Abstract
Two loci encoding subunit III of the cytochrome oxidase (COX) in Oenothera mitochondria have been identified from a cDNA library of mitochondrial transcripts. A 657-bp sequence block upstream from the open reading frame is also present in the two copies of the COX subunit I gene and is presumably involved in homologous sequence rearrangement. The proximal points of sequence rearrangements are located 3 bp upstream from the COX I and 1139 bp upstream from the COX III initiation codons. The 5'-termini of both COX I and COX III mRNAs have been mapped in this common sequence confining the promoter region for the Oenothera mitochondrial COX I and COX III genes to the homologous sequence block.
Keywords: Oenothera mitochondria, mtcDNA, cytochrome oxidase subunits I and III, promoter region
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Birchmeier W., Kohler C. E., Schatz G. Interaction of integral and peripheral membrane proteins: affinity labeling of yeast cytochrome oxidase by modified yeast cytochrome c. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4334–4338. doi: 10.1073/pnas.73.12.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun C. J., Levings C. S. Nucleotide Sequence of the F(1)-ATPase alpha Subunit Gene from Maize Mitochondria. Plant Physiol. 1985 Oct;79(2):571–577. doi: 10.1104/pp.79.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennicke A. Mitochondrial DNA from Oenothera berteriana: PURIFICATION AND PROPERTIES. Plant Physiol. 1980 Jun;65(6):1207–1210. doi: 10.1104/pp.65.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning K. S., RajBhandary U. L. Cytochrome oxidase subunit III gene in Neurospora crassa mitochondria. Location and sequence. J Biol Chem. 1982 May 10;257(9):5253–5256. [PubMed] [Google Scholar]
- Chomyn A., Mariottini P., Cleeter M. W., Ragan C. I., Matsuno-Yagi A., Hatefi Y., Doolittle R. F., Attardi G. Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature. 1985 Apr 18;314(6012):592–597. doi: 10.1038/314592a0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Deng G., Wu R. An improved procedure for utilizing terminal transferase to add homopolymers to the 3' termini of DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4173–4188. doi: 10.1093/nar/9.16.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey R. E., Levings C. S., 3rd, Timothy D. H. Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell. 1986 Feb 14;44(3):439–449. doi: 10.1016/0092-8674(86)90465-4. [DOI] [PubMed] [Google Scholar]
- Dewey R. E., Levings C. S., Timothy D. H. Nucleotide sequence of ATPase subunit 6 gene of maize mitochondria. Plant Physiol. 1985 Nov;79(3):914–919. doi: 10.1104/pp.79.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey R. E., Schuster A. M., Levings C. S., Timothy D. H. Nucleotide sequence of F(0)-ATPase proteolipid (subunit 9) gene of maize mitochondria. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1015–1019. doi: 10.1073/pnas.82.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconet D., Lejeune B., Quetier F., Gray M. W. Evidence for homologous recombination between repeated sequences containing 18S and 5S ribosomal RNA genes in wheat mitochondrial DNA. EMBO J. 1984 Feb;3(2):297–302. doi: 10.1002/j.1460-2075.1984.tb01799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Leaver C. J. The Zea mays mitochondrial gene coding cytochrome oxidase subunit II has an intervening sequence and does not contain TGA codons. Cell. 1981 Nov;26(3 Pt 1):315–323. doi: 10.1016/0092-8674(81)90200-2. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Brennicke A. Cytochrome oxidase subunit II gene in mitochondria of Oenothera has no intron. EMBO J. 1983;2(12):2173–2178. doi: 10.1002/j.1460-2075.1983.tb01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac P. G., Brennicke A., Dunbar S. M., Leaver C. J. The mitochondrial genome of fertile maize (Zea mays L.) contains two copies of the gene encoding the alpha-subunit of the F1-ATPase. Curr Genet. 1985;10(4):321–328. doi: 10.1007/BF00365628. [DOI] [PubMed] [Google Scholar]
- Isaac P. G., Jones V. P., Leaver C. J. The maize cytochrome c oxidase subunit I gene: sequence, expression and rearrangement in cytoplasmic male sterile plants. EMBO J. 1985 Jul;4(7):1617–1623. doi: 10.1002/j.1460-2075.1985.tb03828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B. F., Burger G. A collection of programs for nucleic acid and protein analysis, written in FORTRAN 77 for IBM-PC compatible microcomputers. Nucleic Acids Res. 1986 Jan 10;14(1):455–465. doi: 10.1093/nar/14.1.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver C. J., Harmey M. A. Higher-plant mitochondrial ribosomes contain a 5S ribosomal ribonucleic acid component. Biochem J. 1976 Jul 1;157(1):275–277. doi: 10.1042/bj1570275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale D. M., Hodge T. P., Fauron C. M. The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 1984 Dec 21;12(24):9249–9261. doi: 10.1093/nar/12.24.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna E., Brennicke A. Primary and secondary structure of 26S ribosomal RNA of Oenothera mitochondria. Curr Genet. 1985;9(6):505–515. doi: 10.1007/BF00434055. [DOI] [PubMed] [Google Scholar]
- Mariottini P., Chomyn A., Riley M., Cottrell B., Doolittle R. F., Attardi G. Identification of the polypeptides encoded in the unassigned reading frames 2, 4, 4L, and 5 of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1563–1567. doi: 10.1073/pnas.83.6.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal L., Guillemaut P., Grienenberger J. M., Jeannin G., Weil J. H. Sequence and codon recognition of bean mitochondria and chloroplast tRNAsTrp: evidence for a high degree of homology. Nucleic Acids Res. 1985 Jun 25;13(12):4411–4416. doi: 10.1093/nar/13.12.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Peacock S. L., McIver C. M., Monahan J. J. Transformation of E. coli using homopolymer-linked plasmid chimeras. Biochim Biophys Acta. 1981 Sep 28;655(2):243–250. doi: 10.1016/0005-2787(81)90014-9. [DOI] [PubMed] [Google Scholar]
- Prochaska L. J., Bisson R., Capaldi R. A., Steffens G. C., Buse G. Inhibition of cytochrome c oxidase function by dicyclohexylcarbodiimide. Biochim Biophys Acta. 1981 Sep 14;637(2):360–373. doi: 10.1016/0005-2728(81)90175-4. [DOI] [PubMed] [Google Scholar]
- Roe B. A., Ma D. P., Wilson R. K., Wong J. F. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985 Aug 15;260(17):9759–9774. [PubMed] [Google Scholar]
- Schnare M. N., Gray M. W. 3'-Terminal sequence of wheat mitochondrial 18S ribosomal RNA: further evidence of a eubacterial evolutionary origin. Nucleic Acids Res. 1982 Jul 10;10(13):3921–3932. doi: 10.1093/nar/10.13.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
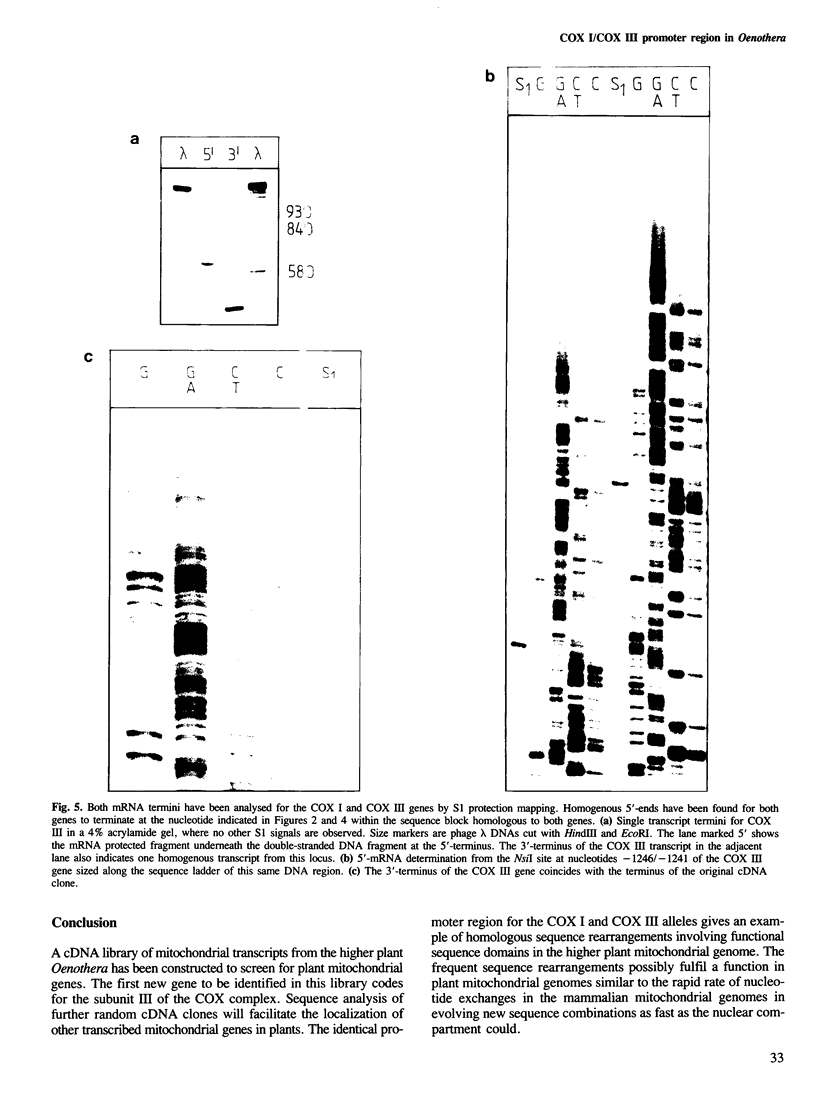
- Schuster W., Hiesel R., Isaac P. G., Leaver C. J., Brennicke A. Transcript termini of messenger RNAs in higher plant mitochondria. Nucleic Acids Res. 1986 Aug 11;14(15):5943–5954. doi: 10.1093/nar/14.15.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippel A. E. Purification and characterization of adenosine triphosphate: ribonucleic acid adenyltransferase from Escherichia coli. Eur J Biochem. 1973 Aug 1;37(1):31–40. doi: 10.1111/j.1432-1033.1973.tb02953.x. [DOI] [PubMed] [Google Scholar]
- Sone N., Kosako T. Evidence for dimer structure of proton-pumping cytochrome c oxidase, an analysis by radiation inactivation. EMBO J. 1986 Jul;5(7):1515–1519. doi: 10.1002/j.1460-2075.1986.tb04391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalenfeld B. E., Tzagoloff A. Assembly of the mitochondrial membrane system. Sequence of the oxi 2 gene of yeast mitochondrial DNA. J Biol Chem. 1980 Jul 10;255(13):6173–6180. [PubMed] [Google Scholar]
- Tzagoloff A., Macino G., Sebald W. Mitochondrial genes and translation products. Annu Rev Biochem. 1979;48:419–441. doi: 10.1146/annurev.bi.48.070179.002223. [DOI] [PubMed] [Google Scholar]
- Van Etten R. A., Walberg M. W., Clayton D. A. Precise localization and nucleotide sequence of the two mouse mitochondrial rRNA genes and three immediately adjacent novel tRNA genes. Cell. 1980 Nov;22(1 Pt 1):157–170. doi: 10.1016/0092-8674(80)90164-6. [DOI] [PubMed] [Google Scholar]
- Ward B. L., Anderson R. S., Bendich A. J. The mitochondrial genome is large and variable in a family of plants (cucurbitaceae). Cell. 1981 Sep;25(3):793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]