ABSTRACT
The in vivo function of microRNAs (miRs) in diabetic retinopathy (DR) and age-related macular degeneration (AMD) remains unclear. We report here that let-7 family members are expressed in retinal and choroidal endothelial cells (ECs). In ECs, overexpression of let-7 by adenovirus represses EC proliferation, migration, and networking in vitro, whereas inhibition of the let-7 family with a locked nucleic acid (LNA)–anti-miR has the opposite effect. Mechanistically, silencing of the let-7 target HMGA2 gene mimics the phenotype of let-7 overexpression in ECs. let-7 transgenic (let-7-Tg) mice show features of nonproliferative DR, including tortuous retinal vessels and defective pericyte coverage. However, these mice develop significantly less choroidal neovascularization (CNV) compared to wild-type controls after laser injury. Consistently, silencing of let-7 in the eye increased laser-induced CNV in wild-type mice. Together, our data establish a causative role of let-7 in nonproliferative diabetic retinopathy and a repressive function of let-7 in pathological angiogenesis, suggesting distinct implications of let-7 in the pathogenesis of DR and AMD.
KEYWORDS: let-7, endothelial cells, angiogenesis, diabetic retinopathy, choroidal, neovascularization, AMD
INTRODUCTION
The retina has been an excellent model for studying the mechanism of angiogenesis. Retinal vascularization in mice begins after birth, when endothelial cells (ECs) sprout from the central retinal artery to the peripheral region and then to the intermediate and deep retinal layers (1, 2). Pathological angiogenesis in the eye is the most common cause of blindness at all ages and underlies conditions such as retinopathy of prematurity in children, diabetic retinopathy (DR) in young adults, and age-related macular degeneration (AMD) in the elderly. DR is the leading cause of blindness in working-age adults (18 to 64 years old), with a prevalence of 5.4% (∼7.7 million people) in the United States (3). Early-stage DR (or nonproliferative DR [NPDR]) is characterized by pericyte loss, basement membrane thickening, and EC dysfunction. Proliferative diabetic retinopathy (PDR) develops at the advanced stage and is associated with new blood vessel growth along the retina and the vitreous surface. AMD is a degenerative disease of the retina and the leading cause of blindness among the elderly (4). Neovascular (or wet) AMD, which accounts for the majority of acute vision loss in AMD, is characterized by choroidal neovascularization (CNV), a process involving abnormal growth of blood vessels from the choroid into the retina. Vascular endothelial growth factor (VEGF) is a major cytokine driving neovascularization and vascular permeability during CNV and diabetic edema. Several U.S. Food and Drug Administration-approved anti-VEGF agents are the current mainstay for wet AMD treatment and have been recently approved for the treatment of diabetic macular edema (5–8). Although anti-VEGF agents have markedly improved the clinical outcome of wet AMD, only 30 to 40% of individuals experienced vision improvement after treatment, and some patients do not respond to anti-VEGF therapy at all (9, 10). Therefore, novel and alternative therapeutic approaches are still greatly needed. Moreover, mechanistic study of AMD and DR is still scarce, and many critical questions remain to be answered. For example, what is the mechanism driving the transition from NPDR to PDR? What are common and distinct mechanisms of AMD and DR?
MicroRNAs (miRs) are small noncoding RNAs that posttranscriptionally repress multiple target genes and are implicated in numerous diseases, including vascular diseases (11–13). The in vivo function of miRs in DR and AMD is still largely unknown (14, 15). Several miRs, including let-7, miR-375, miR-9, miR-103/107, miR-143/145, and miR-144, have been shown to regulate insulin production, release, or insulin sensitivity (16–22). A number of miRs have been shown to be significantly regulated in the retinas or retinal ECs (RECs) in streptozotocin-induced or Akita (type I diabetic) DR animal models or in the oxygen-induced retinopathy model (23–32). Particularly, miR-146a and miR-146b are upregulated in the RECs of diabetic rats and act as negative feedback on NF-κB activation to control the inflammatory response (23, 33); miR-200b and miR-126 are downregulated in a DR mouse model and regulate VEGF expression during retinal neovascularization (25, 29); miR-200b downregulation was also found in the retinas of patients with diabetes (25, 34). However, miR-200b was also shown to be significantly upregulated in the retinas of Akita rats and could contribute to increased cell death in DR (26). More recently, using a genetic knockout mouse model, miR-155 was shown to modulate microglial activation and abnormal vessel growth in an oxygen-induced retinopathy model (35). Regarding the role of miRs in AMD, several miRs, including miR-23/miR-27, miR-21, miR-24, and miR-126, have proven to be important regulators of CNV in a laser injury-induced CNV model (36–39). However, the overall in vivo function of miRs related to DR and AMD is still unclear.
The let-7 family genes are the founding members of miRs, with established functions in tissue differentiation and tumor suppression (40, 41). The mouse let-7 miR family consists of 12 gene loci that resolve into nine unique but highly homologous mature miR sequences, which include let-7a, let-7b, let-7d, let-7e, let-c, let-7f, let-7g, let-7i, and miR-98. In humans, the let-7 family also includes miR-202. All family members are believed to have similar functions due to the identical seed region (nucleotides 2 to 7) that are required for miR-target mRNA interaction. To date, there is no genetic knockout study of the let-7 family miRs in vertebrates reported, possibly due to the location of the let-7 family members in multiple loci of the genome. Transgenic overexpression of Lin28a and Lin28b, negative regulators of let-7 biogenesis, promotes an insulin-sensitized state that resists high-fat-diet-induced diabetes, whereas the overexpression of let-7 results in insulin resistance and impaired glucose tolerance (21, 22). Consistently, the inhibition of let-7 expression by locked nucleic acid (LNA)-modified anti-miR resulted in improvement of insulin resistance and blood glucose levels. Regarding let-7 family function in angiogenesis, inhibition of let-7f was shown to reduce sprouting angiogenesis in vitro (42). Based on these reports, we hypothesized that the let-7 gene family plays a role in angiogenesis related to DR and AMD. We provide evidence that the overexpression of let-7 in vivo leads to features of NPDR, including tortuous retinal vessels and defective pericyte coverage. However, this phenotype did not progress to the PDR phenotype. Consistent with these findings, we found a repressive function of let-7 in angiogenesis. Overexpression of let-7 represses EC proliferation, migration, and EC networking, whereas inhibition of let-7 has the opposite effect. Knockdown of the let-7 target HMGA2 gene mimics the effect of let-7 overexpression in vitro. Moreover, let-7 transgenic mice show significantly less neovascularization in the choroid after laser injury, whereas the knockdown of let-7 using let-7 anti-miR strongly enhanced CNV. These data indicate that let-7 contributes to NPDR but represses angiogenesis and CNV in vivo.
RESULTS
Expression of let-7 family members in human retinal cells and mouse retinal tissues.
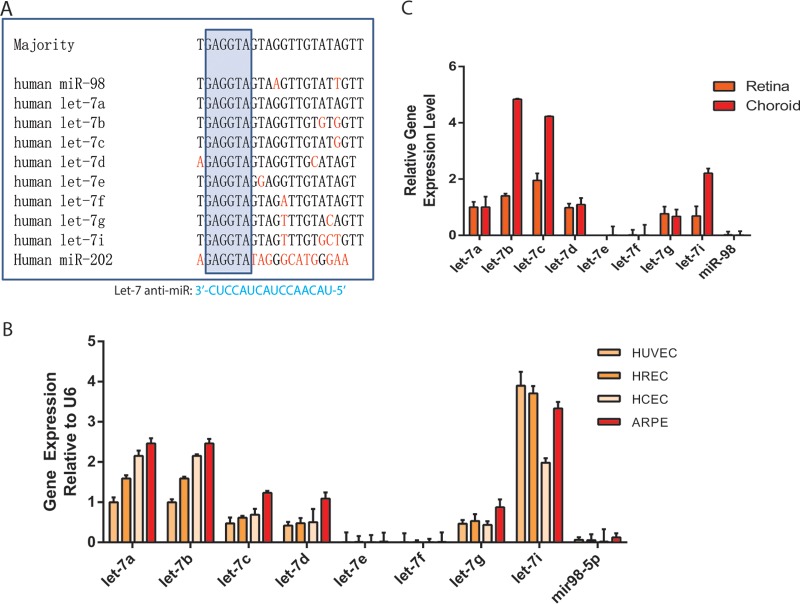
The let-7 family contains 10 unique homologs in humans and 9 homologs in mice (Fig. 1A and data not shown). They are highly expressed in ECs and were shown to play a role in angiogenesis in an in vitro study (42). However, their function in retinal angiogenesis is currently unknown. To study the role of let-7 family in the retinal angiogenesis, the expression of let-7 family members in several retinal cell types, including human retinal ECs (HRECs), human choroidal ECs (HCECs), and human retinal pigment epithelial (RPE) cells (ARPE-19), was compared to that in human umbilical vein ECs (HUVECs) by quantitative reverse transcription-PCR (qRT-PCR). After normalization to U6, the let-7a, let-7b, let-7c, let-7d, let-7g, and let-7i expression was found to be much higher in all four cell types than that of let-7e, let-7f, and miR-98 (Fig. 1B). We also examined the expression of let-7 family members in adult mouse retina and RPE/choroid tissues and found the same 6 let-7 members to be expressed at much higher levels than the other three let-7 members, a finding consistent with their expression pattern in human retinal cells (Fig. 1C). Interestingly, let-7b, let-7c, and let-7i are expressed at >2-fold-higher levels in the RPE/choroid compared to the retina. These results indicate the let-7 family members are expressed in retinal tissues, including retinal ECs, choroid ECs, and RPE cells.
FIG 1.
let-7 expression in retinal cells and tissues. (A) Sequence alignment of the human let-7 family members. The nucleotides that are different from the majority of let-7 family members are labeled in red. The box indicates the seed region (nucleotides 2 to 8). A 16-mer LNA–anti-miR designed to silence let-7 members with complementarity to nucleotides 2 to 17 of let-7a and let-7c is shown below. (B) Relative expression level of let-7 family members in HUVECs, HRECs, HCECs, and ARPE cells as detected by qPCR. (C) Relative expression level of let-7 family members in the whole retina or RPE/choroid from 1-month-old wild-type C57BL6 mice. U6 served as a normalization control for miR qPCRs.
Regulation of EC angiogenic behavior by let-7.
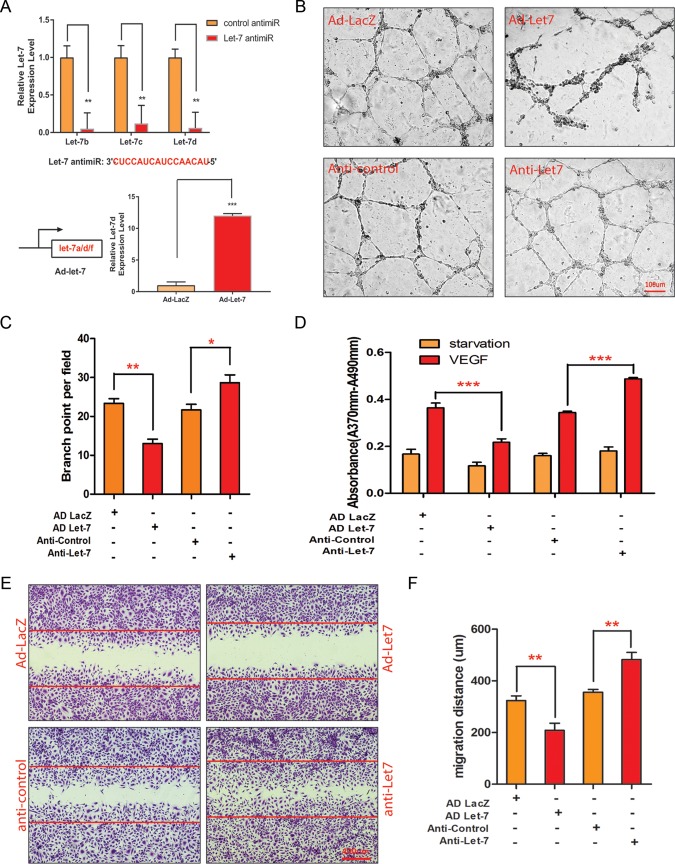
For let-7 functional study, a specific LNA-modified anti-miR has been designed to silence all let-7 family members to circumvent the functional redundancy of the let-7 family members (Fig. 1A) (22). Overexpression of let-7 was achieved by an adenovirus expressing the let-7a/d/f cluster (Ad-let-7). The efficacy of the let-7 silencing by anti-miR and let-7 overexpression by adenovirus in HUVECs was confirmed by qRT-PCR (Fig. 2A).
FIG 2.
Regulation of angiogenic activities by let-7. (A) Relative expression of let-7 family members let-7b, let-7c, and let-7d in HUVECs after let-7 anti-miR transfection as detected by qPCR. U6 served as a normalization control. An anti-let-7 sequence is shown. **, P < 0.01. The relative expression of let-7d after Ad-let-7a/d/f or Ad-LacZ infection in HUVECs was detected by qPCR. U6 served as a loading control. ***, P < 0.001. (B) Representative pictures of in vitro Matrigel assays after Ad-let-7 or Ad-LacZ infection or after anti-let-7 or control anti-miR transfection in HUVECs. (C) Quantification of branching point per field in panel B. (D) Quantification of VEGF-induced proliferation indicated by BrdU incorporation after Ad-let-7 and anti-let-7 transfection in HUVECs. ***, P < 0.001. (E) Representative pictures of migration assays in response to VEGF treatment after Ad-let-7 and anti-let-7 transfection in HUVECs. (F) Quantification of the migration distance (in micrometers) depicted in panel E.
To define the function of let-7 in angiogenesis, an in vitro Matrigel network formation assay was performed with HUVECs transfected with control and let-7 anti-miRs, respectively, or infected with adenovirus overexpressing let-7 or a LacZ control. Cultured ECs form primary vascular tubular network at 6 to 18 h after seeding on Matrigel. Overexpression of let-7 by adenovirus repressed tube formation, as quantified by a significantly reduced number of branch points (23.3 ± 1.2 in control versus 10.3 ± 1.6 in Ad-let-7, n = 3, P < 0.0034), while silencing of let-7 with the LNA–anti-miR significantly enhanced the formation of tubular structures (21.7 ± 1.5 in control versus 28.3 ± 12.0 in anti-let-7, n = 3, P < 0.049) (Fig. 2B and C). Angiogenesis is a multistep process in which EC proliferation and migration play a key role. To examine the cellular mechanism of let-7 in regulating angiogenesis, a bromodeoxyuridine (BrdU) incorporation assay and a scratch wound assay were performed to test the effect of let-7 overexpression or silencing on EC proliferation and migration. Under the basal starvation condition, either let-7 overexpression or silencing had no significant effect on EC proliferation, as shown by similar levels of BrdU incorporation (Fig. 2D). However, let-7 overexpression strongly repressed EC proliferation in response to VEGF stimulation (absorbance = 0.36 ± 0.01 in Ad-LacZ versus 0.22 ± 0.009 in Ad-let-7, n = 3, P < 0.0006), whereas silencing of let-7 with LNA–anti-miR significantly increased EC proliferation (absorbance = 0.34 ± 0.003 in anti-control versus 0.48 ± 0.003 in anti-let-7, n = 3, P < 0.0001). In the scratch wound cell migration assay, VEGF-stimulated ECs are allowed to migrate into the wounded region. The HUVECs were treated with 5-fluorouracil to rule out the influence of EC proliferation on migration. Overexpression of let-7 significantly inhibited EC migration in response to VEGF (migration distance = 324 ± 10 μm in Ad-LacZ control versus 209 ± 15 μm in Ad-let-7, n = 3, P < 0.004), whereas let-7 LNA anti-miR drastically enhanced EC migration (migration distance = 356 ± 6 μm in anti-control versus 482 ± 16 μm in anti-let-7, n = 3, P < 0.002) (Fig. 2E and F). These data indicate that let-7 is sufficient and necessary to repress EC proliferation, migration, and EC networking in vitro.
Silencing of the let-7 target HMGA2 gene suppresses EC angiogenic behavior.
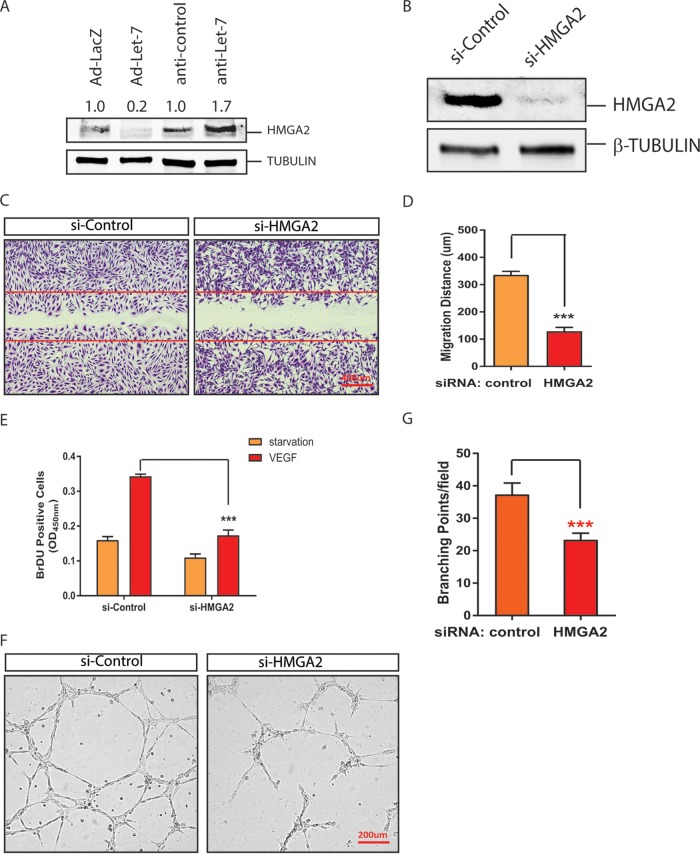
let-7 family members have been shown to target multiple genes and pathways to regulate cell growth and invasion (43). To determine the mechanism whereby let-7 regulates angiogenesis, we focused on one of the key let-7 target genes: the high-mobility-group A2 (HMGA2) gene (44–46). HMGA2 is a nonhistone chromatin protein and an architectural transcription factor that is important for tissue growth and tumorigenesis (47, 48). To determine whether let-7 targets HMGA2 in ECs, the expression of HMGA2 protein was examined after adenovirus overexpression of let-7 and anti-miR let-7 silencing, respectively. HMGA2 protein levels were suppressed by let-7 overexpression and significantly upregulated by let-7 silencing (Fig. 3A). To test whether silencing of HMGA2 mimics the angiogenic phenotype of let-7 overexpression, HMGA2 was silenced with a specific small interfering RNA (siRNA) (Fig. 3B). As shown in Fig. 3C to G, HMGA2 silencing repressed EC proliferation and migration in response to VEGF and network formation in a Matrigel assay, suggesting that HMGA2 at least partially mediates the angiogenic effects of let-7 in ECs.
FIG 3.
Regulation of EC angiogenic activities by let-7 target HMGA2 gene. (A) Representative Western blot analysis showing regulation of HMGA2 protein level by let-7 silencing or overexpression in HUVEC cells. Tubulin was used as a loading control. The band density is indicated. (B) Silencing of HMGA2 by specific siRNAs in ECs, as demonstrated by Western blotting. Tubulin was used as a loading control. (C) EC migration in response to VEGF treatment after HMGA2 knockdown with siRNAs. (D) Quantification of migration distance (in micrometers). ***, P < 0.001. (E) Quantification of VEGF-induced proliferation indicated by BrdU incorporation after HMGA2 silencing in HUVECs. ***, P < 0.001. (F) Representative pictures of in vitro Matrigel assays after HMGA2 silencing in HUVECs. (G) Quantification of branching point per field in panel F. ***, P < 0.001.
Repression of retinal vascular development in mice overexpressing let-7.
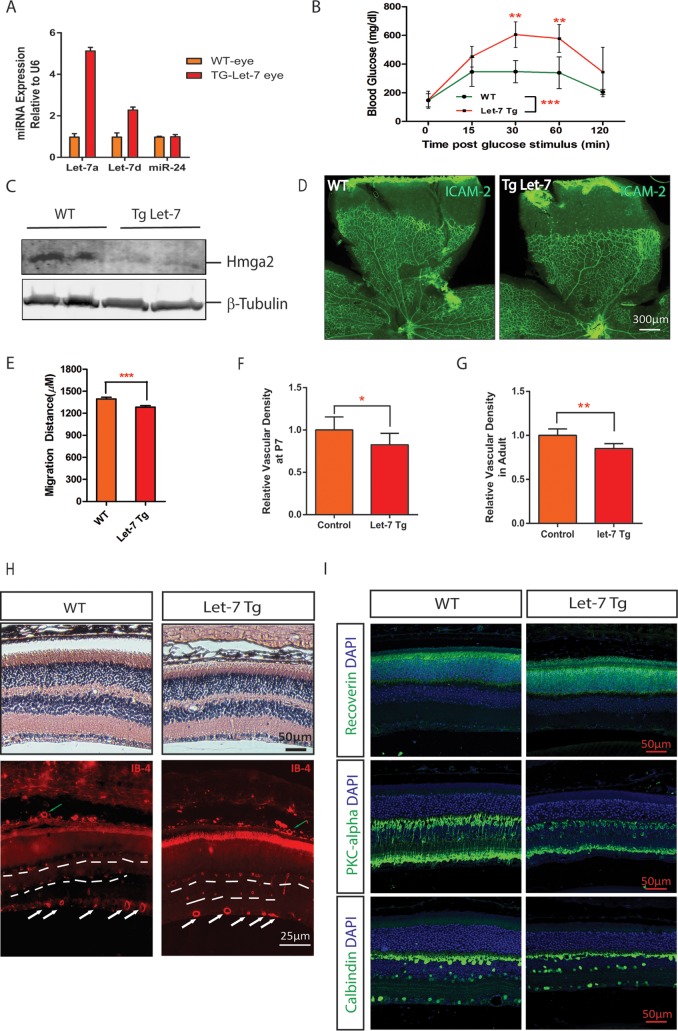
To study the function of let-7 in retinal development and retinal angiogenesis in vivo, we took advantage of the let-7-transgenic (let-7-Tg) mice we had generated previously (22). In these mice, let-7 (a, d, and f) expression was turned on when the mice expressing inactive let-7 transgene were crossed to transgenic mice overexpressing CAG-Cre. In the let-7-Tg mice, let-7a was upregulated by ∼5-fold and let-7d by ∼2-fold in the retina (Fig. 4A). Consistent with the previous report, let-7-Tg mice showed significantly reduced body weight, elevated glucose levels, and an impaired glucose tolerance response compared to the littermate wild-type (WT) control mice (Fig. 4B and data not shown) (22). Consistently, the downregulation of let-7 target protein HMGA2 was confirmed in the retinas of the let-7-Tg mice (relative band density = 0.9 ± 0.1 in WT and 0.45 ± 0.05 in let-7-Tg, **, P < 0.01) (Fig. 4C). Retinal vascularization in mice begins after birth when vessels sprout from the central retinal artery to the periphery region. To study the retinal vascular development phenotype in the let-7-Tg mice, we stained the retinal vasculature with ICAM-2, a marker for retinal endothelial cells. As shown in Fig. 4B, the P6 retina vasculature appeared normal in let-7-Tg mice compared to the WT littermates (Fig. 4D). However, detailed quantification revealed that the retinal vessel sprouting distance and retinal vessel density were mildly but significantly reduced in let-7-Tg mice compared to WT controls (1,396 ± 22.5 μm (n = 40) in control versus 1,284 ± 20.5 μm in let-7-Tg mice (n = 35), P < 0.0005) (Fig. 4E and F). Of note, the sizes of the retinas in let-7-Tg mice were comparable to those of WT controls. The decrease in retinal vascular density persisted in adult let-7-Tg mice, as shown by quantification of the superficial layer of the retinal vasculature (Fig. 4G). By isolectin-B4 staining of vessels in the retinal sections, the deep-layer and intermediate vessels (shown by dashed lines), as well as the choroid vessels (shown by green arrows), appeared normal in adult let-7-Tg mice, indicating that the patterning of retinal and choroid vessels was not affected in let-7-Tg mice (Fig. 4H). Moreover, retinal structures appeared normal in let-7-Tg mice by hematoxylin-and-eosin (H&E) staining. By immunostaining, photoreceptor cells labeled by antibody to recoverin, bipolar cells labeled by antibody to protein kinase Cα (PKCα), and horizontal cells and retinal ganglion cells labeled by antibody to calbindin appeared to be normal in let-7-Tg mice (Fig. 4I). These data suggest that let-7 overexpression mildly represses retinal vascular development without affecting the patterning of the major retinal cell types.
FIG 4.
Delayed retinal vascular development in let-7 transgenic mice. (A) qPCR results showing let-7a and let-7d expression in the retinas of WT and let-7-Tg mice. miR-24 served as a control. (B) let-7-Tg mice show impaired glucose tolerance in let-7-Tg mice. The blood glucose level at 0 to 120 min after glucose stimulation was measured. Statistics were derived from three wild-type and three let-7 transgenic mice. **, P < 0.01; ***, P < 0.001. (C) Downregulation of let-7 target protein Hmga2 in the retinas of let-7-Tg mice, as shown by Western blotting. β-Tubulin was used as a loading control. Band density analyses were performed from three WT and three let-7 transgenic mice (band density = 0.9 ± 0.1 in WT versus 0.45 ± 0.05 in let-7-Tg; **, P < 0.01). (D) Flat-mount ICAM-2 staining of retinal vasculature in WT and let-7-Tg mice. (E) Quantification of retinal vessel sprouting distance in panel C. ***, P < 0.001. (F) Quantification of the retinal vessel densities shown in panel C. *, P < 0.05. (G) Quantification of the retinal vessel densities in adult WT and let-7-Tg mice. **, P < 0.01. (H) H&E staining and isolectin-B4 staining of retinal sections in WT and let-7-Tg mice. White arrows indicate the superficial retinal vessels, green arrows indicate the choroidal vessels, and dashed lines ondicate the intermediate and deep layers of retinal vessels. Note that the rupture in WT retinas in the H&E staining images is a histological artifact. (I) Antibody staining of retinal sections in WT and let-7-Tg mice. The antibodies used included recoverin (a marker of photoreceptor cells), PKC-alpha (a marker of bipolar cells), and calbindin (a marker of ganglion cells). DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the nuclei. Scale bar, 50 μm.
let-7-Tg mice develop features of NPDR.
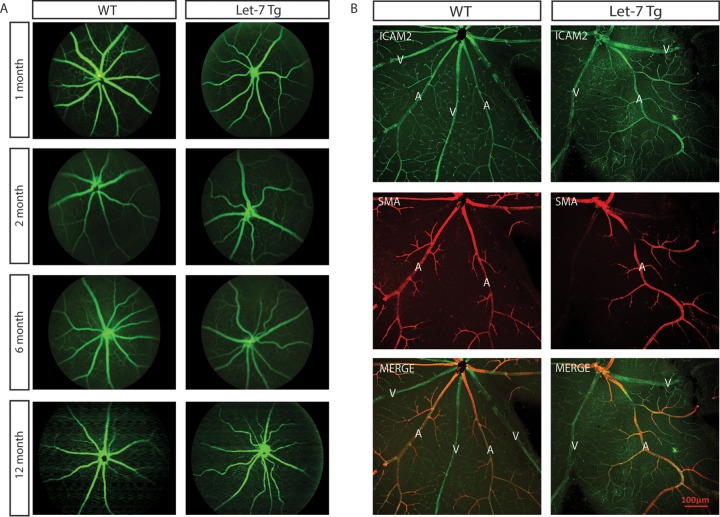
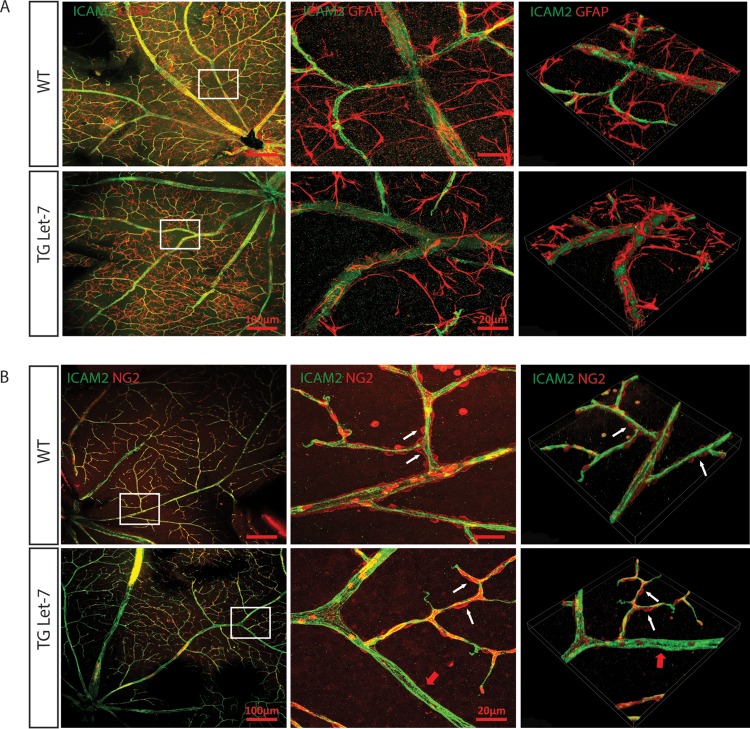
Since let-7-Tg mice showed diabetic phenotypes, we tested whether let-7-Tg mice develop the diabetic retinopathy phenotype (22). The retinal vasculature was monitored by fluorescence angiography from 1 month to 1 year. As shown in Fig. 5A, let-7-Tg mice started to exhibit vessel tortuosity at ∼2 months, which persisted at all later stages tested. As determined by fluorescence angiography, the retinal vessels were not leaky in let-7-Tg mice compared to the WT littermates, suggesting no retinal neovascularization in let-7-Tg mice. The retinas were also stained with an antibody against ICAM2 and an antibody against smooth muscle α-actin (α-SMA) (Fig. 5B). ICAM2 staining confirmed that the retinal vessel patterning is normal. α-SMA staining confirmed that the tortuous vessels are all arteries and not veins. To further characterize the other retinal cell types involved in retinal vascular function in let-7-Tg mice, retinal astrocytes and pericytes were stained with antibodies to glial fibrillary acidic protein (GFAP) and neural/glial antigen 2 (NG2), respectively (Fig. 6). No difference was observed in astrocytes, as shown by GFAP staining. However, patchier pericyte coverage was observed in the let-7-Tg mice, suggesting defective pericyte coverage in let-7-Tg mice (pericyte loss is indicated by the red arrows in the let-7-Tg retina in Fig. 6B). Taken together, retinal vessel tortuosity and defective pericyte coverage without neovascularization suggest features of NPDR in the let-7-Tg mice.
FIG 5.
let-7 transgenic mice show tortuous retinal vessels. (A) Representative angiogram pictures shown tortuous retinal vessels in let-7-Tg mice aged 2 to 12 months but not in WT littermates. (B) ICAM-2 and α-SMA staining of 4-month-old WT and let-7-Tg retinas. Normal smooth muscle coverage but tortuous arteries were observed in let-7 transgenic retinas.
FIG 6.
let-7 transgenic mice have normal astrocyte coverage but defective pericyte coverage. (A) Normal astrocyte coverage in 4-month-old let-7-Tg mice compared to WT mice, as shown by ICAM-2 and GFAP costaining. The images in the middle panels represent the boxed areas in the left panels. The images in the right panels are three-dimensional constructions of the representative confocal images. (B) Defective pericyte coverage in 4-month-old let-7-Tg mice compared to WT mice, as shown by ICAM-2 and NG2 costaining. The images in the middle panels represent the boxed areas in the left panels. The images in the right panels are three-dimensional constructions of the representative confocal images. White arrows indicate the pericyte coverage in the arterioles, while red arrows indicate the lack of pericyte coverage in some regions of the let-7-Tg mice.
Regulation of choroidal neovascularization by let-7.
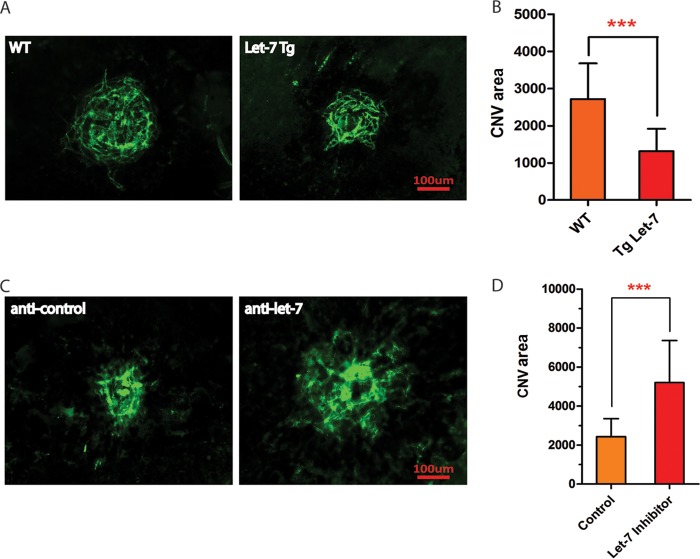
To test the involvement of let-7 in pathological angiogenesis, a laser-induced CNV mouse model, a classic model for CNV in neovascular AMD, was adopted (49). Laser injury was performed in let-7-Tg mice and WT control littermates, and the neovessels that grew out of the choroid were stained with ICAM2 after flat mount at 14 days postinjury. CNV area was significantly less in let-7-Tg mice compared to WT controls (WT = 2,719 ± 195.6 μm2, n = 24; let-7-Tg = 1,314 ± 142.0 μm2 [n = 18, P < 0.0001]) (Fig. 7A and B). To further examine whether laser-induced CNV is enhanced by let-7 silencing, LNA–anti-let-7 or control anti-miR was subretinally injected into WT mice, and CNV was quantified (36). Compared to the control anti-miR, let-7 anti-miR significantly promoted laser-induced CNV, as shown by ICAM2 staining and quantification (control anti-miR = 2,432 ± 264.5 μm2, n = 12; let-7 anti-miR = 5,196 ± 652.3 μm2 [n = 11. P < 0.0006]) (Fig. 7C and D). These results demonstrate that let-7 is sufficient and necessary to repress neovascularization in vivo.
FIG 7.
let-7 regulates pathological angiogenesis in mice. (A) Representative confocal images of ICAM-2 staining showing repression of laser-induced CNV in let-7-Tg mice compared to the controls. Scale bar, 100 μm. (D) Quantification of CNV area (μm2). The P value was calculated from the CNV area from 24 WT and 18 let-7 transgenic mice. ***, P < 0.001. (C) Representative confocal images of ICAM-2 staining showing an increase in laser-induced CNV by LNA–anti-let-7 compared to a scramble control. Scale bar, 100 μm. (B) Quantification of CNV area (μm2). The P value is calculated from the CNV area from 12 control anti-miR-injected and 11 LNA–anti-let-7-injected mice. ***, P < 0.001.
DISCUSSION
Our results reveal an important function for let-7 in diabetic retinopathy and ocular angiogenesis. We found that (1) let-7 is sufficient and necessary to repress the proliferation, migration, and networking of ECs in vitro; (2) silencing of the let-7 target HMGA2 gene mimics the antiangiogenic function of let-7 in ECs; (3) let-7-Tg mice develop features of NPDR, including tortuous retinal vessels and defective pericyte coverage; and (4) let-7-Tg mice show reduced CNV, while silencing of let-7 enhances CNV in vivo. These data establish an important role for let-7 in repressing pathological angiogenesis, suggesting therapeutic implications in angiogenesis-related diseases. let-7 also contributes to NPDR without progressing to PDR. our studies raise caution to design a general let-7-based therapeutic approach in disease settings.
let-7 in angiogenesis.
let-7 is a known regulator of tissue terminal differentiation and a tumor suppressor (50). However, its function in angiogenesis remains unclear. By using adenovirus to overexpress let-7a/d/f and let-7 anti-miR to silence multiple members of the let-7 family, we found that let-7 overexpression is sufficient to repress EC proliferation, migration, and networking, while silencing of let-7 has the opposite effects. The let-7 overexpression effects are similar to when its target HMGA2 gene is silenced. Hence, it is likely that HMGA2 at least partially contributes to the angiogenic phenotype of let-7 in angiogenesis. The HMGA2 gene is an established key let-7 target gene in several different cell types (44–46). Consistent with our results in EC cells, HMGA2 has been shown to be required and sufficient to modulate proliferation, migration, and angiogenesis in endothelial progenitor cells (EPCs) in vitro and the angiogenic capability of bone marrow cells (BMCs) in vivo (51). In an independent study, knockdown of let-7f was shown to repress angiogenesis in a spheroid sprouting assay, suggesting that let-7f may promote angiogenesis (42). The different conclusion from our studies may be a consequence of the different system used or the specific function of let-7f. In our case, to circumvent the potential redundancy of let-7 family members, the let-7 LNA–anti-miR was designed to silence multiple members of the let-7 family. Our in vitro results are consistent with our in vivo laser injury experiments showing that laser-induced CNV is significantly reduced in let-7-Tg mice but increased when let-7 is silenced in the eye. Interestingly, in the let-7-Tg mice, we did not observe a strong effect of let-7 in retinal vascular development, although the retinal vessel density was reduced in adult let-7-Tg mice compared to WT controls. These findings suggest that let-7 regulates pathological angiogenesis with minimal effect on developmental angiogenesis. In this regard, it would be interesting to test the effect of let-7 overexpression in tumor angiogenesis because of the documented function of let-7 as a tumor suppressor.
let-7 in diabetic retinopathy.
let-7 has been shown to regulate glucose metabolism and insulin sensitivity (21, 22). Consistently, let-7 targets are enriched in genes that contain single-nucleotide polymorphisms associated with type 2 diabetes (21). Our study suggests that let-7 is involved in diabetes-associated retinal diseases. We show that let-7-Tg mice show features of DR, which include tortuous retinal vessels and defective pericyte coverage. No obvious difference was observed in smooth muscle cells and astrocytes. In these mice, retinal neovascularization and blood vessel leakage was not observed, suggesting that let-7 overexpression leads to a nonproliferative DR phenotype without progressing to PDR. One possible reason that PDR was not observed in let-7-Tg mice is that let-7 has a repressive role in angiogenesis. To our knowledge, our study provides the first in vivo evidence that a miR is causative for NPDR. Our data that let-7 overexpression leads to NPDR but has a repressive role in CNV suggests different mechanisms for NPDR and wet AMD. One caveat of the study is that the let-7-Tg mice examined here are global transgenic mice; therefore, it is unclear which cell type is responsible for the NPDR phenotype in the let-7-Tg mice. It would also be interesting to characterize the let-7-TG phenotype in the aged mice to determine whether they progress to PDR eventually. In addition to the HMGA2 gene, additional let-7 target genes involved in angiogenesis and NPDR await to be discovered.
Therapeutic implications.
Our data that let-7 contributes to diabetic retinopathy but represses pathological angiogenesis has important therapeutic implications. LNA-modified anti-let-7, which has been shown to prevent impaired glucose tolerance and improve insulin sensitivity, could be beneficial for DR, whereas let-7 overexpression may have implications in CNV in AMD and pathological angiogenesis. Cautions should be taken when applying these technologies because let-7 overexpression may lead to DR, while let-7 silencing may increase pathological angiogenesis. Future studies are needed unveil the cell-type-specific or tissue-specific function of let-7 in DR and pathological angiogenesis to develop tissue-specific let-7-based therapeutic approaches for DR and angiogenesis-related diseases.
MATERIALS AND METHODS
Animal.
Animal studies were conducted in accordance with the ARVO statement for the use of animals in ophthalmic and vision research and were approved by the Institutional Animal Care and Use Committees at the Tulane University. let-7 transgenic mice were used as reported previously (22).
Fluorescein angiography.
Fluorescein angiography was performed using a Micron III Fundus camera connected to the Image system (Phoenix Research Labs, Pleasanton, CA). Briefly, fluorescein dye (0.1 ml/kg of 10% fluorescein sodium) was injected intraperitoneally, and multiple pictures were taken at 30 to 300 s after injection of the dye.
Laser-induced CNV.
Laser-induced CNV was induced in 6- to 8-week-old male C57BL/6J mice or let-7-Tg mice as described using a murine Micron III system (39). If necessary, animals were injected subretinally with 1 μl of a 200-ng/μl solution of LNA-modified let-7 anti-miR or scramble control anti-miR (Exiqon, Vedbaek, Denmark) after laser photocoagulation. Two more injections were performed at days 5 and 9 after injury. At 14 days after laser injury, the treated eyes were fixed in 4% paraformaldehyde and subjected to flat-mount ICAM-2 staining. Images of the CNV or retinal vasculature were captured by using s Nikon Eclipse Ti-S/L100 inverted microscope with fluorescence and cameras or a Nikon A1 laser scanning confocal microscope, and the CNV volumes were quantified using NIH ImageJ software (52).
Cell culture, siRNA, mimic inhibitor transfection, and adenovirus infection.
HUVECs (American Type Culture Collection ([ATCC]) were grown in EC growth medium EGM-2 (Lonza). HCECs and HRECs were kindly provided by Ashwath Jayagopal from Vanderbilt University and grown in EGM2 media (Lonza). ARPE-19 (ATCC) cells were growth in DMEM–F-12 (HyClone) media with 10% fetal bovine serum (FBS). For VEGF treatment, HUVECs were starved with EC basal medium 2 with 0.1% FBS overnight and then treated with VEGF (20 ng/ml) for the indicated periods of time. LNA–anti-miRs for let-7 were synthesized from Exiqon. The sequence for let-7 LNA–anti-miR is 5′-UACAACCUACUACCUC-3′. siRNA transfection in cell culture was performed as described previously (39). siRNAs for si-HMGA2 were purchased from Sigma. The sequences for the siRNAs were as follows: si-HMGA2, 5′-TCCGATATAGAAACCTATC-dTdT-3′ and 5′-GATAGGTTTCTATATCGGA-dTdT-3′. Generation of and infection with adenoviruses were performed as described previously (37). DNA fragments containing mouse let-7a, -d, and -f were cloned into the pShuttle-CMV vector. The derived vector was then recombined into adenovirus vectors and was used to generate adenovirus. Adeno-LacZ virus was used as a control for the experiments. Normally, viruses with a multiplicity of infection of 50 to 100 were used for infecting ECs.
Proliferation, network formation, and scratch wound assays.
EC proliferation, network formation, and scratch wound assays were performed using HUVECs as described previously (39). For the cell proliferation assay, approximately 2 × 103 transfected HUVECs were seeded in 96-well plates. After starvation with 0.1% serum overnight, the cells were stimulated with VEGF-A at 20 ng/ml for 20 h and then subjected to BrdU labeling for 4 h. DNA synthesis as determined by BrdU incorporation was quantified using a commercial enzyme-linked immunosorbent assay kit from Roche according to the manufacturer's instructions. For the scratch wound assay, a scratch wound was made using a 200-μl pipette tip in control or experimental treated HUVEC monolayers before VEGF stimulation (20 ng/ml). Then, 1 μM 5-fluorouracil (Sigma) was added to the cells right after the scratch wound to block cell proliferation. Postscratch EC migration was scored 14 h after the wound scratch. For the in vitro angiogenesis assay, the cells were harvested at 3 days after microRNA inhibitor or siRNA transfection with Lipofectamine RNAiMAX reagent (Invitrogen) for RNA or Matrigel assay and branch point analysis as described previously (37).
RNA, Western blot, and immunofluorescence analyses.
Total RNA was isolated from cell lines and tissues using TRIzol reagent (Invitrogen). miR real-time qRT-PCR was performed using qScript cDNA synthesis and the microRNA quantification system (Quanta Biosciences). The primers for microRNA real-time PCRs were purchased from Quanta Biosciences. For Western blot analysis, protein lysates were resolved by SDS-PAGE and blotted according to standard procedures. The antibodies used were HMGA2 (Cell Signaling) and β-tubulin (Abcam) antibodies. For visualization of the retinal vasculature, retinas were dissected from pups or adult mice and stained with Alexa Fluor 594-conjugated isolectin-B4 (Molecular Probes) or ICAM-2 for the retinal vasculature GFAP for astrocytes, and/or NG2 for pericytes. Immunofluorescence experiments were performed according to standard protocols. After treatment with 1% Triton X-100 in phosphate-buffered saline (PBS), samples were incubated in PBS containing 4% goat serum for 30 min. The samples were then incubated with primary antibodies overnight at 4°C, followed by incubation with the appropriate secondary antibodies. Antibodies used for immunofluorescence included ICAM2 (BD Pharmingen), NG2 (Millipore), GFAP (Millipore), calbindin (Sigma), PKC-alpha (Sigma), recoverin (Millipore), and SM-alpha (Sigma).
Statistics.
Student's t tests were used to determine statistically significant differences between groups. P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank Yiping Chen and Frank Jones for encouragement, Eric Olson from University of Texas Southwestern Medical Center in Dallas for providing the let-7 transgenic mice, and Ashwath Jayagopal from Vanderbilt University for providing HCECs and HRECs for the experiments.
S.W. was supported by a startup fund from Tulane University, NIH grants EY021862 and EY026069, a career development award from the Research to Prevent Blindness foundation, and a BrightFocus Foundation Award in Age-related Macular Degeneration. Q.Z. was supported by an American Heart Association Southeast Affiliate Postdoctoral fellowship.
REFERENCES
- 1.Saint-Geniez M, D'Amore PA. 2004. Development and pathology of the hyaloid, choroidal and retinal vasculature. Int J Dev Biol 48:1045–1058. doi: 10.1387/ijdb.041895ms. [DOI] [PubMed] [Google Scholar]
- 2.Stahl A, Connor KM, Sapieha P, Chen J, Dennison RJ, Krah NM, Seaward MR, Willett KL, Aderman CM, Guerin KI, Hua J, Lofqvist C, Hellstrom A, Smith LE. 2010. The mouse retina as an angiogenesis model. Invest Ophthalmol Vis Sci 51:2813–2826. doi: 10.1167/iovs.10-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kempen JH, O'Colmam BJ, Leske C, Haffner SM, Klein R, Moss SE, Taylor HR, Hamman RF, West SK, Wang JJ, Congdon NG, Friedman DS, Grp EDPR . 2004. The prevalence of diabetic retinopathy among adults in the United States. Arch Ophthalmol 122:552–563. doi: 10.1001/archopht.122.4.552. [DOI] [PubMed] [Google Scholar]
- 4.Jager RD, Mieler WF, Miller JW. 2008. Age-related macular degeneration. N Engl J Med 358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 5.Brown DM, Kaiser PK, Michels M, Soubrane G, Heier JS, Kim RY, Sy JP, Schneider S, Group AS. 2006. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med 355:1432–1444. doi: 10.1056/NEJMoa062655. [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeld PJ, Brown DM, Heier JS, Boyer DS, Kaiser PK, Chung CY, Kim RY, ANCHOR Study Group . 2006. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med 355:1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 7.Zampros I, Praidou A, Brazitikos P, Ekonomidis P, Androudi S. 2012. Antivascular endothelial growth factor agents for neovascular age-related macular degeneration. J Ophthalmol 2012:319728. doi: 10.1155/2012/319728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardlik R, Fusekova I. 2015. Pharmacologic therapy for diabetic retinopathy. Semin Ophthalmol 30:252–263. doi: 10.3109/08820538.2013.859280. [DOI] [PubMed] [Google Scholar]
- 9.Folk JC, Stone EM. 2010. Ranibizumab therapy for neovascular age-related macular degeneration. N Engl J Med 363:1648–1655. doi: 10.1056/NEJMct1000495. [DOI] [PubMed] [Google Scholar]
- 10.Kruger Falk M, Kemp H, Sorensen TL. 2013. Four-year treatment results of neovascular age-related macular degeneration with ranibizumab and causes for discontinuation of treatment. Am J Ophthalmol 155:89–95. doi: 10.1016/j.ajo.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 11.Small EM, Olson EN. 2011. Pervasive roles of microRNAs in cardiovascular biology. Nature 469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garofalo M, Croce CM. 2011. microRNAs: Master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol 51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 13.Santulli G. 2016. MicroRNAs and endothelial (Dys) function. J Cell Physiol 231:1638–1644. doi: 10.1002/jcp.25276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mastropasqua R, Toto L, Cipollone F, Santovito D, Carpineto P, Mastropasqua L. 2014. Role of microRNAs in the modulation of diabetic retinopathy. Prog Retinal Eye Res 43:92–107. doi: 10.1016/j.preteyeres.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Wang SS, Koster KM, He YG, Zhou QB. 2012. miRNAs as potential therapeutic targets for age-related macular degeneration. Future Med Chem 4:277–287. doi: 10.4155/fmc.11.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. 2011. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 17.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma XS, MacDonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 18.Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. 2006. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem 281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 19.Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Bottger T, Braun T, Seibler J, Bruning JC. 2011. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol 13:434-U208. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 20.Karolina DS, Armugam A, Tavintharan S, Wong MTK, Lim SC, Sum CF, Jeyaseelan K. 2011. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One 2011:6. doi: 10.1371/annotation/698b7123-174f-4a09-95c9-fd6f5017d622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Altshuler D, Daley GQ, Consortium D, Investigators M. 2011. The Lin28/let-7 axis regulates glucose metabolism. Cell 147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frost RJ, Olson EN. 2011. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A 108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacs B, Lumayag S, Cowan C, Xu SB. 2011. microRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci 52:4402–4409. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- 24.Wu JH, Gao Y, Ren AJ, Zhao SH, Zhong M, Peng YJ, Shen W, Jing M, Liu L. 2012. Altered MicroRNA expression profiles in retinas with diabetic retinopathy. Ophthalmic Res 47:195–201. doi: 10.1159/000331992. [DOI] [PubMed] [Google Scholar]
- 25.McArthur K, Feng BA, Wu YX, Chen SL, Chakrabarti S. 2011. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes 60:1314–1323. doi: 10.2337/db10-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray AR, Chen Q, Takahashi Y, Zhou KK, Park K, Ma JX. 2013. MicroRNA-200b downregulates oxidation resistance 1 (Oxr1) expression in the retina of type 1 diabetes model. Invest Ophthalmol Vis Sci 54:1689–1697. doi: 10.1167/iovs.12-10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng BA, Chen SL, McArthur K, Wu YX, Sen S, Ding QM, Feldman RD, Chakrabarti S. 2011. miR-146a-mediated extracellular matrix protein production in chronic diabetes complications. Diabetes 60:2975–2984. doi: 10.2337/db11-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva VA, Polesskaya A, Sousa TA, Correa VM, Andre ND, Reis RI, Kettelhut IC, Harel-Bellan A, De Lucca FL. 2011. Expression and cellular localization of microRNA-29b and RAX, an activator of the RNA-dependent protein kinase (PKR), in the retina of streptozotocin-induced diabetic rats. Mol Vis 17:2228–2240. [PMC free article] [PubMed] [Google Scholar]
- 29.Bai YY, Bai X, Wang ZY, Zhang XF, Ruan CG, Miao JC. 2011. MicroRNA-126 inhibits ischemia-induced retinal neovascularization via regulating angiogenic growth factors. Exp Mol Pathol 91:471–477. doi: 10.1016/j.yexmp.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Ye PP, Liu J, He FY, Xu W, Yao K. 2014. Hypoxia-induced deregulation of miR-126 and its regulative effect on VEGF and MMP-9 expression. Int J Med Sci 11:17–23. doi: 10.7150/ijms.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortuza R, Feng B, Chakrabarti S. 2014. miR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia 57:1037–1046. doi: 10.1007/s00125-014-3197-9. [DOI] [PubMed] [Google Scholar]
- 32.Shen JK, Yang XR, Xie B, Chen YJ, Swaim M, Hackett SF, Campochiaro PA. 2008. MicroRNAs regulate ocular neovascularization. Mol Ther 16:1208–1216. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowan C, Muraleedharan CK, O'Donnell JJ, Singh PK, Lum H, Kumar A, Xu SB. 2014. MicroRNA-146 inhibits thrombin-induced NF-κB activation and subsequent inflammatory responses in human retinal endothelial cells. Invest Ophthalmol Vis Sci 55:4944–4951. doi: 10.1167/iovs.13-13631. [DOI] [PubMed] [Google Scholar]
- 34.Long J, Wang Y, Wang W, Chang BH, Danesh FR. 2010. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem 285:23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan L, Lee S, Lazzaro DR, Aranda J, Grant MB, Chaqour B. 2015. Single and compound knockouts of MicroRNA (miRNA)-155 and its angiogenic gene target CCN1 in mice alter vascular and neovascular growth in the retina via resident microglia. J Biol Chem 290:23264–23281. doi: 10.1074/jbc.M115.646950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. 2011. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23∼27∼24 clusters. Proc Natl Acad Sci U S A 108:8287–8292. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Q, Anderson C, Zhang H, Li X, Inglis F, Jayagopal A, Wang S. 2014. Repression of choroidal neovascularization through actin cytoskeleton pathways by microRNA-24. Mol Ther 22:378–389. doi: 10.1038/mt.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabatel C, Malvaux L, Bovy N, Deroanne C, Lambert V, Gonzalez ML, Colige A, Rakic JM, Noel A, Martial JA, Struman I. 2011. MicroRNA-21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS One 6:e16979. doi: 10.1371/journal.pone.0016979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q, Anderson C, Hanus J, Zhao F, Ma J, Yoshimura A, Wang S. 2016. Strand and cell type-specific function of microRNA-126 in angiogenesis. Mol Ther 24:1823–1835. doi: 10.1038/mt.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 41.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. 2010. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer 17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 42.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. 2007. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res 101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 43.Barh D, Malhotra R, Ravi B, Sindhurani P. 2010. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol 17:70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YS, Dutta A. 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayr C, Hemann MT, Bartel DP. 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. 2008. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A 105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X, Benson KF, Ashar HR, Chada K. 1995. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 48.Zaidi MR, Okada Y, Chada KK. 2006. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res 66:7453–7459. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- 49.Ryan SJ. 1982. Subretinal neovascularization. Natural history of an experimental model. Arch Ophthalmol 100:1804–1809. [DOI] [PubMed] [Google Scholar]
- 50.Roush S, Slack FJ. 2008. The let-7 family of microRNAs. Trends Cell Biol 18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Zhu SK, Deng SM, Ma Q, Zhang TF, Jia CL, Zhuo DG, Yang FL, Wei JQ, Wang LY, Dykxhoorn DM, Hare JM, Goldschmidt-Clermont PJ, Dong CM. 2013. MicroRNA-10A* and MicroRNA-21 modulate endothelial progenitor cell senescence via suppressing high-mobility group A2. Circ Res 112:152-+. doi: 10.1161/CIRCRESAHA.112.280016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]