Abstract
Fluorine is ubiquitous and the most active non-metal element in nature. While many microorganisms have developed fluoride resistance as a result of the widespread and prolonged application of oral hygiene products, the mechanisms used by these organisms to overcome fluoride toxicity are incompletely understood. In this study, a fluoride-resistant strain, Enterobacter cloacae FRM, was identified which could grow well at a fluoride concentration of 4,000 mg/L. According to comparative genomics, transcriptome under fluoride stress, and sequence analyses of two fluoride-resistant fosmid clones, the genomic island GI3 was found to be important for fluoride resistance. The result of quantitative RT-PCR indicated that six genes on GI3, ppaC, uspA, eno, gpmA, crcB, and orf5249, which encode a fluoride transporter, fluoride-inhibited enzymes, and a universal stress protein, reside in an operon and are transcribed into two mRNAs activated by fluoride with a fluoride riboswitch. The results of knockout and complementation experiments indicated that these genes work together to provide high fluoride resistance to E. cloacae FRM. This study clarified the resistance mechanism of this high fluoride-resistant organism and has expanded our understanding of the biological effects of fluoride.
Introduction
Fluoride is ubiquitous in nature and is commonly used as an additive in water and oral hygiene products because of its strong anticaries function1, 2. At high concentration, fluoride exhibits toxicity against bacteria, fungi, plants, and animals. Many microorganisms have developed resistance to fluoride as a result of its widespread and prolonged application3, 4. However, the exact mechanisms used by fluoride-resistant microorganisms to overcome the toxicity are incompletely understood.
Currently, fluoride resistance is thought to be mediated by sensor and mitigation systems, which have been explained previously5. This research group found that the conserved RNA structure identified by bioinformatics in many species of bacteria and archaea6 functions as a fluoride-sensing riboswitch. These fluoride riboswitches regulate the expression of downstream genes to alleviate the deleterious effects of fluoride. Two of the most common genes associated with fluoride riboswitches are crcB and eriC F. The crcB gene of Escherichia coli and the eriC F gene of Pseudomonas syringae function as fluoride transporters5. Additional studies have also implicated the proteins CrcB and EriCF in fluoride resistance. CLCF has been shown to function as a fluoride channel, exporting fluoride to protect E. coli against its toxicity7. Fluoride-resistant Streptococcus mutans, a causative agent of dental caries, which appears to lack a fluoride riboswitch8 encodes two homologs of the EriCF protein. A single-nucleotide mutation was identified in the promoter of these two eriC F genes9 and this mutation was found to confer fluoride resistance on S. mutans by increasing promoter activity and upregulating the expression of downstream fluoride antiporter-coding genes10. Moreover, these eriC F genes were confirmed to confer fluoride resistance in S. mutans 11. It has also been shown that either EriCF or CrcB plays a role in fluoride resistance in oral Streptococci 12. In these studies, the highest resistance to fluoride was observed at 600 mg/L.
Our laboratory has isolated a bacterium, Enterobacter cloacae FRM with very high fluoride resistance. Although environmental levels of fluoride are typically found in the 10–100 μM range, this bacterium can grow at 4,000 mg/L (210 mM) fluoride. Given that such high fluoride resistance is not common among fluoride-resistant microorganisms, we decided to investigate the mechanism of resistance. To this aim, the genome of the strain was sequenced, the transcriptome under fluoride conditions was analyzed by RNA sequencing, and a fosmid library was constructed to survey fluoride resistance-related genes. The expression characteristics of these genes were analyzed, and their contributions to fluoride resistance were confirmed with a series of knockout and complementation experiments. An operon containing six genes (ppaC, uspA, eno, gpmA, crcB, and orf5249) in the genomic island GI3 was identified and shown to be important for the survival of E. cloacae FRM in high concentration of fluoride.
Results
E. cloacae FRM exhibits high fluoride resistance
A fluoride-tolerant bacterial strain was isolated in our laboratory. 16S rDNA sequence analysis revealed that this strain belongs to the genus Enterobacter, showing the closest similarity to the Enterobacter strains. Based on nucleotide homology, this strain was assigned as Enterobacter cloacae FRM (GenBank Accession Number: CP019889, CP019890, and CP019891), with 100% identity with its nearest homolog species, E. cloacae ECNIH2 (GenBank Accession Number: CP008823).
To determine the level of fluoride resistance of E. cloacae FRM, we examined the growth of E. cloacae FRM, as well as other seven E. cloacae strains (E. cloacae ACCC 10453, ACCC 11690, ACCC0 1620, ACCC 11688, ATCC 7256, CMCC 45301, and NCTC 9394) in liquid medium with various fluoride concentrations. E. cloacae FRM could be grown in the presence of fluoride at concentrations up to 4,000 mg/L (Fig. 1), while the growth of the seven other E. cloacae strains was inhibited in the presence of 1,500 mg/L fluoride (Fig. 1). These data indicated that E. cloacae FRM exhibited greater fluoride resistance than many other E. cloacae strains.
Figure 1.
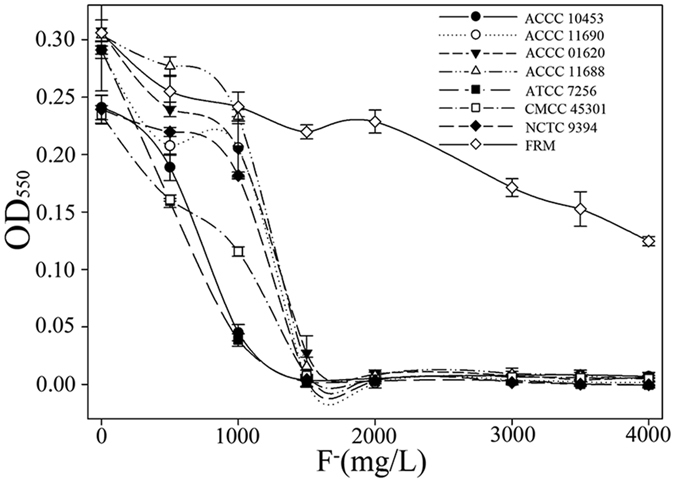
Growth of Enterobacter cloacae strains (FRM, ACCC 10453, ACCC 11690, ACCC0 1620, ACCC 11688, ATCC 7256, CMCC 45301, and NCTC 9394) at varying concentrations of fluoride.
Genomic analysis of E. cloacae FRM
To elucidate the mechanism of fluoride resistance of E. cloacae FRM, the genome of E. cloacae FRM was sequenced and found to consist of a single chromosome (4,899,400 bp and 4,661 genes) and two plasmids, designated as p1 (124,734 bp and 160 genes) and p2 (112,186 bp and 135 genes). Three genomic islands were also identified within the chromosome with IslandViewer 3 (GI1, GI2, and GI3) (Fig. S1).
Ortholog analysis of the genome of E. cloacae FRM and the seven other Enterobacteriaceae strains identified 527 orthologous genes with the RSD algorithm. Based on these genes, a phylogenetic tree was constructed with Phylip software (Fig. S2), which revealed that the closest relative of E. cloacae FRM is E. cloacae NCTC 9394. Comparative analysis of the genome sequences of E. cloacae FRM and E. cloacae NCTC 9394 indicated that the genome of E. cloacae NCTC 9394 did not contain the three genomic islands or the plasmids. As the fluoride resistance of E. cloacae NCTC 9394 was very low (Fig. 1), we hypothesized that the genes involved in fluoride resistance may be located on the genomic islands or plasmids.
The third genomic island, GI3, is important for the fluoride resistance of E. cloacae FRM
To determine whether the genomic islands or plasmids contribute to the fluoride resistance of E. cloacae FRM, we eliminated the three genomic islands and the p1 plasmid. The resulting strains were verified by PCR using specific primers (Fig. S3) and their fluoride resistance was examined (Fig. 2). E. cloacae FRM-Δp1, which has lost the p1 plasmid, grew well on Bulk plates with and without fluoride, similar to the wild-type E. cloacae FRM strain, whereas E. cloacae FRM-Δp1ΔIs123, which has lost the p1 plasmid and the three genomic islands, could only be grown on Bulk plates without fluoride, similar to the E. coli control strain (Fig. 2a). In liquid media, the MIC of fluoride for both E. cloacae FRM-Δp1ΔIs123 and E. coli was 1,500 mg/L, while E. cloacae FRM-Δp1 grew well even at 4,000 mg/L fluoride, similar to E. cloacae FRM (Fig. 2b). These data indicated that the elimination of plasmid p1 does not influence the fluoride resistance of E. cloacae FRM. However, fluoride resistance was dramatically reduced when the three genomic islands were eliminated. Thus, we concluded that the genomic islands, rather than the p1 plasmid, contribute to the fluoride resistance of E. cloacae FRM.
Figure 2.
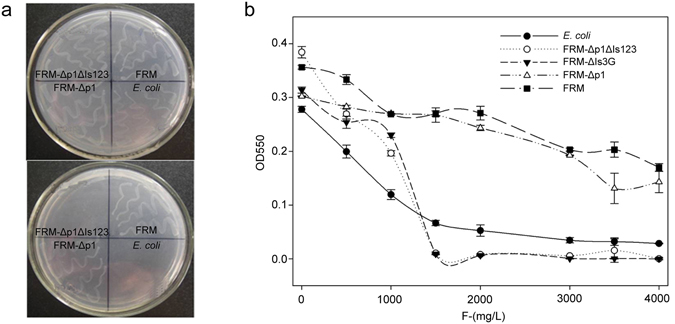
Examination of the fluoride resistance of E. cloacae FRM-Δp1ΔIs123 (elimination of plasmid p1 and the three genomic islands) and E. cloacae FRM-Δp1 (elimination of plasmid p1). (a) Four strains were streaked onto Burk plates without fluoride (upper) and with 1,000 mg/L fluoride (lower). (b) The minimum inhibitory concentration (MIC) of fluoride for E. cloacae FRM, E. cloacae FRM-Δp1ΔIs123, E. cloacae FRM-Δp1, and E. cloacae FRM-ΔIs3G.
The transcriptome of E. cloacae FRM following fluoride ion induction was analyzed by RNA sequencing. Reads per kilobase per million reads values were used to measure transcript abundance. As shown in Table 1, the expression level of some of the genes found on the third genomic island was greatly influenced by the presence of fluoride. For example, fluoride induction increased the expression of crcB (orf5251) 7-fold. The transcripts of the enolase gene eno (orf5255) and the universal stress protein gene uspA (orf5256) were increased 176- and 120-fold, respectively.
Table 1.
Transcriptome annotation of the Enterobacter cloacae FRM genomic island GI3.
| Gene | location | RPKM | Fold change | Annotation | ||
|---|---|---|---|---|---|---|
| Start | End | untreated | induced | |||
| orf05247 | 4068647 | 4069594 | 25.05 | 23.67 | 0.9 | Efflux transporter, RND family, MFP subunit |
| orf05248 | 4069591 | 4072650 | 18.57 | 25.7 | 1.4 | RND efflux transporter |
| orf05249 | 4072819 | 4073079 | 7.47 | 115.91 | 15.5 | Divalent cation transporting ATPase |
| orf05251 | 4073189 | 4073449 | 20.47 | 166.01 | 8.1 | Protein CrcB homolog |
| orf05252 | 4073598 | 4074080 | 28.62 | 281.65 | 9.8 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase |
| orf05255 | 4074611 | 4075897 | 19.97 | 3533.52 | 176.9 | Enolase |
| orf05256 | 4075913 | 4076341 | 15.02 | 1814.37 | 120.8 | Universal stress protein A |
| orf05257 | 4076345 | 4077307 | 16.38 | 2721.34 | 166.1 | Manganese-dependent inorganic pyrophosphatase |
| orf05258 | 4077805 | 4078017 | 156.12 | 280.39 | 1.8 | Putative uncharacterized protein |
| orf05259 | 4078317 | 4078484 | 72.2 | 116.17 | 1.6 | hypothetical protein |
We also constructed a fosmid library from genomic DNA of E. cloacae FRM in E. coli and screened the clones with fluoride resistance to determine the genes that contribute to fluoride resistance. Among the total of 276 clones, only two clones, F1 and F2, could be grown on Bulk plates with fluoride at a concentration of 700 mg/L. The fosmid DNA isolated from F1 and F2 was sequenced. Sequence analyses of the two fosmids revealed that both contained the third genomic island, GI3.
For further confirmation of the importance of GI3, the 4,489-bp GI3 fragment (designated as Is3G), which contains six genes (from orf5249 to orf5257, Fig. 3a) whose expression was increased by fluoride induction, was deleted and the fluoride resistance of this knockout strain, E. cloacae FRM-ΔIs3G, was evaluated. The fluoride MIC for E. cloacae FRM-ΔIs3G was 1,500 mg/L, the same as E. cloacae FRM-Δp1ΔIs123 and E. coli (Fig. 2b). Deletion of the Is3G fragment of E. cloacae FRM resulted in a loss of fluoride resistance. Our results suggested that the genomic island GI3, in particular the 4,489-bp Is3G fragment, is important for the fluoride resistance of E. cloacae FRM.
Figure 3.
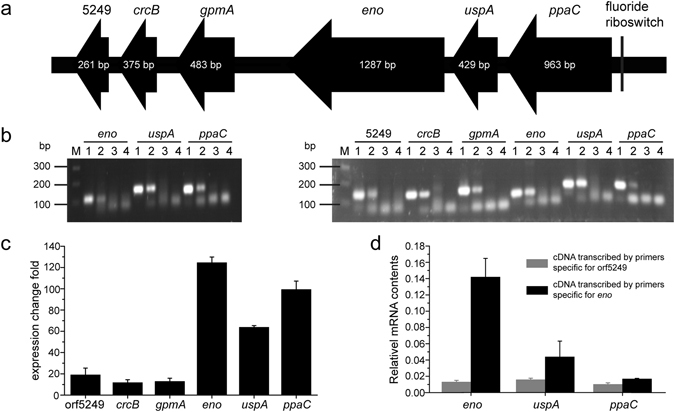
Genetic organization and expression of the Is3G fragment. (a) Schematic representation of the Is3G fragment loci. Thick black line, chromosomal DNA; filled arrows, six open reading frames with corresponding gene size; dotted lines, complementary DNA (cDNA) transcribed with gene-specific primers for orf5249 and eno in the 3′ direction. (b) Analyses of reverse transcription (RT)-PCR products (left, cDNA transcribed with gene-specific primers for orf5249; right, cDNA transcribed with gene-specific primers for eno). Lanes: M, 100-base-pair (bp) DNA ladder; 1, PCR positive control using E. cloacae FRM genomic DNA as a template; 2, RT-PCR using E. cloacae FRM cDNA as a template; 3, negative control using E. cloacae FRM RNA as a template; 4, PCR negative control without template. (c) Transcriptional analysis of the genes orf5249, crcB, gpmA, eno, uspA, and ppaC in E. cloacae FRM cultured with 1,000 mg/L fluoride in comparison with culture in the absence of fluoride using quantitative RT-PCR. Transcript levels of the tested genes were normalized to the 16 S rRNA gene. (d) The relative transcription levels of the genes eno, uspA, and ppaC. The 16 S rRNA gene was used as an internal control for normalization.
The six Is3G genes are transcribed together and controlled by a fluoride riboswitch
To clarify the function of the indispensable Is3G fragment in fluoride resistance, the genetic composition and function of the genes located on this fragment were analyzed. We identified six genes on the Is3G fragment. The chromosomal organization of the six genes is shown in Fig. 3a and detailed annotation information is shown in Table 1. Some of the genes may be involved in fluoride resistance, such as crcB, which has been reported to encode a fluoride transporter protein that reduces the concentration of fluoride in cells5, and the gene uspA, which encodes the universal stress protein A that may be produced in response to a variety of stress conditions13. Two other gene products, enolase and pyrophosphatase, are essential for glycolytic metabolism and nucleic acid synthesis. These enzymes are inhibited by fluoride14, 15.
We scanned the Is3G fragment against the Rfam database16 to search for homologs of known RNAs and found a fluoride riboswitch (Rfam accession number RF01734) upstream of the gene ppaC (Fig. 3a), which regulates the expression of downstream genes5. This suggested that the expression of these six genes may be regulated by the riboswitch. To confirm this, we analyzed the expression of these genes. From the analysis of transcriptome, we found that the mRNA levels of the six genes were low in total RNA isolated from E. cloacae FRM cultured in the absence of fluoride. However, the mRNA levels were significantly increased in total RNA isolated from E. cloacae FRM cultured in the presence of fluoride. Fluoride induction increased expression of the genes ppaC, uspA, and eno by approximately 100-fold, whereas the expression of gpmA, crcB, and orf5249 was increased approximately 10-fold (Table 1). These results were confirmed by qRT-PCR (Fig. 3c). Given the large differences in expression induced by fluoride between the upstream and downstream genes, we hypothesized that ppaC, uspA, and eno are transcribed as one operon and gpmA, crcB, and orf5249 are transcribed as another operon. To confirm this, RT-PCR using total RNA extracted from E. cloacae FRM cultured in Burk medium with 1,000 mg/L fluoride was performed. The RT-PCR results, using an eno-specific antisense primer for reverse transcription, showed that ppaC, uspA, and eno are indeed co-transcribed (Fig. 3b, left). However, the genes ppaC, uspA, and eno were also detected in cDNA transcribed with an orf5249-specific antisense primer (Fig. 3b, right). This result suggested that the three downstream genes (gpmA, crcB, and orf5249) are also transcribed together with the three upstream genes (ppaC, uspA, and eno). However, the mRNA expression of the three upstream genes was greater than the expression of the downstream genes, which suggested that the mRNA of the upstream genes may not only be located on a single transcript. To verify this, qRT-PCR using cDNA transcribed with primers specific for orf5249 and the eno gene as the template was performed to compare the relative mRNA levels of the three upstream genes. The data showed that the levels of the three upstream genes in the cDNA transcribed with primers specific for the eno gene were greater than in the cDNA transcribed with primers specific for orf5249 (Fig. 3d). This suggested that there are two transcripts: one, which contains all six genes, and a second, which only contains the three upstream genes. Overall, these results indicated that the six consecutive genes reside in an operon and are transcribed into two mRNAs that are activated by fluoride through a fluoride riboswitch.
An operon consisting of six genes confers high fluoride resistance
To determine the genes responsible for high fluoride resistance, we knocked out the genes in this operon to examine their contributions to fluoride resistance. As the crcB gene has been reported to be involved in fluoride resistance5 and the uspA gene is involved in physiological processes under conditions of stress, we first knocked out the crcB and uspA genes to determine their roles in E. cloacae FRM fluoride resistance. The knockout strains were verified by PCR (Fig. S4a and b). Deleting crcB and uspA did not cause a complete loss of fluoride resistance in E. cloacae FRM, as the deletion of uspA (E. cloacae FRM-ΔuspA), crcB (E. cloacae FRM-ΔcrcB), and uspA and crcB together (E. cloacae FRM-ΔCU) still permitted growth on Bulk plates with 1,000 mg/L fluoride (Fig. 4a). The MIC for fluoride for all of these strains was 3,000 mg/L (Fig. 4c). These results suggested that there are additional genes involved in fluoride resistance.
Figure 4.
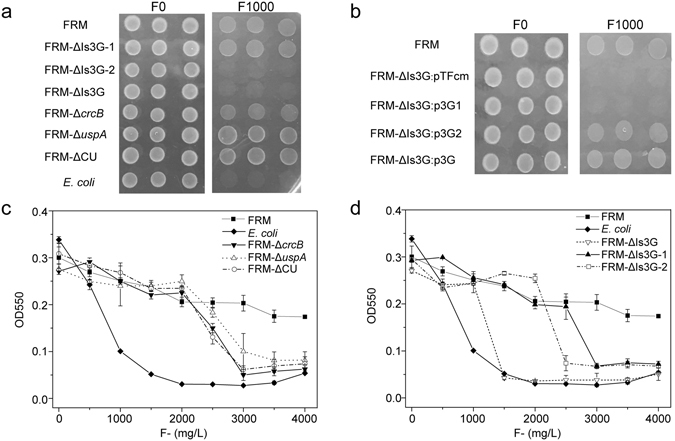
Examination of the fluoride resistance of E. cloacae FRM and knockout strains. (a) Eight strains were spotted onto Burk plates without fluoride (left) and with 1,000 mg/L fluoride (right). E. cloacae FRM-ΔIs3G: deletion of the Is3G fragment; E. cloacae FRM-ΔIs3G-1: deletion of orf5249, crcB, and gpmA; E. cloacae FRM-ΔIs3G-2: deletion of eno, uspA, and ppaC; E. cloacae FRM-ΔcrcB: deletion of crcB; E. cloacae FRM-ΔuspA: deletion of uspA; E. cloacae FRM-ΔCU: deletion of crcB and uspA. (b) Rescue of fluoride toxicity by the fragments Is3G, Is3G-1, and Is3G-2 in E. cloacae FRM-ΔIs3G. (c) The MIC of fluoride for E. cloacae FRM, E. cloacae FRM-ΔcrcB, E. cloacae FRM-ΔuspA, E. cloacae FRM-ΔCU, and E. coli. (d) The MIC of fluoride for E. cloacae FRM, E. cloacae FRM-ΔIs3G, E. cloacae FRM-ΔIs3G-1, E. cloacae FRM-ΔIs3G-2, and E. coli.
Next, we deleted the fragment Is3G-1, which contains the genes orf5249, crcB, and gpmA, and the fragment Is3G-2, which contains the genes eno, uspA and ppaC, to make the knockout strains E. cloacae FRM-ΔIs3G-1 and E. cloacae FRM-ΔIs3G-2 respectively. These strains were verified by PCR (Fig. S4c) and their fluoride resistance was evaluated. Only E. cloacae FRM-ΔIs3G-2 and E. cloacae FRM-ΔIs3G could not be grown on Bulk plates with 1,000 mg/L fluoride (Fig. 4a). In liquid medium, only E. cloacae FRM-ΔIs3G showed weak fluoride resistance, similar to E. coli. The MIC for E. cloacae FRM-ΔIs3G-2 was 2,500 mg/L, lower than the MIC for E. cloacae FRM-ΔIs3G-1, ΔuspA, ΔcrcB, and ΔCU (Fig. 4c and d). These results indicated that although deletion of these genes may influence fluoride resistance, only deletion of the whole Is3G fragment eliminates the high fluoride resistance of E. cloacae FRM.
To confirm the above results, we evaluated the recovery of fluoride resistance of the Is3G fragment. We transformed the plasmids containing the fragments Is3G, Is3G-1, and Is3G-2 into E. cloacae FRM-ΔIs3G to generate E. cloacae FRM-ΔIs3G:p3G, E. cloacae FRM-ΔIs3G:p3G1, and E. cloacae FRM-ΔIs3G:p3G2 respectively, and examined the fluoride resistance of these strains. As shown in Fig. 4b, the fragments Is3G and Is3G-2 allowed E. cloacae FRM-ΔIs3G to grow on Burk plates with 1,000 mg/L fluoride, similar to the wild-type E. cloacae FRM strain. Overall, these results indicated that all six genes found in the operon contribute to the high fluoride resistance in E. cloacae FRM.
Discussion
Our research showed that an operon consisting of six genes is involved in the fluoride resistance of E. cloacae FRM. However, many microorganisms have been reported to use only fluoride transporters, such as the CrcB protein and a fluoride-specific subtype of chloride channels EriCF, to overcome fluoride toxicity5, 7, 10–12. These fluoride-resistant microorganisms do not survive extended exposure to 200 mM fluoride, indicating that the expression of fluoride channels alone is not sufficient to overcome this high concentration of fluoride. This was confirmed by our data: deletion of the crcB gene did not destroy the fluoride resistance of E. cloacae FRM. As E. cloacae FRM can tolerate a concentration of fluoride as high as 4,000 mg/L, and there are few microorganisms that can survive such a high concentration of fluoride, it was not surprising to find that E. cloacae FRM uses six different genes, including crcB, to overcome the toxicity of high fluoride levels.
We determined that the fluoride resistance-related operon was fluoride-induced and controlled by a fluoride riboswitch located on the promoter. The fluoride riboswitch and associated proteins have been reported as mitigation systems for fluoride toxicity that exist pervasively among bacteria and archaea5. The genes associated with the fluoride riboswitch have been analyzed by bioinformatics and found to encode proteins of diverse function5, 6. Our research provides the first experimental evidence linking additional fluoride riboswitch-associated genes to fluoride resistance. According to the function of these genes and their indispensable role in fluoride resistance, the mechanism of fluoride resistance for E. cloacae FRM could be clarified. Fluoride is detected by this riboswitch, which triggers the expression of the six genes that can help E. cloacae FRM to mitigate fluoride toxicity by expelling the anion from cells using the fluoride transporter (CrcB), by utilizing UspA and its role in universal stress adaptation, and by producing additional copies of enzymes (phosphoglycerate mutase, enolase, and pyrophosphatase) that are inhibited by fluoride.
The six genes of the fluoride resistance-related operon were transcribed into two separate transcripts. In addition to the transcript containing the six genes, we also identified a shorter transcript that contained only the three upstream genes, ppaC, uspA, and eno. This expression pattern revealed the precise effects that fluoride has on E. cloacae FRM. Additional transcripts of ppaC and eno suggested that fluoride may primarily influence glycolytic metabolism and nucleic acid synthesis by inhibited the key enzymes, enolase and pyrophosphatase, encoded by the eno and ppaC genes14, 15. The enzyme phosphoglycerate mutase, which is encoded by the pgmA gene, is also involved in glycolytic metabolism, but whether it is inhibited by fluoride has not yet been reported. Our data suggested that this enzyme may also be inhibited by fluoride, but this role may not be as important as the role of enolase in glycolytic metabolism.
Two transposases, orf5235 and orf5264, were also identified at the genomic island GI3, which are located upstream and downstream of the fluoride resistance operon, respectively. The identification of these transposases indicated that this unit of fluoride resistance was acquired by horizontal gene transfer (HGT)17. Thus, other bacteria could receive this genetic material through the same process. HGT may contribute to the spread of this fluoride resistance operon through the exchange of genetic material across genera, which will likely increase the number of fluoride-resistant bacteria.
Overall, our identification of an operon involved in fluoride resistance in E. cloacae FRM clarified the resistance mechanism and has expanded our understanding of the biological effects of fluoride.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S1. E. cloacae Agricultural Culture Collection of China (ACCC) 10453, E. cloacae ACCC 11690, E. cloacae ACCC 01620, and E. cloacae ACCC 11688 were purchased from the ACCC. E. cloacae ATCC 7256, E. cloacae CMCC 45301, and E. cloacae NCTC 9394 were purchased from China General Microbiological Culture Collection Center (CGMCC). E. coli Top 10 (TIANGEN Biotech, Beijing, China) was used as a cloning host. E. coli or E. cloacae were grown in lysogeny broth (LB) or modified Burk mineral medium [0.8 g/L KH2PO4, 0.2 g/L K2HPO4, 1 g/L (NH4)2SO4, 0.2 g/L MgSO4·7H2O, and 1 g/L yeast extract (Difco)] at 37 °C. NaF was used as fluoride. Ampicillin (100 mg/L), kanamycin (50 mg/L), or chloramphenicol (25 mg/L) was added as necessary.
Evaluation of fluoride resistance
To evaluate the growth on plates with 1,000 mg/L fluoride, an overnight culture suspension was spotted or streaked on a plate and incubated at 37 °C for 24 h. To evaluate the growth in liquid medium, the minimum inhibitory concentration (MIC) of fluoride was determined. Burk broth (800 μL) with different concentrations of fluoride was dispensed into a 96-well (12 × 8) microtiter plate (96 × 1.5-mL wells) with a multi-channel micropipette (rows A to H: 0 mg/L, 500 mg/L, 1,000 mg/L, 1,500 mg/L, 2,000 mg/L, 3,000 mg/L, 3,500 mg/L, and 4,000 mg/L). Single colonies of the test strains were inoculated into 3 mL of LB broth and cultured overnight. A sample of this suspension (1%) was inoculated in 50 mL of LB broth and grown at 37 °C in an incubator at 200 rpm for 6 h. The test culture (15 μL) was then inoculated into each well of the prepared 96-well plate. After 20 h at 37 °C and 200 rpm, 200 μL of the cell suspension was transferred into a 96-well plate and the turbidity at OD550 was measured.
Genome sequencing and analysis
Two Illumina libraries [0.5 kilobases (kb) and 3 kb] and Roche 454 paired-end (8 kb insert) whole shotgun libraries were constructed for E. cloacae FRM. Short reads generated from the Illumina paired-end library were assembled using Velvet 1.118. Contigs were then joined into scaffolds with Illumina meta-pair reads and 454 paired-end reads. The gaps between the contigs were closed by PCR amplification and primer walking with an ABI 3730 sequencer. The transfer RNA genes were predicted with tRNAscan-SE19. The ribosomal RNA (rRNA) genes were identified with a basic local alignment search tool (BLAST) search against Rfam16 and rRNA gene sequences from TAC125. The open reading frames were identified with GLIMMER 3.020. The predicted open reading frames were annotated by similarity searches against databases of nonredundant protein sequences from the National Center for Biotechnology Information, clusters of orthologous groups of proteins21, and InterPro22. The annotation of open reading frames was manually curated with Artemis23. The island in the genome of E. cloacae FRM was determined with the program IslandViewer 324.
Comparative genomics and phylogenetic analysis
The following genome sequences were used in genome comparisons with E. cloacae FRM: E. cloacae NCTC 9394 (accession number FP929040), E. cloacae SCF1 (accession number CP002272), E. cloacae ATCC 13047 (accession numbers CP001918, CP001919, and CP001920), E. asburiae LF7a (accession numbers CP003026, CP003027, and CP003028), E. sp. 638 (accession numbers CP000653 and CP000654), E. aerogenes KCTC 2190 (accession number CP002824), Escherichia fergusonii ATCC 35469 (accession number CU928158 CU928144), and Escherichia coli str. K–12 substr. MG1655 (accession number U00096). The sets of orthologous protein-coding genes were defined as mutual fully transitive reciprocal protein BLAST25 hits (with E-value < 10−4)26. Co-ortholog groups were identified by the reciprocal smallest distance (RSD)27. The nucleic acid sequence of each ortholog group was aligned using the CLUSTALW program, version 1.8328. The phylogenetic tree of the orthologous genes was constructed as a neighbor-joining (NJ) tree29 with the NEIGHBOR program from the PHYLIP package 3.6730. Bootstrap values were constructed using the CONSENSE program30 from 100 reproduced trees. The CONSENSE program was used to obtain the bootstrap values and generate the final phylogenetic tree.
RNA sequencing and transcriptome analysis
An overnight culture of E. cloacae FRM was diluted 1:1,000 in Bulk medium in the presence (1,000 mg/L) and absence of fluoride, and the cultures were grown at 37 °C and 200 rpm to exponential phase (OD550 of approximately 0.6). Total RNA from both groups was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Isolated RNA samples were used for first-strand complementary DNA (cDNA) synthesis using random hexamers and Superscript II reverse transcriptase. Following end repair and the addition of a 3′-dA overhang, the cDNA was ligated to an Illumina paired-end adapter oligonucleotide mix and was size-selected for enrichment for 200-base-pair (bp) fragments by gel purification. After 16 PCR cycles, the libraries were sequenced using Illumina GAIIx. TopHat was used to map mRNA reads to the genome, and Cufflinks was used to calculate the expected fragments per kilobase of transcript per million mapped reads as expression values for each transcript.
Elimination of plasmids and genetic islands
Plasmid and genetic island elimination was performed using sodium dodecyl sulfate (SDS, 0.6%) and a high temperature (42 °C) as previously described31. Single colonies were selected for on Burk plates without or with 1,000 mg/L fluoride and 100 μg/mL ampicillin to screen for the p1 plasmid and genetic island-eliminated strains. Primers p1-F and p1-R (Table S2) were used to identify the ampicillin-resistant gene located on the p1 plasmid. The primer pairs Is1-F and Is1-R, Is2-F and Is2-R, and Is3-F and Is3-R (Table S2) were used to detect the three genetic islands (Fig. S3).
Construction and screening of an E. cloacae FRM genomic fosmid library
High-molecular-weight genomic DNA from E. cloacae FRM was extracted by the cetyl trimethylammonium bromide method32, and used to construct a fosmid library with the CopyControl Fosmid Library Production kit (vector pCC1FOS; Epicentre, Madison, WI, USA), according to the manufacturer’s instructions. A total of 276 clones were used to construct the library in three 96-well plates. To screen for clones with fluoride resistance, overnight cultures of the 276 clones were spotted onto Burk plates with 700 mg/L fluoride. fosmid DNA was isolated from the positive fosmid clones by alkaline lysis33 and sequenced from both ends on an ABI 3730 DNA analyzer with the pCC1 sequencing primers pCC1-F and pCC1-R (Table S2). The results were aligned with the genome sequence of E. cloacae FRM to confirm the sequence of the inserted fragments.
Reverse transcription (RT)-polymerase chain reaction (PCR) and quantitative RT-PCR (qRT-PCR)
Total RNA was isolated as mentioned above in RNA sequencing. cDNA samples were prepared using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) with gene-specific primers. An RT reaction using the primers 5249-RT and eno-RT (Table S2) as orf5249 and eno-specific antisense primers was performed, and the resulting cDNA was used as a template in PCR reactions for the detection of six genes with the following primers: 5249-F and 5249-R, crcB-F and crcB-R, gpmA-F and gpmA-R, eno-F and eno-R, uspA-F and uspA-R, and ppaC-F and ppaC-R (Table S2). qRT-PCR was performed using the SYBR Green Real-time PCR Master Mix Plus (Toyobo, Osaka, Japan) according to the manufacture’s instructions. 16 S rRNA (amplified with the primers 16SF and 16SR) was used as an internal control. All reactions were performed in biological triplicate, and the normalized fold changes of the relative expression ratio were quantified by the 2−ΔΔCT method34.
Construction of E. cloacae FRM knockout mutants
E. cloacae FRM knockout mutants were constructed by the CRISPR-Cas9 method, using the plasmids pCas (Addgene plasmid #62225) and pTargetF (Addgene plasmid #62226) as previously described35. Briefly, the chloramphenicol resistance (Cmr) gene (cat) amplified from pB1H1 with the primers CM-F and CM-R (Table S2) was inserted into the XhoI/MluI-digested pTargetF to replace the spectinamycin resistance gene (aadA), generating the plasmid pTFcm. To knockout the genes of interest, upstream and downstream fragments of each gene were amplified separately from E. cloacae FRM genomic DNA with the corresponding primers UF/UR and DF/DR (Table S2), and single guide RNA (sgRNA) containing a targeting N20 sequence of the loci of interest was amplified from pTFcm using corresponding SPF primers with the common reverse primer sgRNA-R. These three fragments were combined using an overlap PCR with SPF and DR primers, digested with SpeI/SalI, and inserted into the SpeI/SalI-digested pTFcm to construct the knockout plasmids (Table S1). These plasmids were then transformed into E. cloacae FRM harboring pCas and the deletion strains were screened as previously described35. Gene knockouts were verified by colony PCR with primers which were positioned ~600 bp upstream and downstream of the target genes (Fig. S4).
Complementation
The fragments Is3G, Is3G-1, and Is3G-2 were PCR-amplified from E. cloacae FRM genomic DNA with the primer pairs 5249 L and 5257 R, 5249 L and 5252 R, and 5253 L and 5257 R (Table S2), and digested with HindIII and SacI. These fragments were inserted into the HindIII/SacI-digested pTFcm to create p3G, p3G1, and p3G2 (Table S1). The plasmids were verified by DNA sequencing and transformed into E. cloacae FRM-ΔIs3G.
Data availability
The E. cloacae FRM chromosome and plasmid p1 and p2 sequences have been deposited in GenBank under accession numbers CP019889, CP019890, and CP019891.
Electronic supplementary material
Acknowledgements
We are grateful to Prof. Sheng Yang for the plasmids pCas and pTargetF. This work was supported by the National High Technology Research and Development Program of China (863 Program, 2013AA102804).
Author Contributions
X.L., J.T., N.W., and Y.F. conceived and supervised the study. X.L., J.T., and N.W. designed research and drafted the manuscript. X.L., J.T., L.L., T.Z., and X.Y. performed experiments and analyzed data. X.L., J.T., X.C. and N.W. provided technical assistance. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Xiaoqing Liu and Jian Tian contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06988-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bin Yao, Email: yaobin@caas.cn.
Ningfeng Wu, Email: wuningfeng@caas.cn.
References
- 1.Walsh T, et al. Fluoride toothpastes of different concentrations for preventing dental caries in children and adolescents. The Cochrane database of systematic reviews, CD007868. 2010 doi: 10.1002/14651858.CD007868.pub2. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton, I. R. Biochemical effects of fluoride on oral bacteria. Journal of dental research69 Spec No., 660–667, discussion 682–663 (1990). [DOI] [PubMed]
- 3.Streckfuss JL, et al. Fluoride resistance and adherence of selected strains of Streptococcus mutans to smooth surfaces after exposure to fluoride. Journal of dental research. 1980;59:151–158. doi: 10.1177/00220345800590021501. [DOI] [PubMed] [Google Scholar]
- 4.Van Loveren C, Spitz LM, Buijs JF, Ten Cate JM, Eisenberg AD. In vitro demineralization of enamel by F-sensitive and F-resistant mutans streptococci in the presence of 0, 0.05, or 0.5 mmol/L NaF. Journal of dental research. 1991;70:1491–1496. doi: 10.1177/00220345910700120401. [DOI] [PubMed] [Google Scholar]
- 5.Baker JL, et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberg Z, et al. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biology. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stockbridge RB, et al. Fluoride resistance and transport by riboswitch-controlled CLC antiporters. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15289–15294. doi: 10.1073/pnas.1210896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breaker RR. New insight on the response of bacteria to fluoride. Caries research. 2012;46:78–81. doi: 10.1159/000336397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao Y, et al. Identification and functional analysis of genome mutations in a fluoride-resistant Streptococcus mutans strain. PLoS One. 2015;10:e0122630. doi: 10.1371/journal.pone.0122630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao Y, et al. A Single nucleotide change in the promoter mutp enhances fluoride resistance of Streptococcus mutans. Antimicrob Agents Chemother. 2016;60:7509–7512. doi: 10.1128/AAC.01366-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murata, T. & Hanada, N. Contribution of chloride channel permease to fluoride resistance in Streptococcus mutans. FEMS Microbiol Lett363, 10.1093/femsle/fnw101 (2016). [DOI] [PubMed]
- 12.Men X, Shibata Y, Takeshita T, Yamashita Y. Identification of anion channels responsible for fluoride resistance in oral Streptococci. PLoS One. 2016;11:e0165900. doi: 10.1371/journal.pone.0165900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kvint K, Nachin L, Diez A, Nyström T. The bacterial universal stress protein: function and regulation. Current opinion in microbiology. 2003;6:140–145. doi: 10.1016/S1369-5274(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 14.Samygina VR, et al. Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies. Journal of molecular biology. 2007;366:1305–1317. doi: 10.1016/j.jmb.2006.11.082. [DOI] [PubMed] [Google Scholar]
- 15.Marquis RE, Clock SA, Mota-Meira M. Fluoride and organic weak acids as modulators of microbial physiology. Fems Microbiol. Rev. 2003;26:493–510. doi: 10.1111/j.1574-6976.2003.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. Fems Microbiol. Rev. 2011;35:957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- 18.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome research. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatusov RL, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apweiler R, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 24.Dhillon BK, et al. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 2015;43:W104–108. doi: 10.1093/nar/gkv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhaxybayeva O, Gogarten JP. Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics. 2002;3:4. doi: 10.1186/1471-2164-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- 28.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein J. Mathematics vs. Evolution: Mathematical Evolutionary Theory. Science. 1989;246:941–942. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- 31.Zhao H, et al. Global transcriptional and phenotypic analyses of Escherichia coli O157:H7 strain Xuzhou21 and its pO157_Sal cured mutant. PLoS One. 2013;8:e65466. doi: 10.1371/journal.pone.0065466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, K. Preparation of genomic DNA from bacteria. Current protocols in molecular biology, 2.4. 1–2.4. 5 (1987). [DOI] [PubMed]
- 33.Bimboim H, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic acids research. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Jiang Y, et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The E. cloacae FRM chromosome and plasmid p1 and p2 sequences have been deposited in GenBank under accession numbers CP019889, CP019890, and CP019891.


