Abstract
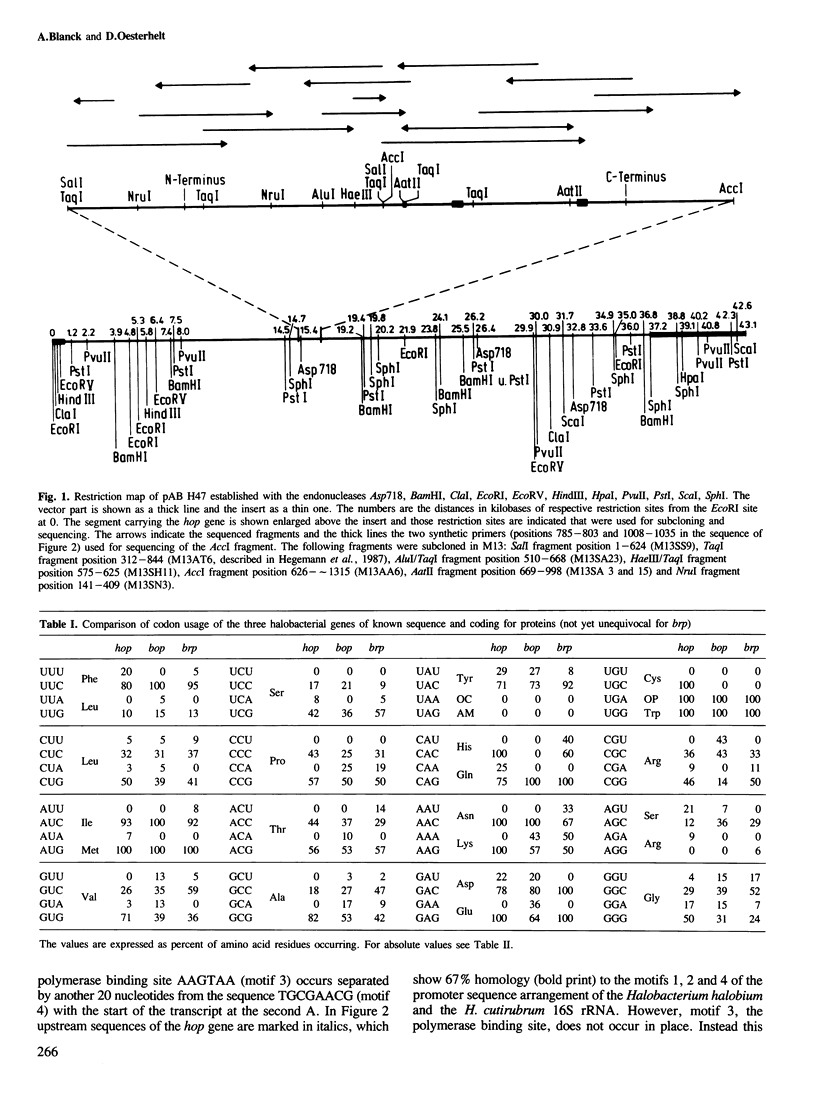
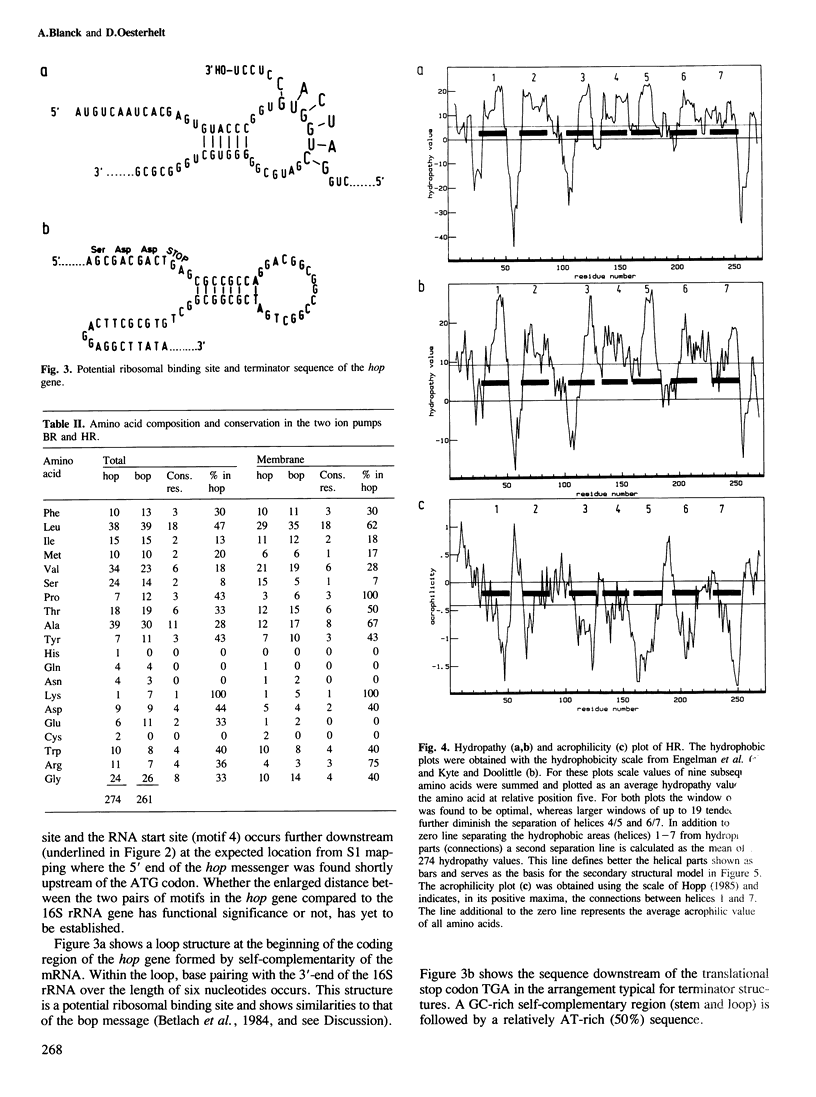
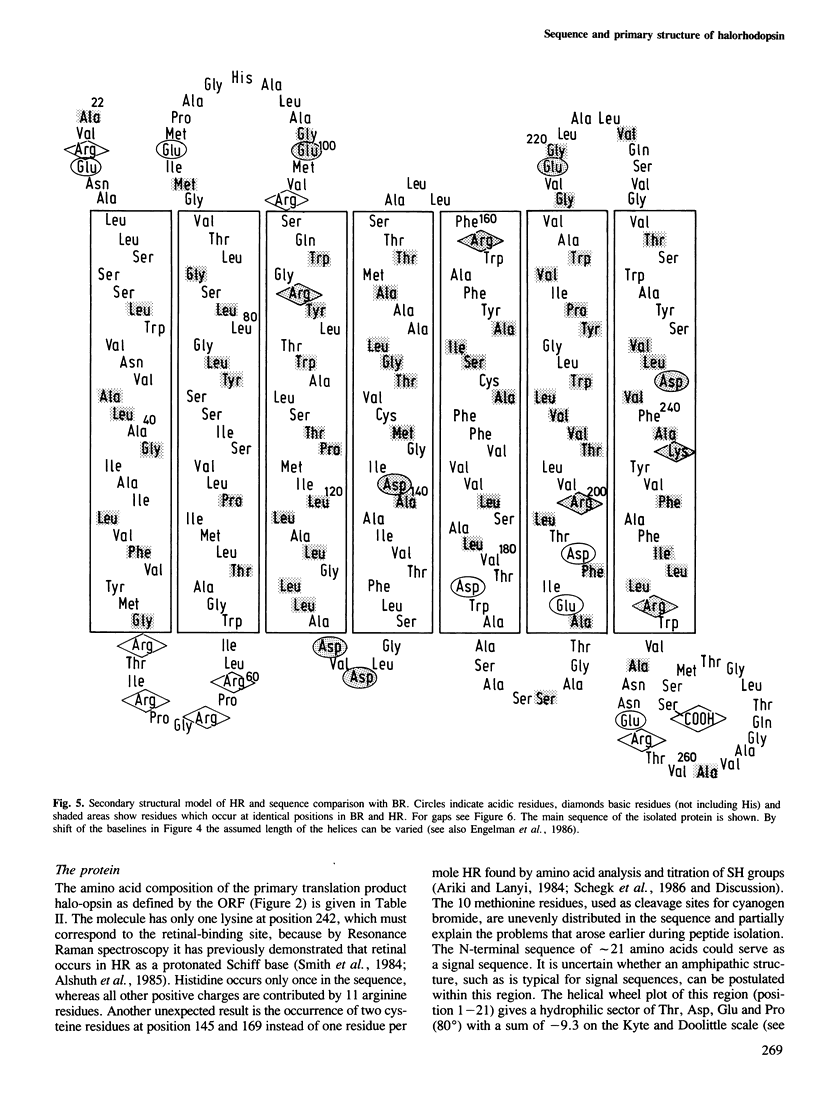
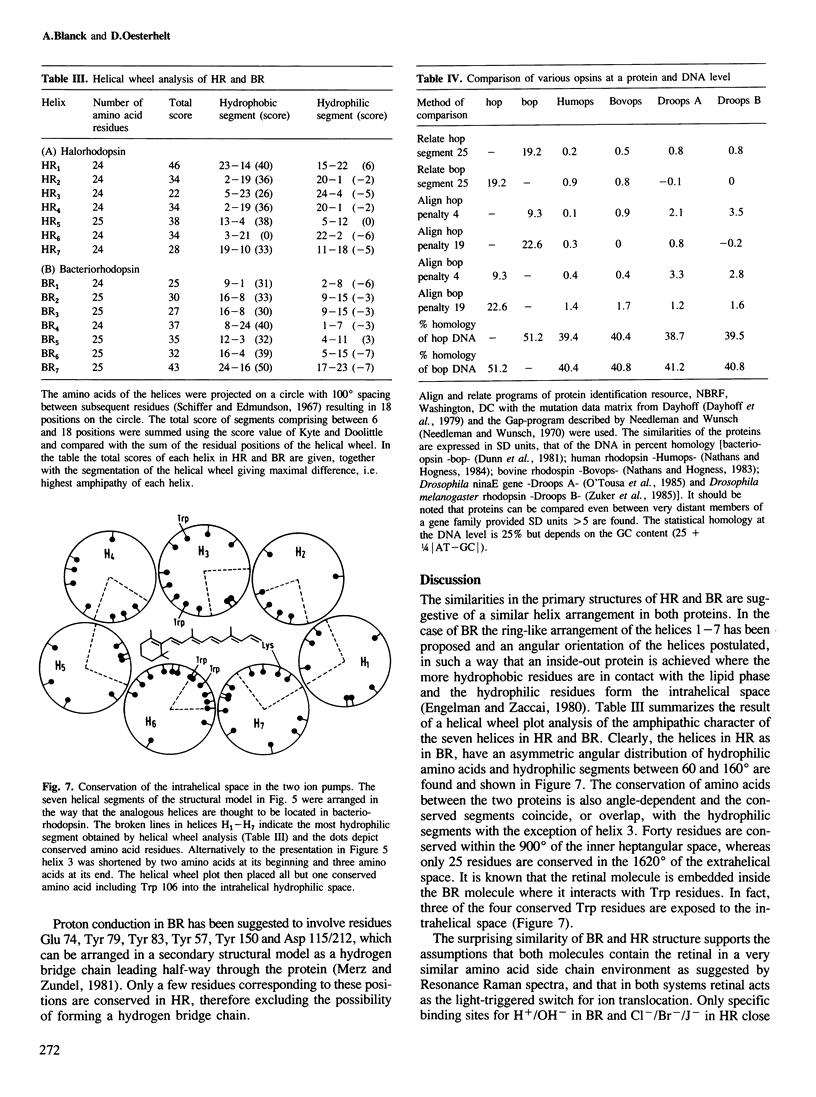
The gene for the protein moiety of the light-driven chloride pump halorhodopsin (HR), hop gene, was sequenced and the primary structure of the protein derived thereof. The gene has a GC content of 67% and codes for 274 amino acids. A promoter structure, resembling that of the halobacterial 16S rRNA genes, is present and both a terminating stem and a loop sequence is found downstream of the TGA stop codon. A ribosomal binding site is located within the translated region. The HR protein moiety is processed at the amino terminus, as well as the carboxy terminus, yielding a dominant species of calculated Mr 26 961. Seven transmembrane helical parts of the protein are defined by hydropathy and acrophilicity calculations. Comparision with the bacteriorhodopsin (BR) structure reveals a conservation of 36% of amino acid residues in the transmembrane part and 19% in the connecting loops at both surfaces. The most conspicuous conserved amino acids are the retinal-binding Lys residue, four Trp residues (eventually interacting with retinal), two Asp residues (providing possibly the negative charge environment of retinal) and three Pro residues of unknown function. No significant homology with the opsins of eucaryotes was found. Helical wheel analysis shows that HR is an inside-out protein with the majority of conserved amino acid residues inside the circle of the seven transmembrane helices. It is postulated that the intrahelical spaces, which could be gated by the retinal moiety, are the physical entities for translocation of protons in BR and chloride ions in HR. Retinal, by its cis−trans isomerization, serves as a switch connecting the ion-specific binding sites in both proteins.
Keywords: halorhodopsin, chloride pump, gene sequence, primary structure, amino acid conservation
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ariki M., Lanyi J. K. Evidence for a sulfhydryl group near the retinal-binding site of halorhodopsin. J Biol Chem. 1984 Mar 25;259(6):3504–3510. [PubMed] [Google Scholar]
- Ariki M., Schobert B., Lanyi J. K. Effects of arginine modification on the photocycle of halorhodopsin. Arch Biochem Biophys. 1986 Aug 1;248(2):532–539. doi: 10.1016/0003-9861(86)90506-0. [DOI] [PubMed] [Google Scholar]
- Betlach M., Friedman J., Boyer H. W., Pfeifer F. Characterization of a halobacterial gene affecting bacterio-opsin gene expression. Nucleic Acids Res. 1984 Oct 25;12(20):7949–7959. doi: 10.1093/nar/12.20.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Dassarma S., Rajbhandary U. L., Khorana H. G. Bacterio-opsin mRNA in wild-type and bacterio-opsin-deficient Halobacterium halobium strains. Proc Natl Acad Sci U S A. 1984 Jan;81(1):125–129. doi: 10.1073/pnas.81.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P. P. Multiple promoters for the transcription of the ribosomal RNA gene cluster in Halobacterium cutirubrum. J Mol Biol. 1985 Nov 20;186(2):457–461. doi: 10.1016/0022-2836(85)90117-2. [DOI] [PubMed] [Google Scholar]
- Dunn R., McCoy J., Simsek M., Majumdar A., Chang S. H., Rajbhandary U. L., Khorana H. G. The bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6744–6748. doi: 10.1073/pnas.78.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Zaccai G. Bacteriorhodopsin is an inside-out protein. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5894–5898. doi: 10.1073/pnas.77.10.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann P., Blanck A., Vogelsang-Wenke H., Lottspeich F., Oesterhelt D. The halo-opsin gene. I. Identification and isolation. EMBO J. 1987 Jan;6(1):259–264. doi: 10.1002/j.1460-2075.1987.tb04748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Jap B. K., Kong S. H. Secondary structure of halorhodopsin. Biochemistry. 1986 Jan 28;25(2):502–505. doi: 10.1021/bi00350a034. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Kühn H. Light-dependent phosphorylation of rhodopsin in living frogs. Nature. 1974 Aug 16;250(467):588–590. doi: 10.1038/250588a0. [DOI] [PubMed] [Google Scholar]
- Mankin A. S., Teterina N. L., Rubtsov P. M., Baratova L. A., Kagramanova V. K. Putative promoter region of rRNA operon from archaebacterium Halobacterium halobium. Nucleic Acids Res. 1984 Aug 24;12(16):6537–6546. doi: 10.1093/nar/12.16.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz H., Zundel G. Proton conduction in bacteriorhodopsin via a hydrogen-bonded chain with large proton polarizability. Biochem Biophys Res Commun. 1981 Jul 30;101(2):540–546. doi: 10.1016/0006-291x(81)91293-6. [DOI] [PubMed] [Google Scholar]
- Michel H., Weyer K. A., Gruenberg H., Dunger I., Oesterhelt D., Lottspeich F. The 'light' and 'medium' subunits of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the genes, nucleotide and amino acid sequence. EMBO J. 1986 Jun;5(6):1149–1158. doi: 10.1002/j.1460-2075.1986.tb04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J., Hogness D. S. Isolation and nucleotide sequence of the gene encoding human rhodopsin. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4851–4855. doi: 10.1073/pnas.81.15.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J., Hogness D. S. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983 Oct;34(3):807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- O'Tousa J. E., Baehr W., Martin R. L., Hirsh J., Pak W. L., Applebury M. L. The Drosophila ninaE gene encodes an opsin. Cell. 1985 Apr;40(4):839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA Rhodopsin and bacteriorhodopsin: structure-function relationships. FEBS Lett. 1982 Nov 8;148(2):179–191. doi: 10.1016/0014-5793(82)80805-3. [DOI] [PubMed] [Google Scholar]
- Polland H. J., Franz M. A., Zinth W., Kaiser W., Hegemann P., Oesterhelt D. Picosecond events in the photochemical cycle of the light-driven chloride-pump halorhodopsin. Biophys J. 1985 Jan;47(1):55–59. doi: 10.1016/S0006-3495(85)83876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal R., Cothran M., Espinoza B., Wall K. A., Bernard M. Light activates the reaction of bacteriorhodopsin aspartic acid-115 with dicyclohexylcarbodiimide. Biochemistry. 1985 Jul 30;24(16):4275–4279. doi: 10.1021/bi00337a004. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert B., Lanyi J. K., Oesterhelt D. Effects of anion binding on the deprotonation reactions of halorhodopsin. J Biol Chem. 1986 Feb 25;261(6):2690–2696. [PubMed] [Google Scholar]
- Smith S. O., Marvin M. J., Bogomolni R. A., Mathies R. A. Structure of the retinal chromophore in the hR578 form of halorhodopsin. J Biol Chem. 1984 Oct 25;259(20):12326–12329. [PubMed] [Google Scholar]
- Stryer L. Cyclic GMP cascade of vision. Annu Rev Neurosci. 1986;9:87–119. doi: 10.1146/annurev.ne.09.030186.000511. [DOI] [PubMed] [Google Scholar]
- Zuker C. S., Cowman A. F., Rubin G. M. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985 Apr;40(4):851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]



