ABSTRACT
Despite intensive investigations, mechanisms of liver cancer are not known. Here, we identified an important step of liver cancer, which is the neutralization of tumor suppressor activities of an RNA binding protein, CUGBP1. The translational activity of CUGBP1 is activated by dephosphorylation at Ser302. We generated CUGBP1-S302A knock-in mice and found that the reduction of translational activity of CUGBP1 causes development of a fatty liver phenotype in young S302A mice. Examination of liver cancer in diethylnitrosamine (DEN)-treated CUGBP1-S302A mice showed these mice develop much more severe liver cancer that is associated with elimination of the mutant CUGBP1. Searching for mechanisms of this elimination, we found that the oncoprotein gankyrin (Gank) preferentially binds to and triggers degradation of dephosphorylated CUGBP1 (de-ph-S302-CUGBP1) or S302A mutant CUGBP1. To test the role of Gank in degradation of CUGBP1, we generated mice with liver-specific deletion of Gank. In these mice, the tumor suppressor isoform of CUGBP1 is protected from Gank-mediated degradation. Consistent with reduction of CUGBP1 in animal models, CUGBP1 is reduced in patients with pediatric liver cancer. Thus, this work presents evidence that de-ph-S302-CUGBP1 is a tumor suppressor protein and that the Gank-UPS-mediated reduction of CUGBP1 is a key event in the development of liver cancer.
KEYWORDS: CUGBP1, HCC, HBL, gankyrin, C/EBPβ, phosphorylation, C/EBP
INTRODUCTION
Quiescent livers express up to 20 tumor suppressor proteins (1); however, they are eliminated by different mechanisms under conditions of liver cancer. Liver cancer remains the fifth most common cancer and the third most common cause of cancer-related death in the world (2). The mechanisms of cancer-related elimination or reduction of tumor suppressor proteins include transcriptional repression of the genes, degradation of proteins, and posttranslational modifications that change biological activities of these proteins (1, 3, 4). A number of recent papers showed that certain proteins work as oncoproteins and as tumor suppressors depending on cell environment, target genes, and posttranslational modifications (5–12). In this regard, Dreijerink et al. have recently shown that menin tumor suppressor protein displays its tumor suppression activity in a variety of cancer cells; however, it possesses oncogenic activities in breast tumorigenesis (5). Another recent report revealed that the Kruppel-associated box zinc finger protein ZNF224 can behave as an oncoprotein and as a tumor suppressor protein and that these opposite activities of ZNF224 are controlled by specific protein-protein interactions (6). In addition to the opposite changes of the activities of tumor suppressor proteins and oncogenes, certain microRNAs display cell type-specific oncogenic or tumor suppression activities. Hickey et al. recently demonstrated that microRNA 181a (miRNA181a) is upregulated by the C/EBPα 30-kDa protein and functions as a tumor suppressor in the hematopoietic system (7). However, later studies demonstrated that this miRNA also has oncogenic activities in the myeloid background (8, 9). Similar to miRNA181a, miRNA22 first was shown to display oncogenic activity (10), but further studies in several tissue culture systems and in vivo clearly demonstrated its tumor suppressor activities (11, 12). More detailed information for the opposite functions of miRNAs can be found in a recent review (12).
CUG triplet repeat binding protein 1 (CUGBP1) is an RNA binding protein that has been shown to be an important regulator of liver homeostasis and involved in several types of cancer (3, 13). It displays its functions mainly through regulation of splicing, stability, and translation of mRNAs (13). Biological activities of CUGBP1 are regulated on several levels, including posttranslational modifications such as phosphorylation. Our previous work has shown that cyclin D1/3-cdk4 phosphorylates CUGBP1 at Ser302, leading to the activation of translation of C/EBPβ and histone deacetylase 1 (HDAC1) by binding to the 5′ region of both of these mRNAs and delivering the eukaryotic initiation factor 2 (eIF2) translational complex to initiate translation of these mRNAs (14–16). This leads to an accumulation of C/EBPβ-HDAC1 complexes which epigenetically silence downstream gene targets (15–17). The CUGBP1-C/EBPβ-HDAC1 pathway is involved in the regulation of several liver functions, including development of the aging phenotype (13), regulation of liver proliferation (17), and development of fatty liver diseases (18). In this paper, we examined the role of posttranslational modifications of CUGBP1 in liver cancer by generation of a CUGBP1-S302A knock-in mouse model, which mimics the unphosphorylated state of CUGBP1. These studies surprisingly showed that the unphosphorylated isoform of CUGBP1 is in fact a tumor suppressor which is eliminated during development of liver cancer in the majority of human hepatocellular carcinoma and hepatoblastoma samples, while the phosphorylated isoform of CUGBP1 is elevated only in rare cases of hepatoblastoma.
Liver cancer uses several mechanisms of elimination of tumor suppressor proteins (TSPs), one of which is the degradation of TSPs by the oncoprotein gankyrin (Gank). Gank is a small non-ATP subunit of 26S proteasome which is increased in hepatocellular carcinoma (3, 4, 19, 20). It has previously been shown that the age-associated development of liver cancer is mediated by activation of Gank (4) and that development of liver cancer in animal models of carcinogenesis involves activation of Gank (21–23). Here, we describe the role of CUGBP1-S302 phosphorylation in liver biology and cancer and provide evidence that CUGBP1 is both a tumor suppressor and target of Gank-mediated degradation when unphosphorylated at this residue.
RESULTS
Generation of CUGBP1 S302A knock-in mice with reduced translational activity of CUGBP1.
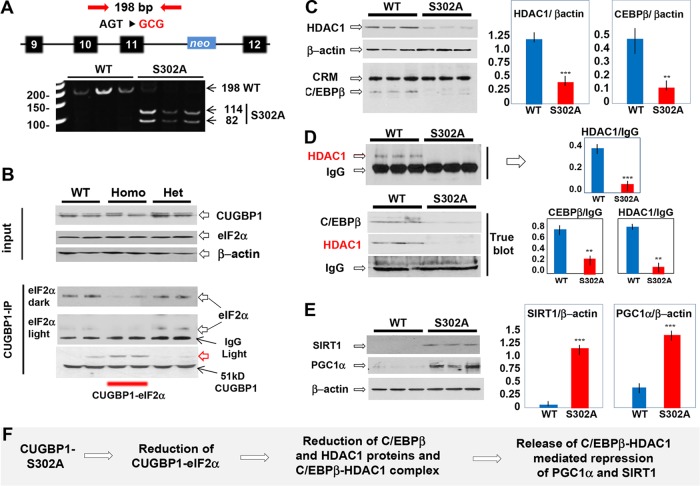
We have reported that translational activity of CUGBP1 is involved in repression of p53 and p21 genes during diethylnitrosamine (DEN)-mediated cancer (16, 24), suggesting that CUGBP1 promotes liver cancer through its translational activity. We also found that phosphorylation of CUGBP1 at Ser302 is changed after DEN treatments (25). To test the role of translational activity of CUGBP1 in cancer, we generated CUGBP1-S302A knock-in mice. We have replaced endogenous CUGBP1 with mutant S302A (Fig. 1A). We first examined translational activities of CUGBP1 by measuring cytoplasmic CUGBP1-eIF2 complexes and found that amounts of CUGBP1-eIF2 complexes are reduced in homozygous S302A mice (Fig. 1B). We next determined amounts of CUGBP1 in CUGBP1 immunoprecipitation (IP); as can be seen in Fig. 1B (bottom), amounts of CUGBP1 are identical in all IPs. We identified an immunoreactive additional isoform of CUGBP1 with a mass of around 60 kDa (Fig. 1B, red arrow). Our studies have revealed that this isoform represents CUGBP1-ubiquitin conjugates. Since the CUGBP1-eIF2 complexes positively regulate translation of several mRNAs, we examined levels of these proteins and found that translational targets of CUGBP1-eIF2 complexes C/EBPβ and HDAC1 are also reduced (Fig. 1C). We next tested formation of C/EBPβ-HDAC1 repressor complexes and levels of its targets, PGC1α and SIRT1. We performed coimmunoprecipitation (co-IP) studies using two IP protocols: regular and TrueBlot co-IP reagents. The use of the TrueBlot system is highly important, since this system reduces IgG signals and is more accurate. Both protocols revealed that C/EBPβ-HDAC1 complexes are reduced in S302A mice (Fig. 1D). Western blotting showed that the reduction of C/EBPβ-HDAC1 complexes resulted in derepression of SIRT1 and PGC1α (Fig. 1E). These studies showed that we have generated knock-in mice in which the translational activity of CUGBP1 is inhibited and known downstream translational targets are reduced (Fig. 1F).
FIG 1.
Generation of CUGBP1-S302A mice, which have reduced translational activity of CUGBP1. (A) The mutation of the S302 codon creates a restriction site for EcoRI, used for genotyping. (B) Levels of cytoplasmic CUGBP1 and eIF2α in livers of 2-month-old WT, heterozygous (Het), and homozygous (Homo) S302A mice determined by Western blotting. For CUGBP1 IP, CUGBP1 was immunoprecipitated and eIF2α was examined in these IPs. The upper image shows input proteins, and the bottom image shows an examination of eIF2α and CUGBP1 in CUGBP1 IPs. The red arrow shows a new immune-reactive CUGBP1 form in homozygous CUGBP1-S302A mice. Dark and light panels indicate dark and light exposures of the filter. IgG light indicates light chains of IgG. (C, left) Examination of downstream targets of CUGBP1-eIF2 complex in livers of S302A mice by Western blotting. Cross-reactive material (CRM) shows loading of protein. (Right) Protein levels of HDAC1 and C/EBPβ were calculated as ratios to β-actin. (D) Examination of C/EBPβ-HDAC1 complexes in livers of S302A mice using regular reagents for co-IP (upper) and TrueBlot reagents (bottom). Bar graphs on the right show quantifications of Western blotting signals for HDAC1 and C/EBPβ as ratios to IgG. (E) Examination of downstream targets of C/EBPβ-HDAC1 complexes, SIRT1, and PGC1α in livers of S302A mice. Images on the left show results of Western blotting; bar graphs show levels of these proteins as ratios to β-actin. (F) A summary of the investigations of translational pathways of CUGBP1 in livers of CUGBP1-S302A mice. **, P < 0.01; ***, P < 0.005.
Livers of CUGBP1-S302A mice are characterized by changes of liver morphology and liver functions.
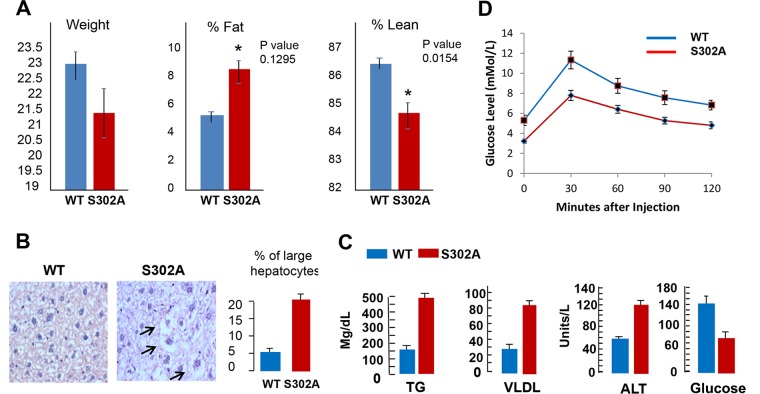
General examination showed that S302A mice have reduced weight and increased fat and are less lean (Fig. 2A). Since we found that S302A mice have increased fat and because livers of S302A mice showed significant changes in three key regulators of liver functions (C/EBPβ, SIRT1, and PGC1α), we examined liver morphology in these mice. Hematoxylin and eosin (H&E) staining of livers of 2-month-old mice revealed that S302A mice have a significant portion of hepatocytes (around 20%) with increased size, and these hepatocytes have accumulation of fat droplets in cytoplasm, suggesting that they have developed fatty liver characteristics (Fig. 2B). In agreement with these results, examination of blood parameters showed elevation of triglycerides (TG), alanine aminotransferase (ALT), and very-low-density lipoproteins (VLDL) in the serum of S302A mice (Fig. 2C). We also observed a reduction of glucose in serum of S302A mice (Fig. 2C), while other blood parameters were identical in wild-type (WT) and S302A mice (data not shown). Given the fatty liver phenotype and alteration of glucose, we next performed a glucose tolerance test. Figure 2D shows that although total levels of glucose are lower in S302A mice at all time points, lowering of glucose levels after injection of glucose has the same kinetics in WT and S302A mice.
FIG 2.
S302A mutation changes liver morphology and liver functions. (A) Examination of body weight and levels of total fat and leanness in CUGBP1-S302A mice. *, P < 0.05. (B) Livers of S302A mice have increased numbers of enlarged hepatocytes with accumulated fat droplets. H&E staining was performed with 2-month-old WT and S302A mice. Bar graphs show percentages of large hepatocytes containing fat droplets. (C) S302A mice have increased triglyceride, VLDL, and ALT levels in blood serum, but levels of glucose are reduced. (D) Glucose tolerance test.
Livers of CUGBP1-S302A mice develop fatty liver phenotype at 2 months of age.
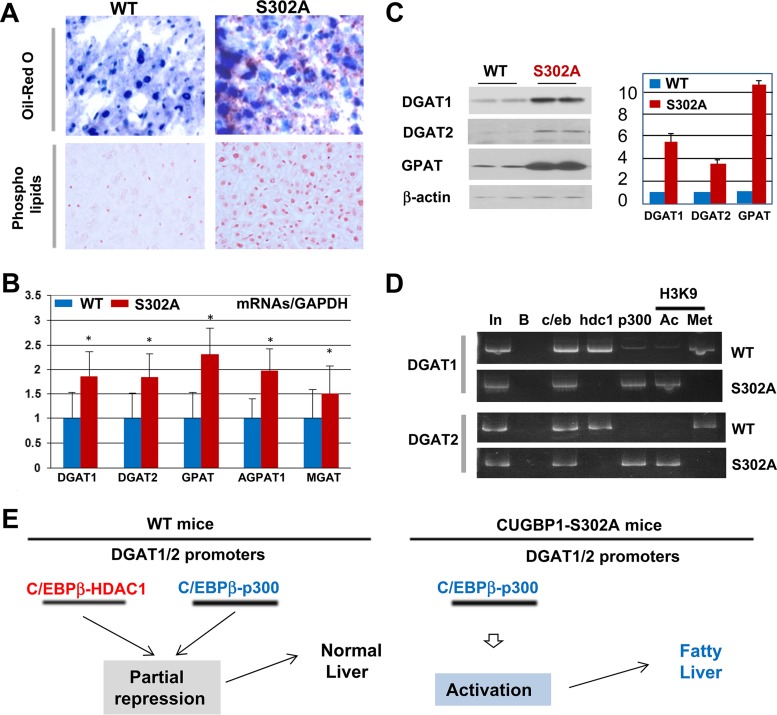
H&E staining suggested that livers of S302A mice contain increased amounts of fat droplets (Fig. 2B). Therefore, we performed two additional tests for fatty liver phenotype: Oil Red O and phospholipid staining. These approaches revealed that S302A mice develop a fatty liver phenotype at 2 months of age (Fig. 3A). We next determined molecular mechanisms that are involved in development of fatty liver in S302A mice. Our previous studies of fatty liver diseases have shown that promoter regions of five enzymes of TG synthesis, GPAT, AGPAT1, MGAT, DGAT1, and DGAT2, contain one or two high-affinity binding sites for C/EBP family proteins (18). Since livers of S302A mice have dramatically reduced C/EBPβ-HDAC1 complex repressors, we hypothesized that the reduction of these complexes eliminates partial repression of the promoters of enzymes of TG synthesis. To test this hypothesis, we examined levels of corresponding mRNAs. Figure 3B shows that amounts of all of these mRNAs are 1.5- to 2-fold higher in livers of S302A mice than in livers of WT mice. Western blotting revealed that protein levels of GPAT, DGAT1, and DGAT2 are also elevated in livers of S302A mice (Fig. 3C). To directly test if the reduction of C/EBPβ-HDAC1 complexes is involved in derepression of the enzymes of TG synthesis, we performed chromatin immunoprecipitation (ChIP) assay with DGAT1 and DGAT2 promoters and examined occupation of these promoters by C/EBPβ-HDAC1 and C/EBPβ-p300 complexes. Although both of these complexes are detected on the promoters, amounts of CEBPβ-HDAC1 complexes are much higher in WT mice (Fig. 3D). However, in livers of S302A mice C/EBPβ-HDAC1 complexes are almost undetectable, while C/EBPβ-p300 complexes are abundant on the DGAT1/2 promoters. We also examined the status of the promoters by determining histone H3 modifications at lysine 9 (K9). As shown in Fig. 3D, H3K9 is mainly methylated on the DGAT1/2 promoters in WT mice; however, it is acetylated on these promoters in S302A mice. This pattern of alterations clearly demonstrates that the DGAT1 and DGAT2 promoters are activated in livers of S302A mice. Figure 3E summarizes studies of fatty liver phenotype in S302A mice. Our data suggest that livers of S302A mice develop a fatty liver phenotype due to derepression of the enzymes of TG synthesis.
FIG 3.
CUGBP1-dependent reduction of C/EBPβ-HDAC1 complexes leads to development of fatty liver phenotype in S302A mice. (A) Livers were stained with Oil Red O and with antibodies to phospholipids. (B) RT-PCR of 5 enzymes of triglyceride synthesis. *, P < 0.05. (C) Protein levels of DGAT1, DGAT2, and GPAT were determined in WT and S302A mice by Western blotting (images on the left) and calculated as ratios to β-actin (bar graphs). (D) ChIP analyses of the promoters of enzymes of TG synthesis, DGAT1 and DGAT2. In, 1/100 input; B, beads; c/eb, C/EBPβ; hdc1, HDAC1; Ac and Met, histone H3K9 acetylated or trimethylated. (E) A diagram showing derepression (activation) of enzymes of TG synthesis.
CUGBP1-S302A mice develop more severe liver cancer under conditions of DEN-mediated carcinogenesis.
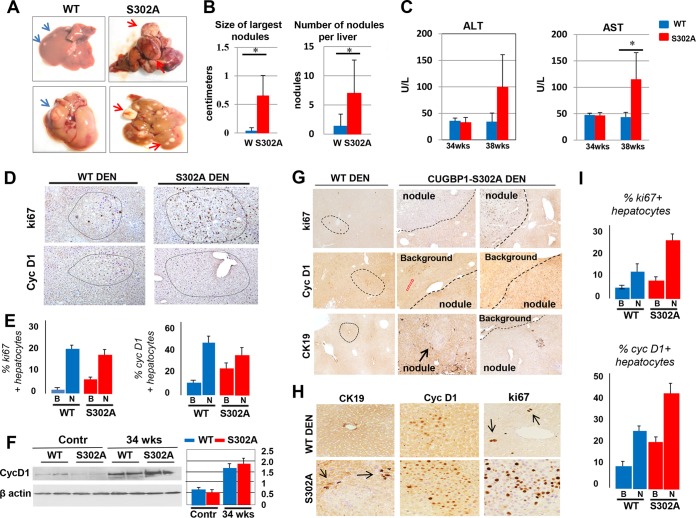
We next examined development of liver cancer in WT and S302A mice under DEN-mediated carcinogenesis. Mice were injected with DEN, and development of liver cancer was examined at 30, 34, and 38 weeks. No liver cancer was observed at 30 weeks in both groups. Given the fatty liver phenotype of S302A mice (Fig. 2 and 3), we expected that the development of liver cancer might be accelerated in S302A mice. In agreement with this suggestion, we observed a more severe phenotype of liver cancer in S302A mice than in WT mice at 34 and 38 weeks, with the most dramatic differences at 38 weeks. A typical picture of WT and S302A livers is shown in Fig. 4A. Calculations of size and number of nodules per liver showed an elevation of liver cancer in S302A mice (Fig. 4B). Additionally, levels of ALT and aspartate aminotransferase (AST) are significantly higher in serum of S302A mice at 38 weeks after DEN injections (Fig. 4C). We next performed immunostaining of the livers with known markers of liver cancer and proliferation: ki67, cyclin D1, and CK19. These analyses showed a moderate difference at 34 weeks but revealed a dramatic difference at 38 weeks after DEN injections. Similar to 38-week DEN treatments, the number and size of nodules are much smaller at 34 weeks in WT mice than those in S302A livers. Typical pictures of ki67 and cyclin D1 staining of 34-week-treated mice is shown in Fig. 4D. We examined distribution of ki67-positive and cyclin D1-positive hepatocytes in the background and tumor nodule sections of WT and S302A livers. These calculations showed about 2-fold higher numbers of positive hepatocytes in background regions of S302A mice compared to those of WT mice. However, the percentages of the positive cells in tumor nodules do not differ significantly (Fig. 4E). To confirm results of immunostaining, we performed Western blotting with antibodies to cyclin D1. Figure 4F shows that levels of cyclin D1 are slightly higher in S302A mice. Examination of ki67- and cyclin D1-positive hepatocytes at 38 weeks showed a more dramatic difference in the size and number of tumor nodules (Fig. 4G). First, ki67-, cyclin D1-, and CK19-positive tumor foci are much larger and contained significantly higher numbers of proliferating hepatocytes (Fig. 4G and H). Second, while proliferating hepatocytes of WT mice are detected mainly in tumor nodules, S302A mice contain ki67-, cyclin D1-, and CK19-positive hepatocytes in both tumor nodules and background (adjacent) regions of the liver (Fig. 4J). Lastly, in WT mice, staining for cyclin D1 showed equal staining in nucleus and in cytoplasm, while livers of S302A mice contain mainly nuclear cyclin D1 (Fig. 4H). Therefore, we conclude that S302A mice develop more severe liver cancer than WT mice under conditions of DEN-mediated carcinogenesis at both 34 and 38 weeks; however, blood parameters, size and number of tumor nodules, and distribution of proliferating hepatocytes showed much more significant differences at 38 weeks. Therefore, we further focused our studies on mice treated for 38 weeks.
FIG 4.
CUGBP1-S302A mice develop more severe liver cancer under conditions of DEN-mediated carcinogenesis. (A) Typical images of livers at 38 weeks after DEN injections. Blue and red arrows show tumor nodules in livers. (B) Bar graphs show the size and number of nodules per liver. (C) Serum levels of ALT and AST in WT and S302A mice at 34 and 38 weeks after DEN injection. (D) Typical images of ki67 and cyclin D1 staining of livers of DEN-treated WT and S302A mice at 34 weeks after DEN injections. Circles show tumor nodules. (E) Calculations of percentages of ki67- and cyclin D1-positive hepatocytes in background (B) and tumor nodule (N) sections of WT and S302A mice at 34 weeks after DEN injections. (F) Examination of cyclin D1 in WT and S302A livers at 34 weeks after DEN injections by Western blotting. Contr, age-matched control mice not treated with DEN. Bar graphs show levels of cyclin D1 as ratios to β-actin. (G) Typical images of staining of livers of DEN-treated WT and S302A mice with markers of liver cancer ki67, cyclin D1, and CK19. Positions of tumor nodules and background regions are shown. The red arrow indicates an area of liver with high levels of cyclin D1 expression within background tissue. (H) Images of staining with markers of liver cancer under higher magnifications. Black arrows in ki67-positive WT DEN indicate areas of proliferating hepatocytes with mitotic figures. (I) Percentages of ki67- and cyclin D1-positive hepatocytes in livers of WT and S302A mice at 38 weeks after DEN injections. *, P < 0.05.
Mutant CUGBP1-S302A is reduced in livers of mice treated with DEN.
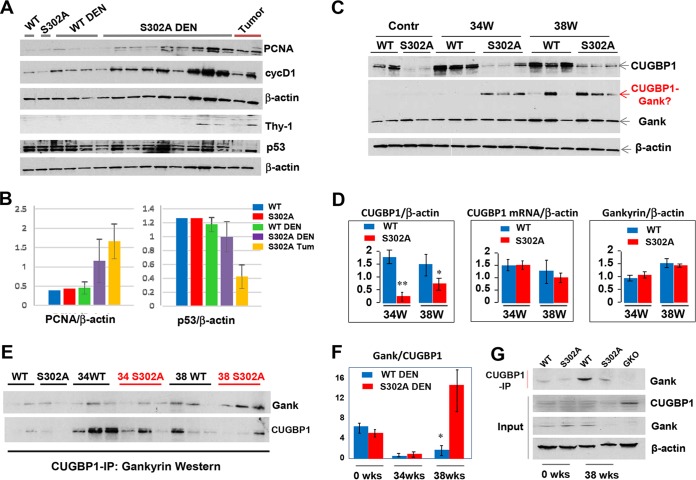
We next performed examination of cell cycle proteins and CUGBP1 using Western blot analyses on DEN-treated and untreated WT and S302A mice 38 weeks after treatment. Consistent with immunostaining, levels of cell cycle proteins cyclin D1 and PCNA are higher in S302A-DEN-treated mice than in WT-DEN-treated mice (Fig. 5A and B). Since p53 is a key tumor suppressor protein, we examined expression of p53 in DEN-treated WT and S302A mice. Western blotting did not detect significant differences in p53 between WT mice and background regions of DEN-treated S392A mice; however, tumor nodule sections of S302A mice showed 2- to 3-fold reductions of p53 (Fig. 5A and B). Together with the higher elevation of cyclin D1 and PCNA, this reduction of p53 confirms that tumor nodules of S302A show more severe liver cancer. One of the key events in the development of liver cancer is the appearance of tumor initiating cells or cancer stem cells. Therefore, we examined expression of a marker of stem cells, Thy-1, and found that it is detectable in livers of DEN-treated S302A mice (Fig. 3A). Interestingly, Thy-1 is elevated in all tumor nodules of S302A mice; while some background regions have elevation of Thy-1, others do not. We think that this difference might reflect the proximity of the background from tumor nodules; therefore, background sections in close proximity to tumor nodules are positive for Thy-1. Our previous examination of CUGBP1 in cytoplasm showed very minor or no differences between WT and S302A livers (Fig. 1B). However, examination of CUGBP1 in nuclear extracts surprisingly found a significant reduction of CUGBP1 in livers of unchallenged S302A mice and in livers of mice after DEN treatment (Fig. 5C and D). However, levels of CUGBP1 mRNA are only slightly reduced at 38 weeks (Fig. 5D). These results suggested that the mutant CUGBP1 is degraded in DEN-treated mice. Previous studies have shown that the oncoprotein Gank is elevated in liver cancer and triggers degradation of tumor suppressor proteins (3, 4, 20, 21). Therefore, we have examined the hypothesis that the reduction of CUGBP1 is associated with elevation of Gank in liver cancer. We found that Gank is elevated in both WT and S302A mice after DEN injection in terms of the levels of mRNA (Fig. 6B) and protein (Fig. 6A and B and 5C and D). We observed the appearance of an extra band on Gank Western blotting with a molecular mass close to that of CUGBP1 and correlated with reduction of CUGBP1 (Fig. 5C). We suggested that this additional band is a strong Gank-CUGBP1 complex; therefore, we examined if Gank interacts with CUGBP1 using a co-IP approach. Figure 5E and F shows that despite low levels of CUGBP1, amounts of Gank in CUGBP1 IP are higher in DEN-treated S302A mice than in WT mice. Quantitation of Gank in CUGBP1 IPs showed 4- to 5-fold larger amounts of Gank (Fig. 5F). To determine the specificity of this interaction, we included livers from Gank liver-specific knockout (GLKO) mice, which were generated recently in our laboratory. Co-IP studies with WT and S302A and GLKO mice confirmed that CUGBP1 specifically interacts with Gank (Fig. 5G).
FIG 5.
Levels of the mutant CUGBP1-S302A protein are dramatically reduced in DEN-treated S302A mice. (A) Western blots with antibodies to PCNA, cyclin D1, Thy-1, and p53 using nuclear extracts of livers from WT and S302A mice 38 weeks after DEN injection. The tumor lane shows protein extracts isolated from big external tumor nodules. (B) Quantification of PCNA and p53 protein levels as ratios to β-actin. (C) Western blots of livers of WT and S302A mice (control and DEN-treated) with antibodies to CUGBP1 and Gank. The red arrow shows the position of Gank immune-reactive protein, the molecular mass of which corresponds to the sum of CUGBP1 and Gank. (D) Bar graphs show calculations of levels of CUGBP1 protein (left) and CUGBP1 mRNA (right) as ratios to β-actin. (E) CUGBP1 was immunoprecipitated from protein extracts of untreated and DEN-treated mice (shown on the top). The IPs were probed with antibodies to Gank and then reprobed with antibodies to CUGBP1. (F) Levels of Gank in CUGBP1 IPs were calculated as ratios to CUGBP1. (G) CUGBP1 was immunoprecipitated from protein extracts of livers of mice shown on the top. The IPs were probed with antibodies to Gank. Input shows Western blotting of CUGBP1 and Gank. The final lane shows CUGBP1 IP from livers of GLKO mice as a control. *, P < 0.05; **, P < 0.01.
FIG 6.
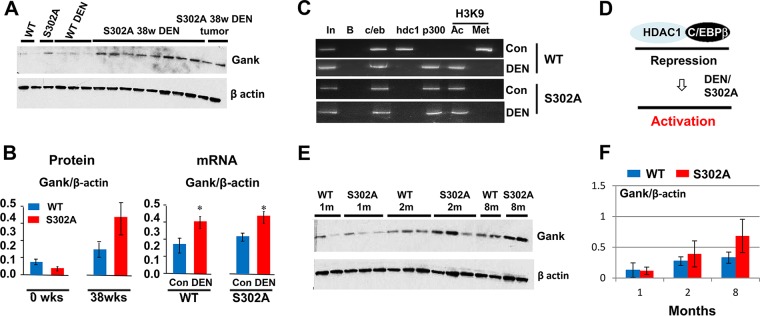
Gank is upregulated in livers of CUGBP1 S302A mice due to derepression of the Gank promoter. (A) Western blot of Gank in livers of S302A mice 38 weeks after DEN injection. The filter was reprobed with antibodies to β-actin. (B, left) Quantification of protein levels of CUGBP1 as a ratio to β-actin. (Right) Levels of Gank mRNA were determined by qRT-PCR and calculated as ratios to β-actin mRNA. *, P < 0.05. (C) ChIP. C/EBPβ, HDAC1, p300, histone H3K9met, and histone H3K9Ac were immunoprecipitated from chromatin solutions, and the IPs were used for isolation of DNA and PCR with primers covering the C/EBP site within the Gank promoter. (D) Schematic showing the proposed mechanism of Gank promoter activation. (E) Western blotting showing that S302A mice have increased Gank expression at ages of 1 to 8 months (m). (F) Quantification of Gank protein levels as ratios to β-actin.
Feedback regulation of Gank and S302A-CUGBP1.
Given the reduction of S302A-CUGBP1 in DEN-mediated liver cancer and the strong interaction of Gank with S302A-CUGBP1, we suggested that Gank eliminates the tumor suppressor isoform of CUGBP1 in liver cancer. To test this hypothesis, we examined the expression of Gank in WT and S302A mice treated with DEN. Figure 6A and B (left) shows that protein levels of Gank are elevated in S302A livers to much higher degrees than in WT mice. Although Gank mRNA was increased by DEN treatments, there were no significant differences in mRNA expression between WT and S302A mice under DEN treatment (Fig. 6B, right). S302A mice have reduced C/EBPβ-HDAC1 complexes (Fig. 1), and the reduction of these complexes leads to derepression of promoters of DGAT1/2 genes (Fig. 3). Given this observation and our previous results showing that the Gank promoter is partially repressed in WT mice by C/EBPβ-HDAC1 complex (22), we asked if the reduction of these complexes in S302A mice is involved in the activation of Gank. ChIP assay with Gank promoter revealed that C/EBPβ-HDAC1 complexes occupy the promoter in livers of WT mice but not in livers of CUGBP1-S302A mice. Examination of histone H3 revealed that, similar to DGAT1/2 promoters, changes of complexes leads to the derepression of the Gank promoter in S302A mice (Fig. 6C). DEN treatment leads to elimination of HDAC1-C/EBPβ complexes from the Gank promoter, while activators of transcription (C/EBPβ-p300 complexes) are increased on the Gank promoter in WT mice after DEN treatments and in S302A mice. These alterations of the chromatin structure lead to the activation of the Gank promoter in WT mice and, to a higher degree, in livers of S302A mice (Fig. 6C and D). We next asked if activation of Gank takes place during spontaneous development of liver cancer, which is frequently observed in old mice. Western blotting showed that the levels of Gank are elevated during aging in both WT and S302A mice; however, elevation of Gank is higher in S302A mice (Fig. 6E and F).
Gank causes degradation of the S302A-CUGBP1 mutant, while the CUGBP1-S302D mutant is resistant to Gank-mediated degradation.
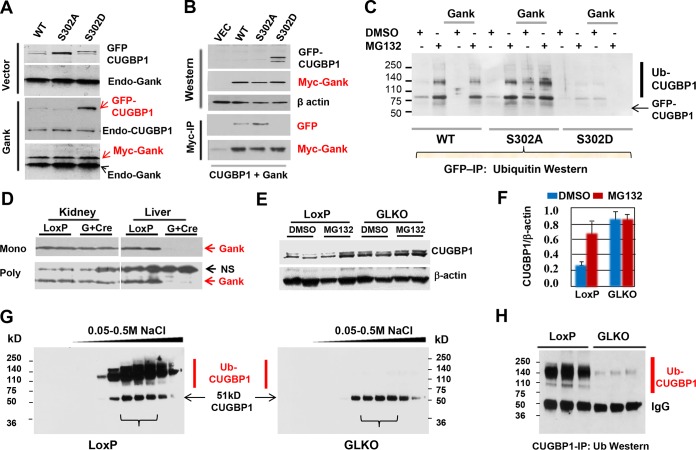
To directly address the role of Gank in degradation of S302A-CUGBP1, we have performed experiments in Hepa-1c17 hepatoma cells using vectors expressing green fluorescent protein (GFP)-WT CUGBP1, GFP-CUGBP1-S302A and GFP-CUGBP1-S302D mutants, and Gank fused to the c-myc epitope. CUGBP1 proteins were cotransfected with Gank (or empty vector) into Hepa-1c17 cells, and levels of CUGBP1 proteins were examined. Examination of protein levels of GFP-linked CUGBP1 revealed that amounts of the WT and S302A mutant are dramatically reduced in cells cotransfected with c-myc–Gank compared to amounts of proteins cotransfected with GFP vector only (Fig. 7A). However, the levels of S302D mutant are not changed by Gank. These results demonstrated that Gank triggers degradation of WT and CUGBP1-S302A but not CUGBP1-S302D mutant. We next examined the interactions of these CUGBP1 proteins with Gank and found that Gank strongly interacts with WT and S302A CUGBP1 proteins; however, the interaction with S302D mutant was not detected (Fig. 7B). The differences between degradation of WT and S302A mutant, if they exist, likely are not significant, since the majority of WT CUGBP1 is not phosphorylated at Ser302 due to overexpression. We next treated transfected cells with the proteasome inhibitor, MG132, and measured amounts of GFP-CUGBP1-Ub conjugates. Inhibition of proteasome leads to the accumulation of Ub-GFP-CUGBP1 conjugates in cells transfected with WT plus Gank and S302A Gank plasmids, while GFP-CUGBP1-Ub conjugates are very weak in cells cotransfected with GFP-CUGBP1-302D mutant (Fig. 7C). Thus, Gank triggers degradation of CUGBP1-S302A and unphosphorylated WT CUGBP1 through the UPS system.
FIG 7.
Gank triggers degradation of dephosphorylated CUGBP1 and S302A-CUGBP1 in cultured cells and in livers. (A) GFP-linked WT and S302A and S302D mutant CUGBP1 proteins were cotransfected with empty vector or with plasmid coding for c-myc-tagged Gank in Hepa-1c17 cells, and levels of GFP-CUGBP1 proteins were determined by Western blotting with antibodies to GFP and with antibodies to CUGBP1. The membranes were reprobed with antibodies to Gank. Positions of endogenous Gank, CUGBP1, and c-myc-tagged Gank are shown. (B) c-myc–Gank was cotransfected with GFP-linked CUGBP1 proteins and immunoprecipitated with antibodies to c-myc tag. The IPs were probed with antibodies to GFP and c-myc tag. Upper panels (Western) show protein input. (C) GFP-linked WT and S302A and S302D mutant CUGBP1 were cotransfected with Gank, and cells were treated with DMSO or with inhibitor of proteasome MG132. CUGBP1 was immunoprecipitated with antibodies to GFP and probed with antibodies to ubiquitin. Positions of GFP-CUGBP1 and ubiquitinated conjugates are shown. (D) Examination of protein level of Gank in GLKO mice by Western blotting with monoclonal (Mono; upper) and polyclonal (Poly; lower) antibodies to Gank. Protein extracts were isolated from kidney and liver of LoxP and GLKO mice and probed with antibodies to Gank. NS, nonspecific band which serves as a loading control. (E) Western blot of CUGBP1 with nuclear extracts isolated from livers of GLKO and LoxP control mice treated with MG132. (F) Quantification of CUGBP1 protein levels as a ratio to β-actin. (G) Examination of ion-exchange chromatography (IEC) fractions of protein extracts isolated from LoxP and GLKO mice by Western blotting with antibodies to CUGBP1. Linear gradient of NaCl is shown on the top. (H) Examination of CUGBP1 IPs from three fractions of IEC by Western blotting with antibodies to ubiquitin.
Liver-specific deletion of Gank in mice increases levels of CUGBP1 by blocking UPS-mediated degradation of CUGBP1.
We next examined if Gank causes ubiquitination and degradation of CUGBP1 in livers. For this, we used GLKO mice that were generated recently in our laboratory. We are currently investigating Gank LKO mice in detail. In this work, we present the part of these studies related to the Gank-mediated regulation of CUGBP1. Figure 7D shows that these animals do not express Gank in the liver; however, Gank is not affected in kidney of liver-specific Gank LKO mice. To examine if Gank causes ubiquitination of CUGBP1 in liver, we have treated control (LoxP) and Gank LKO mice with proteasome inhibitor MG132 and examined levels of CUGBP1 in the nucleus and accumulation of ubiquitin-CUGBP1 conjugates in cytoplasm. Inhibition of proteasome by MG132 in LoxP mice leads to the 3- to 4-fold elevation of CUGBP1, while in Gank LKO mice, the basal levels of CUGBP1 are higher and are not increased by treatments with MG132 (Fig. 7E and F). For detection of CUGBP1-Ub conjugates, we concentrated cytoplasmic proteins on an ion-exchange column (UnoQ; Bio-Rad) followed by elution with a sharp gradient of NaCl. Figure 7G shows the Western blotting of UnoQ fractions with antibodies to CUGBP1. In LoxP mice treated with MG132, we have detected a strong smear region located above the 51-kDa CUGBP1. However, these signals are very weak in Gank LKO mice treated with MG132. To determine if these additional high-molecular-mass conjugates of CUGBP1 are ubiquitinated CUGBP1, we immunoprecipitated CUGBP1 from UnoQ fractions and performed Western blotting with antibodies to ubiquitin. Figure 7H shows that Ub-CUGBP1 conjugates are highly abundant in LoxP livers treated with MG132 and that these conjugates are dramatically reduced in Gank LKO mice. Taken together, examination of Gank LKO mice revealed that Gank triggers degradation of CUGBP1 in the liver through the UPS system.
Tumor suppressor activity of CUGBP1 is associated with repression of multiple pathways of liver cancer.
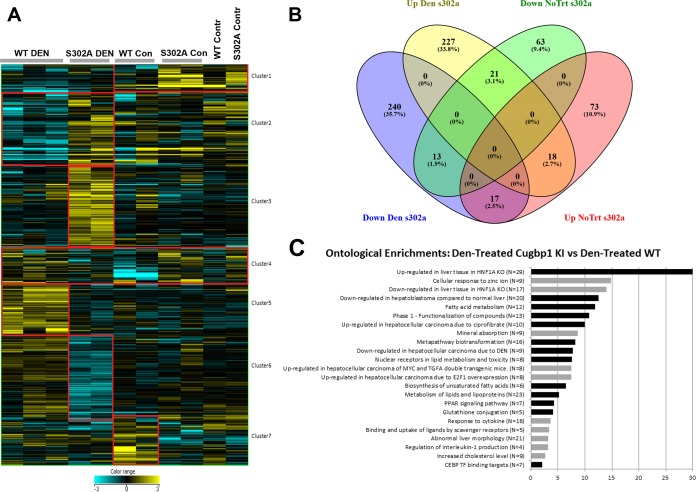
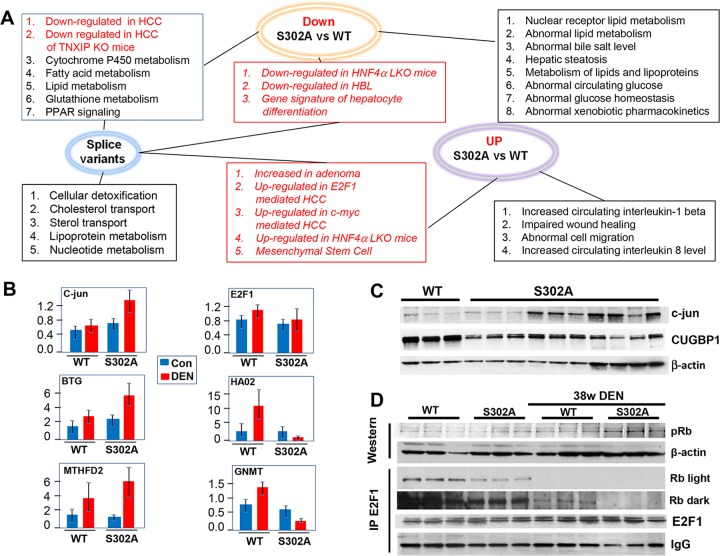
Our results have identified Gank as one of the targets of CUGBP1. We also showed that CUGBP1-dependent inhibition of Gank contributes to tumor suppressor activity of S302A-CUGBP1. This inhibition is mediated by translational activity of CUGBP1, which takes place in cytoplasm. However, CUGBP1 is a multifunctional protein which displays three major functions: regulation of splicing, regulation of translation, and regulation of stability of mRNAs. We observed complex alterations of CUGBP1 in S302A mice, including loss of translational activity (cytoplasmic function) and reduction of the protein in the nucleus, which suggests that targets of splicing activity of CUGBP1 also are affected. Given these alterations, one should assume that there are many downstream pathways which contribute to accelerated cancer in livers of S302A mice. We have performed transcriptome sequencing (RNA-Seq) in livers of control and DEN-treated (38 weeks) WT and S302A mice. Since CUGBP1 regulates translation, stability, and splicing of mRNAs, we have analyzed alterations in levels of mRNAs and in splicing. We have detected significant alterations of 7 clusters of genes (Fig. 8A). Gene lists were overlapped between DEN-treated S302A versus DEN-treated WT mice and control S302A versus control WT mice using a Venn diagram to determine the level of independence of the genotype effect in treated and untreated mice (Fig. 8B). We next determined ontological changes in signaling pathways by comparing RNA-Seq results of DEN-treated WT and S302A mice. Figure 8C represents a summary of pathways enriched for genes up- and downregulated in S302A livers compared to DEN-treated WT mice. As shown, several cancer-related pathways are up- or downregulated in S302A liver to a much higher degree than in WT mice. Interestingly, the directions of changes are in agreement with positive or negative roles of these pathways in liver cancer.
FIG 8.
Alterations of signaling pathways in WT and S302A livers after DEN treatments. (A) Heat map of alterations in expression of five cluster genes. (B) Diagram showing overlap in genes of each cluster. (C) Ontological pathways differentially altered in livers of DEN-treated S302A mice compared with DEN-treated WT mice.
To understand the contribution of splicing activity of CUGBP1 and the activities of CUGBP1 that regulate translation/stability of mRNAs to liver cancer, we have performed comparisons of differential splice variants and levels of mRNAs in WT and S302A DEN-treated mice. Figure 9A shows overlaps of downregulated and upregulated mRNAs with mRNAs that have alterations in splicing. As shown, there are several pathways in DEN-treated S302A mice which are affected by alteration of splicing only. Those mainly include pathways of cellular detoxification, lipoprotein metabolism, and nucleotide metabolism. There are also alterations of distinct pathways which are associated only with down- or upregulation of mRNAs. Importantly, the majority of cancer-related pathways in DEN-treated S302A mice include alterations in both splicing and in levels of mRNAs (Fig. 9A, red boxes). We have further verified the differences in expression of several mRNAs using quantitative reverse transcription-PCR (qRT-PCR) (Fig. 9B). As shown, BTG3 and MTHFD2 mRNAs are elevated by DEN to a higher degree in S302A mice. Additionally, a very different pattern of HAO2 and GNMT mRNA expression was detected for WT and S302A DEN-treated mice. While in WT mice DEN treatment elevates levels of these mRNAs, S302A livers have a dramatic reduction of these mRNAs after DEN treatments (Fig. 9B). Taken together, RNA-Seq analyses revealed that the mutation of CUGBP1 at Ser302 to Ala and the subsequent reduction of the CUGBP1 protein make mice more sensitive to development of liver cancer due to alterations in expression of downstream mRNAs on the levels of splicing and mRNA expression.
FIG 9.
Tumor suppressor activity of CUGBP1 is associated with repression of multiple pathways of liver cancer for the levels of splicing and expression of mRNAs. (A) Analysis of RNA-Seq data shows lists of altered gene pathways regarding the levels of splice variants and mRNA in livers of DEN-treated CUGBP1 S302A mice versus livers of DEN-treated WT mice. Pathways in red are associated with liver cancer. (B) Confirmation of changes of levels of mRNAs by qRT-PCR. (C) Western blot of c-jun protein. The filter was first probed with antibodies to c-jun and then reprobed with antibodies to CUGBP1 and β-actin. (D) Western blotting with antibodies to ph-S780-Rb. Nuclear extracts of untreated and DEN-treated (38 weeks) mice were used. The three lower images show results of co-IP studies. E2F1 was immunoprecipitated and Rb and E2F1 were examined in these IPs. The image labeled Rb dark shows a dark exposure of Rb membrane to detect Rb in IPs from DEN-treated mice. The bottom image shows IgG signals on the same filter.
E2F1 signaling and c-jun are activated to a higher degree in livers of DEN-treated CUGBP1-S302A mice.
Among signaling pathways which are elevated in S302A mice compared to WT mice, RNA-Seq analysis identified pathways which are upregulated in human and animal models of hepatocellular carcinoma (HCC). These critical pathways include stronger activation of c-myc signaling, E2F1 signaling, and HNF1A signaling, as well as activation of c-jun mRNA. We further investigated E2F1-Rb pathways and c-jun, since the first pathway is usually regulated by posttranslational alterations, while c-jun mRNA is a known target of CUGBP1 and CUGBP1 destabilizes this mRNA, leading to repression of c-jun (26, 27). In agreement with previous reports, c-jun mRNA was elevated to a higher degree in DEN-treated S302A mice (Fig. 9B). We next examined the levels of c-jun protein and found that this target of CUGBP1 is also elevated in DEN-treated S302A mice compared to that in WT animals (Fig. 9C). Examination of levels of E2F1 mRNA showed no significant alterations in E2F1 expression (Fig. 9B). In agreement with mRNA levels, the IP-Western approach showed no significant differences for the E2F1 protein levels (Fig. 9D). Therefore, we further examined if E2F1 is activated by release from the complexes with Rb protein. The rationale for this hypothesis was that we detected the activation of cyclin D1 in S302A mice to a much higher degree (Fig. 4). Since cyclin D1 activates cdk4, which phosphorylates Rb at Ser780 and disrupts E2F1-Rb complexes, we suggested that this mechanism is activated in S302A mice to a higher degree. Therefore, we examined the phosphorylation status of Rb and E2F1-Rb complexes in WT and S302A mice treated with DEN. Rb is phosphorylated in both WT and S302A mice; however, the degree of phosphorylation is much higher in S302A livers (Fig. 9D). Co-IP revealed that amounts of Rb-E2F1 complexes are reduced in quiescent livers of S302A mice compared to amounts in WT mice, while levels of E2F1 are not different. Moreover, examination of these complexes in DEN-treated mice showed that, in both groups, these complexes are reduced; however, the reduction in livers of S302A mice is more significant. Thus, these studies demonstrated that CUGBP1 represses the E2F1-dependent pathway of liver cancer via support of E2F1-Rb complexes. Taken together, these studies confirmed data of RNA-Seq analysis and showed specific CUGBP1-dependent pathways which activate E2F1 signaling and c-jun to a higher degree in S302A mice.
Gank-CUGBP1 pathway of liver cancer is a signature of pediatric hepatocellular carcinoma and HBL as well as of spontaneous liver cancer in animal models.
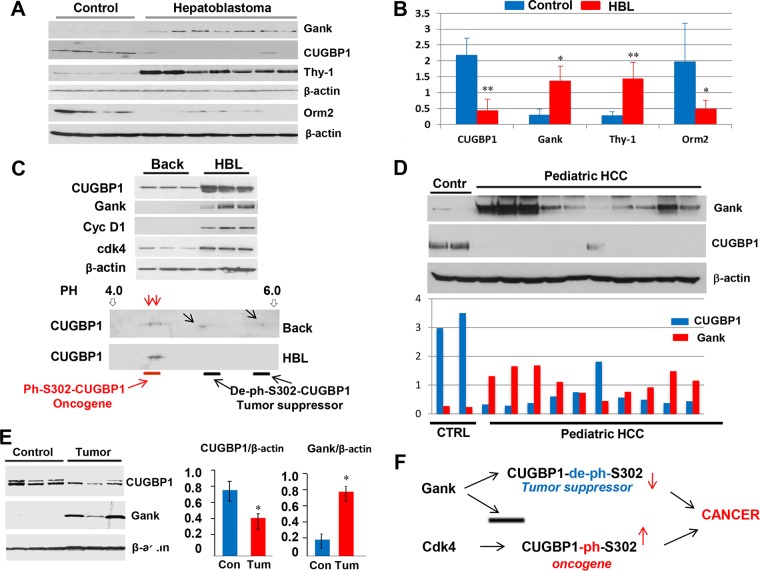
DEN-mediated liver cancer in mice is a very powerful protocol for liver cancer; however, it is initiated by treatments of mice with the chemical and might not reflect the pathways of spontaneous liver cancer in humans. To investigate the relevance of the Gank-CUGBP1 axis to human liver cancer, we have examined expression of Gank and CUGBP1 in human pediatric hepatoblastoma and hepatocellular carcinomas. The detailed examination of 54 hepatoblastoma (HBL) samples is described in our separate paper, which is currently under review (35). As one can see in Fig. 10A and B, the levels of Gank are elevated in representative samples, while CUGBP1 levels are reduced. Examination of the CUGBP1-dependent marker of hepatocyte Orm2 and CUGBP1-dependent marker of stem cells, Thy-1, revealed that Orm2 is reduced while Thy-1 is dramatically elevated in HBL samples, suggesting that the Gank-mediated reduction of CUGBP1 is critical for the development of HBL.
FIG 10.
Gank-CUGBP1 axis is a signature of pediatric human liver cancer. (A) Western blotting was performed with protein extracts isolated from 4 control and 7 HBL samples. (B) Levels of CUGBP1, Gank, Thy-1, and Orm2 were determined as ratios to β-actin. (C, upper) Western blotting of three HBL samples and background regions from corresponding HBL samples with antibodies to CUGBP1, Gank, cyclin D1, and cdk4. (Bottom) 2D gel electrophoresis of proteins from background and tumor sections of patients with high levels of CUGBP1. (D) Western blotting with protein extracts isolated from 10 HCC samples. Bar graphs show levels of CUGBP1 and Gank as ratios to β-actin. (E) Images on the left show Western blotting, and bar graphs on the right show levels of CUGBP1 and Gank calculated as ratios to β-actin. (F) A diagram showing the hypothetical role of CUGBP1 in liver cancer (see the text). *, P < 0.05; **, P < 0.01.
Our results confirm that dephosphorylated CUGBP1 is a tumor suppressor protein while phosphorylated CUGBP1 is not. In the course of studies of HBL samples, we have identified a small group in which CUGBP1 levels are upregulated despite high levels of Gank. Figure 10C (top) shows examination of three HBL samples with higher levels of CUGBP1. We next asked why and how CUGBP1 is not degraded by Gank, which is also elevated in these HBL samples. We found that, in addition to elevation of CUGBP1 and Gank, these HBL samples have elevated levels of cyclin D1 and cdk4. Cyclin D1-cdk4 has been shown to phosphorylate CUGBP1 at Ser302 (5). Given our data from cultured cells (Fig. 7), we suggested that CUGBP1 is resistant to Gank due to phosphorylation by cdk4. To test this suggestion, we examined CUGBP1 isoforms using two-dimensional (2D) gel electrophoresis. In background sections of HBL livers, CUGBP1 exists in 4 isoforms; two of them are hyperphosphorylated and two are less or not phosphorylated (Fig. 10C, bottom). In tumor sections, only two hyperphosphorylated forms are observed. Based on our previous observations (35), we suggest that these isoforms display oncogenic activities.
We next examined the Gank-CUGBP1 pathway in 10 pediatric hepatocellular carcinoma samples. Western blotting showed that 9 HCC samples have significant elevation of Gank and a dramatic reduction of CUGBP1 (Fig. 10D). One sample showed a minor elevation of Gank and a minor reduction of CUGBP1. To further translate our observations in human cancer to animal models, we asked if spontaneous development of liver cancer in mice involves the Gank-CUGBP1 axis. It has been shown previously that around 25 to 30% of mice develop liver cancer at an age of 24 months (21). We have examined CUGBP1-Gank pathways in these liver tumors and found that CUGBP1 is reduced while Gank is dramatically elevated in spontaneously developed liver cancer (Fig. 10E). Thus, our investigations revealed that the Gank-CUGBP1 pathway is involved in the development of human liver cancer and spontaneous liver cancer.
DISCUSSION
Despite the significant progress with studies of liver cancer, the critical/causal events in development of liver cancer are not known. RNA binding protein CUGBP1 previously has been implicated in the regulation of multiple processes in the liver, including liver differentiation after birth (15), liver proliferation after surgery (17), liver recovery after injury (24), and development of fatty liver disease (18). CUGBP1 displays its functions mainly through three activities: regulation of splicing, stability, and translation of mRNAs. Since translational activity of CUGBP1 depends on phosphorylation at Ser302, we generated CUGBP1-S302A mice, which exhibit a loss of CUGBP1 translational activity, with the goal to examine the role of this activity in liver cancer and in liver biology. Examination of liver phenotype found that translational activity of CUGBP1 (which occurs in the cytoplasm) is involved in the epigenetic control of liver functions and inhibits development of the fatty liver phenotype. These studies have identified a new function of CUGBP1, which is the regulation of body homeostasis. Several previous reports found that CUGBP1 is elevated in patients with HCC and in mouse liver cancer mediated by DEN (15, 28, 29). In addition, a recent report showed that CUGBP1 is involved in the development of liver fibrosis, which is one of the critical steps of the development of liver cancer in humans (30). These studies strongly suggested that CUGBP1 displays oncogenic activities. In agreement with this prediction, CUGBP1-dependent accumulation of C/EBPβ-HDAC1 complexes leads to the repression of tumor suppressors p53, p21, and C/EBPα (15, 16). Since previous studies reported the elevation of CUGBP1 in HCC from adult patients (28, 29) and showed oncogenic activities of CUGBP1, we expected that liver cancer was inhibited in S302A mice. However, we found that S302A mice have more severe liver cancer under DEN-mediated carcinogenesis. Our studies of human pediatric liver cancer, spontaneously developed HCC in mice, and DEN-mediated liver cancer revealed that CUGBP1 is reduced in nuclei of these three types of liver cancer. It is important to note that only the Ser302 de-ph isoform of CUGBP1 displays tumor suppressor activities, but the phosphorylated form of CUGBP1 does not have this activity and actually works as an oncoprotein (Fig. 10F). We think that the dephosphorylation of CUGBP1 at Ser302 is the result of a balance between cdk4 and unknown kinases since, in several situations, CUGBP1 is dephosphorylated in livers with high activity of cdk4. Thus, our work presents an example where posttranslational modifications of RNA binding proteins determine if the proteins work as oncoproteins or as tumor suppressors. CUGBP1 regulates translation, splicing, and stability of many mRNAs, and our RNA-Seq assay identified alterations of many of these targets in S302A mice.
How is de-ph-Ser302-CUGBP1 reduced in liver cancer? We could not detect significant changes of CUGBP1 mRNA levels in liver cancer. Instead, our work revealed that CUGBP1 is reduced at the level of protein degradation. Using tissue culture systems and Gank LKO mice, we found that Gank interacts with the S302A-CUGBP1 mutant and causes ubiquitination and subsequent degradation of CUGBP1 in the liver during development of liver cancer. Previous studies and our recent work showed that Gank is elevated in all types of liver cancer (20–24, 26–34), suggesting that this is the main mechanism that causes degradation of CUGBP1. Our results show that the Gank-dependent ubiquitination of CUGBP1 is mainly observed in cytoplasm; however, CUGBP1 is mainly reduced in the nucleus. This suggests that there is an additional level of regulation which includes inhibition of the translocation of CUGBP1 from cytoplasm to nucleus, as has been shown in previous studies in which Gank retains NF-κB in the cytoplasm (34). Interestingly, our studies revealed that there is a feedback regulation of Gank by CUGBP1 when CUGBP1 is phosphorylated at S302 and works as a positive regulator of translation of C/EBPβ and HDAC1. ChIP studies showed that the CUGBP1-dependent C/EBPβ-HDAC1 complexes occupy the Gank promoter and partially repress Gank. Our work is clinically important, since a recent paper has identified a small drug (cjoc42) which inhibits activities of Gank (25). Therefore, future studies will show if the pharmaceutical inhibition of Gank rescues the de-ph-S302-CUGBP1 isoform and protects against or treats liver cancer.
MATERIALS AND METHODS
Animals.
Experiments with animals were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine (protocol AN-1439) and by the IACUC at Cincinnati Children's Hospital (protocol IACUC2014-0042). CUGBP1 S302A KI mice were generated by replacement of WT CUGBP1 with the mutant (Fig. 1A). Blood serum was sent to the Center for Comparative Medicine pathology core at Baylor College of Medicine for ALT and AST assessment. In order to understand the role of Gank on regulation of CUGBP1, we created a liver-specific Gank knockout (Gank LKO) mouse model using the cre-lox system. Mice expressing the cre recombinase protein driven by the albumin promoter were crossed with mice that had loxP sequences flanking exons 2 to 4 of the Gank gene. The resulting offspring had the Gank gene excised only in cells expressing albumin.
Diethylnitrosamine treatment.
Three-week-old male mice were injected with DEN at 50 μg/g body weight. Livers were collected 30 to 38 weeks later and stored at −80°C or prepared for histology in 4% formaldehyde. Five to 7 mice were used for each time point.
Normal, background, and HBL pediatric liver samples.
In these studies we analyzed 6 normal, 51 HBL, and 10 pediatric HCC liver samples, which were obtained from the Cincinnati Children's Hospital Medical Center Bio Bank repository. Samples were obtained from patients between the ages of 0.01 months and 6 years of age over the course of 10 years. All samples were pathologically confirmed through histological analysis prior to further testing.
Histology.
Collected liver tissue was taken from the left lobe and fixed in 4% formaldehyde. Mice were injected intraperitoneally (i.p.) with 66.5 mg/kg bromodeoxyuridine (BrdU) 2 h before sacrifice. BrdU incorporation was measured using a commercially available kit (93-3943; Invitrogen).
ChIP.
Chromatin was prepared and immunoprecipitated using the ChIP-it express enzymatic kit (active motif; 53009). Precipitated DNA was amplified using Gank promoter-specific forward primer ACATGGCAACTCACACTGTCT and reverse primer GTAATGGCAATTACCATCGAGA and run on a 6% polyacrylamide gel.
Treatments of mice with MG132 and examination of CUGBP1-ubiquitin conjugates.
Mice were i.p. injected with 10 μg/g body weight of MG132, and livers were collected 7 h later. To concentrate proteins, cytoplasmic extracts were isolated and loaded on a UnoQ ion-exchange column (Bio-Red). The proteins were eluted by a sharp gradient of NaCl. To determine if CUGBP1 forms conjugates with ubiquitin, CUGBP1 was immunoprecipitated from ion-exchange chromatography fractions, and the IPs were separated by gel electrophoresis and probed with antibodies to ubiquitin.
Cell culture experiments.
Hepa-1c17 (ECACC no. 95090613) cells were grown in αMEM plus GlutaMAX without nucleosides (Fisher) with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were cotransfected with WT, S302A, or S302D GFP-CUGBP1 plasmid and myc-Gank plasmid using GenJet transfection reagent (SignaGen). Following 24 h of transfection, cells were treated with 40 μM MG132 or dimethyl sulfoxide for 4 h immediately before collection of cell lysates.
RNA-Seq.
RNA sequencing libraries were prepared using an Illumina TruSeq RNA preparation kit and sequenced on an Illumina HiSeq 2500 using paired-end, 100-bp reads (Illumina, San Diego, CA). Following removal of primers and barcodes, reads were aligned using mm10 annotations, produced by UCSC, and quantified using Kallisto, which accurately quantifies read abundances (in transcripts per million) through pseudoalignment. All statistical analysis was performed in GeneSpring 13.0. Raw counts were thresholded at 1, normalized using a 75th percentile shift, and baselined to the median of all samples (n = 31,253 transcripts). A filtration was applied to ensure analysis of reasonably expressed transcripts, requiring at least two reads in >50% of samples under at least one experimental condition (n = 10,427 transcripts). Differential expression was assessed between treatment and genotype conditions using a 2-way analysis of variance (ANOVA), with a significance cutoff of a fold change of >1.5 (n = 672 transcripts across two comparisons). Splice variant analysis was performed using AltAnalyze, which identifies alternative exons/introns using de novo isoform comparisons and also incorporates splicing predictions generated by UCSC. Specifically, a splicing index algorithm was used to identify alternative algorithms with a P value requirement of <0.05. Alternate splice events were visually assessed using sashimi plots generated by the Integrated Genome Viewer due to the high frequency of false positives in similar analyses (between 40 and 70% false positives detected).
Real-time qRT-PCR.
Total RNA was isolated from mouse and human livers using an RNeasy plus minikit (Qiagen). cDNA was synthesized with 2 μg of total RNA using a high-capacity cDNA reverse transcription kit (ThermoFisher). cDNA was diluted five times with diethyl pyrocarbonate-treated water and subsequently used for RT-PCR assays with the TaqMan gene expression system (Applied Biosystems). Gene expression analysis was performed using the TaqMan universal PCR master mix (Applied Biosystems) in a total volume of 10 μl containing 5 μl master mix, 1.5 μl water, 3 μl cDNA template, and 0.5 μl of the gene-specific TaqMan assay probe mixture. The cycling profile was 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, as recommended by the manufacturer. TaqMan probe mixtures were purchased from Applied Biosystems. The following probes were used: E2F1, Mm00432939_m1; β-actin, Mm02619580_g1; CUGBP1 (CELF1), Mm04279608_m1; BTG3, Mm01975968-g1; HAO2, Mm00469507_m1; MTHD2, Mm00485276_m1; GNMT, Mm00494688_m1. Levels of all mRNAs were normalized to β-actin.
Western blotting.
Nuclear and cytoplasmic extracts were prepared as previously described (21, 24). Lysates were separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting. The following antibodies were used: CUGBP1 (sc-20003; Santa Cruz), C/EBPβ (sc-150; Santa Cruz), C/EBPα (sc-61; Santa Cruz), Gank (12985S; Cell Signaling Technologies), cyclin D1 (RM-9104-S1; ThermoFisher), Rb (sc-50; Santa Cruz), E2F1 (sc-193; Santa Cruz), pRb (3590S; Cell Signaling Technologies), c-jun (9165; Cell Signaling Technologies), PGC1α (sc-13067; Santa Cruz), SIRT1, DGAT1 (sc-32861; Santa Cruz), DGAT2 (sc-66859; Santa Cruz), GPAT, ubiquitin (3933S; Cell Signaling Technologies), β-actin (A5316; Sigma), PCNA (sc-7907; Santa Cruz), Thy-1 (sc-9163; Santa Cruz), and Orm2 (11199-1-AP; Proteintech) antibodies. Membranes were then incubated with the corresponding secondary horseradish peroxidase-coupled antibodies, sc-2031 and sc-2313 (Santa Cruz) or 18-8817-31 and 18-8816-33 (Rockland).
Statistical analysis.
All values are presented as means ± standard deviations (SD). A Student t test was applied to pairwise comparison for normally distributed data.
Accession number(s).
RNA sequencing data may be accessed in NCBI's Gene Expression Omnibus under accession number GSE83240.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01DK102597 and R01CA159942 (N.T.), by NIH grants AR052791 and AR064488 (L.T.), and by Internal Development Funds from CCHMC (N.T. and L.T.).
REFERENCES
- 1.Martin J, Dufour JF. 2008. Tumor suppressor and hepatocellular carcinoma. World J Gastroenterol 14:1720–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oishi N, Yamashita T, Kaneko S. 2014. Molecular biology of liver cancer stem cells. Liver Cancer 3:71–84. doi: 10.1159/000343863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iakova P, Timchenko L, Timchenko NA. 2011. Intracellular signaling and hepatocellular carcinoma. Semin Cancer Biol 21:28–34. doi: 10.1016/j.semcancer.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timchenko NA, Lewis K. 2015. Elimination of tumor suppressor proteins during liver carcinogenesis. Cancer Studies Mol Med 1:27–38. doi: 10.17140/CSMMOJ-1-104. [DOI] [Google Scholar]
- 5.Dreijerink KM, Groner AC, Vos ES, Font-Tello A, Gu L, Chi D, Reyes J, Cook J, Lim E, Lin CY, de Laat W, Rao PK, Long HW, Brown M. 2017. Enhancer-mediated oncogenic function of the menin tumor suppressor in breast cancer. Cell Rep 18:2359–2372. doi: 10.1016/j.celrep.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesaro E, Sodaro G, Montano G, Grosso M, Lupo A, Costanzo P. 2017. The complex role of the ZNF224 transcription factor in cancer. Adv Protein Chem Struct Biol 107:191–222. doi: 10.1016/bs.apcsb.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Hickey CJ, Schwind S, Radomska HS, Dorrance AM, Santhanam R, Mishra A, Wu YZ, Alachkar H, Maharry K, Nicolet D, Mrózek K, Walker A, Eiring AM, Whitman SP, Becker H, Perrotti D, Wu LC, Zhao X, Fehniger TA, Vij R, Byrd JC, Blum W, Lee LJ, Caligiuri MA, Bloomfield CD, Garzon R, Marcucci G. 2013. Lenalidomide-mediated enhanced translation of C/EBPα-p30 protein up-regulates expression of the antileukemic microRNA-181a in acute myeloid leukemia. Blood 121:159–169. doi: 10.1182/blood-2012-05-428573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bräuer-Hartmann D, Hartmann JU, Wurm AA, Gerloff D, Katzerke C, Verga Falzacappa MV, Pelicci PG, Müller-Tidow C, Tenen DG, Niederwieser D, Behre G. 2015. PML/RARα-regulated miR-181a/b cluster targets the tumor suppressor RASSF1A in acute promyelocytic leukemia. Cancer Res 75:3411–3424. doi: 10.1158/0008-5472.CAN-14-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su R, Lin HS, Zhang XH, Yin XL, Ning HM, Liu B, Zhai PF, Gong JN, Shen C, Song L, Chen J, Wang F, Zhao HL, Ma YN, Yu J, Zhang JW. 2015. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene 34:3226–3239. doi: 10.1038/onc.2014.274. [DOI] [PubMed] [Google Scholar]
- 10.Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, Delwel R, Pandolfi PP. 2013. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X, Hu C, Arnovitz S, Bugno J, Yu M, Zuo Z, Chen P, Huang H, Ulrich B, Gurbuxani S, Weng H, Strong J, Wang Y, Li Y, Salat J, Li S, Elkahloun AG, Yang Y, Neilly MB, Larson RA, Le Beau MM, Herold T, Bohlander SK, Liu PP, Zhang J, Li Z, He C, Jin J, Hong S, Chen J. 2016. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat Commun 7:11452. doi: 10.1038/ncomms11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wurm AA, Tenen DG, Behre G. 2017. The Janus-faced nature of miR-22 in hematopoiesis: is it an oncogenic tumor suppressor or rather a tumor-suppressive oncogene? PLoS Genet 13:e1006505. doi: 10.1371/journal.pgen.1006505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones K, Timchenko L, Timchenko NA. 2012. The role of CUGBP1 in age-dependent changes of liver functions. Ageing Res Rev 11:442–449. doi: 10.1016/j.arr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timchenko LT, Salisbury E, Wang G-L, Nguyen H, Albrecht JH, Hershey JWB, Timchenko NA. 2006. Age-specific CUGBP1-eIF2 complex increases translation of CCAAT/enhancer-binding protein beta in old liver. J Biol Chem 281:32806–32819. doi: 10.1074/jbc.M605701200. [DOI] [PubMed] [Google Scholar]
- 15.Wang G-L, Salisbury E, Shi X, Timchenko LT, Medrano EE, Timchenko NA. 2008. HDAC1 promotes liver proliferation in young mice via interaction with C/EBPβ. J Biol Chem 283:26179–26187. doi: 10.1074/jbc.M803545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin J, Iakova P, Jiang Y, Lewis K, Sullivan E, Jawanmardi NN, Donehower L, Timchenko L, Timchenko NA. 2013. Transcriptional and translational regulation of C/EBPβ-HDAC1 protein complexes controls different levels of p53, SIRT1, and PGC1α proteins at the early and late stages of liver cancer. J Biol Chem 288:14451–14462. doi: 10.1074/jbc.M113.460840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin J, Hong IH, Lewis K, Iakova P, Breaux M, Jiang Y, Sullivan E, Jawanmardi N, Timchenko L, Timchenko NA. 2015. Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration in mice. Hepatology 61:315–325. doi: 10.1002/hep.27295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J, Iakova P, Breaux M, Sullivan E, Jawanmardi N, Chen DN, Chen D, Jiang Y, Medrano EE, Timchenko NA. 2013. Increased expression of enzymes of triglyceride synthesis is essential for the development of hepatic steatosis. Cell Rep 3:831–843. doi: 10.1016/j.celrep.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson SP. 2008. Hepatocellular carcinoma and the ubiquitin-proteasome system. Biochim Biophys Acta 1782:775–784. doi: 10.1016/j.bbadis.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Higashitsuji H, Itoh K, Nagao T, Dawson S, Nonoguchi K, Kido T, Mayer RJ, Arii S, Fujita J. 2000. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med 6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 21.Wang G-L, Shi X, Haefliger S, Jin J, Major A, Iakova P, Finegold M, Timchenko NA. 2010. Elimination of C/EBPalpha through the ubiquitin-proteasome system promotes the development of liver cancer in mice. J Clin Investig 120:2549–2562. doi: 10.1172/JCI41933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Iakova P, Jin J, Sullivan E, Sharin V, Hong I-H, Anakk S, Major A, Darlington G, Finegold M, Moore D, Timchenko NA. 2013. Farnesoid X receptor inhibits gankyrin in mouse livers and prevents development of liver cancer. Hepatology 57:1098–1106. doi: 10.1002/hep.26146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim IK. 2002. Spectrum of molecular changes during hepatocarcinogenesis induced by DEN and other chemicals in Fischer 344 male rats. Mech Ageing Dev 123:1665–1680. doi: 10.1016/S0047-6374(02)00087-8. [DOI] [PubMed] [Google Scholar]
- 24.Hong IH, Lewis K, Iakova P, Jin J, Sullivan E, Jawanmardi N, Timchenko L, Timchenko NA. 2014. Age-associated change of C/EBP family proteins causes severe liver injury and acceleration of liver proliferation after CCl4 treatments. J Biol Chem 289:1106–1118. doi: 10.1074/jbc.M113.526780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chattopadhyay A, O'Connor CJ, Zhang F, Galvagnion C, Galloway WRJD, Tan YS, Stokes JE, Rahman T, Verma C, Spring DR, Itzhaki LS. 2016. Discovery of a small-molecule binder of the oncoprotein gankyrin that modulates gankyrin activity in the cell. Sci Rep 6:23732. doi: 10.1038/srep23732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. 2008. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell 29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HH, Gorospe M. 2008. GU-rich RNA: expanding CUGBP1 function, broadening mRNA turnover. Mol Cell 29:151–152. doi: 10.1016/j.molcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Huang H, Yuan B, Luo T, Li J, Qin X. 2014. Suppression of CUGBP1 inhibits growth of hepatocellular carcinoma cells. Clin Investig Med 37:10–19. doi: 10.25011/cim.v37i1.20864. [DOI] [PubMed] [Google Scholar]
- 29.Chettouh H, Fartoux L, Aoudjehane L, Wendum D, Clapron A, Chrtien Y, Rey C, Scatton O, Soubrane O, Conti F, Praz F, Housset C, Rosmorduc O, Desbois-Mouthon C. 2013. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res 73:3974–3986. doi: 10.1158/0008-5472.CAN-12-3824. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, Wu X, Ma Y, Shao F, Tan Y, Tan T, Gu L, Zhou Y, Sun B, Sun Y, Wu X, Xu Q. 2016. CUG-binding protein 1 regulates HSC activation and liver fibrogenesis. Nat Commun 7:13498. doi: 10.1038/ncomms13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng T, Hong X, Wang J, Pei T, Liang Y, Yin D, Song R, Song X, Lu Z, Qi S, Liu J, Sun B, Xie C, Pan S, Li Y, Luo X, Li S, Fang X, Bhatta N, Jiang H, Liu L. 2014. Gankyrin promotes tumor growth and metastasis through activation of IL-6/STAT3 signaling in human cholangiocarcinoma. Hepatology 59:935–946. doi: 10.1002/hep.26705. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Higashitsuji H, Higashitsuji H, Itoh K, Sakurai T, Koike K, Hirota K, Fukumoto M, Fujita J. 2013. Overexpression of gankyrin in hepatocytes induces hemangioma by suppressing factor inhibiting hypoxia-inducible factor-1 (FIH-1) and activating hypoxia-inducible factor-1. Biochem Biophys Res Commun 432:22–27. doi: 10.1016/j.bbrc.2013.01.093. [DOI] [PubMed] [Google Scholar]
- 33.Yang C, Tan Y-X, Yang G-Z, Zhang J, Pan Y-F, Liu C, Fu J, Chen Y, Ding ZW, Dong LW, Wang HY. 2016. Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma. J Exp Med 213:859–875. doi: 10.1084/jem.20151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Li HH, Fu J, Wang XF, Ren Bin Y, Dong LW, Tang SH, Liu SQ, Wu MC, Wang HY. 2007. Oncoprotein p28 GANK binds to RelA and retains NF-kappaB in the cytoplasm through nuclear export. Cell Res 17:1020–1029. doi: 10.1038/cr.2007.99. [DOI] [PubMed] [Google Scholar]
- 35.Valanejad L, Lewis K, Wright M, Jiang Y, D'Souza A, Karns R, Sheridan R, Gupta A, Bove K, Witte D, Geller J, Tiao G, Nelson DL, Timchenko L, Timchenko N. 23 May 2017. FXR-gankyrin axis is involved in development of pediatric liver cancer. Carcinogenesis doi: 10.1093/carcin/bgx050. [DOI] [PMC free article] [PubMed] [Google Scholar]