPCR) was then performed using a Mini Opticon Real-Time PCR System (BioRad, Hercules, CA). The genes examined encode for endothelial cell-cell junctions [Vascular endothelial-cadherin (VE-cadherin; CDH5)], mesenchymal transformation [alpha-smooth muscle actin: (α-SMA, ACTA2; Snail)], pro-inflammatory cell-surface receptors [vascular cell adhesion molecule 1 (VCAM-1) and intercellular cell adhesion molecule 1 (ICAM-1)],28 and collagen type I alpha 2 (COL1A2) fibrosis marker. Activated fibroblasts are characterized by the gene expression of mesenchymal-like markers (α-SMA), pro-inflammatory markers (VCAM-1 and ICAM-1), and COL1A2.29,30 For cell-GAG interaction studies, the genes examined are for cell-surface binding to CS (EMR2) and HA (CD44). For activated fibroblast, GAG production genes that were examined include CS production (CSPG4) and HA production (HAS3). Gene expression values were quantified using the ΔΔCt method with glyceraldehydes-3-phosphate dehydrogenase (GAPDH) as a housekeeping gene. Primers used for RT-qPCR are listed in supplementary material Table I.
D. Invasion and immunocytochemistry
HUVEC invasion and immunocytochemistry staining were previously described.19 After 48 h under static or shear conditions, HUVEC on 3D collagen-only and collagen + GAG gels were fixed and stained for endothelial cell tight junction protein (VE-cadherin) and pro-EndMT marker (α-SMA).
The MDA-MB231 BCC spheroids were stained for Ki-67 proliferation marker (1:100 v/v in PBS, mouse anti-human, Santa Cruz Biotechnology, Dallas, TX, USA) and allowed to incubate overnight at 4 °C. After incubation, cells were washed and 1:100 dilutions of secondary antibody (Alexafluor® 488, Invitrogen, Grand Island, NY) were added for 2 h in the dark at room temperature (RT). Alexafluor 488 was used for Ki-67 so that there would not be an overlap with the CellTrace Far Red 633 that stained the cancer cells' cytoplasm. Fluorescent images were obtained by sandwiching the gels between a glass slide and coverslip (24 × 40 mm micro cover glass No. 1 SUPERSLIP, VWR® International, Radnor, PA) using a Leica TCS SP5 confocal imaging system with a 63× water-immersion objective. Five confocal images from each condition were captured and analyzed using a MATLAB (Mathworks, Natick, MA) custom written code to quantify the fluorescent intensity of α-SMA protein expression.
E. Flow cytometry
HUVEC were grown in the 3D collagen gel for 48 h and then fixed in situ with 4% paraformaldehyde. The 3D collagen gels were then enzymatically digested with prepared papainase solution. The papainase protocol was adapted from Smeriglio et al.,31 where phosphate-buffer with EDTA (PBE) was prepared by dissolving 7.1 g of sodium phosphate dibasic (Na2HPO4) and 1.6 g of ethylenediaminetetraacetic acid disodium salt (EDTA-Na2) in 500 ml of diluted water with pH adjusted to 6.5 and filtered through a 0.22 mm filter (Corning, Teterboro, NJ, USA). Lastly, 0.035 g of L-cysteine was dissolved in 20 ml of PBE buffer. The solution was then re-filtered and 100 μl of sterile papain enzyme was added. Finally, the prepared papain enzyme was added to each 3D gel and incubated at 37 °C for 16 h.
Following the completion of hydrogel degradation, cells were collected and suspended in PBS. For cell proliferation studies, a fluorescence-activated cell sorting (FACS) Aria Cell Sorter (BD Biosciences, San Jose, CA, USA) with 488 nm and 637 nm fluorescence filters was used to detect the CellTrace Far Red 633 stained spheroids and CellTrace CSFE green 488 stained HUVEC. Data were analyzed with FCS 6 express software (De Novo™, Glendale, CA).
F. Image analysis
The confocal images were analyzed using Lecia LAS X software version 3.0 (Buffalo Grove, IL, USA).
G. Statistics
All data were analyzed with a one-way analysis of variance (ANOVA), a two-way ANOVA, or an unpaired student t-test n ≥ 3, p < 0.05 using Graphpad Prism version 6 for windows (GraphPad Software, San Diego, CA).
III. RESULTS AND DISCUSSION
A. HUVEC only model
1. HUVEC immunocytochemistry for vascular endothelial-cadherin (VE-cadherin), gap junction width quantification, alpha-smooth muscle actin (α-SMA) expression, and HUVEC invasion
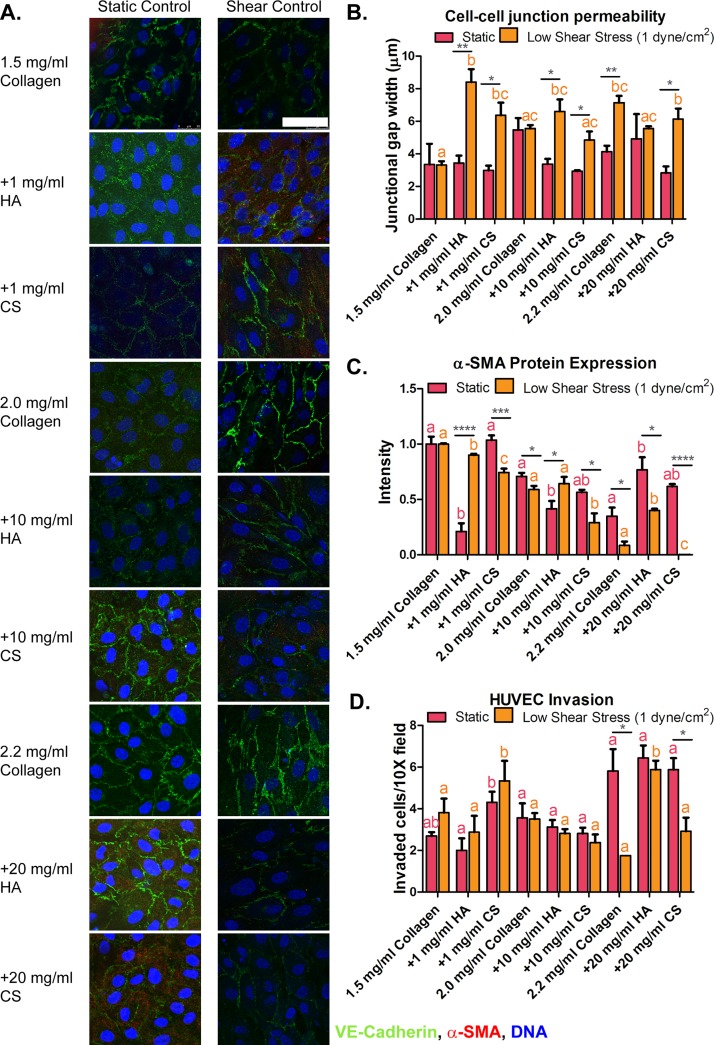
Confocal images of HUVEC captured 48 h after exposure to static conditions and low shear stress cultured on collagen + GAG gels were compared to those of their corresponding collagen-only stiffness controls. Confocal images of HUVEC protein markers VE-cadherin and α-SMA expression are shown in Fig. 1(a). HUVEC exposed to altered ECM stiffness and composition with and without shear stress demonstrated the expression of cell-cell junction marker VE-cadherin throughout all samples, as shown in Fig. 1(a). We further measured the gap width of these cell-cell junctions, which was previously described19 and quantitative results are shown in Fig. 1(b). The gap width was increased in HUVEC cultured onto 2.2 mg/ml collagen-only control stiffness gels exposed to low shear stress when compared to the 1.5 mg/ml collagen-only control under static conditions (3.35 ± 1.27 and 7.14 ± 0.42 μm, respectively). Furthermore, an increased gap junction width was observed in HUVEC cultured onto collagen + GAG gels exposed to low shear stress when compared to the 1.5 mg/ml collagen-only control under static conditions [as shown in Fig. 1(b)]. An increased gap width suggests that HUVEC are more likely to undergo EndMT.
FIG. 1.
Protein expression and invasion of human umbilical vein endothelial cells (HUVEC) seeded onto 3D hydrogels. (a) HUVEC immunocytochemistry images for alpha-smooth muscle actin (α-SMA; red), vascular endothelial-cadherin (VE-cadherin; green), and cell DNA (blue) on three-dimensional (3D) collagen-only or collagen + glycosaminoglycans [GAGs, chondroitin sulfate (CS) or hyaluronic acid (HA)] gels after 48 h of exposure to static and flow-induced 1 dyne/cm2 shear stress. (b) Gap junction width measurements of HUVEC seeded onto 3D collagen-only or collagen + GAGs hydrogels exposed to static and 1 dyne/cm2 shear stress conditions. (c) Fluorescence intensity analysis of mesenchymal marker (α-SMA) protein expression in HUVEC. (d) HUVEC invasion into the extracellular matrix of cells cultured on collagen-only controls and collagen-GAG exposed to static conditions and 1 dyne/cm2 shear stress. Fluorescence intensity analysis of mesenchymal marker α-SMA protein expression in HUVEC. Data shown are mean ± SEM, n ≥ 3 culture wells. All static or all fluidic conditions were analyzed with a one-way ANOVA with Tukey's post-test. Tukey groups are shown for statistically significant results (p < 0.05). Identical extracellular matrix conditions exposed to static or shear conditions were analyzed with an unpaired Student's t-test. Bars connected with * represent statistical significance with an unpaired Student's t-test (p < 0.05). Scale bar = 50 μm.
We examined the characteristics of HUVEC by analyzing the protein expression of the pro-EndMT marker α-SMA from the confocal images. Collagen + GAG samples were compared to their corresponding static collagen-only stiffness controls, as described by Dahal et al.,21 to make it possible to determine if stiffness or GAG content induces EndMT. For the lowest gel concentration, there was a significant difference in GAG containing gels (1.5 mg/ml collagen + 1 mg/ml HA gels under static and 1.5 mg/ml collagen + 1 mg/ml CS gels under shear conditions) when compared to 1.5 mg/ml collagen-only stiffness control gels. For moderate gel concentrations, similar intensity profiles of α-SMA marker expression were obtained. For the stiffest gel concentration under low shear stress conditions, α-SMA marker expression decreased in the collagen + GAG (20 mg/ml of HA and CS concentrations, respectively) samples when compared to the 2.2 mg/ml collagen-only stiffness control. These results suggest that α-SMA expression was the lowest in HUVEC exposed to low steady shear stress and stiff ECM conditions. Increased expression of pro-EndMT marker protein (α-SMA) suggests that the endothelial cells are ready to invade into the altered 3D collagen-only and collagen + GAGs gels; however, applying a low shear stress to the HUVEC reduces α-SMA expression, as shown by the results in Fig. 1(c). These findings are similar to in vivo conditions where cell-cell junctions are produced on adjacent cells and only form when cells are fully confluent. Continuous staining of VE-cadherin is a physical marker of endothelium function.32 Previous studies have observed that altered ECM properties mediate how a cell senses and perceives external forces, which results in altered cell behavior.26,33–40
Collagen + GAG samples were compared to their corresponding static collagen-only stiffness controls by Dahal et al.21 to determine if stiffness or GAG content induces invasion in HUVEC. There was no significant difference between the GAG samples and their stiffness control collagen-only gels, except for the cases of 1 mg/ml CS and its 1.5 mg/ml stiffness control under static conditions. The overall increased invasion observed in stiff and high GAG concentration gels [as shown in Fig. 1(d)] may be due in part to increased gel stiffness, fibril alignment, and pore size. It was previously reported by Provenzano et al.41 that a dense collagen microenvironment enhanced mammary tumor formation and metastasis. They observed 3 stages of tumor-associated collagen signature (TACS) in transgenic mice. For TACS-1, the presence of increased collagen concentration surrounding tumors was indicated by increased signal intensity in a region near the tumor. In TACS-2, they observed straightened collagen fibers stretched around the tumor and TACS-3 identified aligned collagen fibers that facilitated local invasion. Overall, they found that tumors in collagen-dense tissues had more collagen signals, enhanced collagen fibril realignment, and increased local invasion.
Invasion assays allow for the quantification of the number of HUVEC that have transformed to the mesenchymal phenotype and have invaded 60 μm into the collagen-only or collagen + GAG hydrogels. An illustration of the invasion assay and a confocal image of an invaded HUVEC are shown in supplementary material, Fig. 4. Under static conditions, HUVEC cultured on 2.2 mg/ml collagen-only control and collagen + 20 mg/ml GAG (20 mg/ml of HA or CS) gels invaded significantly more (5.81 ± 2.09, 6.43 ± 1.19, and 5.87 ± 1.10 invaded cells/field, respectively) when compared to 1.5 mg/ml collagen-only gels (2.68 ± 0.37 invaded cells/field). Shear stress decreased HUVEC invasion. HUVEC exposed to 1 dyne/cm2 shear stress and cultured on 2.2 mg/ml collagen-only control or collagen + 20 mg/ml GAG (20 mg/ml HA or CS) gels showed a decreased invasion rate to 2.25 ± 0.50, 5.87 ± 0.85, and 2.91 ± 1.12 invaded cells/field, respectively. The number of invaded HUVEC seeded on collagen + 10 mg/ml GAG (10 mg/ml HA or CS) gels was found to be similar to cells exposed to 1.5 mg/ml collagen-only control gels with and without shear exposure. The number of invaded HUVEC seeded on collagen + 1 mg/ml GAG (1 mg/ml HA or CS) under static or shear conditions was statistically different (for HA Fluidic: 2.88 ± 1.56 and Static: 2.00 ± 1.16 invaded cells/field, and for CS, Fluidic: 5.33 ± 1.66 and Static: 4.31 ± 1.01 invaded cells/field). These results show that 2.2 mg/ml collagen-only gels and collagen + 20 mg/ml GAG (20 mg/ml HA or CS) gels resulted in the most mesenchymally transformed and invaded HUVEC. Low shear stress attenuated the invasion of HUVEC on stiff (2.2 mg/ml collagen-only control) or 1.5 mg/ml collagen + 20 mg/ml GAG gels. HUVEC invasion results are shown in Fig. 1(d).
The addition of GAGs and increased collagen concentrations of the gels alter gel stiffness and fibril alignment but not pore size. Collagen I gels similar to those used in this study have been found to have tunable structural and mechanical properties. Yang et al.42 fabricated 1.0, 3.0, and 4.0 mg/ml collagen gels that gelled at 37 °C. They found that stiffness increased with increasing collagen concentration, but the least stiff gel and the stiffest gel had similar pore sizes. When they embedded tumor spheroids in their gel, they found that invasion distances in the 1.0 and 4.0 mg/ml collagen gels were similar even though the stiffness of the gels differed. Similarities in pore sizes between the least stiff and the stiffest collagen gels may explain why similar HUVEC invasion distances were observed in both these ECM alterations. These results also demonstrate that there is a correlation between invasion distance and pore size. In another study, Yang et al.43 investigated altered ECM compositions that were composed of acid-solubilized (AS) or pepsin-treated (PT) collagen and GAG (CS or HA) and found that collagen + CS gels induced changes in the storage and loss moduli of AS gels when compared to PT gels. It was also observed that CS induced fibril bundling, increased pore size, and inhibited invasion in the AS collagen network and not in the PT collagen networks. On the other hand, they also observed that the presence of HA reduced pore sizes and facilitated cell invasion. These previous studies highlight the importance of considering the biomechanical effect of changes in the collagen network structure induced by ECM components such as GAGs when examining cell invasion in a composite microenvironment.
2. Downregulation of the endothelial marker and upregulation of pro-inflammatory and mesenchymal markers in HUVEC cultured on collagen + GAG gels and exposed to low fluid-induced shear stress conditions
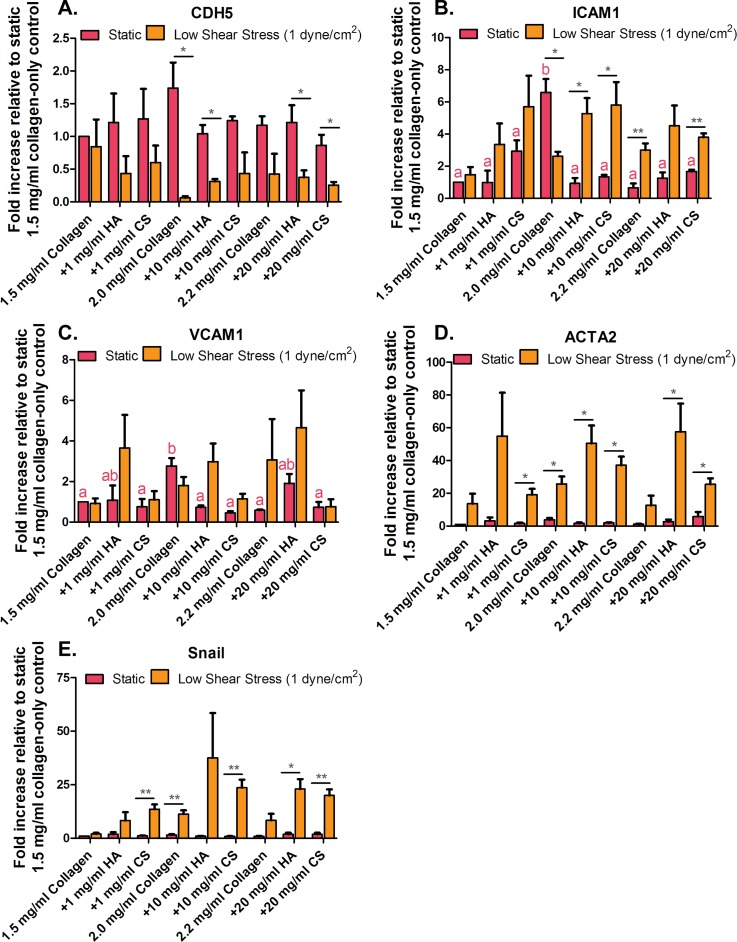
For RT-qPCR characterization, we examined endothelial marker VE-cadherin (CDH5), pro-inflammatory markers (VCAM-1 and ICAM-1), and pro-EndMT markers (ACTA2 and Snail) gene expression in HUVEC under static conditions and low steady shear flow (as shown in Fig. 2). Gene expression for all conditions was compared to static 1.5 mg/ml collagen-only stiffness control conditions. VE-cadherin/CDH5 gene expression in HUVEC cultured on collagen-only stiffness controls for corresponding collagen + GAG gels was the same when cultured under static conditions. However, HUVEC exposed to low shear conditions statistically downregulated CDH5 expression when cultured on 2.0 mg/ml collagen-only, collagen + 10 mg/ml HA, 2.2 mg/ml collagen-only control, and collagen + GAG (HA and CS) gels compared to 1.5 mg/ml collagen-only (0.06 ± 0.04, 0.31 ± 0.06, 0.42 ± 0.53, 0.7 ± 0.18, 0.25 ± 0.08, and 1.00 ± 0.00 fold, respectively). Low shear stress conditions increased pro-inflammatory gene expression in HUVEC cultured on collagen + 20 mg/ml GAG (HA and CS) compared to 1.5 mg/ml collagen-only static control conditions (4.50 ± 1.26, 3.79 ± 0.24, and 1.00 ± 0.00 fold for ICAM-1, respectively). For mesenchymal marker gene expression, low shear stress conditions had a similar effect to the pro-inflammatory markers for collagen + 20 mg/ml GAGs (HA and CS) conditions, where ACTA2 gene expression was upregulated when compared to 1.5 mg/ml collagen-only static control conditions (57.54 ± 17.18, 25.46 ± 3.66 fold, respectively). These data suggest that ECM composition induction of EndMT is not only collagen stiffness dependent but also GAG concentration dependent. This is observed from the increased pro-inflammatory and mesenchymal markers expression with increased GAG (HA and CS) concentration. Thus, the GAG concentration in the ECM has a prominent role in cells undergoing EndMT. The addition of shear stress exposure with these GAG conditions accelerated the EndMT process in HUVEC. However, HUVEC undergoing EndMT do not similarly express pro-inflammatory markers. When examining pro-EndMT behavior, no significant difference was quantified phenotypically in HUVEC exposed to 2.0 mg/ml collagen-only control gels under the low shear stress and static conditions. However, when examining pro-inflammatory (ICAM1 and VCAM1) genes in 2.0 mg/ml collagen-only control under the low shear stress condition, it is lower than that under the static condition. These results may suggest that low shear stress protects the cells from inflammation, which is why we observed more myofibroblastic-like characteristics in static conditions than under low shear stress. Overall, cells exposed to biophysical cues, including low shear stress and the presence of GAG in the ECM, resulted in increased pro-inflammatory and mesenchymal markers gene expression compared to the endothelial marker, VE-cadherin/CDH5, except for the moderate stiffness collagen-only control.
FIG. 2.
Human umbilical vein endothelial cells (HUVEC) inflammatory and pro-EndMT gene expression. (a) Vascular endothelial-cadherin (VE-cadherin; CDH5), (b) pro-inflammatory cell receptor intercellular adhesion molecule-1 (ICAM-1), and (c) vascular cell adhesion molecule-1 (VCAM-1). (d) Endothelial to mesenchymal transformation (EndMT) markers, actin, alpha 2 (ACTA2) and (e) Snail. Error bars show mean ± SEM, n = 3 culture wells. All static or all fluidic conditions were analyzed with a one-way ANOVA with Tukey's post-test. Tukey groups are shown for statistically significant results (p < 0.05). Identical extracellular matrix conditions exposed to static or shear conditions were analyzed with an unpaired Student's t-test. Bars connected with * represent statistical significance with an unpaired Student's t-test (p < 0.05).
These gene expression results suggest that the presence of GAGs in the ECM induces HUVEC to undergo EndMT. Additionally, HUVEC exposure to low shear stress perturbs the EC monolayer and induces the cells to undergo EndMT as seen by the increased expression of pro-inflammatory and pro-EndMT markers. These data indicate that GAGs and low shear stress may act synergistically to induce EndMT in HUVEC; however, it is also dependent on ECM stiffness. For example, low and high stiffness gels exhibited more phenotypic and genotypic EndMT characteristics (Figs. 1 and 2). This induced transformation, owing to the biomechanical cues from shear stress and ECM properties, mediates mechanotransducers such as adherens junctions, cell-surface pro-inflammatory, and cell motility markers. In this study, the cell-cell bond width was examined. Previous EndMT studies by Dahal et al. have also shown that decreased cell-ECM bond strength induced EndMT in EC.21
3. Low fluid-induced shear stress and GAGs regulate cell-surface receptors for matrix binding and ECM production gene expression levels in HUVEC
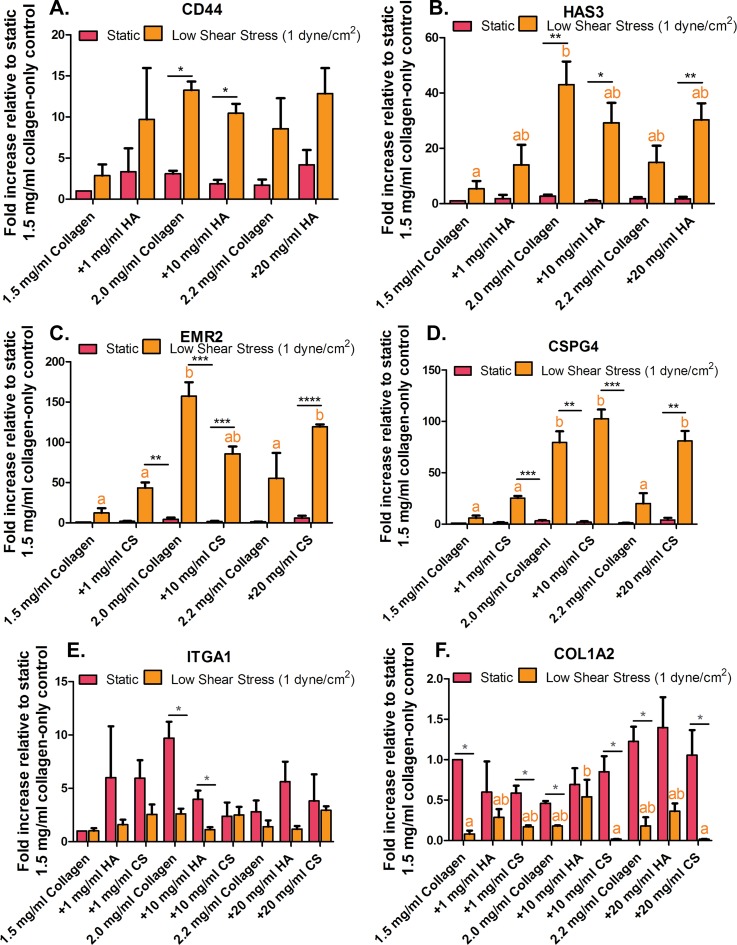
Myofibroblasts generated from EndMT secrete ECM proteins involved in wound repair, which are prominent in tissue fibrosis of many organs.14,44,45 Gene expression levels for cell-surface receptors to GAGs (HA; CD44 and CS; EMR2) and collagen (ITGA1) were examined. Additionally, new GAGs and collagen synthesis (HA: HAS3, CS: CSPG4, collagen: COL1A2) gene expression levels were studied. Results for cell-matrix binding and the new production of ECM proteins are shown in Fig. 3.
FIG. 3.
Human umbilical vein endothelial cells (HUVEC) glycosaminoglycan (GAG) and collagen cell-surface receptors and extracellular matrix (ECM) remodeling gene expression. (a) Cell-surface receptor for hyaluronic acid (HA; CD44) and (b) production of HA (HAS3). (c) Cell-surface receptor for chondritic sulfate (CS; EMR2) and (d) production of CS (CSPG4). (e) Collagen 1 subunit integrin receptor (ITGA1) and (f) fibrosis marker (COL1A2). Error bars show mean ± SEM, n = 3 culture wells. All static or all fluidic conditions were analyzed with a one-way ANOVA with Tukey's post-test. Tukey groups are shown for statistically significant results (p < 0.05). Identical extracellular matrix conditions exposed to static or shear conditions were analyzed with an unpaired Student's t-test. Bars connected with * represent statistical significance with an unpaired Student's t-test (p < 0.05).
Cell-surface receptors for GAGs (HA; CD44 and CS; EMR2) were found to be significantly upregulated in HUVEC exposed to low fluid-induced shear stress conditions when compared to static conditions [Figs. 3(a) and 3(c)]. The various ECM compositions had no effect on the GAG cell binding receptor gene expression when cells were exposed to static conditions. Similar results were quantified for GAG synthesis [HA: HAS3 and CS: CSPG4, Figs. 3(b) and 3(d)]. These data suggest that shear stress mediates GAG-cell binding and GAG production. HA binding receptor, CD44, can selectively bind to different growth factors and function as signal receptor targeting mesenchymal cells and has been previously found to impact the direction of cell-cell communication.46,47 It is also known that the CS cell-surface binding receptor, EMR2, is a member of adhesive-(G protein)-coupled receptors, which are involved in tumorigenesis and have been found to be upregulated in invasive breast carcinomas.48,49 HAS3 appears to promote processes leading to tumor growth, invasion, and metastasis.50–53 CSPG4 has been previously found to be involved in the formation of cell surface ligands in aggressive breast cancer cells.13,54,55 The freshly synthesized HA and CS bind to aggregating proteoglycans that contribute to the assembly of pericellular matrix and thus take possession in controlling growth factors and cytokines secretion into the stroma environment.56–58 The upregulation of these gene markers may be due to the disease-prone culture conditions that the HUVEC were exposed to, which are low (pathological) shear stress and collagen + GAG ECM.
The gene for HA production is hyaluronan synthases 3 (HAS3) and for CS is chondroitin sulfate proteoglycan 4 (CSPG4). Many of these ECM components and their cell-surface receptors such as those for GAGs (HA: CD44 and CS: EMR2) and cell-collagen integrin binding alpha 1 (ITGA1) that can facilitate growth factor signaling are frequently overproduced in cancer.59 EMR2 is also known as an adhesion G protein-coupled receptor E2 (ADGRE2) and has been found to be involved in tumorigenesis.48,49 Additionally, ECM deposition of GAGs (HA: HAS3, CS: CSPG4) and collagen can alter ECM biomechanical properties. These biochemical and biomechanical properties of the ECM can transmit to biological signals that allow cells to sense and interact with other stroma cells in the microenvironments. This process occurs through the use of various signaling transduction cascades originating from the cell surface to the nucleus and results in protein and gene expression and cell invasion. For example, changes in ECM biomechanical and biochemical properties may facilitate cancer cell migration toward the endothelium. Mesenchymally transformed endothelial cells may also interact with stroma cancer cells, resulting in the generation of activated fibroblasts, increased cell migration rate and distance, and cancer cell proliferation.
In Figs. 3(e) and 3(f), the expression of α1 integrin subunit ITGA1 and fibrosis marker COL1A2 in HUVEC cultured on collagen-only stiffness control gels and corresponding stiff collagen + GAG gels indicates that altered ECM properties induce the formation of activated fibroblasts. ITGA1 is part of the α1β1 integrin and is primarily expressed in mesenchymal cells. Its expression has been linked to fibroblast proliferation and collagen synthesis regulation.60 ITGA1 and COL1A2 were both found to be upregulated in HUVEC exposed to static conditions and altered ECM compositions and stiffness. From Fig. 3(f), it was found that COL1A2 collagen synthesis marker was downregulated in HUVEC exposed to shear stress conditions on the 2.2 mg/ml collagen-only gels and collagen + 20 mg/ml GAG (20 mg/ml HA and CS) gels by 0.17 ± 0.10, 0.36 ± 0.9, and 0.01 ± 0.00 fold, respectively. However, COL1A2 was upregulated in HUVEC exposed static conditions by 1.22 ± 0.18, 1.39 ± 0.37, and 1.055 ± 0.31 fold for 2.2 mg/ml collagen-only and collagen + GAGs (20 mg/ml HA and CS), respectively. These data suggest that shear does not play a role in promoting fibrosis but may inhibit the generation of myofibroblastic-like cells. The cells exposed to shear stress and altered ECM composition and stiffness have not formed myofibroblastic-like characteristics even though they have undergone EndMT. On the contrary, the EC that have undergone EndMT under static conditions cultured on collagen-only (with various collagen concentrations) gels and collagen + GAGs (HA and CS at various concentrations) produced more collagen compared to shear conditions. Collagen I production has been shown to increase during tumor formation.61 However, shear stress may inhibit ECM remodeling.
B. HUVEC-BCC model
1. Low steady shear stress decreases spheroid size
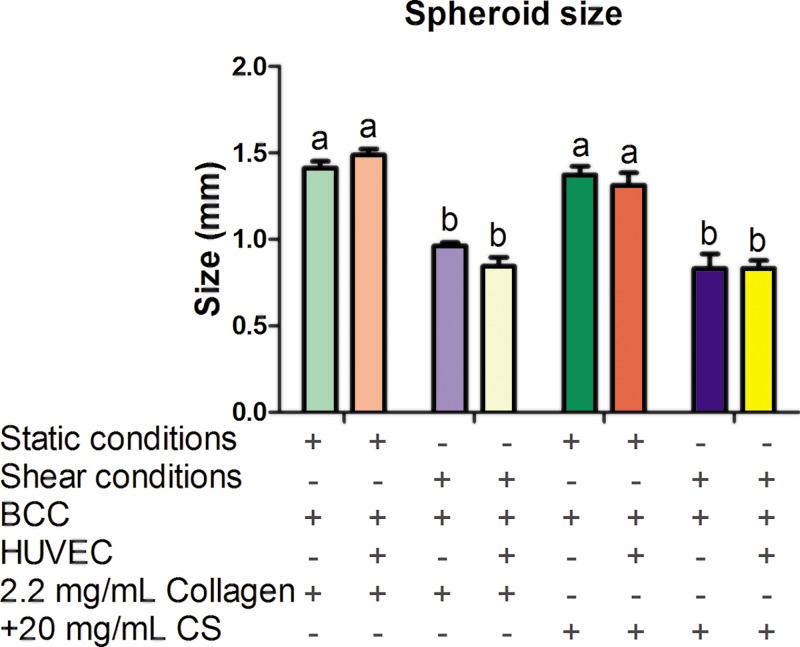
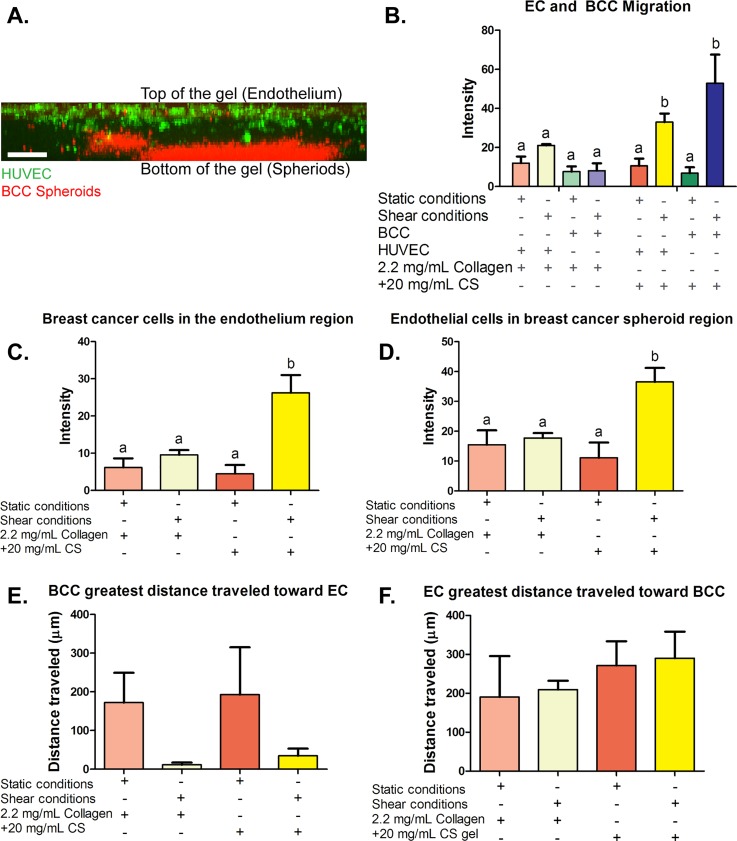
The condition yielding the largest amount of observed EndMT was 1.5 mg/ml collagen mixed with 20 mg/ml CS (from the study of the role of shear stress and altered ECM composition in EndMT, results are shown in supplementary material, Table III). For the HUVEC-BCC model, breast cancer spheroids were embedded in both a 2.2 mg/ml collagen-only stiffness control and 1.5 mg/ml collagen + 20 mg/ml CS ECM environment under static and shear conditions. In addition, the spheroids were embedded in collagen-only and collagen + GAG gels and then co-cultured with HUVEC, again under static and shear stress conditions. Second, spheroids were embedded in collagen-only and collagen + GAG gels and then co-cultured with HUVEC under static and shear stress conditions. The original average size of the formed spheroids was 0.56 ± 0.04 mm. After 48 h, the spheroids-only embedded in 2.2 mg/ml collagen-only gels statistically increased in size when exposed to static and shear conditions (Static: 1.41 ± 0.17 mm and Shear: 0.97 ± 0.12 mm). Interestingly, when spheroids were co-cultured with HUVEC and exposed to shear stress conditions in collagen + GAG gels, there was a significant difference in spheroid size when compared to static co-culture conditions (Static: 1.31 ± 0.25 and Shear: 0.80 ± 0.14 mm, results shown in Fig. 4). Overall, shear stress decreased spheroid size in both the mono-culture and co-culture when embedded in collagen-only and collagen + GAG gels. ECM composition and stiffness had no significant effect on spheroid size when comparing mono-culture and co-culture results. Reduction in spheroid size by shear stress could be related to transport of oxygen, nutrients, or cell metabolic wastes. Both convective and diffusive transport processes are important in providing nutrients, the removal of wastes, and the delivery of drugs. Within the ECM, transport is largely by diffusion. For macromolecules, diffusion in tissue is impacted by enzyme degradation such as collagenase and hyaluronidase. A previous study using fluorescein isothiocyanate (FITC)-conjugated dextran found that increased penetration of drug macromolecules into the tumor tissue is facilitated by collagenase and hyaluronidase after 18 h.62 Furthermore, in this study, high gene expression of cell surface markers for GAGs (EMR2 and CD44) in HUVEC exposed to low shear and altered ECM composition and stiffness was observed. It has been suggested that GAG cell surface markers can form a protective coating around the cell, therefore limiting the diffusion of drug molecules to the cancer cell surface.63 It would also be a possible explanation as to why spheroid size increased in static conditions when embedded in altered collagen and GAGs concentration.
FIG. 4.
Breast cancer spheroid size. Spheroid size was measured following embedding in three-dimensional (3D) collagen-only or collagen + chondroitin sulfate (CS) gels and after 48 h of exposure to static or shear stress conditions. Error bars show mean ± SEM, n ≥ 8 spheroids and n = 2 biological samples. Bars that do not share any letters are significantly different according to a two-way ANOVA with Tukey's post-hoc test (p < 0.05).
Regarding the result of shear stress reducing the size of spheroid-only embedded in collagen gels and collagen + CS gels (shown in Fig. 4), one possible explanation may be due to the diffusion of molecules from the cell culture medium into the collagen hydrogel leading to an increase in the number of BCC separating from the spheroids and migrating towards the endothelium. Another explanation may be that BCC migrated up towards the top of the gels for oxygen supply. In the case of shear limited spheroid size in spheroids embedded in stiff gels co-cultured with HUVEC exposed to these altered ECM compositions, this may be due in part to the cell-cell crosstalk and soluble factors secreted by ECs. Shear stress has been shown to activate signaling pathways (MAPK), leading to upregulation of cell-cell adhesion molecules, growth factors, and gene and protein expression in EC.64,65 Increased spheroid size suggests that the BCCs are more likely to migrate and proliferate. This highlights the need for the use of microfluidic devices because static conditions make for a more pathological-like environment.
2. HUVEC and BCC migrated significantly when cultured in collagen + 20 mg/ml CS hydrogels and when exposed to shear stress
We examined HUVEC and BCC migration through the stiff ECM when cultured in 2.2 mg/ml collagen-only and collagen + 20 mg/ml CS gels. Figure 5(a) shows a side view confocal image of the 3D gel with CellTrace live staining of HUVEC on top of the gel (green) and CellTrace live staining of BCC spheroids (red) embedded in collagen + 20 mg/ml CS at the bottom of the gel. BCC and EC migration through the ECM was quantified by measuring fluorescence intensity in the middle of the gel. The green fluorescently labeled HUVEC exposed to shear conditions cultured on spheroids embedded in collagen + CS gels resulted in increased intensity when compared to HUVEC co-cultured with spheroids in 2.2 mg/ml collagen-only control. Similar results were observed for spheroids embedded in collagen + CS co-cultured with HUVEC exposed to shear conditions. These data depict that shear stress moderates BCC and HUVEC invasion in collagen + CS ECM composition [results quantified in Fig. 5(b)]. Additionally, more BCC were found to be present in the endothelium region as well as more HUVEC were observed to be present inside the breast cancer spheroid region [results are shown in Figs. 5(c) and 5(d)]. The fact that shear stress significantly increased the HUVEC-BCC interaction can be correlated with the reduced tumor spheroid size described earlier. Fluid shear stress has been suggested to act as a disruptor for HUVEC surface receptors which in turn may promote cancer cell adhesion to the endothelium, resulting in decreased spheroid size when co-cultured with EC. Figures 5(e) and 5(f) demonstrate the results of the quantification of the greatest distance HUVEC and BCC have traveled from their primary region. The length of the cell that traveled the greatest distance is quantified in Fig. 6 in the supplementary material. There was no significant difference in distance traveled and cell length, indicating that the distance traveled by cells was consistent across all the gels with the same stiffness. The data suggest that shear, GAGs (CS) and the presence of HUVEC result in the greatest cell migration and interaction.
FIG. 5.
Migration and migration distance quantification of breast cancer cells (BCC) and human umbilical vein endothelial cells (HUVEC). (a) Side view confocal image of the three-dimensional (3D) hydrogels with live cell staining. Live cell stain of human umbilical vein endothelial cells (HUVEC) on top of the gels (green) and live cell staining of spheroids (red) embedded in a collagen hydrogel. (b) BCC embedded in three-dimensional (3D) collagen-only gels and collagen +20 mg/ml chondroitin sulfate (CS) gels after 48 h of exposure to static or shear stress conditions migration toward the endothelium. (c) Fluorescent intensity quantification of BCC stained with CellTrace Far Red in the endothelium region and (d) fluorescent intensity quantification of EC stained with CellTrace Green in the spheroid region. (e) and (f) Greatest distance breast cancer and endothelial cells traveled in 2.2. mg/ml collagen-only or collagen + 20 mg/ml chondroitin sulfate (CS) gels after 48 h of exposure to static or shear stress conditions. (e) Quantification of the greatest distance BCC traveled. (f) Quantification of the greatest distance HUVEC traveled. Error bars show mean ± SEM, n = 6 confocal images from 2 biological samples. All conditions were analyzed with a one-way ANOVA with Tukey's post-test. Tukey groups are shown for statistically significant results (p < 0.05). Scale bar = 250 μm.
3. HUVEC-BCC interactions
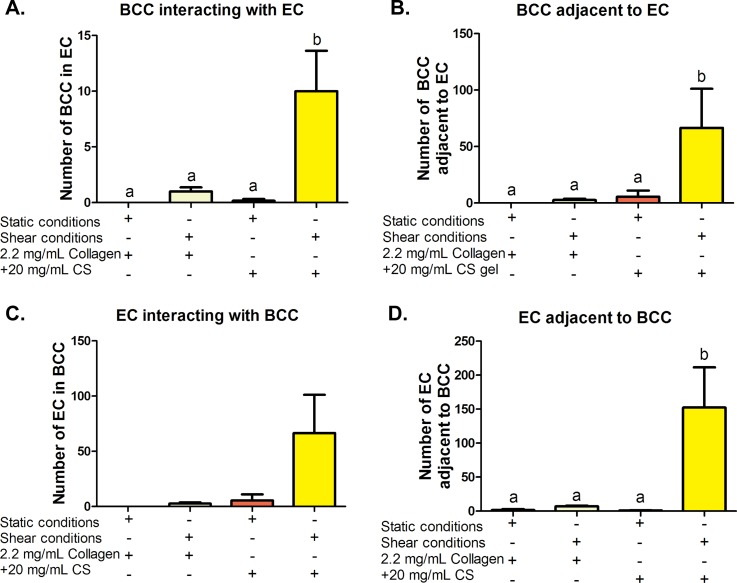
When BCC separated from spheroids and migrated towards the endothelium, interactions occurred along the way that were detected and quantified. Figure 6 shows results of migrated BCC interacting with invading EC and the endothelium. Figure 6(a) shows the quantified data of the number of BCC in a group of HUVEC, (b) number of BCCs physically touching or adjacent to HUVEC, (c) likewise HUVEC count inside a group of BCC, and (d) number of EC adjacent to BCC. It was determined that cells co-cultured in collagen + CS gels exposed to shear stress had the highest interactions between two different cell types. Statistically, 10.00 ± 3.64 BCC were inside a group of EC when exposed to shear stress and collagen + CS ECM conditions while there were only 0.16 ± 0.16 BCC inside a group of EC under the static collagen + CS conditions. For 2.2 mg/ml collagen-only stiffness controls, 1.00 ± 0.36 BCC were counted under shear while none was observed under the static conditions. These results suggest that shear stress and collagen + CS ECM conditions induce interactions between BCC and EC. Increased HUVEC-BCC interaction may be due in part to the upregulation of GAG cell surface receptors (EMR2 for CS and CD44 for HA) that was described previously. Many of the ECM components and their receptors such as hyaluronic acid and CD44 have been found to facilitate the signaling of growth factors which are frequently overproduced in cancer.12 For example, hyaluronic acid receptor CD44 can selectively bind to growth factors and act as a signal co-receptor or a presenter and help determine cell-cell communication.46,66 BCC can then form direct contacts (adhere to) EC. Thus, abnormal changes in ECM composition and stiffness can alter ECM biochemical properties and can potentially influence oncogenic effects of various growth factor signaling pathways, leading to alteration of stromal cell behaviors and transformation.
FIG. 6.
Quantification of breast cancer cell (BCC)-human umbilical vein endothelial cell (HUVEC) interactions. (a) The number of BCC inside a group of HUVEC and (b) physically touching HUVEC and likewise (c) HUVEC count inside a group of BCC and (d) number of HUVEC touching BCC. Error bars show SEM, n = 6 confocal images from 2 biological samples. Bars that do not share any letters are significantly different according to a two-way ANOVA with Tukey's post-hoc test (p < 0.05).
4. Co-culture of endothelial-breast spheroids resulted in increased breast cancer proliferation in 2.2 mg/ml collagen-only hydrogels
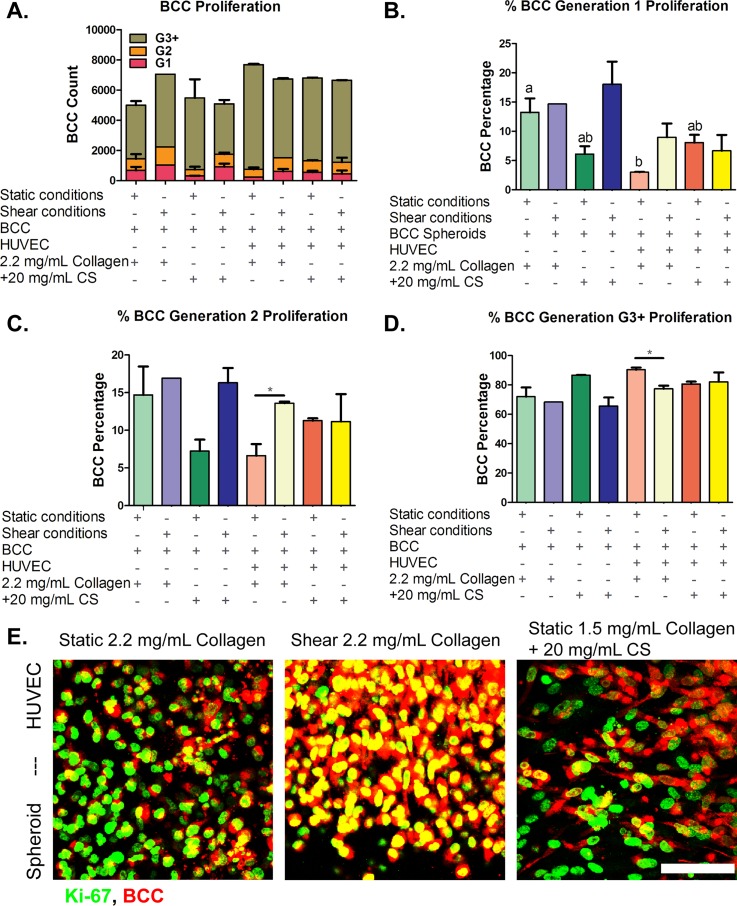
CellTrace Far Red BCC proliferation dye was used for in vitro labeling of cells to trace multiple generations using dye dilution by flow cytometry techniques. BCC proliferation analysis by dye dilution was gated to a positive control (cells + dye) and negative control (cells-only) to distinguish fluorescently labeled cells from auto-fluorescence after several cell divisions. For flow cytometry, analysis of 10 000 events was detected for each sample. These events represent BCC that were fluorescently tagged and have proliferated into generation 1, generation 2, and generation 3 and higher. The analysis examined the number of BCC that have proliferated at the various conditions (altered ECM stiffness and composition and exposed to shear stress and static conditions) in mono-culture (spheroids-only) and co-culture (spheroids-HUVEC) over a 48 h-period [results are shown in Fig. 7(a)]. A higher percentage of G1 BCC is considered favorable as the cancer cells have proliferated less, while a higher percentage of G2 and G3 BCC is a more pathogenic result.
FIG. 7.
Breast cancer cells (BCC) proliferation. BCC were cultured for 48 h with human umbilical vein endothelial cells in three-dimensional collagen or collagen + chondroitin sulfate (CS) gels for 48 h under static or shear conditions. (a) Total cell count for all generations. Cell generations are noted as (b) G1, (c) G2, and (d) G3 and higher. (e) Ki-67 proliferation marker immunocytochemistry confocal microscopy images of BCC. BCC spheroids embedded in 2.2 mg/ml collagen-only stiffness control cultured with human umbilical vein endothelial cells (HUVEC) on top of the gels and exposed to (Left) static conditions and (Middle) shear stress conditions. (Right) BCC spheroids embedded in 1.5 mg/ml collagen + 20 mg/ml CS cultured with HUVEC on top of the gels and exposed to static conditions. BCC were stained with CellTrace Far Red and Ki-67 proliferation marker (green). Bar Scale = 50 μm. Error bars show mean ± SEM, n = 2 biological samples. All static or all fluidic conditions were analyzed with a one-way ANOVA with Tukey's post-test. Tukey groups are shown for statistically significant results (p < 0.05). Identical extracellular matrix conditions exposed to static or shear conditions were analyzed with an unpaired Student's t-test. Bars connected with * represent statistical significance with an unpaired Student's t-test (p < 0.05). Scale bar = 50 μm.
Due to our BCC isolation method and gating range, the number of events/cell counts could vary from condition to condition so we evaluated the percentage of cell proliferation at every generation. For generation 1, there was no statistically significant difference in BCC proliferation for all shear conditions [Fig. 7(b)]. For static conditions, the highest percentage of G1 cells was in stiff collagen-only without the presence of HUVEC [Fig. 7(b)]. The lowest percentage of G1 cells was measured to be in static conditions and collagen + 20 mg/ml CS gels (6.11 ± 1.90%).
For generation 2, there were no significant statistical differences between all sheared and all static samples. The quantitative results are shown in Fig. 7(c). A t-test between identical ECM conditions with or without shear showed that there was a significantly higher percentage of G2 cells in sheared 2.2 mg/ml collagen gels with HUVEC when compared to 2.2 mg/ml collagen with HUVEC under static conditions. This shows that shear flow helps to prevent cancer cell proliferation in stiff collagen-only gels.
For generations 3 and higher [Fig. 7(d)], there were no statistically significant results for all shear or all static results according to a one-way ANOVA (p > 0.05). An unpaired student's t-test was also performed on conditions that shared identical ECM and HUVEC conditions but differed by static or shear. BCC embedded in 2.2 mg/ml collagen-only gels with HUVEC and exposed to shear had significantly less G3+ cells when compared to BCC cultured in 2.2 mg/ml collagen-only gels with HUVEC under static conditions [77.41 ± 3.00% and 90.37 ± 1.46%, respectively, shown in Fig. 7(d)]. BCC cultured in collagen + 20 mg/ml CS gels exposed to static conditions also showed a low expression of G3 cells (80.65 ± 2.34%). In the stiff (2.2 mg/ml) collagen + HUVEC, static condition it was shown in Figs. 1–3 that HUVEC underwent EndMT and expressed activated fibroblast-like markers such as upregulation of COL1A2 gene expression, and these conditions significantly affected BCC proliferation. To confirm these results, we used immunofluorescence to stain the BCC for Ki-67, a cell marker for proliferation. It was observed that BCC in collagen + 20 mg/ml CS gels exposed to static conditions showed elongated morphologies compared to the other proliferated samples, shown in Fig. 7(e). The results suggest that BCC proliferation is arbitrated by mesenchymally transformed EC. This aggressive population of cancer cells was quantified in conditions with the highest mesenchymally transformed EC indicated by an upregulation of activated fibroblast-like marker (COL1A2–fibrosis marker). For BCC proliferation, it was observed that BCC embedded in 2.2 mg/ml collagen-only stiffness control gels under static conditions resulted in the lowest number of cells in generations 1 and 2; however, in generations 3 and higher, the BCC cultured in those activated fibroblasts conditions have proliferated the most among all the samples. These data suggest that activated fibroblasts generated in stiff ECM (2.2 mg/ml collagen-only gels) under static conditions facilitated BCC proliferation and may play a role in cancer metastasis. This is important for understanding the complex role of tissue properties associated with poor prognosis of the disease, which can be difficult to isolate in vivo.
IV. CONCLUSIONS
In this study, an in vitro 3D culture microfluidic device was utilized to recreate a tumor vasculature environment for investigating the role of solid tumor, endothelial cells, and surrounding tissue interactions. It was found that the interplay of low fluid-induced shear stress and altered ECM composition (changes in collagen and GAG concentrations, and stiffness) on EndMT resulted in increased cell invasion and protein and gene expression of mesenchymal-like (α-SMA, Snail, and COL1A2) markers. Additionally, gene expression of HUVEC surface receptors for pro-inflammatory and GAG binding and GAG productions markers were upregulated in shear and altered ECM conditions. Cancer cell spheroids were embedded under the experimental conditions that were found to produce the most EndMT, which results in increased interaction of cancer cells and mesenchymally transformed endothelial cells. Furthermore, these pathological-like conditions lead to increased cancer cell migration rate and distance and cancer cell proliferation. The results show that the tumor stroma environment generates activated fibroblasts through EndMT and that these cells, in turn, can promote cancer cell metastasis. This novel model represents the first critical step towards recapitulating well known cancer-endothelial cell interactions in a stiff tumor stroma microenvironment. Additionally, fluidic conditions help elucidate the capabilities of both physical and biological parameters involved in altering cellular behavior and functionality, offering an important tool to advance the knowledge about tumor progression and the development of more effective treatment methodologies.
V. SUPPLEMENTARY MATERIAL
See supplementary material for additional experimental data.
ACKNOWLEDGMENTS
This research was supported by the National Science Foundation Grant No. CMMI 1436173 and the Clifford D. Clark Graduate Fellowship (SGM).
References
- 1. Weir H. K., Thompson T. D., Soman A., Moller B., and Leadbetter S., Cancer 121, 1827 (2015). 10.1002/cncr.29258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray F. and Møller B., Nat. Rev. Cancer 6, 63 (2006). 10.1038/nrc1781 [DOI] [PubMed] [Google Scholar]
- 3. Mariotto A. B., Yabroff K. R., Shao Y., Feuer E. J., and Brown M. L., J. Natl. Cancer Inst. 103, 117 (2011). 10.1093/jnci/djq495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schroer A. K. and Merryman W. D., J. Cell Sci. 128, 1865 (2015). 10.1242/jcs.162891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frangogiannis N. G., Nat. Rev. Cardiol. 11, 255 (2014). 10.1038/nrcardio.2014.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xing F., Saidou J., and Watabe K., Front. Biosci. 15, 166 (2010). 10.2741/3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zeisberg E. M., Potenta S., Xie L., Zeisberg M., and Kalluri R., Cancer Res. 67, 10123 (2007). 10.1158/0008-5472.CAN-07-3127 [DOI] [PubMed] [Google Scholar]
- 8. Wong A. D. and Searson P. C., Cancer Res. 74, 4937 (2014). 10.1158/0008-5472.CAN-14-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zervantonakis I. K., Hughes-Alford S. K., Charest J. L., Condeelis J. S., Gertler F. B., and Kamm R. D., Proc. Natl. Acad. Sci. U.S.A. 109, 13515 (2012). 10.1073/pnas.1210182109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cox T. R. and Erler J. T., Dis. Models & Mech. 4, 165 (2011). 10.1242/dmm.004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moonen J. R. A. J., Lee E. S., Schmidt M., Maleszewska M., Koerts J. A., Brouwer L. A., Van Kooten T. G., Van Luyn M. J. A., Zeebregts C. J., Krenning G., and Harmsen M. C., Cardiovasc. Res. 108, 377 (2015). 10.1093/cvr/cvv175 [DOI] [PubMed] [Google Scholar]
- 12. Whatcott C. J., Han H., Posner R. G., Hostetter G., and Von Hoff D. D., Cancer Discovery 1, 291 (2011). 10.1158/2159-8290.CD-11-0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prinz R. D., Willis C. M., Viloria-Petit A., and Klüppel M., Genet. Mol. Res. 10, 3901 (2011). 10.4238/2011.December.8.9 [DOI] [PubMed] [Google Scholar]
- 14. Zeisberg E. M., Tarnavski O., Zeisberg M., Dorfman A. L., McMullen J. R., Gustafsson E., Chandraker A., Yuan X., Pu W. T., Roberts A. B., Neilson E. G., Sayegh M. H., Izumo S., and Kalluri R., Nat. Med. 13, 952 (2007). 10.1038/nm1613 [DOI] [PubMed] [Google Scholar]
- 15. Chang S.-F., Chang C. A., Lee D.-Y., Lee P.-L., Yeh Y.-M., Yeh C.-R., Cheng C.-K., Chien S., and Chiu J.-J., Proc. Natl. Acad. Sci. U.S.A. 105, 3927 (2008). 10.1073/pnas.0712353105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rizvia I., Gurkanb U. A., Tasoglub S., Alagica N., Cellia J. P., Mensaha L. B., Maia Z., Demircib U., and Hasana T., Proc. Natl. Acad. Sci. U.S.A. 110, E1974 (2013). 10.1073/pnas.1216989110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Polacheck W. J., Charest J. L., and Kamm R. D., Proc. Natl. Acad. Sci. U.S.A. 108, 11115 (2011). 10.1073/pnas.1103581108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shields J. D., Fleury M. E., Yong C., Tomei A. A., Randolph G. J., and Swartz M. A., Cancer Cell 11, 526 (2007). 10.1016/j.ccr.2007.04.020 [DOI] [PubMed] [Google Scholar]
- 19. Mina S. G., Wang W., Cao Q., Huang P., Murray B. T., and Mahler G. J., RSC Adv. 6, 85457 (2016). 10.1039/C6RA16607E [DOI] [Google Scholar]
- 20. Schneider C. A., Rasband W. S., and Eliceiri K. W., Nat. Methods 9, 671 (2012). 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dahal S., Huang P., Murray B. T., and Mahler G. J., “Endothelial to mesenchymal transformation is induced by altered extracellular matrix in aortic valve endothelial cells,” J. Biomed. Mater. Res., Part A (published online). 10.1002/jbm.a.36133 [DOI] [PubMed]
- 22. Engler A. J., Sen S., Sweeney H. L., and Discher D. E., Cell 126, 677 (2006). 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- 23. Cross V. L., Zheng Y., Choi N. W., Verbridge S. S., Bryan A., Bonassar L. J., Fischbach C., and Stroock A. D., Biomaterials 31, 8596 (2011). 10.1016/j.biomaterials.2010.07.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sieminski A., Hebbel R., and Gooch K., Exp. Cell Res. 297, 574 (2004). 10.1016/j.yexcr.2004.03.035 [DOI] [PubMed] [Google Scholar]
- 25. Levental I., Georges P. C., and Janmey P. A., Soft Matter 3, 299 (2007). 10.1039/B610522J [DOI] [PubMed] [Google Scholar]
- 26. Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D., Hammer D. A., and Weaver V. M., Cancer Cell 8, 241 (2005). 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- 27. Denis M., Gregory A., Bayat M., Fazzio R. T., Whaley D. H., Ghosh K., Shah S., Fatemi M., and Alizad A., PLoS One 11, e0165003 (2016). 10.1371/journal.pone.0165003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amaya R., Pierides A., and Tarbell J. M., PLoS One 10, e0129952 (2015). 10.1371/journal.pone.0129952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalluri R. and Zeisberg M., Nat. Rev. Cancer 6, 392 (2006). 10.1038/nrc1877 [DOI] [PubMed] [Google Scholar]
- 30. Small E. M., Thatcher J. E., Sutherland L. B., Kinoshita H., Gerard R. D., Richardson J. A., Dimaio J. M., Sadek H., Kuwahara K., and Olson E. N., Circ. Res. 107, 294 (2010). 10.1161/CIRCRESAHA.110.223172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smeriglio P., Lai J. H., Yang F., and Bhutani N., J. Visualized Exp. 104, e53085 (2015). 10.3791/53085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hordijk P. L., Anthony E., Mul F. P. J., Rientsma R., Oomen L. C. J. M., and Roos D., J. Cell Sci. 112, 1915 (1999). [DOI] [PubMed] [Google Scholar]
- 33. Gehler S., Baldassarre M., Lad Y., Leight J. L., Wozniak M. A., Riching K. M., Eliceiri K. W., Weaver V. M., Calderwood D. A., and Keely P. J., Mol. Biol. Cell 20, 3224 (2009). 10.1091/mbc.E08-12-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopez J., Mouw J., and Weaver V., Oncogene 27, 6981 (2008). 10.1038/onc.2008.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DuFort C. C., Paszek M. J., and Weaver V. M., Nat. Rev. Mol. Cell Biol. 12, 308 (2011). 10.1038/nrm3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fernandez-Gonzalez R., de Matos Simoes S., Röper J. C., Eaton S., and Zallen J. A., Dev. Cell 17, 736 (2009). 10.1016/j.devcel.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Montell D. J., Science 322, 1502 (2008). 10.1126/science.1164073 [DOI] [PubMed] [Google Scholar]
- 38. Hoon J. L., Tan M. H., and Koh C.-G., Cells 5, 17 (2016). 10.3390/cells5020017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sit S.-T. and Manser E., J. Cell Sci. 124, 679 (2011). 10.1242/jcs.064964 [DOI] [PubMed] [Google Scholar]
- 40. Ridley A. J., J. Cell Sci. 114, 2713 (2001). [DOI] [PubMed] [Google Scholar]
- 41. Provenzano P. P., Inman D. R., Eliceiri K. W., Knittel J. G., Yan L., Rueden C. T., White J. G., and Keely P. J., BMC Med. 6, 11 (2008). 10.1186/1741-7015-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Y. L., Motte S., and Kaufman L. J., Biomaterials 31, 5678 (2010). 10.1016/j.biomaterials.2010.03.039 [DOI] [PubMed] [Google Scholar]
- 43. Yang Y., Sun C., Wilhelm M. E., Fox L. J., Zhu J., and Kaufman L. J., Biomaterials 32, 7932 (2011). 10.1016/j.biomaterials.2011.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Van De Water L., Varney S., and Tomasek J. J., Adv. Wound Care 2, 122 (2013). 10.1089/wound.2012.0393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ghatak S., Maytin E. V., MacK J. A., Hascall V. C., Atanelishvili I., Moreno Rodriguez R., Markwald R. R., and Misra S., Int. J. Cell Biol. 2015, 834893. 10.1155/2015/834893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu P., Takai K., Weaver V. M., and Werb Z., Cold Spring Harbor Perspect. Biol. 3, a005058 (2011). 10.1101/cshperspect.a005058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sherman L., Wainwright D., Ponta H., and Herrlich P., Genes Dev. 12, 1058 (1998). 10.1101/gad.12.7.1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davies J. Q., Lin H. H., Stacey M., Yona S., Chang G. W., Gordon S., Hamann J., Campo L., Han C., Chan P., and Fox S. B., Oncol. Rep. 25, 619 (2011). 10.3892/or.2010.1117 [DOI] [PubMed] [Google Scholar]
- 49. Langenhan T., Aust G., and Hamann J., Sci. Signaling 6, re3 (2013). 10.1126/scisignal.2003825 [DOI] [PubMed] [Google Scholar]
- 50. Lai E., Singh R., Teng B., Zhao Y., Sharratt E., Howell G., Rajput A., and Dunn K. B., Dis. Colon Rectum 53, 475 (2010). 10.1007/DCR.0b013e3181c87084 [DOI] [PubMed] [Google Scholar]
- 51. Young T. W., Mei F. C., Yang G., Thompson-lanza J. A., Liu J., and Cheng X., Cancer Res. 64, 4577 (2004). 10.1158/0008-5472.CAN-04-0222 [DOI] [PubMed] [Google Scholar]
- 52. Bullard K. M., Kim H. R., Wheeler M. A., Wilson C. M., Neudauer C. L., Simpson M. A., and McCarthy J. B., Int. J. Cancer 107, 739 (2003). 10.1002/ijc.11475 [DOI] [PubMed] [Google Scholar]
- 53. Itano N., Sawai T., Miyaishi O., and Kimata K., Cancer Res. 59, 2499 (1999). [PubMed] [Google Scholar]
- 54. Van Sinderen M., Cuman C., Winship A., Menkhorst E., and Dimitriadis E., Placenta 34, 907 (2013). 10.1016/j.placenta.2013.07.065 [DOI] [PubMed] [Google Scholar]
- 55. Cooney C. A., Jousheghany F., Yao-Borengasser A., Phanavanh B., Gomes T., Kieber-Emmons A. M., Siegel E. R., Suva L. J., Ferrone S., Kieber-Emmons T., and Monzavi-Karbassi B., Breast Cancer Res. 13, R58 (2011). 10.1186/bcr2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kodama J., Hasengaowa, Kusumoto T., Seki N., Matsuo T., Nakamura K., Hongo A., and Hiramatsu Y., Eur. J. Cancer 43, 1460 (2007). 10.1016/j.ejca.2007.02.007 [DOI] [PubMed] [Google Scholar]
- 57. Sakko A. J., Ricciardelli C., Mayne K., Tilley W. D., LeBaron R. G., and Horsfall D. J., Cancer Res. 61, 926 (2001). [PubMed] [Google Scholar]
- 58. Kultti A., Li X., Jiang P., Thompson C. B., Frost G. I., and Shepard H. M., Cancers 4, 873 (2012). 10.3390/cancers4030873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Slevin M., Krupinski J., Gaffney J., Matou S., West D., Delisser H., Savani R. C., and Kumar S., Matrix Biol. 26, 58 (2007). 10.1016/j.matbio.2006.08.261 [DOI] [PubMed] [Google Scholar]
- 60. Leitinger B. and Hohenester E., Matrix Biol. 26, 146 (2007). 10.1016/j.matbio.2006.10.007 [DOI] [PubMed] [Google Scholar]
- 61. Zhu G. G., Risteli L., Mäkinen M., Risteli J., Kauppila A., and Stenbäck F., Cancer 75, 1010 (1995). [DOI] [PubMed] [Google Scholar]
- 62. Eikenes L., Tufto I., Schnell E. A., Bjørkøy A., and Davies C. D. L., Anticancer Res. 30, 359 (2010). [PubMed] [Google Scholar]
- 63. Mitchell M. J. and King M. R., Am. J. Physiol. Cell Physiol. 306, C89 (2014). 10.1152/ajpcell.00285.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chlen S., Li S., and Shyy J. Y.-J., Hypertension 31(2), 162 (1998). 10.1161/01.HYP.31.1.162 [DOI] [PubMed] [Google Scholar]
- 65. Gimbrone M. A., Nagel T., and Topper J. N., J. Clin. Invest. 99, 1809 (1997). 10.1172/JCI119346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lu P., Weaver V. M., and Werb Z., J. Cell Biol. 196, 395 (2012). 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for additional experimental data.