Abstract
Finger millet grains contain exceptionally high levels of calcium which is much higher compared to other cereals and millets. Since calcium is an important macronutrient in human diet, it is necessary to explore the molecular basis of calcium accumulation in the seeds of finger millet. CIPK is a calcium sensor gene, having role in activating Ca2+ exchanger protein by interaction with CBL proteins. To know the role of EcCIPK24 gene in seed Ca2+ accumulation, sequence is retrieved from the transcriptome data of two finger millet genotypes GP1 (low Ca2+) and GP45 (high Ca2+), and the expression was determined through qRT-PCR. The higher expression was found in root, shoot, leaf and developing spike tissue of GP45 compared to GP1; structural analysis showed difference of nine SNPs and one extra beta sheet domain as well as differences in vacuolar localization was predicted; besides, the variation in amino acid composition among both the genotypes was also investigated. Molecular modeling and docking studies revealed that both EcCBL4 and EcCBL10 showed strong binding affinity with EcCIPK24 (GP1) compared to EcCIPK24 (GP45). It indicates a genotypic structural variation, which not only affects the affinity but also calcium transport efficiency after interaction of CIPK-CBL with calcium exchanger (EcCAX1b) to pull calcium in the vacuole. Based on the expression and in silico study, it can be suggested that by activating EcCAX1b protein, EcCIPK24 plays an important role in high seed Ca2+ accumulation.
Keywords: Finger millet, Transcriptome data, CIPK24, CBL, Modeling, Docking
Introduction
Finger millet (Eleusine coracana), an allotetraploid (4X) and annual robust grass, is mainly grown as a grain cereal in the semi-arid tropics and subtropics of the world under rain-fed conditions (Fakrudin et al. 2004). As compared to other cereals, calcium (Ca2+) content is very high in finger millet grains that varies from 100 to 450 mg/100 g of seed, and is amazingly 10–30 times higher than that found in the grains of rice and wheat (National Research Council 1996; Panwar et al. 2010). It can be used in formulating diets for pregnant and lactating women and growing children. Also, high calcium supplements can help in controlling osteoporosis occurring during menopause (Kumar et al. 2012). Molecular characterization of finger millet genotypes revealed that high calcium accumulation in finger millet grain is mainly genetically determined and less is environmentally influenced (Panwar et al. 2010; Kumar et al. 2012; Nath et al. 2013; Singh et al. 2014a).
Ca2+ signaling and transport gene family has very important role in seed Ca2+ accumulation (Kumar et al. 2015a, b; Singh et al. 2014b, 2015; Sharma et al. 2017). The role of Ca2+ exchanger protein is well studied and also used for making transgenic plant for higher seed Ca2+ accumulation but study about its regulatory sensor protein, especially CIPK (CBL-interaction protein kinase) is not well studied. CIPK(s) is a type of Ca sensor protein, playing an important role in regulation of ion transport, especially Ca exchanger protein. The structure of CIPKs is related to sucrose non-fermenting kinase (SNF1) from yeast and AMP-activated protein kinase (AMPK) from animals (Hrabak et al. 2003). The typical CIPK consists of the conserved N-terminal SNF1-type kinase domain (24 amino acid), which is fused, via a junction domain, to a highly variable C-terminal regulatory domain (Batistic and Kudla 2004). This domain is required for interaction with CBL proteins (Albrecht et al. 2001).
It is noteworthy that, during salinity stress, CIPK24 appears to target other ion transporters at the tonoplast including an H+ pump and a Ca2+/H+ exchanger (Verslues et al. 2007). CIPK24 interacts with CBL10 to the vacuolar membrane (Kim et al. 2007). The CIPK24/CAX1 interaction is Ca2+ dependent and SOS2 must be recruited to the tonoplast to activate cation exchanger (CAX) (Cheng et al. 2004). Although the role of CIPK24 in CAX activation during stresses is well defined, but its role in Ca2+ accumulation by activating CAX is not studied so far. Therefore, it was speculated that there might be specific interactions between CIPK24 and CBL complexes with vacuolar calcium exchanger (CAX) in the seed that regulate action of CAX to pump calcium into vacuole of developing seed.
In the present investigation, attempts were made to characterize and define the role of CIPK24 gene homologue of finger millet to explore its role in high grain calcium accumulation. Transcript profiling of CIPK24 gene homologue was done in developing spikes for investigating its role in differential expression in finger millet genotypes differing in grain calcium content. Full-length structural annotation of CIPK24 was done from the developing spike transcriptome data of two finger millet genotype. In silico characterization and interaction studies of CIPK24 gene homologues isolated from finger millet genotypes with CBLs of finger millet were performed using bioinformatics tools.
Materials and methods
Isolation and molecular cloning of partial sequence of CIPK24 gene from finger millet
Primer designing
The homologs of CIPK24 gene of rice, sorghum and maize were retrieved from NCBI database (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignment of these sequences was done using MEGA5 to determine the conserved regions, and these conserved regions were subjected to Primer3 tool (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) to obtained primers for expression analysis using qRT-PCR (Tamura et al. 2011).
Genomic DNA extraction and PCR amplification
In this investigation, CTAB method of DNA isolation described by Murray and Thompson (1980) was applied for obtaining good quality genomic DNA which was used as template DNA for subsequent PCR amplification. PCR amplification was performed using 50–100 ng of template DNA, 30 ng of primer, 0.1 mM dNTPs, 1.5 U Taq DNA polymerase (Bangalore Genei Pvt. Bangalore, India), 1X PCR buffer (10 Mm Tris pH 8.0, 50 mM KCL and 1.8 mM MgCl2) in volume of 25 μl. PCR amplified products of all the primers were subjected to gel electrophoresis and were documented using Alpha Imager 1200TM (Alpha Innotech Corporation, USA).
Cloning and sequencing of partial CIPK24 gene in pGEMT vector
The amplified product was analyzed on agarose gel and the expected size amplicon was gel; eluted using QIAquick Gel Extraction Kit (Qiagen, USA) and cloned in pGEM-Teasy vector (Promega, USA) as per the kit instructions. Putative cloned CIPK24 gene was sequenced using M13 universal Primer present in pGEM-Teasy vector. The confirmation of CIPK24 sequence was done through BLAST analysis.
Transcript profiling of EcCIPK24 gene in vegetative and reproductive tissues
Plant materials
Seeds of finger millet genotypes were collected from Uttarakhand and obtained from Uttarakhand University of Horticulture and Forestry, Ranichauri, India. Two genotypes of finger millet GP1 (low) and GP45 (high) were selected in the present study due to the difference in their total grain calcium contents (Panwar et al. 2010).
Surface sterilized finger millet seeds were germinated on wet paper and were planted on commercial soil mix. Plants were grown at 37 °C in a glass house. The developing spikes of different stages (S1 to S4) were cut off, frozen in liquid nitrogen and kept at 80 °C until further use.
Preparation of RNA and first strand cDNA synthesis
Total RNA was isolated from different developmental stages of finger millet using total RNA isolation iRIS system from IHBT Palampur. Total RNA was treated with RNase free DNaseI according to manufacturer’s instruction (Fermentas, Germany). The first strand cDNA was synthesized with 2 μg of purified total RNA (pre-treated with DNase I) using the RT-PCR system (Promega, USA) according to the manufacturer’s protocol.
Quantitative real-time PCR
Real-time polymerase chain reaction (RT-PCR) was used to quantitatively determine the expression profile of the EcCIPK24 genes in vegetative and reproductive tissues of finger millet genotypes. Gene-specific primers were designed to amplify the specific cDNA fragment of gene. Tubulin (CX265249) was used as internal control. RT-PCR was performed in the reaction volume of 20 µl containing 2.5 × Real Master Mix SYBR ROX/20 × SYBR solution, 100 ng of cDNA, 100 nM of forward and reverse primers. The ratio of the target band intensity to the tubulin was used to investigate the relative expression level of the target gene.
Isolation of full-length EcCIPK24 gene from finger millet transcriptome data
Partial nucleotide sequence of EcCIPK24 obtained after sequencing was used as a query sequence to perform local BLAST search against our local assembled transcriptome database (TSA accession SRR1151079 and SRR1151080) constructed from transcript sequencing of spike tissues of low and high finger millet genotypes. The contigs sequences showed maximum similarity with CIPK24 of finger millet were retrieved and assembled as well as its further characterizations were done through computational approaches.
SNP analysis of EcCIPK24 of both genotypes was performed using clustal W alignment tool. Domain analysis of EcCIPK24 gene homologue sequences of both genotypes were performed using SMART on line tool (http://www.smart.co.in) with default parameters. Motif analyses of sequences of both genotypes were done with default parameter using MEME (http://meme.nbcr.net) (Schultz et al. 1998; Bailey et al. 2009). ProtParam analysis was done using EcCIPK24 gene homologue sequences of both genotypes. The protein secondary structure prediction, GOR algorithm based tool GORIV (http://abs.cit.nih.gov/gor/) was used to determined the amino acid residues involved in the formation of helix, sheet and turn (Sen et al. 2005). Secondary structures of proteins were predicted using EcCIPK24 gene homologue sequences of both genotypes with default parameter. Prediction of protein sub-cellular localization of EcCIPK24 sequences of GP1 and GP45 was done using CELLO V. 2.5 sub-cellular localization predictor tool (http://cello.life.nctu.edu.tw/).
Protein structure prediction, evaluation and validation
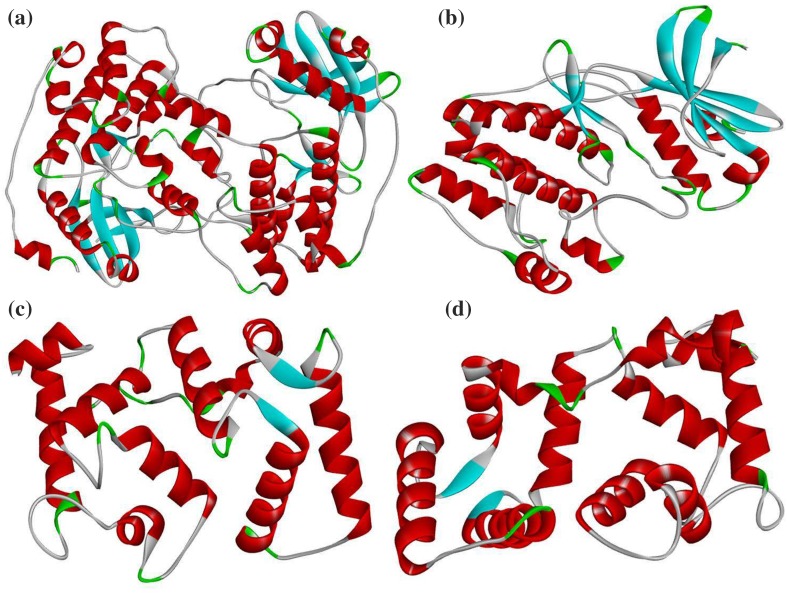
Protein sequence of EcCBL4, EcCBL10 and EcCIPK24 (GP1 and GP45) was subjected to BLASTp against PDB database (http://www.rcsb.org/pdb/home/home.do) for identification of suitable template as well as methods for modeling of 3D model (Altschul et al. 1990). In addition to BLASTp search, SWISS-MODEL was employed to model 3D structure of target protein sequences (Arnold et al. 2006). The structural refinements through energy minimization of each predicted models were performed by SPDB viewer (http://spdbv.vital-it.ch/refs.html). RAMPAGE server (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) was used to analyze the stereochemical quality of structure coordinates of the predicted protein models through Ramachandran plot analysis. The overall quality analysis of each models was done by ProSA (Protein Structure Analysis) and ProQ (Protein Quality Predictor) (Wiederstein and Sippl 2007; Wallner and Elofsson 2003), and DS Visualizer was used for the visualization of 3D models (Fig. 4) (Pathak et al. 2016).
Fig. 4.
Modeled 3D structures of finger millet proteins involved in calcium accumulation a EcCIPK24 (GP45), b EcCIPK24 (GP1), c EcCBL4 and d EcCBL10
Protein–protein docking
The refined predicted structures of EcCIPK24 (GP1 and GP45) were taken as receptor and the structures of EcCBL4 and EcCBL10 were considered as ligands for the protein–protein docking studies. EcCBL4 and EcCBL10 were docked with EcCIPK24 protein of the both high and low calcium genotype using ClusPro (https://cluspro.bu.edu/home.php) server at default parameter. The best docked conformation was taken to analyze the interactions, which are responsible for regulation of the biological processes on the basis of their binding free energy. The docked complex files were visualized and analyzed by PyMol (Fig. 5) (DeLano 2002; Comeau et al. 2004; Pathak et al. 2013).
Fig. 5.
Protein–protein docking; a EcCIPK24 (GP45)(cyan)–EcCBL10(red), b EcCIPK24(GP1)(magenta)–EcCBL10(red), c EcCIPK24(GP45)(cyan)–EcCBL4(green), d EcCIPK24(GP1)(magenta)–EcCBL4(green)
Results and discussion
Finger millet, an under-utilized cereal crop, contains exceptionally high amounts of calcium in grains. Since calcium is an important macronutrient in human diet, it requires immediate attention to understand the molecular basis of high calcium accumulation in seed of finger millet. It has been reported in our lab that the seeds of high calcium containing finger millet genotype are having higher level expression of Ec CAX1 gene while compared with low calcium containing genotype of finger millet (Mirza et al. 2014; Singh et al. 2015). Further, it has also been reported that CIPK24 gene regulate vacuolar CAX1 transporter that leads to pull cytosolic calcium in vacuole. CIPK24 activates transport protein after interaction with CBL proteins, viz. CBL4, CBL10, etc. (Chinnusamy et al. 2005; Kim et al. 2007; Quan et al. 2007). This gives an indication regarding the role of CIPK24 not only in signaling but also in regulation of calcium transport efficiency across the membranes in seed. The present section deals with isolation and characterization of EcCIPK24 gene in grain Ca2+ accumulation.
Isolation and molecular cloning
A prominent single band with expected size 0.25 kb was consistently observed in genomic DNA of finger millet after PCR amplification with set of primer. It showed conserved sequences in pairwise alignment using Clustal W online tool and closely related in phylogenetic tree using neighbor joining (NJ) method for all cereals CIPK24 gene homologue. The PCR amplicon of 0.25 kb was eluted from the gel and purified using QIAquick Gel Extraction Kit (Qiagen, USA), Subsequently, cloned in pGEM T-Easy vector (Promega, USA) as per the kit instructions. Putative cloned EcCIPK24 gene was sequenced at DNA sequencing facility, University of Delhi, South Campus. The sequence was confirmed by homology alignment with CIPK24 of other cereals and used for fetching the contigs from transcriptome data of low and high calcium containing genotype of finger millet.
Expression studies of EcCIPK24 gene in vegetative and reproductive tissues
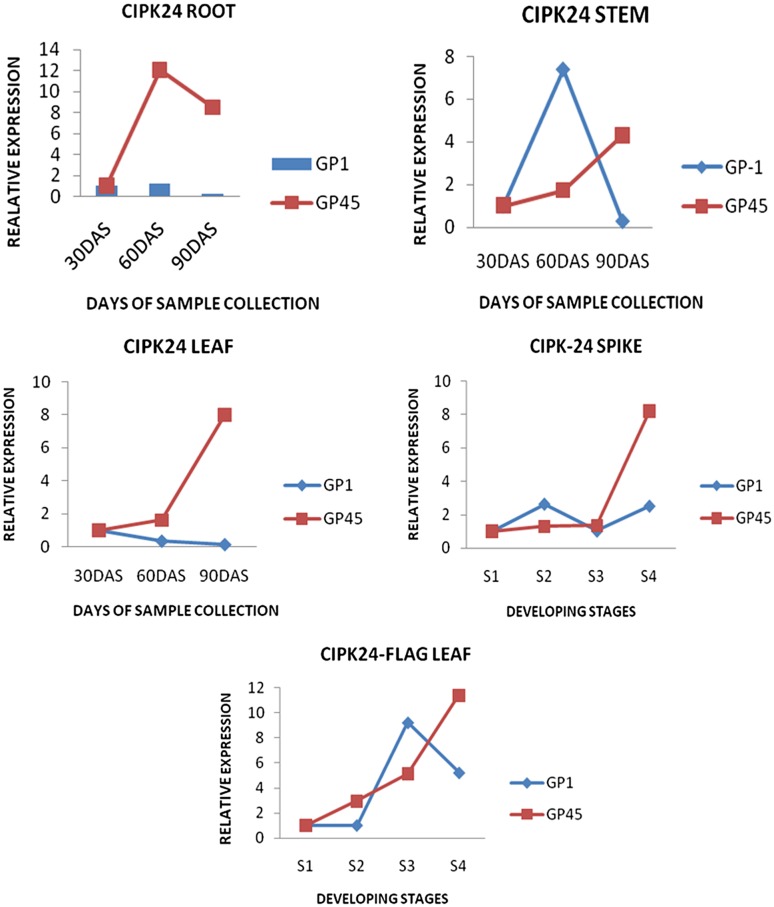
The expression pattern of EcCIPK24 gene homologues was investigated in two finger millet genotypes (GP1 and GP45). The abundance of specific mRNA was investigated in root, stem and third leaf at three different vegetative stages, viz. 30, 60, 90 DAS (day after sowing). The expression pattern of EcCIPK24 gene was also studied in flag leaf and developing spike at four defined reproductive stages, viz., S1, S2, S3 and S4. The relative expression of gene within genotypes and among genotypes was investigated.
The increased expression of EcCIPK24 gene was observed in root, stem and leaf tissues of GP45 genotype compared to GP1 genotype. It was noticed that the expression of EcCIPK24 gene was more in GP45 at 60 DAS (root, leaf tissue) and 90 DAS (in root stem and leaf tissue) but less in GP1 at 60 DAS in stem.
The expression of EcCIPK24 gene in GP45 genotype increased continuously in flag leaf tissue and developing spike tissues from S1 to S4 stages (Fig. 1). However, in case of spike tissues of GP1 genotype the expression increased 2.7-fold at S2 stage, and decreased at S3 then it increased 2.5-fold at S4 stage. In flag leaves expression continuously increased from S1 to S4 stage in GP45, while in GP-1 it drops at S4 stage. The expression of EcCIPK24 gene is in same increasing pattern in GP45, while decreased in spike and flag leaves tissues of GP1 genotype.
Fig. 1.
Relative expression of EcCIPK24 transcripts in root, stem, leaf tissue at 30, 60 and 90 DAS and in developing stages of Spike, flag leaf among genotypes GP1 (Low calcium genotype) and GP45 (High calcium genotype). DAS days after sowing
Previous studies conducted in our lab suggested that the expression of EcCAX1 is higher in developing spikes of GP45 genotype, as compared to GP1. The similar expression patterns of EcCIPK24 gene were recorded in both vegetative as well as reproductive tissues of GP45 and GP1 genotypes, which indicate the co-regulation of both the genes (Mirza et al. 2014). The result is in agreement with study that CIPK24 after interaction with CBL10 regulate the activity of CAX protein (Cheng et al. 2004; Verslues et al. 2007; Kim et al. 2007).
In silico analysis of EcCIPK24 gene homologue with other cereal CIPK24 gene homologue
The contig sequence of EcCIPK24 and interactive sequences (EcCBL4, EcCBL10) was retrieved by performing local BLAST against transcriptome local database of finger millet and reassembled by SeqMan Pro gene analysis package (DNASTAR Inc., Madison, WI, USA).
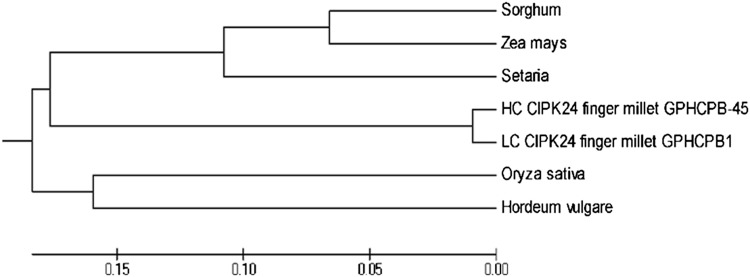
Finger millet CIPK24 homologue showed more than 90% similarity with available cereal CIPK gene homologues, among them with Oryza sativa CIPK24 gene homologue gave highest similarity. The result showed that retrieved CIPK24 gene homologue of finger millet has more conserved sequence among cereals. The result of phylogenetic analysis showed that CIPK24 gene homologue of both finger millet genotypes was situated in two sub–sub-cluster of a distinct sub-cluster (Fig. 2).
Fig. 2.
Phylogenetic tree of CIPK24 genes of cereals constructed using neighbor joining method showing relationship with EcCIPK (GP1 and GP45)
Comparative analysis of SNPs, amino acid composition, and other physico-chemical properties of EcCIPK24 homologues
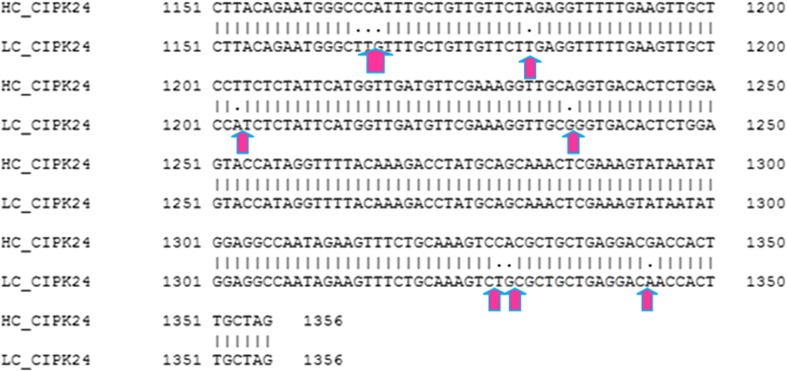
Nine SNPs were detected in EcCIPK24 gene sequence of both genotypes using clustal W (Fig. 3). Domain analysis showed that both sequences have S TKc kinase domain with E value 1.23e-105, which is more conserved at C-terminal and variable at N-terminal. The domain starts from the amino acid sequences from 14 and End at 267. Motif analyses of sequences of both genotypes showed the presence of three motifs using MEME (Bailey et al. 2009). Results of ProtParam analysis are summarized in Tables 1 and 2.
Fig. 3.
SNP analysis of EcCIPK24 gene in GP1 and GP45 genotypes
Table 1.
Comparison of physico-chemical properties of EcCIPK24 sequences isolated from two finger millet genotypes differing grain calcium contents
| S. No. | Physico-chemical properties | High calcium genotype (GP45) | Low calcium genotype (GP1) |
|---|---|---|---|
| 1. | Molecular weight | 50860.3 | 50846.3 |
| 2. | Iso electric point | 7.64 | 7.64 |
| 3. | Number of amino acid | 451 | 451 |
| 4. | Sub-cellular localization | 2.456 (cytoplasmic) | 2.357 (cytoplasmic) |
| 0.044 (vacuolar) | 0.043 (vacuolar) | ||
| 5. | Total number of atoms | 7189 | 7190 |
| 6. | Instability index | 33.20 | 32.78 |
| 7. | Aliphatic index | 92.31 | 93.39 |
| 8. | Grand average of hydropath city (GRAVY) | −0.230 | −0.213 |
| 9. | Helix | 296 | 296 |
| 10. | Sheet | 160 | 161 |
| 11. | Turns | 47 | 47 |
Table 2.
Comparison of amino acid composition of EcCIPK24 sequences isolated from two finger millet genotypes differing grain calcium contents
| S. No. | Amino acid composition | CIPK24 of GP45 (%) | CIPK24 of GP1 (%) |
|---|---|---|---|
| 1. | Ala (A) | 7.1 | 7.3 |
| 2. | Arg (R) | 6.9 | 6.9 |
| 3. | Asn (N) | 3.3 | 3.3 |
| 4. | Asp (D) | 6.0 | 6.0 |
| 5. | Cys (C) | 1.1 | 1.1 |
| 6. | Gln (Q) | 2.7 | 2.7 |
| 7. | Glu (E) | 7.1 | 7.1 |
| 8. | Gly (G) | 6.9 | 6.9 |
| 9. | His (H) | 2.0 | 2.0 |
| 10. | Ile (I) | 7.1 | 7.1 |
| 11. | Leu (L) | 9.3 | 9.5 |
| 12. | Lys (K) | 6.4 | 6.4 |
| 13. | Met (M) | 2.4 | 2.4 |
| 14. | Phe (F) | 4.0 | 4.0 |
| 15. | Pro (P) | 3.3 | 3.1 |
| 16. | Ser (S) | 6.9 | 6.9 |
| 17. | Thr (T) | 5.3 | 5.1 |
| 18. | Trp (W) | 0.9 | 0.9 |
| 19. | Tyr (Y) | 4.0 | 4.0 |
| 20. | Val (V) | 7.3 | 7.3 |
| 21. | Pyl (O) | 0.0 | 0.0 |
| 22. | Sec (U) | 0.0 | 0.0 |
Molecular weight 50860.3 Dalton of EcCIPK24 sequence (GP45) was found to be higher than EcCIPK24 sequence (GP1) 50846.3 Dalton. Isoelectric point and number of amino acids in two sequences are similar with values 7.64 and 451, respectively. Though the number of amino acids is same but change in amino acid composition is observed in both sequences of EcCIPK24. Secondary structure of EcCIPK24 (GP1) showed one extra sheet (161) than that of EcCIPK24 (GP45) (160) using GORIV. The results showed cytoplasmic localization of CIPK24 sequences of both genotypes having 2.456 and 2.357 as an index, respectively, which was predicted by CELLO V. 2.5 sub-cellular localization prediction tool (Yu et al. 2006). Interestingly, it was found that vacuolar localization of EcCIPK24 (GP45) was higher than vacuolar localization of EcCIPK24 (GP1), i.e., 0.044 and 0.043, respectively. Results of ProtParam analysis are summarized in Table 2.
3D structure prediction and validation
The homology modeling approach was employed to determine a reasonable 3D structure of these proteins based on the known structure available in Protein Data Bank. The 3D structure modeling of EcCPIK24 (GP45), EcCPIK24 (GP1), EcCBL4 and EcCBL10 was done using SWISS-MODEL (http://swissmodel.expasy.org) (Biasini et al. 2014) showed in Fig. 4. The modeled protein structures were subjected to SPDB Viewer for stabilizing their stereochemical properties through energy minimization. It is a computational technique employed to eliminate the unwanted contacts of the macromolecules for the purpose of structure refinements (Vyas et al. 2012). The refined structures were further subjected to RAMPAGE, ProSA (Protein Structure Analysis), and ProQ (Protein Quality Predictor) to validate its overall quality. It was suggested that the predicted model quality to be acceptable and can be utilized for molecular docking studies. The structural template used for modeling of target sequences and physico-chemical properties of modeled protein structures was shown in Table 3 (Wallner and Elofsson 2003; Wiederstein and Sippl 2007).
Table 3.
Comparative analysis of protein models of EcCIPK24 gene homologues
| S. No. | EcCIPK24 and its interacting proteins of both genotypes | Template | Physico-chemical properties | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of group | Number of atoms | Number of bonds | QMEAN4 | Sequence identity in percent | C_beta interaction energy | Torsion angle energy | |||
| 1 | EcCIPK24 (GP45) | 4dz8.A | 562 | 4485 | 4575 | −3.90 | 70.14 | −2.14 | −2.64 |
| 2 | EcCIPK24 (GP1) | 4dz8.A | 282 | 2248 | 2293 | −2.95 | 70.14 | −0.95 | −2.78 |
| 3. | EcCBL10 (GP45) | 2zfd.A | 184 | 1506 | 1532 | −1.08 | 54.75 | 0.28 | −0.48 |
| 5. | EcCBL4 (GP45) | 2ehb.A | 181 | 1471 | 1496 | −0.80 | 68.32 | −0.24 | −0.33 |
Molecular interaction prediction of target proteins through molecular docking
ClusPro (https://cluspro.bu.edu/home.php) server was used for the molecular docking studies between EcCIPK and EcCBL proteins to investigate its role in calcium accumulation. It is a fully automated, web-based program for the docking of protein structures. It required the 3D co-ordinate files of receptor and ligand protein structure to predict its binding affinity.
EcCBL4 was docked with EcCIPK24 (both GP45 and GP1) with binding energies −860.4 and −915.2 kcal/mol, respectively (Table 4); the probable range of interacting amino acid residues through non-covalent bonding is found at 17–24 and 101–107 of the EcCIPK24 (GP45) with EcCBL4 at 40–50, 61–66 and 194–197. Moreover, 237–247 and 288–292 of EcCIPK24 (GP1) with 137–139 and 189–197 of CBL4 are found to be interacting. EcCBL10 was docked with EcCIPK24 (GP45) and (GP1) with energy values −827.3 and −909.8 kcal/mol, respectively (Table 4), the probable interacting amino acid residues predicted between these proteins at 48–54, 86–88, 105–108 and 167–171 of EcCIPK24 (GP45) with 52–59, 82–90 and 123–126 of EcCBL10. Moreover, 241–247 and 287–290 of EcCIPK24 (GP1) with 175–180 and 226–232 interacted through non-covalent bonding. On the basis of docking energy we have investigated the binding mode of EcCBL proteins, and was predicted that the EcCBL4 has greater affinity with EcCIPK24 proteins compared to CBL10 (Fig. 5).
Table 4.
Protein–protein docking studies: illustrates minimum free binding energy
| S. No. | Protein–protein interactions | Docking energy (KCal/mol) | ||
|---|---|---|---|---|
| Receptor | Ligand | High calcium genotype GP45 | Low calcium genotype GP1 | |
| 1. | EcCIPK24 | EcCBL4 | −860.4 | −915.2 |
| 2. | EcCIPK24 | EcCBL10 | −827.3 | −909.8 |
Based on such protein–protein docking studies, we have found that EcCBL4 might interact with EcCIPK24 at vacuolar membrane and form an EcCIPK24–EcCBL4 complex that might interact and regulate EcCAX1 at vacuolar membrane to pull calcium in vacuole. Probably, this might also be playing a significant role in high accumulation of calcium in finger millet seed.
Conclusions
In the present study, an attempt has been made to isolate partial EcCIPK24 ORF from finger millet, and the eluted product was cloned in pGEM T-Easy vector and sequenced. In silico analysis of full-length EcCIPK24 gene homologues identified from transcriptome of both genotypes showed differences of nine SNPs, one extra beta sheet domain, and vacuolar localization and amino acid composition in their sequences. Docking study suggested that EcCBL4 has stronger binding affinity with EcCIPK24 and might play a significant role in the accumulation of calcium in seeds. Based on such studies, elucidation of a probable pathway for exploring differential accumulation of calcium in finger millet is proposed, which suggest that EcCBL4 gets activated from Ca signature and interact with EcCIPK24 at vacuolar membrane. The EcCIPK24–EcCBL4 complex interacted and regulated EcCAX1 at vacuolar membrane to pull calcium in vacuole. The differences in the structures of EcCIPK24, variation in their interaction with interacting proteins, EcCBL4 and EcCAX1 and differential expression of EcCIPK24, in two genotypes of finger millet genotypes may be a plausible reason for interpreting it for differential accumulation of Ca in finger millet genotypes. Higher expression of EcCIPK24 in high calcium containing genotype and lower affinity of EcCIPK24 with EcCBL4 and EcCBL10 might regulate EcCaX1B at vacuolar membrane to pull calcium in vacuoles in finger millet seeds. Probably, this interacting pathway may also contribute for differential accumulation of calcium in finger millet seed. It can be further validated by establishing the relationship of membrane calcium transport efficiency with EcCIPK–EcCBL interaction as proposed in the present study using comparative genomics and interactomics studies.
Acknowledgements
This work was financially supported by the grant of program mode support for research and development in Agricultural Biotechnology (Grant No. BT/PR7849/AGR/02/374/2006-Part II) and HRD-DBT, Govt. of India. Bioinformatics Distributed Information Sub Centre, G. B. Pant University of Agriculture and Technology, Pantnagar is also duly acknowledged for providing research facilities.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
References
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J. The NAF domain defines a novel protein–protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001;20(5):1051–1063. doi: 10.1093/emboj/20.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O, Kudla J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta. 2004;219(6):915–924. doi: 10.1007/s00425-004-1333-3. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Zhu JK, Hirschi KD. The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem. 2004;279(4):2922–2926. doi: 10.1074/jbc.M309084200. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Jagendorf A, Zhu JK. Understanding and improving salt tolerance in plants. Crop Sci. 2005;45:437–448. doi: 10.2135/cropsci2005.0437. [DOI] [Google Scholar]
- Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 2004;32(suppl 2):W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL molecular graphics system. http://virology.wisc.edu/acp/Classes/DropFolders/Drop660_lectures/2013_660/L01_PyMOL_2013r.pdf
- Fakrudin B, Shashidhar HE, Kulkarni RS, Hittalmani S. Genetic diversity assessment of finger millet, Eleusine coracana (Gaertn), germplasm through RAPD analysis. PGR Newslett. 2004;138:50–54. [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132(2):666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Park SJ, Jang B, Jung CH, et al. Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. 2007;50(3):439–451. doi: 10.1111/j.1365-313X.2007.03057.x. [DOI] [PubMed] [Google Scholar]
- Kumar A, Sharma N, Panwar P, Gupta AK. Use of SSR, RAPD markers and protein profiles based analysis to differentiate Eleusine coracana genotypes differing in their protein content. Mol Biol Rep. 2012;39(4):4949–4960. doi: 10.1007/s11033-011-1291-3. [DOI] [PubMed] [Google Scholar]
- Kumar A, Pathak RK, Gupta SM, Gaur VS, Pandey D. Systems biology for smart crops and agricultural innovation: filling the gaps between genotype and phenotype for complex traits linked with robust agricultural productivity and sustainability. OMICS. 2015;19(10):581–601. doi: 10.1089/omi.2015.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Singh UM, Manohar M, Gaur VS. Calcium transport from source to sink: understanding the mechanism(s) of acquisition, translocation, and accumulation for crop biofortification. Acta Physiol Plant. 2015;37(1):1722. doi: 10.1007/s11738-014-1722-6. [DOI] [Google Scholar]
- Mirza N, Taj G, Arora S, Kumar A. Transcriptional expression analysis of genes involved in regulation of calcium translocation and storage in finger millet (Eleusine coracana L. Gartn.) Gene. 2014;550(2):171–179. doi: 10.1016/j.gene.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath M, Roy P, Shukla A, Kumar A. Spatial distribution and accumulation of calcium in different tissues, developing spikes and seeds of finger millet genotypes. J Plant Nutr. 2013;36(4):539–550. doi: 10.1080/01904167.2012.748072. [DOI] [Google Scholar]
- National Research Council . Lost crops of Africa. Volume 1: grains. Washington: National Academy Press; 1996. [Google Scholar]
- Panwar P, Nath M, Yadav VK, Kumar A. Comparative evaluation of genetic diversity using RAPD, SSR and cytochrome P450 gene based markers with respect to calcium content in finger millet (Eleusine coracana L. Gaertn.) J Genet. 2010;89(2):121–133. doi: 10.1007/s12041-010-0052-8. [DOI] [PubMed] [Google Scholar]
- Pathak RK, Giri P, Taj G, Kumar A. Molecular modeling and docking approach to predict the potential interacting partners involved in various biological processes of MAPK with downstream WRKY transcription factor family in Arabidopsis thaliana. Int J Comput Bioinform In Silico Model. 2013;2:262–268. [Google Scholar]
- Pathak RK, Taj G, Pandey D, Kasana VK, Baunthiyal M, Kumar A. Molecular modeling and docking studies of phytoalexin(s) with pathogenic protein(s) as molecular targets for designing the derivatives with anti-fungal action on Alternaria spp. of Brassica. Plant Omics. 2016;9(3):172. doi: 10.21475/poj.16.09.03.p7654. [DOI] [Google Scholar]
- Quan R, Lin H, Mendosa I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo J, Guo Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007;19:1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen TZ, Jernigan RL, Garnier J, Kloczkowski A. GOR V server for protein secondary structure prediction. Bioinformatics. 2005;21(11):2787–2788. doi: 10.1093/bioinformatics/bti408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Jamra G, Singh UM, Sood S, Kumar A. Calcium biofortification: three pronged molecular approaches for dissecting complex trait of calcium nutrition in finger millet (Eleusine coracana) for devising strategies of enrichment of food crops. Front Plant Sci. 2017;7:2028. doi: 10.3389/fpls.2016.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh UM, Pandey D, Kumar A. Determination of calcium responsiveness towards exogenous application in two genotypes of Eleusine coracana L. differing in their grain calcium content. Acta Physiol Plant. 2014;36(9):2521–2529. doi: 10.1007/s11738-014-1625-6. [DOI] [Google Scholar]
- Singh UM, Chandra M, Shankhdhar SC, Kumar A. Transcriptome wide identification and validation of calcium sensor gene family in the developing spikes of finger millet genotypes for elucidating its role in grain calcium accumulation. PLoS One. 2014;9(8):e103963. doi: 10.1371/journal.pone.0103963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh UM, Metwal M, Singh M, Taj G, Kumar A. Identification and characterization of calcium transporter gene family in finger millet in relation to grain calcium content. Gene. 2015;566(1):37–46. doi: 10.1016/j.gene.2015.04.021. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Batelli G, Grillo S, Agius F, et al. Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol Cell Biol. 2007;27(22):7771–7780. doi: 10.1128/MCB.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas VK, Ukawala RD, Ghate M, Chintha C. Homology modeling a fast tool for drug discovery: current perspectives. Indian J Pharm Sci. 2012;74(1):1. doi: 10.4103/0250-474X.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12(5):1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(suppl 2):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins Struct Funct Bioinf. 2006;64(3):643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]