Abstract
Breast cancer encompasses a group of heterogeneous diseases, each associated with distinct clinical implications. Dozens of molecular biomarkers capable of categorizing tumors into clinically relevant subgroups have been proposed which, though considerably contribute in precision medicine, complicate our understandings toward breast cancer subtyping and its clinical translation. To decipher the networking of markers with diagnostic roles on breast carcinomas, we constructed the diagnostic networks by incorporating 6 publically available gene expression datasets with protein interaction data retrieved from BioGRID on previously identified 1015 genes with breast cancer subtyping roles. The Greedy algorithm and mutual information were used to construct the integrated diagnostic network, resulting in 37 genes enclosing 43 interactions. Four genes, FAM134B, KIF2C, ALCAM, KIF1A, were identified having comparable subtyping efficacies with the initial 1015 genes evaluated by hierarchical clustering and cross validations that deploy support vector machine and k nearest neighbor algorithms. Pathway, Gene Ontology, and proliferation marker enrichment analyses collectively suggest 5 primary cancer hallmarks driving breast cancer differentiation, with those contributing to uncontrolled proliferation being the most prominent. Our results propose a 37-gene integrated diagnostic network implicating 5 cancer hallmarks that drives breast cancer heterogeneity and, in particular, a 4-gene panel with clinical diagnostic translation potential.
Introduction
Despite the considerable contributions of traditional diagnostic and treatment modalities made in the battle against breast cancer, it still remains as the leading cause of women death worldwide1, 2. Though, if diagnosed early and treated appropriately, breast cancer patients have relatively better outcomes than many other types of malignancies, it is difficult to reach accurate diagnosis and optimal therapeutic design given distinct patients’ morphological features and treatment responses3–7. Canonically, breast carcinomas are grouped as luminal (luminal A and B), HER2 positive, and triple negative subtypes based on the status of estrogen receptor (ER), progesterone receptor (PR) and epidermal growth factor receptor 2 (HER2). While luminal tumors respond well to the hormonal therapy Tamoxifen8, and HER2 positive cancers could be properly treated with Herceptin9, triple negative breast cancers do not actively react to any available targeted modalities without severe adverse effects due to, primarily, lack of the three primary surface receptors10–13.
The diverse clinical consequences of breast cancer patients have led to a surge in the exploration of novel biomarkers and subtyping strategies of this complicated disease7. For example, androgen receptor (AR) was used, instead of HER2, to classify ER-PR- breast cancers into ER−PR−AR+, ER−PR−AR− subclasses with distinct clinical features14. The additional use of proliferation markers KI6715 and/or TOP2A16 with the conventional diagnostic modality has led to improved accuracy of identifying luminal A from B tumors. Lots of efforts have been devoted to sub-classify the triple negative group (TNG). While some studies use cytokeratins such as CK5/617–23, CK1422, CK1722, CK8/1819 to differentiate the basal subtype from the rest TNG tumors, some find EGFR17–20, 22, vimentin19, P-cadherin21 or TP6321 effective for this purpose. With the diverse biomarkers identified, the number of breast cancer subtypes varies considerably among studies24. These though contribute in deciphering breast cancer heterogeneity, considerably complicate our understandings toward breast cancer differentiation and hamper their clinical translations.
Biomarkers identified from networks are reported more reproducible than individual ones selected without network information25. An integrated network has been considered useful to integrate multiple levels of high-throughput information and gain comprehensive understandings of cancer related genomic alterations26. Tumor clonal network, by treating tumor as an evolving system and computationally dissecting clones from tumors, has been proposed as an effective tool to gain a ‘whole-system’ view of a tumor for personalized cancer management27, 28. Ever since 2011 when Weinberg brought up the concept of cancer hallmarks, targeting the hallmarks of cancer has been considered as a rational approach to the next-generation cancer therapy29. Accordingly, cancer hallmark network has opened a novel window for predicting patient clinical outcome from a myriad of phenotypic complexities governed by a limited set of organizing principles30. Under this framework, a set of mutations and copy number variations were reported effective in predicting subtype-specific drug targets in breast cancer31; and cancer hallmark-based gene signature sets were identified beneficial in predicting the recurrence and chemotherapy response of stage II colorectal cancer patients32.
Inspired by these previous efforts we, in this paper, focus on identifying genes and hallmarks governing the heterogeneity of breast cancer from the network point of view. For this, we constructed six diagnostic networks by integrating each of 6 publically available gene expression datasets with protein interaction data retrieved from BioGRID33 on 1015 diff-genes previously reported with breast cancer subtyping roles34. Using the Greedy algorithm and mutual information we condensed each of the 6 networks, and merged genes present in at least three networks to preserve as much information as possible with the most succinct number of genes.
The resulting integrated diagnostic network contains 37 genes and 43 interactions, among which four, i.e., FAM134B, KIF2C, ALCAM, KIF1A, were identified with comparable subtyping efficacies with the initial 1015 genes (which were evaluated by hierarchical clustering and leave-one-out cross validations). Pathway, Gene Ontology, and proliferation marker enrichment analyses reveal five critical cancer hallmarks driving the complexity and heterogeneity of breast cancers, which are ‘enabling replicative immortality’, ‘sustaining proliferative signaling’, ‘resisting cell death’, ‘deregulating cellular energetics’, and ‘activating invasion & metastasis’. Our results offer a 4-gene panel with feasible size for clinical translation, and underpin 5 cancer hallmarks and associated pathways for therapeutic design. These not only update our knowledge toward breast cancer complexity and, more importantly, provide practical insights and tools for breast cancer control.
Methods
Construction of the diff-gene protein network
Protein interactions (PPI) of 1015 genes differentiating breast cancer subtypes (diff-genes) proposed in ref. 34 were retrieved from the public database BioGRID (Biology General Repository for Interaction Datasets)33 and used for ‘diff-gene protein network’ construction. BioGRID version 3.4.147 was requested which encompasses 1,421,025 protein and genetic interactions, 27,785 chemical associations and 38,559 post-translational modifications of major modelling organisms from 58,514 papers.
Construction of diagnostic diff-gene networks
Six datasets, GSE70947, GSE15852, GSE20711, GSE65212, GSE18229-GPL887, GSE65194, were retrieved from the GEO database (Gene Expression Omnibus)35 and included in this study. We conducted the analysis using data free of metastasis. Two datasets are comprised of case-control sample pairs (i.e., each pair is consist of one breast cancer tissue sample and its adjacent normal breast tissue), with GSE70947 and GSE15852 each encompassing 148 and 43 sample pairs. GSE20711 contains 88 breast cancer and 2 normal breast tissue samples. We removed cell line and mammoplasty data from GSE65212, GSE18229-GPL887 and GSE65194, and kept 164 (out of 178) samples from GSE65212 (comprised of 153 breast cancer and 11 normal breast tissue samples), 77 (out of 94) samples from GSE18229-GPL887 (including 72 breast cancer and 5 normal tissue samples), and 165 (out of 178) samples from GSE65194 (composed of 153 breast cancer and 12 normal breast tissue samples) for diff-gene network construction.
Differential expression analysis of the diff-genes between breast cancer and normal samples was conducted for each dataset using GEO2R, which is an interactive web tool allowing comparisons between two or among multiple groups of samples in a GEO series using limma R packages on the original submitter-supplied processed data36–38. Diff-genes differentially expressed in breast cancer tissues obtained using each dataset are believed to capture subtyping features and specific to tumor cells. They are considered with more profound diagnostic values and named ‘diagnostic diff-genes’ here.
The p-values of these diagnostic diff-genes were corrected using the Benjamini & Hochberg adjustment method and transformed to paired t-scores using Equation 1,
| 1 |
where denotes the expression level of g th gene in the i th sample and jth experiment, and n i represents the sample size of each sample cohort. The higher the t-score of a given gene is the more significant diagnostic value the gene is associated with.
Construction of diagnostic networks
Each diagnostic diff-gene network was combined with the diff-gene protein network by keeping edges in common, forming six independent diagnostic networks. The Greedy searching strategy based on mutual information was employed to find the most succinct network maintaining the highest accumulated t-score for each diagnostic network using the jActiveModules plugin in Cytoscape39. Mutual information is computed by Equation 2, where a and c each denotes the nodes, x and y each represents the t-scores of a and c, p(x, y) is the probability density function of a and c, p(x) and p(y) are the partial probability density function of a and c, respectively.
| 2 |
The Greedy algorithm is an iterative approach where, in each round, it randomly selects one node (seed), expands the network by adding nodes that raise the overall t-score until no further increase is obtainable. The top 10 sub-networks (ranked by t-scores) were merged after generating ‘n 0’ (the number of genes in the initial network) sub-networks with each gene as the seed, resulting in a network containing ‘n r’ genes (r denotes the r th run). Multiple rounds of the Greedy algorithm were run using ‘n r−1’ nodes as the starting network until n r meets the stopping criterion which was set to approximately 50 here.
Construction of integrated diagnostic network
The overlapping rate was computed for each combination of the six diagnostic networks using Equation 3
| 3 |
where n ranges from 2 to 6, G 1, G 2 and G n each denotes the number of genes in the n th diagnostic network under comparison, and G 1…n denotes the genes in common among the n compared networks. Genes and edges present in at least three diagnostic networks were selected as the integrated diagnostic network.
Identification and evaluation of pivotal diagnostic genes
Connectivity assessment
The degee of each node, i.e., the number of edges each gene connects with its neighbors, was asssessed to measure the importance of each identified diagnostic diff-gene. Genes were categorized into <25%, 25–50%, 50–75%, >75% quantiles of the degree distribution, i.e., genes with 1–12, 13–35, 36–76 or >76 degrees were grouped into distinct classes. BioGRID contains 13369 nodes and 109670 edges after the removal of singletons, with the node degree ranges from 1 to 3576. We computed the percentage of each group of identified diagnostic diff-genes represented in BioGRID (Per i) using Equation 4
| 4 |
where N s,i represents the number of diagnostic diff-genes in level i and N D,i represents the number of genes in BioGRID fell in level i. Permutation test with 1000 runs was conducted to evaluate whether genes in the highly connected group (>75% percentile) are obtained by chance.
The enrichment of the connecitivity for gene j (EC j) from the integrated diagnostic network was computed using (C s,j represents the number of connectivity of gene j in the integrated diagnostic network, and CD,j represents the connectivity of gene j in BioGRID).
Genes whose connectivity is highly enriched in the integrated diagnostic network were considered specific to and crucial for breast cancer diagnosis, and were selected as candidate ‘pivotal diagnostic genes’.
Patient survival association study
Kaplan Meier Plotter40 (http://kmplot.com/analysis/index.php?p=service&cancer=breast), a database containing clinical information and gene expression data on 3951 breast cancer patients, was used to evaluate the clinical association of each candidate pivotal diagnostic gene with breast cancer patient 10-year relapse free survival. Genes without significant association with patient survival were excluded from the pivotal diagnostic gene panel.
Cross validation and hierarchical clustering analysis
Cross validation was used to quantitatively finalize the pivotal diagnostic gene panel and assess its predictive power in breast cancer subtyping according to the status of ER, PR and HER2. Leave-1-out and 10-fold cross validations were used, where support vector machine (SVM) and k-nearest neighbor (KNN) were employed as the kernels. Both SVM and KNN are supervised machine learning methods widely applied in classification. SVM constructs a set of hyperplanes in a high-dimensional space, and the classification is achieved by the hyperplane that has the largest distance to the nearest training data point of any class. KNN classifies an object by taking a vote of its ‘k’ nearest neighbors, and the object is assigned to the class voted by the majority of the ‘k’ neighbors (k = 10 to be consistent with34). The statistics computed from 1000 simulations were reported.
The hierarchical clustering was used to draw heatmaps for the finalized diagnostic gene panel using R (https://www.r-project.org), where the distance matrix and agglomeration method were optimized to produce the optimal results.
We benchmark the predictive power of the pivotal gene panel against that in ref. 34 where GSE24450, TCGA and GSE22220 were used. As FAM134B is missing from GSE22220, we included GSE24450 and TCGA in this study. In addition, we added GSE25055 to generalize the subtyping functionality of the pivotal diagnostic gene panel. GSE24450 and GSE25055 were retrieved from the Gene Expression Omnibus (GEO) database. GSE24450 contains 183 primary breast tumors that were processed and hybridized to Illumina HumanHT-12_V3 Expression BeadChips. GSE25055 data was obtained using Affymetrix Human Genome U133A Array (HG-U133A) and encompasses 300 samples where 10 samples without consensus subtyping between immunohistochemistry marker-based and PAM50 classification were removed (original sample size is 310). TCGA data (level 3) was retrieved from the TCGA portal at http://tcga.cancer.gov/dataportal, which contains 451 samples profiled using Agilent 244 K Custom Gene Expression G4502A-07-3.
Patient tumor sample stratification
We performed tumor sample stratification based on the expression of each of the four pivotal diagnostic genes using GSE24450, TCGA and GSE25055 datasets. Student t test was used to assess the significance of each gene in distinguishing breast cancer subtypes stratified by ER, PR and HER2.
Evaluation of diagnostic genes
Pathway and Gene Oncology enrichment analysis
Enrichment analyses on the pathways and Gene Oncology (GO) of the identified diagnostic genes were performed using Enrichr (http://amp.pharm.mssm.edu/Enrichr/). The performance of the enrichment analysis was evaluated by p-value, adjusted p-value, Z-score and C-score. The p-value is computed from the Fisher exact test which assumes a binominal distribution and independence of genes under test. The adjusted p-value is the p-value corrected from multiple hypotheses testing using the Benjamini-Hochberg method. The Z-score is computed as the deviation from the expected rank, which has been precomputed using Fisher’s exact test for many random input gene lists for each term in the gene set library. Combined score (denoted as ‘C-score’) was computed to assess the enrichment of each pathway or GO term using Equation 5
| 5 |
where C is the C-score, p and Z each refers to the p-value and Z-score, respectively.
A gene set is a group of genes sharing a common biological function and used as the prior biological knowledge to be compared against for the enrichment analysis. Enrichr contains 103 gene sets, with genes covered in each set ranging from 280 to 49238.
In the pathway enrichment analysis, ‘BioCarta_2016’ was chosen as the gene set, where BioCarta is an interactive on-line resource designed for life science research with pathway information retrieval as a featured functionality41. In GO analysis the latest gene ontology annotations (‘GO_2015’) were used as the background.
Cancer proliferation marker enrichment analysis
Enrichment analysis of genes present in the integrated diagnostic network among cancer cell proliferation markers was conducted using Enrichr, where ‘Achilles_fitness_decrease’ was selected as the gene set. The Achilles project performed a genome-scale screen across 216 cancer cell lines for genes required for cancer cell proliferation and/or viability42.
Data availability
The datasets analysed during the current study include 7 gene expression datasets, GSE70947, GSE15852, GSE20711, GSE65212, GSE18229-GPL887, GSE65194, GSE24450, retrieved from GEO (http://www.ncbi.nlm.nih.gov/geo), the level 3 breast cancer patient data downloaded from the TCGA repository (https://cancergenome.nih.gov), protein interaction data obtained from BioGRID (https://thebiogrid.org/), patient gene expression and clinical survival information stored in Kaplan Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=breast), pathways from BioCarta (https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways), gene ontologies from GO database (http://www.geneontology.org/page/download-annotations), and proliferation markers identified from the Achilles project (https://portals.broadinstitute.org/achilles).
Results
The workflow of this study is summarized in Fig. 1.
Figure 1.

Workflow of this project. Each rounded square box represents one dataset, each square box shows one set of results, and each diamond box illustrates one operation together with associated algorithms. Datasets are shown in italic, where ‘Data_1-6’ represents GSE70947, GSE15852, GSE20711, GSE65212, GSE18229-GPL887, GSE65194, GSE24450, ‘Data_7-9’ represents GSE24450, GSE25055, TCGA, and ‘BioGRID’ means the BioGRID database. The primary outputs are highlighted in bold face, ‘6×’ means that 6 sets of networks were generated. Square brackets in each diamond box represent the algorithm or approach used in the operation.
Diff-gene protein network
The diff-gene protein network, constructed by retrieving protein interactions from BioGRID using diff-genes identified in ref. 34, is comprised of 317 edges and 318 nodes and densely connected around two hubs, i.e., APP and ER (Supplementary Figure S1). The number of edges (degree) connected to APP and ER are 100 and 21, respectively. Those of APP and ER from the whole network stored in BioGRID are 2346 and 571, respectively.
Diagnostic diff-gene networks
Six diagnostic diff-gene networks, each formed by mapping clinical gene expression data to the diff-gene protein network, were obtained (Supplementary Figure S2). These networks were named by concatenating the gene expression dataset with ‘PPI’, which represents protein interactions retrieved from BioGRID, by ‘&’. Each network contains, on average, 48 nodes and 53 edges, with detailed information available in Supplementary Table S1.
Integrated diagnostic network
The overlaping rates among diagnostic networks enter the plateau when we start merging them in triplets (Fig. 2), i.e., the double, triple, quadruple, quintuple, and sextuple integated networks contain, on average, 43, 42, 12, 8 and 1 genes, respectively.
Figure 2.

Overlapping rates for different combinations of diagnostic networks. (A,B,C,D,E and F) each denotes the diagnostic network GSE70947&PPI, GSE18229&PPI, GSE15852&PPI, GSE20711&PPI, GSE65194&PPI and GSE65212&PPI, respectively, where the network names are defined as the gene expression dataset concatenated with ‘PPI’ (representing BioGRID) by ‘&’.
We, thus, selected nodes and edges at least present in three diagnostic networks and merged them as the integrated diagonosis network (Fig. 3), which includes 37 genes and 43 interactions. The condensed network preserves the two hubs (APP and ER) of the diff-gene protein network, with the degree being 19 and 6, respectively, for each gene.
Figure 3.

Integrated diagnostic network. This network was obtained by merging nodes and edges present in at least three diagnostic networks.
Genes fell into <25, 25–50, 50–75 and >75 percentile of total degree represent 0.07%, 0.95%, 1.55% and 3.64% of total genes stored in BioGRID. The gene with the highest degree enrichement is FAM134B (33.33%) which together with KIF2C (28.57%), ALCAM (25%) and KIF1A (25%) represent the top 10 percentile degree enrichment among the 37 genes in the integrated diagnostic network.
Pivotal diagnostic gene
Connectivity assessment
The BioGRID database contains 13369 nodes and 109670 edges after removing singletons, with the degree of a single gene ranging from 1 to 3576. In accordance with the <25%, 25–50%, 50–75% and >75% percentile of degrees in the integrated diagnostic network, the number of degrees are classified into four groups, i.e., 1–27, 28–52, 53–111 and >111 degrees, respectively. Genes in the integrated diagnostic network are condensed in the group representing the top 25 percentile degrees, i.e., 3.64% of the total genes from BioGRID in this group as compared with the 1.55%, 0.95%, 0.07% statistics in the lower 25 percentile, 25 to 50 percentile and 50 to 75 percentile groups (Fig. 4). Permutation test with 1000 runs show that the high enrichment (3.64%) of the highly connected group (>75% percentile degree) in the integrated diagnostic network is not obtained by chance (p = 0.005).
Figure 4.

Enrichment of nodes degree in each percentile level for genes in the integrated diagnostic network. The percentile levels were defined as <25, 25–50, 50–75, and >75 percentile of the degrees of each gene in the integrated diagnostic network, which correspond to 1–27, 28–52, 53–111, and >111 number of degrees, respectively. ‘Genes_BioGRID’ represents the number of genes from BioGRID felt into a given percentile level of node degree, and ‘Genes_Selected’ shows that from the integrated diagnostic network.
The connectivity enrichment of each gene in the integrated diagnostic network as compared with the whole protein interaction network from BioGRID ranges from 33.3% (FAM134B) to 0.81% (APP), as listed in Supplementary Table S2. There are two break points, i.e., the 4th and 6th genes, where the connectivity enrichment of the diagnostic genes significantly drops (Supplementary Figure S3). The 3rd and 4th genes share the same connectivity enrichment. We, thereby, consider the top 6, top 5, top 4 and top 3 as candidates in the pivotal gene panel.
Patient survival association study
The top five diagnostic genes are significantly associated with breast cancer 10-year relapse free survival (Fig. 5). FAM134B (p = 7E-08, HR = 0.79), ALCAM (p = 6.7E-10, HR = 0.61), KIF1A (p = 2E-05, HR = 0.79) confer protective effect, and KIF2C (p < 1E-16, HR = 1.69) and KIFC1 (p < 1E-16, HR = 1.69) are risky on patient clinical outcome. No statistical significance was observed for PHLDA1. Thus, we exclude the 6th gene from the candidate gene panel.
Figure 5.
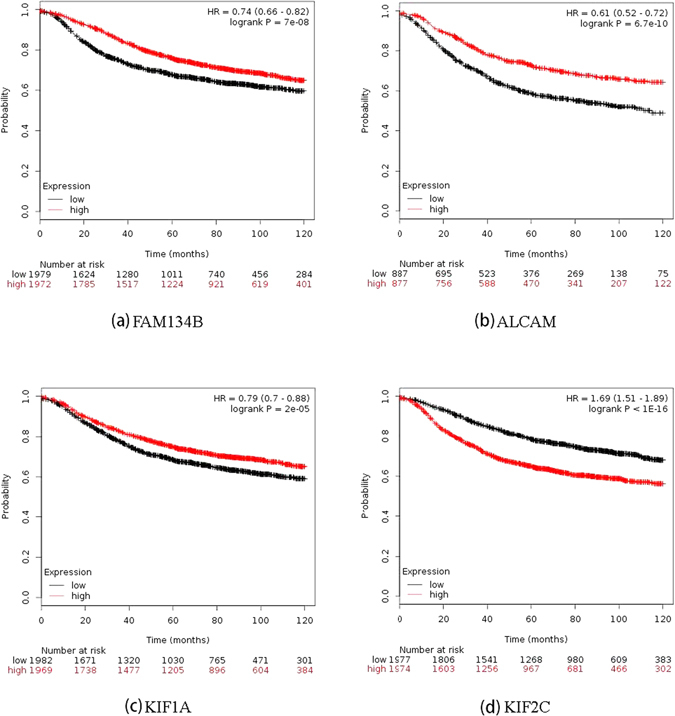
Breast cancer patient 10-year relapse free survival associated with each of the four pivotal diagnostic genes computed from Kaplan Meier Plotter.
Cross validation
Leave-1-out cross-validation results show the maximum prediction power of 74.8% and 76.5% accuracies from 1000 runs as assessed by SVM and KNN (k = 10), respectively, when applied to the GSE24450 data; exhibit 68.1% (SVM) and 67.2% (KNN) accuracies when the TCGA dataset was used; and obtain 89.6% (SVM) and 88.7% (KNN) scores when GSE25055 was used (Table 1). The average behaviors are 67.8% (SVM) and 66.7% (KNN) using GSE24450; 58.6% (SVM) and 56.4% (KNN) using TCGA; and 77.6% (SVM) and 74.6% (KNN) using GSE25055 (Table 1).
Table 1.
Cross-validations of the four pivotal diagnostic genes in differentiating breast cancer subtypes.
| Gene panel | Statistics | GSE24450 | TCGA | GSE25055 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leave-1-out | 10-fold | Leave-1-out | 10-fold | Leave-1-out | 10-fold | ||||||||
| SVM | KNN | SVM | KNN | SVM | KNN | SVM | KNN | SVM | KNN | SVM | KNN | ||
| 3-gene panel | Median | 0.677 | 0.670 | 0.677 | 0.670 | 0.565 | 0.539 | 0.579 | 0.544 | 0.739 | 0.713 | 0.731 | 0.716 |
| Mean | 0.676 | 0.666 | 0.676 | 0.665 | 0.572 | 0.543 | 0.585 | 0.550 | 0.732 | 0.712 | 0.731 | 0.712 | |
| Max | 0.739 | 0.757 | 0.730 | 0.739 | 0.672 | 0.661 | 0.682 | 0.665 | 0.887 | 0.878 | 0.878 | 0.875 | |
| Min | 0.617 | 0.574 | 0.626 | 0.583 | 0.523 | 0.470 | 0.505 | 0.456 | 0.557 | 0.478 | 0.550 | 0.491 | |
| 4-gene panel | Median | 0.678 | 0.670 | 0.678 | 0.670 | 0.582 | 0.561 | 0.591 | 0.562 | 0.783 | 0.748 | 0.775 | 0.760 |
| Mean | 0.678 | 0.667 | 0.678 | 0.667 | 0.586 | 0.564 | 0.596 | 0.561 | 0.776 | 0.746 | 0.776 | 0.757 | |
| Max | 0.748 | 0.765 | 0.748 | 0.765 | 0.681 | 0.672 | 0.690 | 0.682 | 0.896 | 0.887 | 0.889 | 0.878 | |
| Min | 0.626 | 0.591 | 0.635 | 0.591 | 0.525 | 0.479 | 0.522 | 0.483 | 0.565 | 0.478 | 0.587 | 0.528 | |
| 5-gene panel | Median | 0.678 | 0.670 | 0.678 | 0.670 | 0.573 | 0.548 | 0.586 | 0.557 | 0.765 | 0.739 | 0.749 | 0.745 |
| Mean | 0.677 | 0.668 | 0.677 | 0.667 | 0.579 | 0.553 | 0.590 | 0.559 | 0.760 | 0.735 | 0.751 | 0.743 | |
| Max | 0.748 | 0.757 | 0.739 | 0.739 | 0.672 | 0.667 | 0.682 | 0.665 | 0.878 | 0.870 | 0.871 | 0.871 | |
| Min | 0.617 | 0.591 | 0.635 | 0.591 | 0.528 | 0.490 | 0.517 | 0.475 | 0.548 | 0.513 | 0.554 | 0.524 | |
Leave-1-out and 10-fold represent two types of cross-validations used for performance assessment. ‘SVM’ and ‘KNN’ are used as the kernels for cross-validation, which represents support vector machine and k-nearest neighbor classifiers (k = 10), respectively. Statistics of 1000 rounds of iterations are shown.
Using 10-fold cross-validation and as compared with the leave-1-out approach, the same maximum and average prediction power were obtained using GSE24450; similar maximum and average scores were obtained using GSE25055, i.e., 88.9% (SVM) and 87.8% (KNN) for the maximum prediction power and 77.6% (SVM) and 75.7% (KNN) for the average performance; slightly higher performance was observed using TCGA data, i.e., 59.6% (SVM) and 56.1% (KNN) for the average performance, and 69% (SVM) and 68.2% (KNN) for the maximum behavior.
Results using SVM as the kernel are more stable than those using KNN as, in most cases, higher average performance, lower maximum and higher minimum values were obtained using SVM than KNN. 10-fold cross validation behaves better than the leave-1-out approach when data of relatively larger sample size was used. That is, the advantage of SVM over KNN becomes evident when TCGA data was used which encompasses 451 samples whereas GSE24450 and GSE25055 have 183 and 300 samples, respectively.
Most statistics measured for the 4-gene panel outweigh those in the 3-gene and 5-gene panels, though the difference is nuance (Table 1). Using GSE24450 as the discovery set for finalizing the pivotal gene panel, we selected the 10-fold cross validation approach (with SVM being the kernel) to assess the trajectory of the prediction power of the gene panels where one gene from the integrated diagnostic network was added at one time. The results show that 1) having more genes added in the panel, overall, improves the prediction power, and 2) the trajectory undergoes a sharp increase during the first 4 genes followed by a mild recession and relativley long plateau (Supplementary Figure S4). We thus consider the 4-gene panel as pivotal genes for the subsequent analyses and discussions.
Hierarchical clustering analysis
Four subtypes, [ER+|PR+]HER2−, [ER+|PR+]HER2+, [ER−|PR−]HER2+, [ER−|PR−]HER2- (also named TNG), were defined based on the status of ER, PR and HER2, conventionally used in clinic. Using only the four pivotal diagnostic markers, ER- tumors (red and yellow), especially the [ER−|PR−]HER2− cohort (red), could be clearly distinguished from ER+ samples (green and blue) (Fig. 6) using GSE24450, TCGA and GSE25055 datasets.
Figure 6.

Breast cancers from GSE24450, TCGA and GSE25055 clustered by the four pivotal diagnostic genes.
Patient sample stratification
Among the four genes, FAM134B and KIF1A function in differentiating ER positive and ER negative subtypes. The p values are 3.10E-25 (FAM134B) and 3.66E-13 (KIF1A) using TCGA data; 2.71E-10 (FAM134B) and 4.83E-03 (KIF1A) using GSE24450; and 6.33E-03 (FAM134B) and 1.43E-05 (KIF1A) using GSE25055 (Fig. 7). ALCAM and KIF2C could nicely distinguish TNG from the rest. That is, the p values are 1.01E-12 (ALCAM) and 2.24E-21 (KIF2C) using TCGA data; 1.82E-04 (ALCAM) and 1.97E-03 (KIF2C) using GSE24450; and 4.11E-14 (ALCAM) and 2.24E-27 (Fig. 7).
Figure 7.

Breast cancers from TCGA categorized into ‘luminal vs. non-luminal’ or ‘TNG vs. non-TNG’ for each of the four pivotal diagnostic genes.
Diagnostic genes
Pathway enrichment analysis
Genes from the integrated diagnostic network are enriched in 22 pathways obtained from BioCarta41 (Supplementary Figure S5, Supplementary Table S3). The C-score and the p-value decreases and increases dramatically from the 6th enriched pathway (Supplementary Figure S5). The top five pathways are ‘CDK regulation of DNA replication’ (p = 7.15E-05, C-score = 14.54), ‘downregulation of MTA3 in ER-negative breast tumors’ (p = 7.15E-05, C-score = 11.11), ‘role of HER2 in signal transduction and oncology’ (p = 5.37E-03, C-score = 8.72), ‘cyclines and cell cycle regulation’ (p = 4.52E-03, C-score = 6.07) and ‘role of Ran in mitotic spindle regulation’ (p = 1.33E-03, C-score = 5.85) (Table 2). Three out of the 5 pathways are associated with cell cycle, one represents the metastatic feature of ER negative subtype, and one shows the importance of HER2 mediated signaling in differentiating breast cancer subtypes.
Table 2.
Statistics of the top 5 pathways enriched by genes present in the integrated diagnostic network.
| Pathways | Adjusted p-value | Z-score | C-score | Genes |
|---|---|---|---|---|
| CDK Regulation of DNA Replication | 7.15E-05 | −1.5235 | 14.54 | CDT1;MCM5;MCM2 |
| Downregulation of MTA3 in ER-negative Breast Tumors | 7.15E-05 | −1.1643 | 11.11 | HSPB1;ESR1;GAPDH |
| Role of HER2 in Signal Transduction and Oncology | 5.37E-03 | −1.6685 | 8.72 | ERBB3;ESR1 |
| Cyclins and Cell Cycle Regulation | 4.52E-03 | −1.1252 | 6.07 | CCND3;CDKN2A |
| Role of Ran in mitotic spindle regulation | 1.33E-03 | −0.8836 | 5.85 | RANGAP1;AURKA |
Genes from the integrated diagnostic network and enriched in a given pathway are listed accordingly as ‘Genes’.
Gene Ontology enrichment analysis
72 biological processes, 17 cellular components and 14 molecular functions, collectively called GO terms, are enriched by genes from the integrated diagnostic network with adjusted p values below 0.05 (Supplementary Table S4). The top 5 enriched biological processes are ‘mitotic cell cycle’ (adjusted p = 4.85E-09, C-score = 44.12), ‘mitotic cell cycle phase transition’ (adjusted p = 1.27E-05, C-score = 26.19), ‘cell cycle phase transition’ (adjusted p = 1.27E-05, C-score = 26.14), ‘cell division’ (adjusted p = 3.65E-05, C-score = 22.33), and ‘microtubule-based process’ (adjusted p = 2.27E-04, C-score = 20.32), which are all associated with cell cycle and division (Fig. 8). The top 5 enriched cellular components are ‘nucleoplasm’ (adjusted p = 1.12E-04, C-score = 20.37), ‘cytosol’ (adjusted p = 8.33E-05, C-score = 19.70), ‘microtubule cytoskeleton’ (adjusted p = 1.12E-04, C-score = 19.12), ‘perinuclear region of cytoplasm’ (adjusted p = 2.69E-03, C-score = 13.74) and ‘kinesin complex’ (adjusted p = 2.69E-03, C-score = 12.91), which are locations and components involved during mitotic cell division (Fig. 8). Accordingly, the top 5 enriched molecular functions, ‘microtubule binding’ (adjusted p = 2.57E-03, C-score = 14.41), ‘tubulin binding’ (adjusted p = 4.42E-03, C-score = 13.04), ‘ATP binding’ (adjusted p = 4.42E-03, C-score = 12.94), ‘protein kinase binding’ (adjusted p = 5.88E-03, C-score = 12.91) and ‘kinase binding’ (adjusted p = 8.46E-03, C-score = 11.99), convolve the proteins, ATP and kinases required for cell division (Fig. 8).
Figure 8.
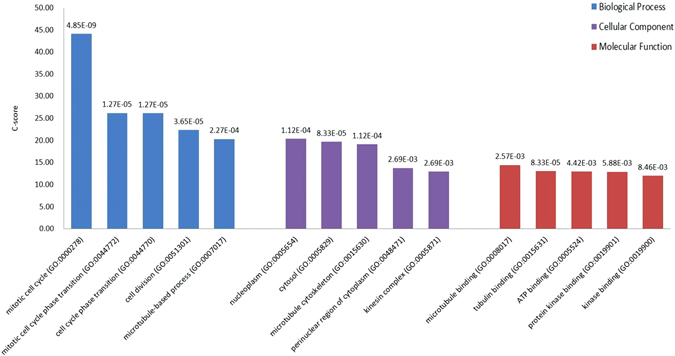
Top 5 enriched GO terms for genes in the integrated diagnostic network.
Cancer proliferation marker enrichment analysis
Out of the 216 cell lines used to screen genes having a context-specific effect on cell proliferation and/or viability in the Achilles project42, 64 are enriched with genes in the integrated diagnostic network (Supplementary Figure S6. The C-score drops considerably and the p value undergoes a sharp increase from the 6th cell line (cell lines are ranked with the decrease of the C-score and increase of the p-value). The top five cell lines are ZR7530, SNU840, HCC2218, NCIH23 and BT474, among which 4 out of 5 are breast or ovary cancers. Genes enriched in these 5 cell lines are TOP2A, HER3, CDC25B, MCM2, TUBB, HNRNPU, and CCND3, where TOP2A, HER3, CC25B and MCM2 appear three times, TUBB and HNRNPU pop up twice and CCND3 is only present in the ovary cell line SNU840 (Table 2). Genes enriched in the top breast cancer cell lines are TOP2A, HER3, CDC25B, MCM2 and TUBB (Table 2).
Discussion
Integrated diagnostic network reveals 4 pivotal genes with diagnostic potential
The integrated diagnostic network preserves the top two hubs of the diff-gene protein network retrieved from BioGRID, i.e., APP and ER (Supplementary Figure S7). It is intuitive that ER dominates the diagnostic network given its prominent roles and canonical use in breast cancer subtyping7. APP, however, is even more promiscuous, which has 4 to 5 times number of edges of ER in the whole protein interaction network of BioGRID or the diff-gene protein network, and the fold drops to 3 when the network is condensed to the integrated diagnostic network. This, on one hand, implicates that the network, once trimed to capture breast cancer heterogeneity, is shifted towards ER-driven and, on the other hand, suggests the critical roles played by APP in mediating carcinogenesis and subtype differentiation. APP has multiple human isoforms due to alternative splicing and encodes a type I transmembrane protien (amyloid precursor protein) expressed in many tisues. APP has been implicated in many cellular processes including hormonal regulation43. In particular, APP has been reported as a primary androgen target gene promoting prostate cancer growth43, and suggested to promote breast cancer proliferation with its immunohistochemical status proposed as a prognostic factor in ER positive breast cancers44; a recent study further unveiled its role in accerlerating the motility of advanced breast tumors, implicating its therapeutic targeting opportunity45.
Genes with degrees over-represented in the integrated diagnostic network are FAM134B, KIF2C, ALCAM and KIF1A, the combined effort of which has shown a comparable subtyping accuracy with the 1015 diff-genes reported in ref. 34 (Fig. 6, Table 1). The leave-one-out cross validations using GSE24450 (namely HEBCS in ref. 34) were reported to be 0.757 and 0.748, respectively, from SVM and KNN in ref. 34, and were 0.75 and 0.77, respectively, in this study; similarly, 0.735 and 0.723 were obtained using TCGA data from SVM and KNN in ref. 34, and 0.67 was observed from both approaches here (Table 1). These results suggest that the four pivotal diagnostic genes capture, if not all, the majority of the subtyping information imbedded in the diff-genes. By varying subtype combinations, we found that FAM134B and KIF1A function best in stratefying cancers according to ER status, and ALCAM and KIF2C act as the identifiers of triple negative cancers; while FAM134B and ALCAM express relatively higher in ER+ or non-TNG subtypes, KIF1A and KIF2C have comparatively lower expression in tumors of these classes (Fig. 7). Expression of these four genes, thus, may offer a succint panel for breast cancer diagnosis in addition to ER, PR and HER2 status. Truly, in accordance with this, patient 10-year relapse free survival analysis of each gene from this panel reveals that over-expression of FAM134B, ALCAM, KIF1A and low-expression of KIF2C each conveys a favorable clinical outcome with statistical significance (Fig. 5). FAM134B encodes an endoplasmic reticulum-anchored autophagy receptor mediating the degration of endoplasmic reticulum46. Its genetic mutation, resulting in decreased FAM134B expression, is a frequent event in the progression of oesophageal squamous cells47 and colorectal cancers48, which is adversely associated with patient clinical and pathological parameters and congruent with the tumor suppressive properties of FAM134B as previously reported48 as well as demonstrated in this study (Figs 6 and 7). ALCAM, the activated leukocyte cell adhesion molecule, has been known involved in cell migration and adhesion49, 50, in accordance with its identified role here in distinguishing TNG breast cancers, featured by high invasiveness, from the rest (Fig. 7). Decreased ALCAM expression has been implicated in poor breast cancer prognosis and promoted metastasis ability49–54, confirming with its tumor suppressive roles observed in Fig. 5 as well as previously suggested55. Impaired ALCAM expression is associated with induced ER+ breast cancer cell apoptosis and autophagy56, and down-regulating ALCAM expression sensitizes ER+ breast cancers to Tamoxifen treatment57, suggesting the therapeutic potential of down-regulating ALCAM in ER+ cancers which is consistent with its relatively higher expression in such tumor subtypes (Fig. 7). Both KIF1A and KIF2C encode members of the kinesin family, whose active movement supports several cellular functions including mitosis58. KIF1A was reported over-expressed in ER- breast cancer cell lines MDA-MB-231 and MDA-MB-468, and contributes to their chemotherapeutic resistance15. Elevated level of KIF2C was found in non-small cell lung cancer cells, which promotes cancer cell migration and could be suppressed by targeting the RAS-RAF-MEK1 pathway59. These not only support our observations on their diagnostic potential (Figs 6 and 7) but also suggest their therapeutic opportunities in cancer control.
Enrichment analysis reveals 5 cancer hallmarks driving breast cancer heterogeneity
The top 5 pathways enriched by genes in the integrated diagnostic network (adjusted p < 0.01) are ‘CDK regulation of DNA replication’, ‘down-regulation of MTA3 in ER negative breast cancers’, ‘role of HER2 in signal transduction’, ‘cyclins and cell cycle regulation’ and ‘role of Ran in mitotic spindle regulation’ (Table 3). These pathways show two prominent phenotypic features dominating breast cancer heterogeneities, i.e., proliferation and metastasis, and imply three cancer hallmarks. That is, three out of the five pathways reflect the ‘enabling replicative immortality’ (cell cycle) hallmark, one is associated with the ‘sustaining proliferative signaling’ (HER2 transduction), and one represents the ‘activating invasion & metastasis’ (MTA3 is metastasis associated 1 family member 3). As MTA3 is an estrogen-regulated gene60 whose promoter region contains an ER binding site, these pathways also consolidate the roles of ER and HER2 in breast cancer subtyping.
Table 3.
Top 5 enriched cell lines from cancer proliferation marker enrichment analysis.
| Name | Type | p-value | Z-score | C-score | Genes |
|---|---|---|---|---|---|
| ZR7530 | breast | 0.023110909 | −1.760811151 | 6.633768881 | TOP2A;HER3;CDC25B;MCM2 |
| SNU840 | ovary | 0.023110909 | −1.713280168 | 6.454698254 | CCND3;TUBB;HNRNPU |
| HCC2218 | breast | 0.023110909 | −1.710427594 | 6.443951326 | TOP2A;HER3;CDC25B;MCM2 |
| NCIH23 | lung | 0.023110909 | −1.67847526 | 6.323572486 | TOP2A;HNRNPU;MCM2 |
| BT474 | breast | 0.035418911 | −1.619832446 | 5.411065504 | HER3;TUBB;CDC25B |
Cell line name, type, p-value, Z-score, C-score and genes enriched in each cell line are provided.
Genes enriched in these 5 pathways are ER, HER3, MCM2, MCM5, CDT1, CCND3, CDKN2A, RANGAP1, AURKA, HSPB1, GAPDH. Genes such as ER and HER3 reflect the proliferative property of breast cancer cells. ER has long been recognized to mediate cell signaling in response to hormonal stimuli and known to drive the proliferative feature of breast cancer cells61. HER3 forms heterodimers with other members of this family, leading to the activation of pathways governing cell proliferation and differentiation. Seven of the 11 genes suggest the vital roles of the G1/S and G2/M check points for ‘enabling replicative immortality’. MCM2 and MCM5 are members of the MCM family of chromatin-binding proteins which, together with CDT1, are involved in DNA replication initiation and up-regulated during the G1/S transition. CCND3 encodes cyclin D3 that forms a complex with CDK4/6, the activity of which is required for the G1/S transition in the cell cycle; and CDKN2A encodes an inhibitor of CDK4. RANGAP1 encodes a protein interacting with Ras-related nuclear protein 1 (Ran), which is phosphorylated by the cyclin B/CDK1 complex (M phase kinase) and plays essential roles during cell mitosis62. AURKA is a cell cycle regulated kinase involved in microtubule formation and/or stabilization at the spindle pole during chromosome segregation and, thus, implicated with fundamental roles during mitosis and meiosis63. HSPB1, a member of the heat shock protein family, is reported to suppress PTEN level and, consequently, leads to reduced apoptosis in human breast cancer cells64, implicating the properties of cancer cells in ‘resisting cell death’. GAPDH encodes the glyceraldehyde-3-phosphate dehydrogenase whose up-regulation is correlated with aberrant gene profiling associated with both glycolysis and gluconeogenesis65. This suggests the Warburg effect, which represents the ‘deregulating cellular energetics’ hallmark.
Almost all genes enriched in the top 5 pathways have been implicated with cancer diagnostic potentials. ER has been canonically used as a clinical routine for breast cancer subtyping7. HER3 overexpression has been observed in diverse human cancers and been reported diagnostic of poor outcome in, e.g., breast cancer66 and melanoma67. MCM2 and MCM5 have been used for the diagnosis of colon cancers68. The prognostic value of CDT1 has been recently evaluated in breast cancer, whose over-expression was observed in tumor cells and significantly associated with poor patient survival69. CCND3 amplification has been proposed as a marker predicting tumor progression in, e.g., breast cancer70 and bladder urothelial carcinoma71. CDKN2A hyper-methylation has been suggested as a predictive factor for unfavorable prognosis of, e.g., colorectal cancer72, 73, rectal cancer74, and adult acute lymphoblastic leukemia patients harboring BCR-ABL1 fusions75. AURKA over-expression is reported strongly associated with tumor grade and proposed with prognostic value for disease progression76. HSPB1 encodes the heat-shock protein 27 which plays crucial roles in tumorigenesis and is reported an independent prognosis marker for malignancies such as lung cancer77. Elevated level of GAPDH positively associated genes is proportional to the malignant stage of various tumors and unfavorable prognosis65.
Gene ontology analysis reveals cell division to be the most enriched cellular event differentiating breast cancer subtypes (Fig. 8). This, together with the 7 out of 11 genes identified from pathway analysis and participating directly in cell cycle, implicate that ‘enabling replicative immortality’ may be one of the driving hallmarks fostering the proliferative feature of breast cancer cells and their differentiation.
Cancer proliferation marker enrichment analysis reveals that 7 genes from the integrated diagnostic network are enriched in cancer cells. Among them, 5 (TOP2A, HER3, CDC25B, MCM2 and TUBB) are from breast cancer cell lines (Table 3) and, in particular, HER2 positive cells (ZR7530 is [ER+PR−]HER2+, HCC2218 and BT474 are [ER−PR−]HER2+78). TOP2A, topoisomerase II alpha, functioning as an enzyme relaxing DNA supercoils, has long been used as a cancer proliferation marker and applied for breast tumor subtyping7. Importantly, abnormal TOP2A expression has been reported associated with increased cancer responsiveness to anthracycline-based chemotherapy79, suggesting its therapeutic implications besides confirmed diagnostic roles. HER3 encodes an EGFR family protein that is used as a prognostic marker in hormone receptor-negative breast cancers including the TNG and the HER2 positive subtype66, 67, and is as critical as HER2 in cell proliferation maintenance80. CDC25B is a member of the CDC25 family of phosphatases that activates the cyclin dependent kinase CDC2 and required for the entry of cells into mitosis. The association between CDC25B expression and cell proliferation is multifaceted: on one hand, CDC25B is up-regulated in multiple tumor types with increased levels correlated with higher proliferation, and its elevated level in the mammary glands has led to accelerated mammary epithelial proliferation that ultimately leads to tumor formation when exposed to the carcinogen DMBA in vivo 81; on the other hand, its tumor suppressive roles and anti-proliferative effect have been reported by several studies81, 82. MCM2 expression is correlated with that of KI67, a widely used proliferation marker in addition to ER, PR and HER2 for breast cancer subtyping in some studies7, and proposed as a sensitive maker of gastric cardiac cancer83. TUBB encodes the beta chain of tubulin, which polymerizes into microtubules that function in many essential cellular processes including mitosis, and thus indicative of cell proliferation. It is reported that targeting tubulin arrests mitosis and inhibits tumor cell proliferation, rendering microtubule-targeted drugs indispensable for the therapy of various cancers84. Some of these proliferation markers have intrinsic connections, so far reported, with HER2 status or expression. For instance, TOP2A aberrations are frequently found in HER2-amplifed breast cancers, accounting for 30–90% of such tumors7, 85. HER3 forms heterodimers with HER2 in downstream signal transduction, and plays a central role in HER2-amplified breast cancers80. CDC25B expression could be induced through HER2 signal transduction in human lung cancer cells86. These, collectively, suggest the importance of ‘sustaining proliferative signaling’ and, in particular, HER2 transduction, in driving the complex morphological and pathological features of breast cancers.
Conclusion
This study constructed an integrated diagnostic network composed of 37 nodes and 43 edges, by using information integrated from 6 publically available gene expression datasets and protein interactions retrieved from BioGRID to trim the 1015 diff-genes previously reported. We identified 4 pivotal diagnostic genes (FAM134B, KIF2C, ALCAM, KIF1A) from this network, which form a largely reduced gene panel preserving comparable subtyping efficacies with the initial 1015 diff-genes. Further pathway, GO, and proliferation marker enrichment analyses of the integrated diagnostic network collectively suggest two carcinogenic transitions governing breast cancer differentiation, i.e., proliferation and metastasis, and five out of 10 cancer hallmarks87, i.e., ‘enabling replicative immortality’ (i.e., cell cycle, especially G1/S and G2/M), ‘sustaining proliferative signaling’ (ER, HER2), ‘resisting cell death’, ‘deregulating cellular energetics’ (aerobic glycolysis), and ‘activating invasion & metastasis’ empowering such processes, with the first two being the most prominent. Our work provides a gene panel of reasonable size with clinical translation potential, and hallmarks driving breast cancer heterogeneities. The pivotal genes and primarily hallmarks (or implicated top pathways) identified may offer novel diagnostic markers or therapeutic targets, alone or in combination with current clinical modalities, for the benefit of breast cancer patients.
Electronic supplementary material
Acknowledgements
This study is funded by the National Natural Science Foundation of China (Grant No. 31471251), Natural Science Foundation of Jiangsu Province (Grant No. BK20161130), the Six Talent Peaks Project in Jiangsu Province (Grant No. SWYY-128), and the Fundamental Research Funds for the Central Universities (JUSRP11507).
Author Contributions
X.-F.D. designed, implemented the study, and drafted the manuscript. T.-Y.H. conducted some analyses and joined in finalizing the manuscript. T.-T.H. helped in formatting. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07189-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Spitale A, Mazzola P, Soldini D, Mazzucchelli L, Bordoni A. Breast cancer classification according to immunohistochemical markers: clinicopathologic features and short-term survival analysis in a population-based study from the South of Switzerland. Annals of oncology. 2009;20:628–635. doi: 10.1093/annonc/mdn675. [DOI] [PubMed] [Google Scholar]
- 2.Berry DA, et al. Effect of screening and adjuvant therapy on mortality from breast cancer. New England Journal of Medicine. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 3.Blows FM, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai XF, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. American Journal of Cancer Research. 2015;5:2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 5.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. New England Journal of Medicine. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 6.Montagna E, et al. Heterogeneity of Triple-Negative Breast Cancer: Histologic Subtyping to Inform the Outcome. Clinical Breast Cancer. 2013;13:31–39. doi: 10.1016/j.clbc.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Dai X, Xiang L, Li T, Bai Z. Cancer Hallmarks, Biomarkers and Breast Cancer Molecular Subtypes. J Cancer. 2016;7:1281–1294. doi: 10.7150/jca.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu FL, Bender W. A proposed mechanism of tamoxifen in breast cancer prevention. Cancer Detect Prev. 2002;26:370–375. doi: 10.1016/S0361-090X(02)00124-1. [DOI] [PubMed] [Google Scholar]
- 9.Murray S. Trastuzumab (Herceptin) and HER2-positive breast cancer. CMAJ. 2006;174:36–37. doi: 10.1503/cmaj.051452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gluz O, et al. Triple-negative breast cancer–current status and future directions. Ann Oncol. 2009;20:1913–1927. doi: 10.1093/annonc/mdp492. [DOI] [PubMed] [Google Scholar]
- 11.Group EBCTC. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. The Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 12.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. Journal of Clinical Oncology. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 13.Dieci MV, Orvieto E, Dominici M, Conte P, Guarneri V. Rare Breast Cancer Subtypes: Histological, Molecular, and Clinical Peculiarities. Oncologist. 2014;19:805–813. doi: 10.1634/theoncologist.2014-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakis S, et al. The androgen receptor as a surrogate marker for molecular apocrine breast cancer subtyping. Breast. 2014;23:234–243. doi: 10.1016/j.breast.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Cheang MC, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen KV, et al. The value of TOP2A gene copy number variation as a biomarker in breast cancer: Update of DBCG trial 89D. Acta Oncol. 2008;47:725–734. doi: 10.1080/02841860801995396. [DOI] [PubMed] [Google Scholar]
- 17.Carey LA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 18.Cheang MC, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 19.Livasy CA, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen TO, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 21.Matos I, Dufloth R, Alvarenga M, Zeferino LC, Schmitt F. p63, cytokeratin 5, and P-cadherin: three molecular markers to distinguish basal phenotype in breast carcinomas. Virchows Arch. 2005;447:688–694. doi: 10.1007/s00428-005-0010-7. [DOI] [PubMed] [Google Scholar]
- 22.Rakha EA, et al. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res. 2009;15:2302–2310. doi: 10.1158/1078-0432.CCR-08-2132. [DOI] [PubMed] [Google Scholar]
- 23.Rakha EA, et al. Breast carcinoma with basal differentiation: a proposal for pathology definition based on basal cytokeratin expression. Histopathology. 2007;50:434–438. doi: 10.1111/j.1365-2559.2007.02638.x. [DOI] [PubMed] [Google Scholar]
- 24.Dai X, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Molecular Systems Biology. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang E. Understanding genomic alterations in cancer genomes using an integrative network approach. Cancer Lett. 2013;340:261–269. doi: 10.1016/j.canlet.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 27.Wang E, et al. Cancer systems biology in the genome sequencing era: part 2, evolutionary dynamics of tumor clonal networks and drug resistance. Semin Cancer Biol. 2013;23:286–292. doi: 10.1016/j.semcancer.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Wang E, et al. Cancer systems biology in the genome sequencing era: part 1, dissecting and modeling of tumor clones and their networks. Semin Cancer Biol. 2013;23:279–285. doi: 10.1016/j.semcancer.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Hainaut P, Plymoth A. Targeting the hallmarks of cancer: towards a rational approach to next-generation cancer therapy. Curr Opin Oncol. 2013;25:50–51. doi: 10.1097/CCO.0b013e32835b651e. [DOI] [PubMed] [Google Scholar]
- 30.Wang E, et al. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol. 2015;30:4–12. doi: 10.1016/j.semcancer.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Zaman N, et al. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell Rep. 2013;5:216–223. doi: 10.1016/j.celrep.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 32.Gao S, et al. Identification and Construction of Combinatory Cancer Hallmark-Based Gene Signature Sets to Predict Recurrence and Chemotherapy Benefit in Stage II Colorectal Cancer. JAMA Oncol. 2016;2:37–45. doi: 10.1001/jamaoncol.2015.3413. [DOI] [PubMed] [Google Scholar]
- 33.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai X, Chen A, Bai Z. Integrative investigation on breast cancer in ER, PR and HER2-defined subgroups using mRNA and miRNA expression profiling. Sci Rep. 2014;4:6566. doi: 10.1038/srep06566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics & Molecular Biology3, Article3 (2004). [DOI] [PubMed]
- 37.Smyth, G. K. limma: Linear Models for Microarray Data. Bioinformatics & Computational Biology Solutions Using R & Bioconductor 397–420 (2005).
- 38.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 39.Barrett T, et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Research. 2007;35:D760–765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szasz AM, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura, D. BioCarta. Biotech Software & Internet Report2 (2001).
- 42.Cowley GS, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takayama K, et al. Amyloid precursor protein is a primary androgen target gene that promotes prostate cancer growth. Cancer Research. 2009;69:137–142. doi: 10.1158/0008-5472.CAN-08-3633. [DOI] [PubMed] [Google Scholar]
- 44.Takagi K, et al. Amyloid precursor protein in human breast cancer: An androgen-induced gene associated with cell proliferation. Cancer Science. 2013;104:1532–1538. doi: 10.1111/cas.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim S, et al. Amyloid-β precursor protein promotes cell proliferation and motility of advanced breast cancer. BMC Cancer. 2014;14:928. doi: 10.1186/1471-2407-14-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khaminets A, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 47.Haque MH, et al. Identification of Novel FAM134B (JK1) Mutations in Oesophageal Squamous Cell Carcinoma. Sci Rep. 2016;6:29173. doi: 10.1038/srep29173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Islam F, et al. Novel FAM134B mutations and their clinicopathological significance in colorectal cancer. Hum Genet. 2017;136:321–337. doi: 10.1007/s00439-017-1760-4. [DOI] [PubMed] [Google Scholar]
- 49.Davies SR, et al. Expression of the cell to cell adhesion molecule, ALCAM, in breast cancer patients and the potential link with skeletal metastasis. Oncology Reports. 2008;19:555. [PubMed] [Google Scholar]
- 50.Burkhardt M, et al. Cytoplasmic overexpression of ALCAM is prognostic of disease progression in breast cancer. Journal of Clinical Pathology. 2006;59:403–409. doi: 10.1136/jcp.2005.028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ihnen M, et al. Protein expression analysis of ALCAM and CEACAM6 in breast cancer metastases reveals significantly increased ALCAM expression in metastases of the skin. Journal of Clinical Pathology. 2011;64:146–152. doi: 10.1136/jcp.2010.082602. [DOI] [PubMed] [Google Scholar]
- 52.Fang T, Mosunjac M, Adams AL, Oforiacquah S. Abstract 696: Loss of ALCAM function as a biologic basis for ethnic disparity in breast cancer. Cancer Research. 2012;72:696–696. doi: 10.1158/0008-5472.CAN-11-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burandt E, et al. Loss of ALCAM expression is linked to adverse phenotype and poor prognosis in breast cancer: a TMA-based immunohistochemical study on 2,197 breast cancer patients. Oncology Reports. 2014;32:2628. doi: 10.3892/or.2014.3523. [DOI] [PubMed] [Google Scholar]
- 54.Hein S, et al. Biological role of Activated Leukocyte Cell Adhesion Molecule (ALCAM) overexpression in breast cancer cell lines and clinical tumour tissue. Breast Cancer Research & Treatment. 2010;129:347–360. doi: 10.1007/s10549-010-1219-y. [DOI] [PubMed] [Google Scholar]
- 55.Davies S, Jiang W. The Role of ALCAM, Activated Leukocyte Cell Adhesion Molecule, in the Aggressive Nature of Breast Cancer Cells, a Potential Connection to Bone Metastasis. 2009;30:1163–1168. [PubMed] [Google Scholar]
- 56.Jezierska A, Matysiak W, Motyl T. ALCAM/CD166 protects breast cancer cells against apoptosis and autophagy. Medical Science Monitor International Medical Journal of Experimental & Clinical Research. 2006;12:BR263–273. [PubMed] [Google Scholar]
- 57.Chen MJ, Cheng YM, Chen CC, Chen YC, Shen CJ. MiR-148a and miR-152 reduce tamoxifen resistance in ER+ breast cancer via downregulating ALCAM. Biochemical & Biophysical Research Communications. 2017;483:840–846. doi: 10.1016/j.bbrc.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 58.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/S0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 59.Zaganjor, E. Regulation by ERK1/2 of Novel Substrates, Kinesins KIF2A and KIF2C. Albanesi Joseph P (2013).
- 60.Gururaj AE, et al. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6670–6675. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J, et al. Evaluation of ER and Ki-67 proliferation index as prognostic factors for survival following neoadjuvant chemotherapy with doxorubicin/docetaxel for locally advanced breast cancer. Cancer Chemother Pharmacol. 2008;61:569–577. doi: 10.1007/s00280-007-0506-8. [DOI] [PubMed] [Google Scholar]
- 62.Swaminathan S, et al. RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly. J Cell Biol. 2004;164:965–971. doi: 10.1083/jcb.200309126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, meiosis and mitosis. Biol Cell. 2004;96:215–229. doi: 10.1016/j.biolcel.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 64.Cayado-Gutierrez N, et al. Downregulation of Hsp27 (HSPB1) in MCF-7 human breast cancer cells induces upregulation of PTEN. Cell Stress Chaperones. 2013;18:243–249. doi: 10.1007/s12192-012-0367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D, Moothart DR, Lowy DR, Qian X. The expression of glyceraldehyde-3-phosphate dehydrogenase associated cell cycle (GACC) genes correlates with cancer stage and poor survival in patients with solid tumors. PLoS One. 2013;8:e61262. doi: 10.1371/journal.pone.0061262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bae SY, et al. HER3 status by immunohistochemistry is correlated with poor prognosis in hormone receptor-negative breast cancer patients. Breast Cancer Research and Treatment. 2013;139:741–750. doi: 10.1007/s10549-013-2570-6. [DOI] [PubMed] [Google Scholar]
- 67.Reschke M, et al. HER3 Is a Determinant for Poor Prognosis in Melanoma. Clinical Cancer Research An Official Journal of the American Association for Cancer Research. 2008;14:5188. doi: 10.1158/1078-0432.CCR-08-0186. [DOI] [PubMed] [Google Scholar]
- 68.Burger M. MCM2 and MCM5 as Prognostic Markers in Colon Cancer: A Worthwhile Approach. Digestive Diseases and Sciences. 2009;54:197–198. doi: 10.1007/s10620-008-0416-6. [DOI] [PubMed] [Google Scholar]
- 69.Mahadevappa, R. et al. The prognostic significance of Cdc6 and Cdt1 in breast cancer. Scientific Reports7, doi:10.1038/s41598-017-00998-9 (2017). [DOI] [PMC free article] [PubMed]
- 70.Azzato EM, et al. Effects of common germline genetic variation in cell cycle control genes on breast cancer survival: results from a population-based cohort. Breast Cancer Research. 2008;10:R47. doi: 10.1186/bcr2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beltran AL, et al. Fluorescence in situ hybridization analysis of CCND3 gene as marker of progression in bladder carcinoma. J Biol Regul Homeost Agents. 2013;27:559–567. [PubMed] [Google Scholar]
- 72.Xing X, et al. The prognostic value of CDKN2A hypermethylation in colorectal cancer: a meta-analysis. British Journal of Cancer. 2013;108:2542–2548. doi: 10.1038/bjc.2013.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maeda K, et al. Hypermethylation of the CDKN2A gene in colorectal cancer is associated with shorter survival. Oncology Reports. 2003;10:935. [PubMed] [Google Scholar]
- 74.Kohonen-Corish MRJ, et al. KRAS mutations and CDKN2A promoter methylation show an interactive adverse effect on survival and predict recurrence of rectal cancer. International Journal of Cancer Journal International Du Cancer. 2014;134:2820–2828. doi: 10.1002/ijc.28619. [DOI] [PubMed] [Google Scholar]
- 75.Iacobucci I, et al. Abstract 311: Loss of the tumor suppressor genes CDKN2A/ARF impairs prognosis in adult BCR-ABL1 positive acute lymphoblastic leukemia (ALL) Cancer Research. 2011;71:311–311. doi: 10.1158/1538-7445.AM2011-311. [DOI] [PubMed] [Google Scholar]
- 76.Rouquier S, Pillaire MJ, Cazaux C, Giorgi D. Expression of the microtubule-associated protein MAP9/ASAP and its partners AURKA and PLK1 in colorectal and breast cancers. Dis Markers. 2014;2014:798170. doi: 10.1155/2014/798170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo H, et al. Functional promoter −1271G >C variant of HSPB1 predicts lung cancer risk and survival. Journal of Clinical Oncology. 2010;28:1928. doi: 10.1200/JCO.2009.24.4954. [DOI] [PubMed] [Google Scholar]
- 78.Dai, X., Cheng, H. Y., Bai, Z. H. & Li, J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer revised (2017). [DOI] [PMC free article] [PubMed]
- 79.Press MF, et al. Alteration of topoisomerase II-alpha gene in human breast cancer: association with responsiveness to anthracycline-based chemotherapy. J Clin Oncol. 2011;29:859–867. doi: 10.1200/JCO.2009.27.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee-Hoeflich ST, et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- 81.Lee, C. C. & Manfredi, J. Abstract 3774: Investigating the role of CDC25B in inhibition of cellular proliferation. Cancer Research75 (2015).
- 82.Varmehziaie S, Manfredi JJ. The dual specificity phosphatase Cdc25B, but not the closely related Cdc25C, is capable of inhibiting cellular proliferation in a manner dependent upon its catalytic activity. Journal of Biological Chemistry. 2007;282:24633–24641. doi: 10.1074/jbc.M703105200. [DOI] [PubMed] [Google Scholar]
- 83.Liu M, et al. MCM2 expression levels predict diagnosis and prognosis in gastric cardiac cancer. Histology & Histopathology. 2013;28:481. doi: 10.14670/HH-28.481. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Z, et al. MT119, a new planar-structured compound, targets the colchicine site of tubulin arresting mitosis and inhibiting tumor cell proliferation. Int J Cancer. 2011;129:214–224. doi: 10.1002/ijc.25661. [DOI] [PubMed] [Google Scholar]
- 85.Ejlertsen B, et al. HER2, TOP2A, and TIMP-1 and responsiveness to adjuvant anthracycline-containing chemotherapy in high-risk breast cancer patients. J Clin Oncol. 2010;28:984–990. doi: 10.1200/JCO.2009.24.1166. [DOI] [PubMed] [Google Scholar]
- 86.Oguri T, Nemoto K, Bansal P, Wipf P, Lazo JS. Induction of Cdc25B expression by epidermal growth factor and transforming growth factor-alpha. Biochem Pharmacol. 2004;68:2221–2227. doi: 10.1016/j.bcp.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 87.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analysed during the current study include 7 gene expression datasets, GSE70947, GSE15852, GSE20711, GSE65212, GSE18229-GPL887, GSE65194, GSE24450, retrieved from GEO (http://www.ncbi.nlm.nih.gov/geo), the level 3 breast cancer patient data downloaded from the TCGA repository (https://cancergenome.nih.gov), protein interaction data obtained from BioGRID (https://thebiogrid.org/), patient gene expression and clinical survival information stored in Kaplan Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=breast), pathways from BioCarta (https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways), gene ontologies from GO database (http://www.geneontology.org/page/download-annotations), and proliferation markers identified from the Achilles project (https://portals.broadinstitute.org/achilles).


