Abstract
A microfluidic device composed of variable numbers of multiconstriction channels is reported in this paper to differentiate a human breast cancer cell line, MDA-MB-231, and a nontumorigenic human breast cell line, MCF-10A. Differences between their mechanical properties were assessed by comparing the effect of single or multiple relaxations on their velocity profiles which is a novel measure of their deformation ability. Videos of the cells were recorded via a microscope using a smartphone, and imported to a tracking software to gain the position information on the cells. Our results indicated that a multiconstriction channel design with five deformation (50 μm in length, 10 μm in width, and 8 μm in height) separated by four relaxation (50 μm in length, 40 μm in width, and 30 μm in height) regions was superior to a single deformation design in differentiating MDA-MB-231 and MCF-10A cells. Velocity profile criteria can achieve a differentiation accuracy around 95% for both MDA-MB-231 and MCF-10A cells.
Keywords: multiconstriction channel, microfluidic cell separation, breast cancer cells, smartphone imaging, velocity profiles, video/image processing, particle tracking
Graphical abstract
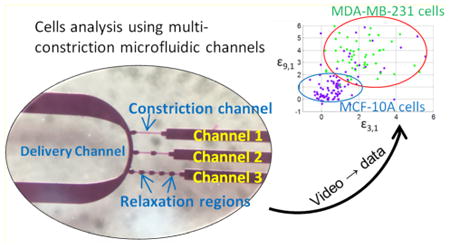
Studies of the mechanical characteristics of single cells have been previously used in cancer research.1,2 Different stages of cancer cells have significant mechanical differences due to the stiffness of their cell membrane.3 Other well-developed methods and techniques based on cell membrane properties, the epithelial cell adhesion molecule (EpCAM) and fluorescence detection for the sorting of cancer cells require specific equipment, such as atomic force microscopy,3,4 costly antibody reagents,5,6 and flow cytometry setups.7–10 Microfluidic chips bring new technologies and methods to both cancer cell analysis and circulating tumor cell (CTC) detection and sorting.11 Size-based microfluidic channels for CTC cancer cell separation are broadly used in CTC studies which sense the different diameters of cancer cells, white blood cells, and red blood cells.12–17 In order to analyze and sort different cancer cells with similar sizes, investigators have chosen to use constriction channels and study the dynamics of cells, usually accompanied by other methods sensitive to both biomechanical and bioelectrical properties to enable identification.18–22
Characterization of the mechanical properties of cancer cells using a microfluidic device with constriction channels has been proven to be an accurate method to differentiate different cell lines.23–27 Metastatic cancer cells have softer cell membranes, as well as deformation ability in microfluidic constriction channels. On the other hand, normal cells have a cytoskeleton with higher mechanical strength, including but not limited to actin filaments, intermediate filaments, and microtubules.23,28,29 Therefore, the deformation time of cancer cells and normal cells has been shown to differ due to their mechanical properties. Many studies have focused on the entry time and transit time of cells in a constriction channel, and analysis has defined criteria typifying specific cell lines.30 By plotting the dynamic parameters of the single cells in constriction channel, different cell lines, including breast cancer cells,3,18,24 lung cancer cells,19,31,32 kidney tumor cells,33 and cervical cancer cell lines,34,35 can be separated into groups in order to achieve cell characterization.36 However, using a single constriction channel or adjustable constriction channels has limited capabilities to distinguish between cancer cells and normal cells. When differentiating various cell lines, scientists studying characterization of cancer cells have added an additional bioelectrical measurement along with the single constriction channel.2,20 At the same time, improvements in the methods used to induce cells' deformation using not only constriction channels, but also lateral displacement array or pillar array17,37 in a microfluidic chamber13,14 have enhanced the resolution of characterizing cells.
This Article reports a multiconstriction multichannel microfluidic device for analyzing velocity profiles of single-cells which provides greater single-cell resolution of normal and cancer cells than that afforded by a single constriction channel studies. Smartphone video-microscopy was used to capture the dynamic motions of cells as each passed through the series of constriction channels. The result was a distinct velocity profile at each stage of deformation in the multiple constriction channels. Compared to single-constriction channels, multiconstriction channels divide the cell deformation into multiple stages, which effectively reveals the deformation process of each cell with higher resolution, and hence a new and simple way to observe the variations in individual cells' velocity profiles. The series of relaxation regions separating the constriction channels causes cells to experience multiple relaxations where cell membrane/shape recovery takes place. The length of relaxation regions are kept as 50 μm constantly, so that the results can focus on the effects of the constriction, the relaxation count and constriction length. The microfluidic channels device was designed with three different configurations to compare the ability to differentiate two different cell lines in each channel. We also compared the effect of adding one or multiple relaxation regions in the constriction channel. The velocity profiles analyses were focused on comparing the initial velocity change due to the deformation at the entrance of each channel and the exiting velocity at the last segment of the channel, where the cells have experienced sufficient deformations. The multiple deformation procedure increases the ability of deformation and recovery of the cells. Based on our experimental results, the ability to differentiate between cancer cells and normal cells in a multiconstriction channel is higher than that achieved with either a single constriction channel or two constriction channels with only one relaxation region. After establishing criteria to examine the difference between initial velocities and final velocities, we successfully differentiated ∼94.3% of the cancer cells from normal cells. Then, four testing samples were used to verify our criteria. The results indicated that the selected criteria for the multiconstriction channels have an accuracy around 95% to tell the difference between cancer cells and normal cells.
Experimental Section
Materials and Methods
Cell Culture and Sample Preparation
We prepared the highly metastatic breast cancer cells MDA-MB-231 and benign breast epithelial cells MCF-10A to represent normal cells with the same concentration of ∼50 000 cells/mL. Both MDA-MB-231 and MCF-10A cells were provided by the Tissue Culture Shared Resources of the Lombardi Comprehensive Cancer Center, Georgetown University in Washington, DC. MDA-MB-231 cells were grown in F12:DMEM (Lonza, Basel, Switzerland) with 10% fetal bovine serum (FBS), 4 mM glutamine and penicillin-streptomycin (100 units per mL). MCF-10A cells were grown in F12:DMEM with penicillin-streptomycin (100 units per mL), 2.5 mM l-glutamine, 20 ng/mL epidermal growth factor (EGF), 0.1 μg/mL cholera toxin, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, and 5% horse serum. The cells were grown in T-25 cm2 culture flasks at 37 °C in a 5% CO2 in air atmosphere until cells were ready for subculture.
The morphology of the cells was observed before trypsinization (Figure 1). The cells were then detached from the flask with trypsin-EDTA solution (trypsin-EDTA solution 10×, Sigma). The MDA-MB-231 cells were trypsinized at 37 °C for 2 min, and MCF-10A cells were trypsinized at 37 °C for 15 min. Before every experiment, the viability of the cells was observed under a microscope using trypan blue (trypan blue solution (0.4%), lot 42K2360, Sigma).38 Both trypan blue and culture medium were warmed to 37 °C in a water bath before usage. The viability of the cells was 100%, as determined by the count using a hemocytometer.
Figure 1.
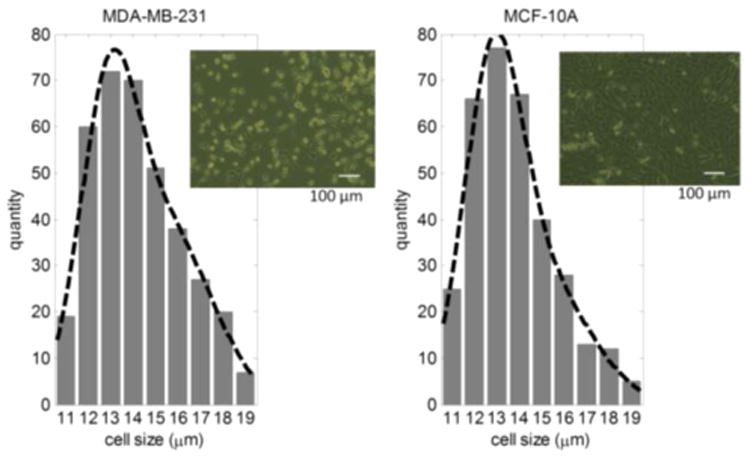
MDA-MB-231 and MCF-10A cell size distribution; cell image before trypsinization.
Microfluidic Channel Fabrication
The multiconstriction channels devices were fabricated on a silicon wafer with two layers of SU-8 (SU-8 3005 and SU-8 3025, MicroChem, Newton, MA) photolithography and polydimethylsiloxane (PDMS) soft-lithography, followed by PDMS and glass bonding after plasma treatment. Three different configurations of channels were connected to a main delivery channel, as shown in the channel configurations in Figure 2. All the constriction channels had a cross section of 8 μm by 8 μm. The first channel contains only one constriction channel with a length of 250 μm; the second channel contains two constriction channels with lengths of 125 μm each, and a relaxation section with a length of 50 μm; the third channel contains five constriction channels with a length of 50 μm each, and relaxation sections of 50 μm between every constriction channel. The detailed fabrication procedures are presented in section 1 of the Supporting Information.
Figure 2.
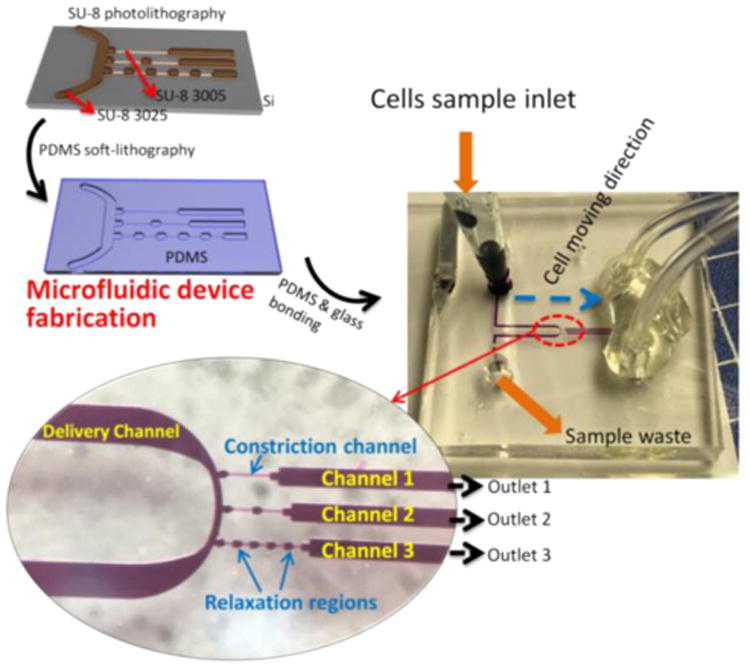
Microfluidic device fabrication processes and experimental setup.
The SU-8 mold consists of two layers with 8 μm of SU-8 3005 and ∼25 μm of SU-8 3025, as illustrated in Figure 2. Tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (TFOCS, Fisher Scientific) was coated on the surface of the molds for the easy release of PDMS. After the mold was prepared, standard PDMS (SYLGARD 184, Dow Corning, Midland, MI) replica molding was conducted to fabricate the microchannels. The PDMS channels were then bonded to a glass slide after air plasma treatment using plasma cleaner (Harrick Plasma, model PDC-001, Ithaca, NY).
Experimental Setup
The microfluidic device was mounted on an inverted microscope (Zeiss Axio Observer, LSM-510, Thornwood, NY) with a lens magnification of 20×. One side of the delivery channel was connected to a reservoir with a cell sample. Three outlets were connected together to a syringe pump to create the same negative pressure. The cell delivery was initiated by applying a one-time negative pressure by another vacuum pump. The cell delivery was maintained by the pressure difference between the cell sample reservoir and the opening of the other side of the delivery channel. This channel dimension is good for cell sizes from 11 to 19 μm. Normal cells (MCF-10A) with 25 μm or larger will completely clog the microchannel.
Instead of a high speed camera, a smartphone's slow motion function was applied to record videos of cell movement with a frame rate of 240 frames per second. The video can be analyzed to extract information about the motion of each cell traveling through the constriction channels. As shown in Figure 2, the single constriction channel is referred to as channel 1; the middle channel with one relaxation at 125 μm is referred to as channel 2; and the five-constriction and four relaxation channel is referred to as channel 3.
Data Collection
The data of the movement of the cells in the three different channels were collected from the smartphone videos. For analysis, only the constriction channels were considered, where all three channels have the same total length of 250 μm in constrictions. The velocity distribution for every 25 μm was plotted in Matlab. As shown in Figure 3, the velocity at each segment was represented as (1)–(10). The velocities in segments (1)–(10) were recorded as V1, V2, V3, …, V10, respectively. Next, the velocity increment between every 25 μm was calculated. The velocity increments between two segments are defined as , where m = 1, 2, 3, …, 8, 9, 10, and n = 1, 2, 3, …, 8, 9, 10, representing 10 segments in the constriction channel.
Figure 3.
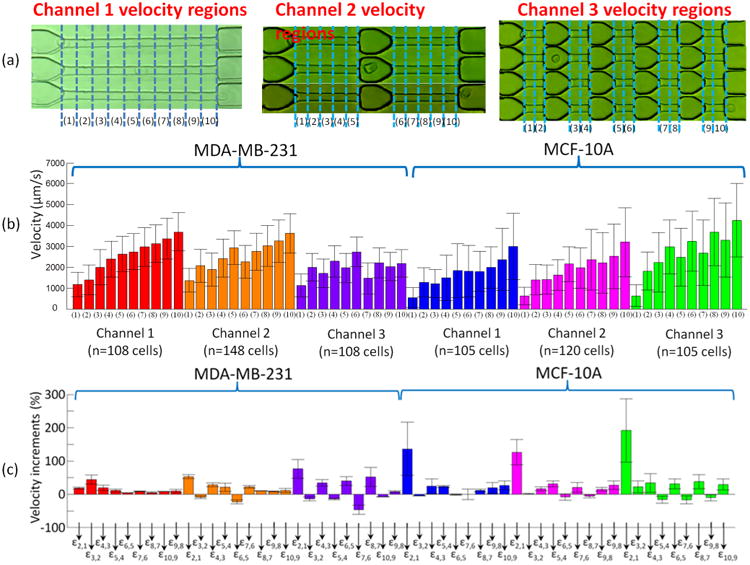
(a) Defining the velocity regions in three channel configurations using segments (1)–(10). (b) Velocity and (c) velocity increments of MDA-MB-231 cells and MCF-10A cells in three different microfluidic channels.
Results
Velocity of the Cells
The velocity and the increments of velocity between every 25 μm of both MDA-MB-231 and MCF-10A in three channels are demonstrated in Figure 3. Each cell line in every channel is represented by a color, and the same color will be used in the single cell analysis.
The initial entry velocity of MDA-MB-231 cells was similar regardless of the number of relaxation regions. This can be seen in Figure 3b by comparing the heights of the red bar (1), orange bar (1), and purple bar (1) into channel 1, channel 2, and channel 3 as shown in the channel configurations in Figure 3b, respectively. Similarly, the initial entry velocity of MCF-10A cells was independent of the number of relaxation regions as can be seen by comparing the heights of blue bar (1), pink bar (1) and green bar (1). These results serve to validate the measurement system because the aperture to each channel type (channel 1, channel 2, and channel 3) is identical. Due to the same dimension of the entrance of constriction channels, the same cell line had a similar entry velocity in channel 1–3.
As seen in Figure 3(b), the initial entry velocity measurement of MDA-MB-231 cells entered segment (1) of each channel twice as fast as MCF-10A cells. Also shown in Figure 3(b), from the initial entry velocity of the MDA-MB-231 cells in channel 1, 2, and 3 (red bar (1), orange bar (1), and purple bar (1), respectively), the average velocity of MDA-MB-231 cells was measured as ∼1.2 × 103 μm/s, with a standard deviation of ∼0.6 × 103 μm/s. Also from Figure 3b, from the initial entry velocity of the MCF-10A cells in channel 1, 2, and 3 (blue bar (1), pink bar (1), and green bar (1), respectively), the average velocity of MCF-10A cells was measured as 594.0 μm/s, with a standard deviation of 473.0 μm/s.
We assume the initial entry velocity
where k represents how many times the velocity of cancer cells is compared to normal cells. Using α = 0.05, the tk in t test is 1.647; therefore, k = 1.92. This means that the probability of the initial velocity of cancer cells is 1.92 times to normal cells is larger than 95%. If k = 2, which means , the possibility p = 79.87%. Similarly, if , the possibility p = 99%. The detailed statistic analysis is presented in section 3 of the Supporting Information.
It is well-recognized that metastatic cancer cells are softer than normal cells 3,26,27 and this has been correlated with faster deformation and transit times in constriction channels.3,22,32,39 Here we have improved the resolution of such studies by analyzing velocity profiles. The initial velocity incremental difference between MDA-MB-231 cells and MCF-10A cells was further analyzed as illustrated by Figure 3c. ε2,1 is defined by the equation: ε2,1 = (V2 − V1)/V1. The error bars in Figure 3c represent the standard deviations defined by the equation: is meaningful in studying large populations of the cells' behavior. As calculated velocity difference (Table 1) from Figure 3c, the average ε2,1 of the MCF-10A cells in channel 1 (blue), channel 2 (pink), and channel 3 (green) is 1.36, 1.26, and 1.91, respectively (Table 1). This shows that after 25 μm of deformation, MCF-10A cells reach a much higher V2 compared to the initial entry velocity V1. However, the ε2,1 of MDA-MB-231 cells was calculated to be 0.18, 0.53, and 0.76, respectively for channels 1, 2, and 3; these data indicated a slight decrease in the velocity of the MDA-MB-231 cells after their initial entry into the channel. This at first seems inconsistent with the “softer” biomechanical nature of the cancer cells, and we investigated this in greater depth.
Table 1. Velocity Difference Calculated by the Velocity Profiles in Figure 3.
| MDA-MB-231 | MCF-10A | |||||
|---|---|---|---|---|---|---|
| channel | 1 | 2 | 3 | 1 | 2 | 3 |
| ε2,1 | 1.36 | 1.26 | 1.91 | 0.18 | 0.53 | 0.76 |
| channel | 2 | 2 | ||||
| ε6,5 | −0.23 | −0.08 | ||||
The average velocity profiles can indicate the heterogeneity of the population of a cell-line. However, we need to analyze the detailed velocity differences in each segment to find the different mechanical characteristics. Channel 2 has one relaxation region between segments (5) and (6) (as illustrated in the “channel 2” in Figure 3c). The velocity has a drop for both MDA-MB-231 cells and MCF-10A cells at V6 compared to V5, as shown in segment (6) of the orange and pink in Figure 3(b). ε6,5 is defined by the equation: ε6,5 = (V6 − V5)/V5. As calculated velocity difference (Table 1) from Figure 3c, the average ε6,5 of the MDA-MB-231 cells and the MCF-10A cells in channel 2 (orange and pink), is −0.23 and −0.08, respectively. This indicated that the velocity of MDA-MB-231 cells had been reduced more than MCF-10A cells. In another word, MCF-10A cells inherited a higher more exiting velocity after the relaxation region between segments (5) and (6).
Channel 3 contains five constriction channels, each 50 μm in length, separated by relaxation sections of 50 μm between every constriction channel. Cells deformed at the entrance of the first constriction channel (defined as segment (1) in the “channel 3” in Figure 3b) and exited at the last segment of the final constriction channel (defined as segment (10) in the “channel 3” in Figure 3b), where the cells have experienced sequential deformations. Cancer cells recovered back to a round cell shape quicker than normal cells in each relaxation segment and we defined this as “deformation performance”. Therefore, the cancer cells showed deformation performance at the entrance to each constriction channel, which resulted in a decreasing velocity. As shown in Figure 3a, the initial velocities (segment (1), 0–25 μm traveling distance in constriction channel) of MDA-MB-231 cells were ∼1100 μm/s (purple bar (1)), and exiting velocities (segment (10), 215–250 μm traveling distance in constriction channel) were ∼2200 μm/s (purple bar (10)). In contrast, normal cells deformed slower initially, but reached a relatively higher velocity after two or three deformations. Normal cells stay in a rod-like shape without fully recovering back to an original spherical shape. The initial velocities of MCF-10A cells were ∼600 μm/s (green bar (1)), and exiting velocities were ∼4100 μm/s (green bar (10)). The geometry shapes of normal cells were almost fixed when the normal cells entered the fourth and fifth constriction channels. The entrance time of cancer cells into the fourth and fifth channel was longer than the normal cells.
Single cell Velocity Analysis
In order to illustrate how well MDA-MB-231 and MCF-10A cells can be differentiated at the single cell level, Figure 4 presents the scatter plots of single cell velocity data in each channel using the same color legend as in Figure 3. Each dot represents the data of one cell.
Figure 4.
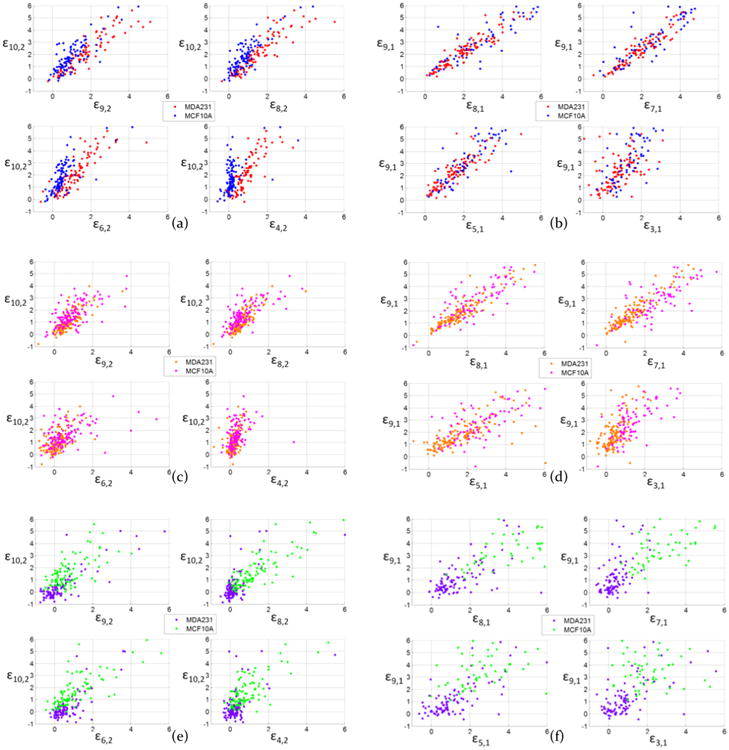
Scatter plot of MDA-MB-231 cells and MCF-10A cells velocity increments of comparing ε10,2 to ε9,2, ε8,2, ε6,2, and ε4,2 in channel 1 (a), channel 2 (c), and channel 3 (e); and comparing ε9,1 to ε8,1, ε7,1, ε5,1, and ε3,1 in channel 1 (b), channel 2 (d), and channel 3 (f).
The scatter plot of the MDA-MB-231 and MCF-10A cells'velocity differences in channel 1, 2, and 3 calculated at differentregions within each channel are shown in Figure 4. The datafrom Channel 1 is the focus in Figure 4a and b. Here it canbeen seen when comparing ε10,2 and ε9,1, (the ending velocity(V9 and V10), respectively) and the entry velocities (V1 and V2),the MDA-MB-231 (red dots) and MCF-10A cells (blue dots)had major overlaps in channel 1. The velocity profiling usingthe channel 1 configuration cannot distinguish between thecancer and normal cells at the single-cell level. About 95% ofthe cancer cells will be recognized as normal cells by thecriterion ε10,2 < 0. Almost no cells have ε10,2 > 0, which meansboth MDA-MB-231 cells and MCF-10A cells have completelydeformed and accelerated during passage through the single,250 μm constriction channel. Compared to ε8,2 and ε6,2 in Figure 4a, the criterion ε4,2 showed a better segregation of redand blue dots into two separate regions. V1 and V2 representthe beginning of the constriction channel, where cells begin todeform. MDA-MB-231 and MCF-10A cells showed a differentdeformation performance during the first 100 μm of channel 1.MDA-MB-231 cells took a shorter time to reach a highervelocity than MCF-10A cells at V4. Therefore, the ε4,2 of MDA-MB-231 cells had a higher value than MCF-10A cells in Figure4a. This phenomenon supports atomic force microscopy(AFM) data showing that MCF-10A cells were stiffer than MDA-MB-231 cells2,3,39, which might account for inability of MCF10A cells to reach higher velocities in a continuous constriction channel of 250 μm.
Channel 2: As shown in Figure 4c and d, the relaxation region between velocity segments (5) and (6) of the constriction channel 2 separated the channel into two 125 μm channels. The scatter plot of MDA-MB-231 cells (orange dots) and MCF-10A cells (pink dots) had major overlap at both in ε10,2 and ε9,1 regions, as well as in different velocity segments, such as ε8,2, ε7,1, ε5,1, and ε3,1. Because the complete deformation of MDA-MB-231 and MCF-10A cells was achieved by around the 100 μm mark within channel 2 (Supporting Information Video), the velocity differences were similar for both of these cell types within each velocity segment. The scatter plot in Figure 4c and d indicated that the channel 2 configuration was not useful in differentiating MDA-MB-231 and MCF-10A cells.
In channel 3, the short relaxation and constriction allowed the cells to continue experiencing a secondary deformation after the first relaxation. As shown in Figure 4e and f, all of the MCF-10A cells (purple dots) had ε10,2 > 0, and the majority of MDA-MB-231 cells (green dots) fell into ε10,2 < 0. By further consideration of ε7,1, more cancer cell dots fell into the region where ε7,1 < 2. V7 was the velocity in the fourth constriction region in channel 3. Therefore, based on the scatter plots of MDA-MB-231 and MCF-10A cells, it is possible using the channel 3 configuration to accurately identify individual single cell from either the MDA-MB-231 or MCF-10A cell line. In channel 1, 2 and 3, the scatter plots with ε2,1 are demonstrated in Supporting Information Figures S1 and S2. The ε4,2 in Figure 4a had a separation between MDA-MB-231 and MCF-10A cells. In channel 1, as plotted in Supporting Information Figure S3, using criterion of ε4,2 > 0.5*ε2,1, the false negative (cancer cells observed as normal cells) rate is 7.4%, and the false positive (normal cells observed as cancer cells) rate is 19%. Using criteria of ε4,2> 0.5*ε2,1 and ε4,2 > 0.3, the false negative rate is 7.4%, and the false positive rate is 19%, which means that the ε4,2 does not further improve the differentiation rate. If we include more criteria, such as criteria II and III, the false negative rate decreased to 5.6%; however, it increases the false positive rate to 21%, so the differentiation results in channel 1 does not have a sufficient balance between false negative and false positive rates.
We also labeled the dots with sequence number from 1–108 (for MDA-MB-231) and 1–105 (for MCF-10A) in Supporting Information Figure S4; using ε10,2, ε8,2, and ε6,2 and in Figure 4a had limitations in further improving the separation results. Some dots that cannot be separated by ε10,2 or ε8,2 but can be separated by ε9,1 and ε7,1. More mechanical alternation of the cells during constriction channels can bring variations in the cells' velocity profiles.
Differentiation Criteria
To better define how normal cells and cancer cells can be differentiated in channel 3, specific criteria are established for separating the different cells based on their velocity profiles and their cell shape changes. Video observations revealed that MDA-MB 231 cells deformed faster than MCF-10A cells. In channel 3, MDA-MB-231 cells recovered their normal spherical shape while in the first and second relaxation regions; in contrast, here, MCF-10A tended to maintain the flattened shape they had acquired while passing through the constriction channels. When MDA-MB-231 cells continued deforming in the third and fourth constriction regions, by virtue of maintaining their flattened shape, MCF-10A cells had a smoother entry and transit in the fourth and fifth constriction regions. We propose that these differences in cell shape recovery are reflective of differing mechanical properties and these contribute to the cell separation seen in the scatter plot of MDA-MB-231 and MCF-10A cells in channel 3, shown in Figure 4e. By using the velocity at the last constriction channel and the first constriction channel, we can differentiate the majority of the cancer cells from normal cells. Based on Figure 4f, further decreasing of the overlapping areas between the MDA-MB-231 and MCF-10A cells can be achieved by adding the additional criteria of ε7,1 < 2. In channel 3, the entrance time of cancer cells into the fourth and fifth channel will take longer than the normal cells. Therefore, the ε7,1 of normal cells will be larger than cancer cells. According to our experiments in channel 3, most MCF-10A cells have ε7,1 > 2, or V7 > 3V1. In summary, we can further distinguish the contrasting mechanical properties between MDA-MB-231 and MCF-10A using the following criteria:
(1) Criterion I:
(2) Criterion II:
(3) Criterion III:
In order to evaluate the sensitivity of the criteria we listed, the receiver operating characteristic (ROC) curve was plotted in Figure 5. The sensitivities of cancer cells in channel 1, 2, and 3 were 0.69, 0.80, and 0.90, respectively; and the sensitivities of normal cells in channel 1, 2, and 3 were 0.76, 0.86, and 0.96, respectively. For channel 3 (green and purple), using criteria II and III listed above had the best differentiation results between cancer cells and normal cells. The ROC curves and sensitivities for criterion I, criterion II, and criterion III are listed in section 6 of the Supporting Information (Figure S5 and Table S2).
Figure 5.
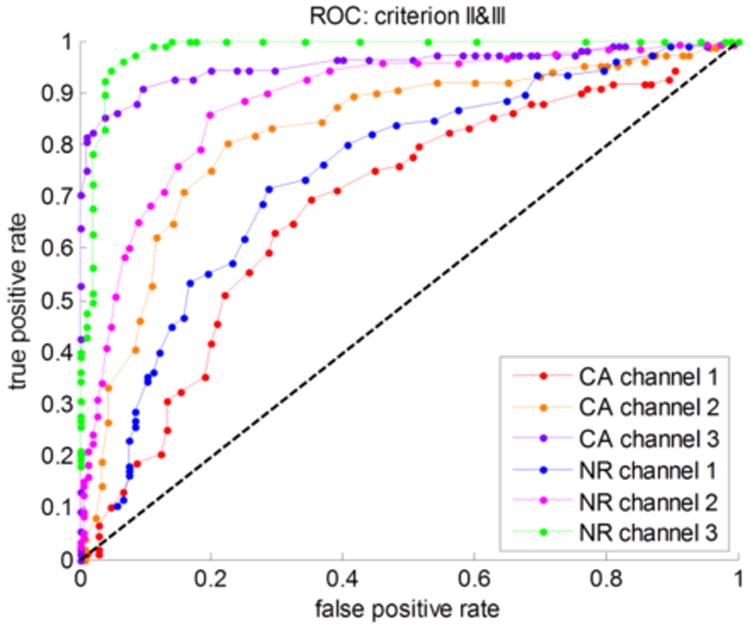
ROC curve of the cancer cells (CA) and normal cells (NR) in channel 1, 2, and 3, respectively.
Based on these differentiation criteria, scatter plots using criteria II, III, and the combination of both II and III was shown in Figure 6. Criterion I was able to differentiate all the normal cells (purple dots with ε10,2 < 0). After labeling the dot of each cancer cell with sequence number 1–108 in scatter plot, we observed the sequence number of each cell. Criterion II with an additional term ε9,1 was able to identify more cancer cells with ε10,2 > 0. Criterion III was similar to criterion II, except that we only used ε8,2 and ε7,1. Criterion III had some overlap of normal cells in ε8,2 < 0 and ε7,1 < 2. Therefore, we combined criteria II and III together, and used scatter plots, displayed in Figure 6c, to help visualize the separation. The normal cells (purple dots) were further apart to the cancer cells (green dots).
Figure 6.
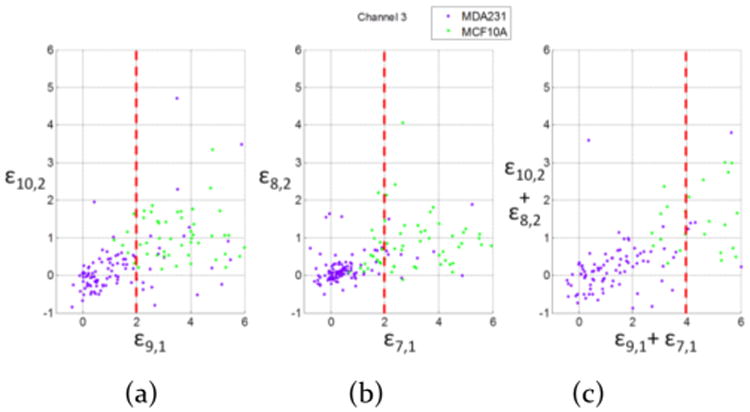
Scatter plot of using criterion II (a), criterion III (b), and combining criteria II and III (c) to differentiate MDA-MB-231 cells and MCF-10A cells in channel 3.
By directly applying ε10,2 > 0, based on the scatter plot data in Figure 6a and bars in Figure 6a, all of the normal cells can be recognized as normal cells (recorded as NR → NR); however, ∼51% of the cancer cells were observed as normal cells (recorded as CA → NR) with ε10,2 > 0. Using criteria II and III, as shown in the scatter plot in Figure 6a and b and bars in Figures 7b and 6c, the identification of normal cells were compromised in order to achieve higher likelihood of cancer cells being observed as either cancer cells (recorded as CA → CA) or as normal cells (CA → NR). After we applied both criteria II and III together, we achieved an acceptable ratio to differentiate cancer cells and normal cells.
Figure 7.
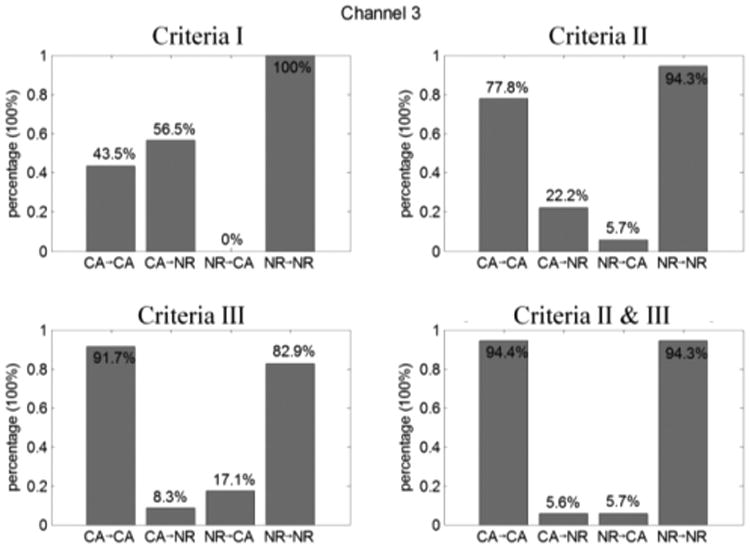
Differentiation rate of cancer cells (CA) and normal cells (NR) in channel 3 using different velocity analysis criteria.
Similarly, we also calculated the observed normal cells and cancer cells from known samples by different criteria in three channels with results shown in Supporting Information Table S-1.
Testing Sample in Channel 3
In order to verify the quality of our differentiation criteria, four blind samples with either cancer cells or normal cells were tested. The data of velocity at each constriction increment in channel 3 was analyzed.
The combination of criteria II and III was applied here to the blind-test samples (s1, s2, s3, and s4). As a result indicated in Figure 8, two samples were observed as cancer cells and the other two samples were observed as normal cells, which agreed to the predicted cells before testing.
Figure 8.
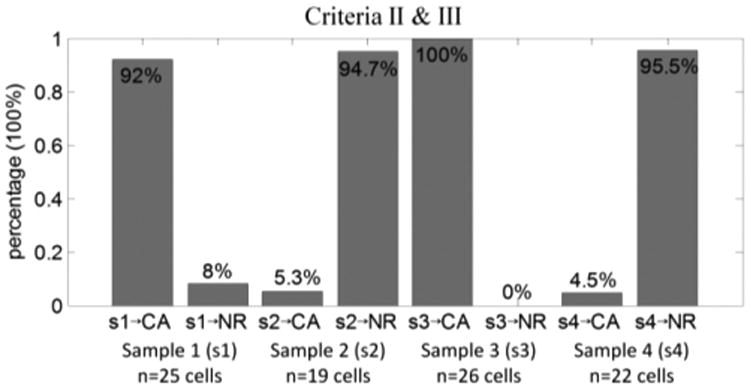
Differentiation ratio of cancer cells (CA) and normal cells (NR) using four blind-testing samples (s1, s2, s3, and s4) in channel 3.
Discussion
The initial deformation of the cells was affected by the mechanical properties of the cells, since the microfluidic channel geometries and experimental conditions were the same. The kinetic behaviors of the cells were directly related to the mechanical properties, such as Young's modulus, cell membrane stiffness, and tension stress of the membrane. These velocity profiles vary between normal cells and cancer cells, because cancer cells have lower membrane stiffness and cytoskeleton strength as documented by previous studies.3,4,40,41 The velocity of the cells depends on fluidic pressure, flow rate, and the geometry of the cells. The cross-section of the microfluidic channels and flow rate are kept constant for both MDA-MB-231 and MCF-10A medium, so that the Reynolds number of both samples can be considered constant. The mechanical characteristics can be revealed by the dynamic behaviors of the cells. The stiffness of the cell membrane dominates the shear force and shear stress between the cell membrane and the microfluidic channel side wall. The Young's modulus of the cells affects the deformation of the cell membrane when the cross-section of the microfluidic channel changes. Both the stiffness of the cell membrane and the Young's modulus of the cells are typical biomarkers used to distinguish cancer cells from normal cells. By studying the average velocity at each segment and the velocity difference between different segments, we can differentiate the different cell types due to their different mechanical characteristics.
The first approaches that used microfluidics for differentiating cells were explored by using a single constriction channel like the channel 1 we presented in our device. The cells experienced a one-time deformation in a single constriction channel, which relied on both the cell size and stiffness. The differentiation merely based on velocity was not reliable due to the variability in cell size which is unavoidable due to at a minimum, cell cycle phase dependent variations in cell size. In a single constriction channel (channel 1), our experiments had a differential ratio of 52.9% for the cancer cells, which was similar to the results of others' experiments with single constriction channel with 8 μm × 8 μm cross section.18 Chen et al., used the elongation and transit time of the cells in single constriction channel to characterize breast cancer cells, with a differential ratio about 57.5%.18 This low recognition rate was another reason why scientists were encouraged to search for new methods to further differentiate different types of cells. Rather than adding electrodes to measure impedance to combine biomechanical and bioelectrical parameters of the cells, we opted to add relaxation regions to separate the single constriction channel into multiple constriction channels to emphasize the biomechanical characteristics of the cells. Our differential ratio for both cancer cells and normal cells can reach more than 94%, which was also verified by four blind-testing samples.
Sample video still shots of MDA-MB-231 and MCF-10A cells passing through channels 1, 2, and 3 appear in the Supporting Information, regenerated from 240 fps videos. From viewing these images, each cell, even within a cell line population, is different and unique. In the single-constriction channel (channel 1), the MDA-MB-231 cells have a higher velocity than the MCF-10A cells. The single-constriction channel with length of 250 μm was separated into 10 segments; however, the 10 segments of the velocity profiles in channel 3 included 5 velocities deformation regions. The two types of cells performed differently in velocity profiles with the five deformation regions, which was suitable in cell type differentiation. In channel 2, both cell types have a decreasing velocity at the relaxation region between 125–150 μm. In channel 3, due to the short constriction region and short relaxation region, MDA-MB-231 cells do not experience a full shape recovery process (deformation performance). Compared to channel 1, channel 3 distinguishes cell types by adding multiple deformation and relaxation regions, which make V7, V5, and V3 more useful in differentiating different cell types. Channel 3 introduced more mechanical alternations for the cells, which increases the membrane mechanical differences between normal cells and cancer cells. Each time the cell deforms, a greater separation is obtained for cells of different metastatic potential. Therefore, based on the velocity profiles generated in constriction regions and relaxation regions, channel 3 had more opportunities to have varying velocity profiles, including velocity increments, for MDA-MB-231 cells and MCF-10A cells. MCF-10A cells in channel 3 have a long deformation time at the entrance, however, they kept the rodlike shape and moved into the succeeding constriction regions with a higher initial velocity. With the continuous acceleration, MCF-10A cells reach a higher final velocity than MDA-MB-231 cells. In other words, once the MCF-10A cells deform, they retain the shape that facilitates their passage. MDA-MB-231 cells show more plasticity in their shape with a net retardation of their velocities.
In our experiments, the effect of multiple relaxation regions are studied in multiple-constriction channels (channel 3) in order to differentiate between cancer cells and normal cells. In future work, the length of relaxation regions can be modified and tested. The cancer cells and normal cells behave differently during the constriction and relaxation regions. The constriction time was determined by the constriction channel length, while the relaxation time was determined by relaxation position and relaxation length. With a longer relaxation region, the cancer cells can recover more after the constriction channels, which may have a recovery that behaves differently compared to the recovered normal cells.
Conclusions
After calculating the velocity increments in the fourth channel and fifth channel compared to the initial velocity in multiple-constriction channels (channel 3), the scatter plot of the single cell velocity data of each MDA-MB-231 (n = 108 cells) and MCF-10A (n = 105 cells) cells showed clear separation of the two cell lines into distinct regions, which successfully differentiated ∼94.4% of the cancer cells from normal cells. Due to the fact that normal cells are stiffer than cancer cells, multiple relaxation regions gave cancer cells more chance to recover into its original shape, and enlarged the dynamic velocity increments along the constriction channels. Our measurement by smart phone video through microscope was reliable. Furthermore, this method has a potential to be developed into a convenient phone application of health examination. Our experimental results indicated that multiconstriction microfluidic channels can be used to differentiate metastatic MDA-MB-231 and MCF-10A cells at the single cell level. For future studies, the quantity of parallel constriction channels can be increased to have higher throughput.
Supplementary Material
Acknowledgments
The authors would like to thank National Institute of Health (NIH) National Cancer Institute R21CA210126 and National Science Foundation (NSF) CBET-1403304 for supporting this research.
Abbreviations
- CTC
circulating tumor cell
- EpCAM
epithelial cell adhesion molecule
- FBS
fetal bovine serum
- EGF
epidermal growth factor
- PDMS
polydimethylsiloxane
- TFOCS
tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane
- AFM
atomic force microscopy
- ROC
receiver operating characteristic
Footnotes
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.6b00823.
Author Contributions: The manuscript was written through contributions of all authors.
Notes: The authors declare no competing financial interest.
References
- 1.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xue C, Wang J, Zhao Y, Chen D, Yue W, Chen J. Constriction channel based single-cell mechanical property characterization. Micromachines. 2015;6(11):1794–1804. [Google Scholar]
- 3.Babahosseini H, Strobl JS, Agah M. Single cell metastatic phenotyping using pulsed nanomechanical indentations. Nanotechnology. 2015;26(35):354004. doi: 10.1088/0957-4484/26/35/354004. [DOI] [PubMed] [Google Scholar]
- 4.Nikkhah M, Strobl JS, De Vita R, Agah M. The cytoskeletal organization of breast carcinoma and fibroblast cells inside three dimensional (3-D) isotropic silicon microstructures. Biomaterials. 2010;31(16):4552–4561. doi: 10.1016/j.biomaterials.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Sheng W, Fan ZH. An ensemble of aptamers and antibodies for multivalent capture of cancer cells. Chem Commun. 2014;50(51):6722–6725. doi: 10.1039/c4cc02002b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Fan ZH. A universal tumor cell isolation method enabled by fibrin-coated microchannels. Analyst. 2016;141(2):563–566. doi: 10.1039/c5an01783a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riethdorf S, Fritsche H, Mx000FC;ller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jx000E4;nicke F, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13(3):920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 8.Zabaglo L, Ormerod MG, Parton M, Ring A, Smith IE, Dowsett M. Cell filtration-laser scanning cytometry for the characterisation of circulating breast cancer cells. Cytometry. 2003;55(2):102–108. doi: 10.1002/cyto.a.10071. [DOI] [PubMed] [Google Scholar]
- 9.Juratli MA, Sarimollaoglu M, Siegel ER, Nedosekin DA, Galanzha EI, Suen JY, Zharov VP. Real-time monitoring of circulating tumor cell release during tumor manipulation using in vivo photoacoustic and fluorescent flow cytometry. Head & neck. 2014;36(8):1207–1215. doi: 10.1002/hed.23439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz I, Cruz JJ, Ramos M, Gx000F3;mez-Alonso A, Adansa JC, Rodríguez C, Orfao A. Evaluation of multiparameter flow cytometry for the detection of breast cancer tumor cells in blood samples. Am J Clin athol. 2005;123(1):66–74. doi: 10.1309/wp3qwkvjfydhhxqd. [DOI] [PubMed] [Google Scholar]
- 11.Bagnall JS, Byun S, Begum S, Miyamoto DT, Hecht VC, Maheswaran S, Stott SL, Toner M, Hynes RO, Manalis SR. Deformability of tumor cells versus blood cells. Sci Rep. 2015;5:18542. doi: 10.1038/srep18542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ, Tai YC. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 2011;13(1):203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohamed H, McCurdy LD, Szarowski DH, Duva S, Turner JN, Caggana M. Development of a rare cell fractionation device: application for cancer detection. IEEE transactions on nanobioscience. 2004;3(4):251–256. doi: 10.1109/tnb.2004.837903. [DOI] [PubMed] [Google Scholar]
- 14.Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto DT, et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12(7):685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riahi R, Gogoi P, Sepehri S, Zhou Y, Handique K, Godsey J, Wang Y. A novel microchannel-based device to capture and analyze circulating tumor cells (CTCs) of breast cancer. Int J Oncol. 2014;44(6):1870–1878. doi: 10.3892/ijo.2014.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee A, Park J, Lim M, Sunkara V, Kim SY, Kim GH, Kim MH, Cho YK. All-in-one centrifugal microfluidic device for size-selective circulating tumor cell isolation with high purity. Anal Chem. 2014;86(22):11349–11356. doi: 10.1021/ac5035049. [DOI] [PubMed] [Google Scholar]
- 17.Loutherback K, D'Silva J, Liu L, Wu A, Austin RH, Sturm JC. Deterministic separation of cancer cells from blood at 10 mL/min. AIP Adv. 2012;2(4):042107. doi: 10.1063/1.4758131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Zheng Y, Tan Q, Shojaei-Baghini E, Zhang YL, Li J, Prasad P, You L, Wu XY, Sun Y. Classification of cell types using a microfluidic device for mechanical and electrical measurement on single cells. Lab Chip. 2011;11(18):3174–3181. doi: 10.1039/c1lc20473d. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Jiang M, Chen D, Zhao X, Xue C, Hao R, Yue W, Wang J, Chen J. Single-Cell Electrical Phenotyping Enabling the Classification of Mouse Tumor Samples. Sci Rep. 2016;6:19487. doi: 10.1038/srep19487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang SB, Zhao Y, Chen D, Lee HC, Luo Y, Chiu TK, Wang J, Chen J, Wu MH. A clogging-free microfluidic platform with an incorporated pneumatically driven membrane-based active valve enabling specific membrane capacitance and cytoplasm conductivity characterization of single cells. Sens Actuators, B. 2014;190:928–936. [Google Scholar]
- 21.Huang SB, Zhao Y, Chen D, Liu SL, Luo Y, Chiu TK, Wang J, Chen J, Wu MH. Classification of cells with membrane staining and/or fixation based on cellular specific membrane capacitance and cytoplasm conductivity. Micromachines. 2015;6(2):163–171. [Google Scholar]
- 22.Zhao Y, Chen D, Luo Y, Chen F, Zhao X, Jiang M, Yue W, Long R, Wang J, Chen J. Simultaneous characterization of instantaneous Young's modulus and specific membrane capacitance of single cells using a microfluidic system. Sensors. 2015;15(2):2763–2773. doi: 10.3390/s150202763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babahosseini H, Strobl JS, Agah M. Microfluidic iterative mechanical characteristics (iMECH) analyzer for single-cell metastatic identification. Anal Methods. 2017;9(5):847–855. doi: 10.1039/C6AY03342C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou HW, Li Q, Lee G, Kumar A, Ong C, Lim CT. Deformability study of breast cancer cells using microfluidics. Biomed Microdevices. 2009;11(3):557–564. doi: 10.1007/s10544-008-9262-8. [DOI] [PubMed] [Google Scholar]
- 25.Mak M, Reinhart-King CA, Erickson D. Elucidating mechanical transition effects of invading cancer cells with a subnucleus-scaled microfluidic serial dimensional modulation device. Lab Chip. 2013;13(3):340–348. doi: 10.1039/c2lc41117b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin D, Deng B, Li J, Cai W, Tu L, Chen J, Wu Q, Wang W. A microfluidic device enabling high-efficiency single cell trapping. Biomicrofluidics. 2015;9(1):014101. doi: 10.1063/1.4905428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan G, Chen PC, Peng WK, Bhagat AA, Ong CJ, Han J. Real-time control of a microfluidic channel for size-independent deformability cytometry. J Micromech Microeng. 2012;22(10):105037. [Google Scholar]
- 28.Hoffman BD, Grashoff C, Schwartz MA. Dynamicmolecular processes mediate cellular mechanotransduction. Nature. 2011;475(7356):316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Nguyen J, Wei Y, Sun Y. Recent advances in microfluidic techniques for single-cell biophysical characterization. Lab Chip. 2013;13(13):2464–2483. doi: 10.1039/c3lc50355k. [DOI] [PubMed] [Google Scholar]
- 30.Khan Z, Vanapalli S. Probing the mechanical properties of brain cancer cells using a microfluidic cell squeezer device. Biomicrofluidics. 2013;7(1):011806. doi: 10.1063/1.4774310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagnall JS, Byun S, Miyamoto DT, Kang JH, Maheswaran S, Stott SL, Toner M, Manalis SR. Deformability-based cell selection with downstream immunofluorescence analysis. Integrative Biology. 2016;8(5):654–664. doi: 10.1039/c5ib00284b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, Chen D, Zhao Y, Wei C, Zhao X, Yue W, Long R, Wang J, Chen J. A constriction channel based microfluidic system enabling continuous characterization of cellular instantaneous Young's modulus. Sens Actuators, B. 2014;202:1183–1189. [Google Scholar]
- 33.Zhao Y, Chen D, Li H, Luo Y, Deng B, Huang SB, Chiu TK, Wu MH, Long R, Hu H, et al. A microfluidic system enabling continuous characterization of specific membrane capacitance and cytoplasm conductivity of single cells in suspension. Biosens Bioelectron. 2013;43:304–307. doi: 10.1016/j.bios.2012.12.035. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Abdelgawad M, Yu L, Shakiba N, Chien WY, Lu Z, Geddie WR, Jewett MA, Sun Y. Electrodeformation for single cell mechanical characterization. J Micromech Microeng. 2011;21(5):054012. [Google Scholar]
- 35.Adamo A, Sharei A, Adamo L, Lee B, Mao S, Jensen KF. Microfluidics-based assessment of cell deformability. Anal Chem. 2012;84(15):6438–6443. doi: 10.1021/ac300264v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byun S, Son S, Amodei D, Cermak N, Shaw J, Kang JH, Hecht VC, Winslow MM, Jacks T, Mallick P, Manalis Sr. Characterizing deformability and surface friction of cancer cells. Proc Natl Acad Sci U S A. 2013;110(19):7580–7585. doi: 10.1073/pnas.1218806110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Zhang W, Huang F, Feng H, Shu W, Xu X, Chen Y. High throughput capture of circulating tumor cells using an integrated microfluidic system. Biosens Bioelectron. 2013;47:113–119. doi: 10.1016/j.bios.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Tang L, Li Z, Lin Y, Li J. In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet–based sensing platform in living cells. Nat Protoc. 2014;9(8):1944–1955. doi: 10.1038/nprot.2014.126. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Li J, Sun Y. Microfluidic approaches for cancer cell detection, characterization, and separation. Lab Chip. 2012;12(10):1753–1767. doi: 10.1039/c2lc21273k. [DOI] [PubMed] [Google Scholar]
- 40.Babahosseini H, Carmichael B, Strobl J, Mahmoodi S, Agah M. Sub-cellular force microscopy in single normal and cancer cells. Biochem Biophys Res Commun. 2015;463(4):587–592. doi: 10.1016/j.bbrc.2015.05.100. [DOI] [PubMed] [Google Scholar]
- 41.Carmichael B, Babahosseini H, Mahmoodi S, Agah M. The fractional viscoelastic response of human breast tissue cells. Phys Biol. 2015;12(4):046001. doi: 10.1088/1478-3975/12/4/046001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


