ABSTRACT
We have detected DNA polymerase beta (Polβ), known as a key nuclear base excision repair (BER) protein, in mitochondrial protein extracts derived from mammalian tissue and cells. Manipulation of the N-terminal sequence affected the amount of Polβ in the mitochondria. Using Polβ fragments, mitochondrion-specific protein partners were identified, with the interactors functioning mainly in DNA maintenance and mitochondrial import. Of particular interest was the identification of the proteins TWINKLE, SSBP1, and TFAM, all of which are mitochondrion-specific DNA effectors and are known to function in the nucleoid. Polβ directly interacted functionally with the mitochondrial helicase TWINKLE. Human kidney cells with Polβ knockout (KO) had higher endogenous mitochondrial DNA (mtDNA) damage. Mitochondrial extracts derived from heterozygous Polβ mouse tissue and KO cells had lower nucleotide incorporation activity. Mouse-derived Polβ null fibroblasts had severely affected metabolic parameters. Indeed, gene knockout of Polβ caused mitochondrial dysfunction, including reduced membrane potential and mitochondrial content. We show that Polβ is a mitochondrial polymerase involved in mtDNA maintenance and is required for mitochondrial homeostasis.
KEYWORDS: DNA polymerase beta, mitochondrial DNA repair, TFAM, base excision repair, mitochondria, mitochondrial health, mutational studies
INTRODUCTION
Cellular DNA repair is critical for genomic stability, and the accumulation of DNA damage has been linked to many debilitating human disorders, including accelerated aging, cancer, and neurodegeneration (reviewed in references 1 and 2). Mammalian cells have two genomes, nuclear and mitochondrial, and both have the ability to replicate, accumulate DNA damage, and propagate mutations. The nucleus contains the vast majority of the mammalian genome and has extensive ability to repair complex bulky adducts, double-strand breaks (DSB), single-strand breaks (SSB), and hundreds of chemical DNA modifications. The ability to effectively repair this breadth of damage is achieved through multiple, often overlapping, DNA repair pathways. In contrast, the repair of mitochondrial DNA (mtDNA) is a more limited version of nuclear DNA (nDNA) repair. Mitochondria lack nucleotide excision repair, and the presence of double-strand break repair is debated (recently reviewed in reference 3). Despite the mitochondria having attenuated DNA repair capabilities compared to the nucleus, the accumulation of mtDNA damage is not without consequence. Ineffective mtDNA maintenance is the underlying cause of many human diseases, including Alpers syndrome and chronic progressive external ophthalmoplegia (CPEO) caused by mutations in mitochondrial polymerase gamma (Polγ) or the TWINKLE helicase (4–6). The accumulation of mtDNA damage has also been linked to normal aging, a phenotype that can be accelerated by mutating the major mitochondrial DNA polymerase, Polγ (7, 8). Indeed, the mitochondrial free radical theory of aging suggests that reactive oxygen species (ROS) produced as a by-product of respiration may cause the gradual accumulation of DNA damage leading to age-related dysfunction (9). Considering the proximity of the mitochondria to ROS production, mtDNA may be preferentially affected. In support, mitochondria are particularly adept at resolving oxidative DNA damage (10). In the mitochondria, as in the nucleus, the majority of oxidative lesions are repaired by the base excision repair (BER) system.
Nuclear and mitochondrial BER are somewhat similar (for a comprehensive review, see reference 11); however, there are distinctions. In the nucleus, there are multiple polymerases, namely, Polβ, Polδ, Polε, Polκ, and Polλ, that can be involved in the different subpathways of BER. These enzymes have overlapping substrate specificity, providing a greater level of genomic protection. In comparison, it is thought that BER in mitochondria relies solely on Polγ. Thus, Polγ supports both replication and repair functions in the mitochondria. As such, Polγ has high replication fidelity; however, this may be detrimental in situations that require the polymerase to bypass a lesion. PrimPol, a highly error prone polymerase involved in translesion DNA synthesis (TLS), has been reported to interact with the major single-stranded binding proteins, replication protein A (RPA) and mitochondrial single-stranded binding protein 1 (SSBP1) (12). Further, another TLS enzyme, polymerase zeta subunit REV3, has very recently been identified in the mitochondria and was shown to protect mtDNA from DNA damage after UV exposure (13). These works demonstrated that there are genomic events in the mitochondria that cannot be resolved by Polγ alone and required further processing from other predominantly nuclear polymerases. Modified mtDNA terminal ends are another form of DNA damage that requires further processing by specialized DNA repair enzymes, many of which have been reported in the mitochondria.
The identification of the DNA terminal modifiers tyrosyl-DNA phosphodiesterase (TDP1) (14), aprataxin (15), and polynucleotide kinase phosphatase (PNKP) (16) in mitochondria is evidence that mtDNA terminal modifications occur with enough frequency that specific repair processes are required. Another common form of DNA terminal modification is the 5′-deoxyribose phosphate (5′-dRP) lesion that can occur as a DNA repair intermediate. The formation of 5′-dRP termini is refractory to DNA ligation and can cause more detrimental protein-DNA adducts if left unrepaired (17). In the nucleus, the majority of 5′-deoxyribophosphodiesterase (5′-dRP lyase) activity comes from the core BER enzyme Polβ, and the activity has been reported to be a rate-limiting step in short-patch BER (SP-BER) (18). Polβ is comprised of a catalytic domain required for nucleotide insertion and an 8-kDa N-terminal 5′-dRP lyase domain (18). Considering that the 5′-dRP lyase activity of Polβ was measured to be 17-fold higher than the same activity in Polγ (19), we sought to reinvestigate the potential role of Polβ in the mitochondria. To date, research has been unable to definitively answer whether Polβ is in mammalian mitochondria. However, a number of parasitic organisms not only have Polβ in the mitochondria but use the enzyme for both replication and repair of mtDNA in the absence of Polγ, evidence that a homolog of mammalian Polβ is able to function in the organelle (20–22). Data obtained by using bovine mitochondrial extracts have found a Polβ-like enzyme in the heart (23); however, purified mitochondrial mouse liver extracts have been negative (24). Using a more comprehensive approach, we investigated the role of Polβ in mtDNA maintenance. Polβ was identified in purified mitochondrial tissue and cell extracts. Polβ interacts with mitochondrial maintenance proteins, transcription factor A mitochondrial (TFAM), TWINKLE, and SSBP1, which is supportive of a role in mtDNA maintenance. Mitochondria derived from samples with a deficiency in Polβ have lower BER capacity, elevated endogenous mtDNA damage, and mitochondrial dysfunction. We report here that Polβ complements Polγ in the maintenance and repair of mtDNA.
RESULTS
DNA Polβ is found in the mitochondria.
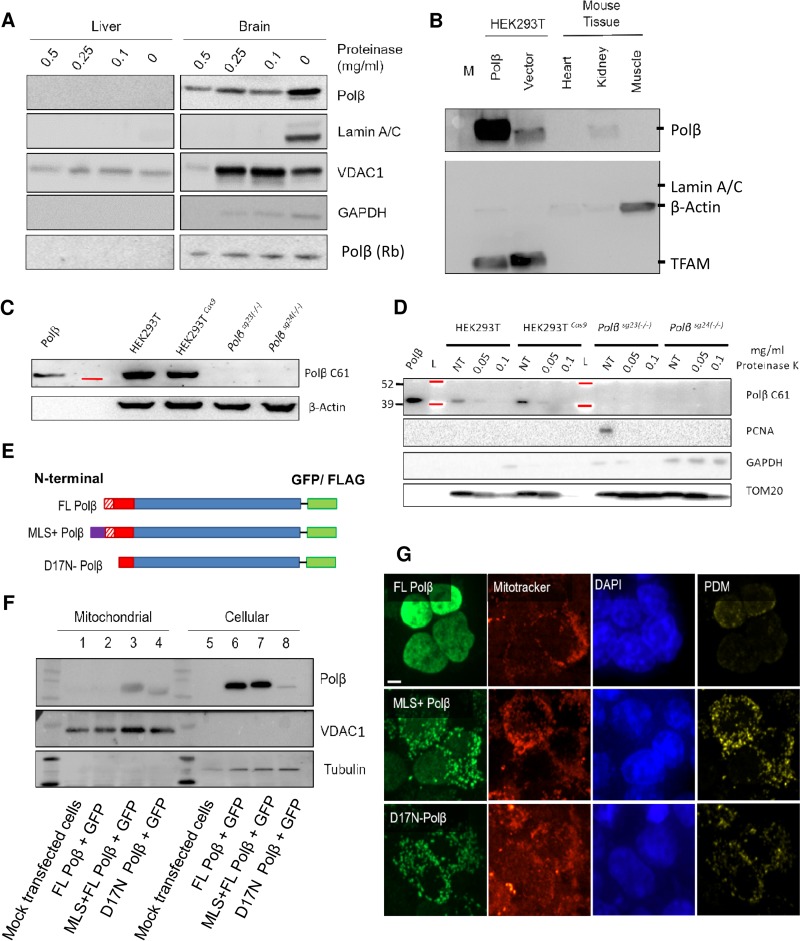
Mice with Polβ heterozygosity have defects in mitochondrion-related pathways in the brain, including oxidative phosphorylation (25). Further, using large-scale mitochondrial extracts from bovine heart, a Polβ-like activity has been reported (23). In contrast to these findings, Hansen and colleagues found that Polβ was not present in mitochondrial extracts derived from mouse liver (24). To reexamine the mitochondrial localization of Polβ, we built upon the research done by Hansen et al. using the same method to extract intact mitochondria from mouse liver. However, we went further, digesting the mitochondria with proteinase to remove outer mitochondrial membrane-adhering proteins. Without proteinase treatment and with overexposure, the Polβ antibody detected a faint band consistent with the size of nuclear Polβ in the mitochondrial liver samples (arrow in Fig. S1A in the supplemental material). With proteinase treatment, nuclear and cytoplasmic contaminations were removed, and the Polβ band from the liver was no longer detected (Fig. 1A, left). This work confirms the previous research reporting that there was no detectable Polβ in liver mitochondria.
FIG 1.
DNA polymerase β is detected in the mitochondria. (A) Mouse mitochondria purified from liver and brain were digested with increasing amounts of proteinase K to remove outer membrane-bound proteins. The liver samples (left) show no Polβ in the mitochondrial compartment. The brain (right) mitochondrial sample revealed Polβ after proteinase digestion, evidence that Polβ was inside the mitochondrial membrane. VDAC1 was used as a mitochondrial outer membrane marker and was digested at 0.5 mg/ml proteinase. Lamin A/C was used as a marker for nuclear contamination. GAPDH shows the extent of cytoplasmic contamination. A second antibody was also used to verify the results [Polβ (Rb)]. Fifty micrograms of protein was loaded. (B) The kidney preparations had a clear Polβ band present. Human kidney cells (HEK293T) were used to determine whether Polβ was found in the mitochondria of human cells and whether the levels could be artificially manipulated. Vector control HEK293T preparations had visible amounts of endogenous Polβ present. Overexpression of Polβ caused elevated levels of mitochondrial Polβ. The TFAM mitochondrial control antibody detected only the human form of the protein. (C) To verify Polβ antibody specificity, HEK293T cells were modified for Polβ knockout using CRISPR/cas9. The far left lane shows the molecular mass of purified Polβ protein. Total cellular protein extracts from clones 23 and 24 have no remaining Polβ. The red line indicates a molecular mass marker at 39 kDa. (D) Mitochondrial extracts from HEK293T/Polβ-KO cells compared to parental and parental+Cas9 cells. Extracts were exposed to either 0.05 or 0.1 mg/ml proteinase K or had treatment (NT). Polβ was detected in the extracts from the parental cell lines but were absent in Polβ-KO lines (clones 23 and 24). Red lines indicate molecular mass markers at 39 and 52 kDa. (E) To visualize Polβ in the mitochondria, HEK293T cells were transfected with either a C-terminal GFP or FLAG full-length (FL) Polβ, a positive control with an MLS sequence from the mitochondrial protein SOD2, and the FL Polβ protein with a 17-aa N-terminal deletion (D17N) (see Materials and Methods for a full description). (F) Immunoblot analysis of modified Polβ-GFP constructs to verify protein localization. Samples 1 to 4, mitochondrial extracts. Samples 5 to 8, whole-cell lysates. Lanes 1 and 5, mock-transfected cells. Lanes 2 and 6, FL Polβ-GFP. Lanes 3 and 7, SOD2 MLS–Polβ-GFP. Lanes 4 and 8, D17N-Polβ-GFP. VDAC1 was used to show mitochondrial enrichment of the samples. Tubulin was used as a cytoplasmic marker. Lanes 1 to 4 contained 10 μg mitochondrial extract per lane, and lanes 5 to 8 contained 2 × 104 cells per lane. (G) Localization of the C-terminal FLAG constructs depicted in panel E by immunofluorescence. The FL Polβ-FLAG construct is detected in the mitochondria (PDM channel). Addition of the SOD2 MLS signal enhanced the PDM signal; however, only with the deletion of the 17-aa N-terminal region (D17N) was there complete loss of nuclear localization and enhanced mitochondrial localization (also see Fig. S1G in the supplemental material). DAPI was used to visualize the nucleus (magnification, ×63, z-stack; scale bar = 20 μm).
In contrast, the Polβ antibody detected bands in all of the proteinase-digested brain mitochondrial preparations (Fig. 1A, right). Significantly, there was a single nuclear Polβ band detected in the liver mitochondria, whereas the brain had two distinct bands for Polβ before proteinase treatment (Fig. 1A and S1B). The weak lower band corresponding to the size of nuclear Polβ (∼39 kDa) was present only before protease digestion. A more prominent, upper band of Polβ remained even at the highest concentration of proteinase, despite almost complete digestion of voltage-dependent anion channel 1 (VDAC1), a mitochondrial outer membrane protein (Fig. 1A). The near complete disappearance of VDAC1 suggests that proteinase K digestion had gone beyond only the outer membrane proteins and had likely also digested inner mitochondrial membrane proteins, hence the reduced signal in the 0.5-mg/ml sample. Of particular note, we achieved similar results with a second alternate host species Polβ antibody binding to a different region of the protein [Fig. 1A, Polβ (Rb)]. Mitochondrial extracts were prepared from other types of tissue to ascertain whether mitochondrial Polβ was found only in brain tissue. These samples were exposed to proteinase treatment at 0.1 mg/ml prior to Western blot analysis. The Polβ antibody also detected a band of the correct, higher-molecular-weight size in the kidney mitochondrial samples but not in heart or muscle (Fig. 1B).
The mouse and human Polβ proteins have nearly identical sequence homology (96% identical [GenBank accession no. NP_002681.1 versus NP_035260.1). Thus, we asked whether the human kidney cell line HEK293T would also have Polβ in the mitochondria and whether the level of the protein in this organelle could be manipulated by overexpression of Polβ (Fig. 1B). We stably transfected HEK293T cells with a vector containing full-length (FL) Polβ plus a C-terminal FLAG tag. The mitochondrial Polβ band was clearly stronger in the overexpressing cell line than in the vector control cell line, which is evidence that human kidney cells have mitochondrial Polβ and Polβ levels in the mitochondria that can be artificially manipulated. This is also direct evidence of the antibody specificity. Pertinently, the antibody bound to endogenous Polβ in the vector alone mitochondrial samples. An antibody that only binds human TFAM confirmed mitochondrial enrichment (Fig. 1B). We also generated Polβ knockouts (KO) in the HEK293T cells using the CRISPR/Cas9 system, targeting the first exon of the Polβ gene. Immunoblot analysis using total cellular extracts from the two KO Polβ cell lines (clones 23 and 24) indeed showed no residual protein remaining (Fig. 1C). To further test the specificity of the Polβ antibody, we extracted mitochondria from the KO cells and probed for the protein. We detected no Polβ protein in the mitochondrial extracts derived from the HEK293T Polβ KO clones (Fig. 1D and S1C and D). Comparing the Polβ bands detected in the two parental lines (wild type [WT] and Cas9 [parental+Cas9]) to purified Polβ protein, we observed that the protein detected in the mitochondria by the antibody is similar in size to Polβ. This antibody was then used to visualize the intracellular localization of the Polβ protein using Polβ KO mouse embryonic fibroblasts (MEFs) (Mβ19tsA) and a control MEF line (Mβ16tsA) (Fig. S1E to G) (26). Polβ protein was detected in the nucleus and surrounding cytoplasm. The mitochondrial nucleoid protein TFAM was used to establish whether the cytoplasmic Polβ signal had any mitochondrial colocalization (Fig. S1F). Indeed, the product of the difference of the mean (PDM) shows regions of significant colocalization between TFAM and Polβ. The Polβ antibody specificity was tested using the Polβ KO MEF line, with limited nonspecific cross-reactivity observed with overexposure of the image (Fig. S1G). We found similar results with other Polβ antibodies we tested (see Materials and Methods). Overall, these results demonstrate that Polβ can localize to the mitochondria and that the amount of protein in the organelle can be manipulated by overexpression of the protein or by KO of the gene.
After our discovery of Polβ in the mitochondria, we next sought to determine how the protein is localized to the organelle. There are numerous DNA repair and maintenance proteins that function in both the nuclear and mitochondrial compartments. Import of these proteins into mitochondria is often facilitated by a mitochondrial localization sequence (MLS). Using the MLS prediction program MitoProt (27), we identified the first 60 amino acids (aa) of Polβ in human and mouse (Fig. S1H) as having the highest prediction of mitochondrial import at a probability of 0.85. Full-length Polβ (FL Polβ) has a second in-frame start codon only 17 aa from the beginning of the protein. Deletion of the first 17 aa from the protein sequence reduced the probability of mitochondrial import from 0.85 to 0.07, suggesting that this sequence is particularly important for mitochondrial transport.
We queried whether the mRNA expression patterns of various Polβ isoforms may predict the extent of mitochondrial localization of the protein within a tissue. There are multiple Polβ isoforms, nine of which are predicted to give a protein product (NCBI). Of these nine, many have truncated C-terminal ends, potentially affecting a putative mitochondrial targeting helix (MTH) (see Fig. S1H) and hence protein import into the mitochondria. We used three separate reverse transcription-PCR (RT-PCR) probes (Fig. S1I) to measure the relative expression of the N-terminal and C-terminal ends and the catalytic domain of Polβ in 13 human tissues. Brain, liver, and testes had high mRNA expression levels of the N-terminal sequence of the gene (Fig. S1J). However, we found no correlation between Polβ protein in the mitochondria and isoform expression levels in the tissues.
To further characterize the N-terminal mitochondrial import sequence of Polβ, we designed vectors overexpressing different versions of Polβ tagged with a green fluorescent protein (GFP) at the C terminus. Specifically, we asked whether we could influence the recruitment of Polβ by manipulating the N terminus (Fig. 1E). We tested two novel Polβ expression constructs in comparison to the FL Polβ-GFP in human kidney cells (HEK293T). The first encoded the MLS derived from SOD2, a mitochondrial inner membrane protein used as a positive control. The second construct had part of the Polβ N terminus removed (designated D17N). Immunoblot analysis was used to verify protein expression (Fig. 1F). Interestingly, the placement of GFP on the C-terminal end of Polβ (unlike what was seen with the FLAG tag [Fig. 1B]) adversely affected mitochondrial localization of the protein (Fig. 1F and S1K). Using FL Polβ-GFP, we were able to detect only limited amounts of Polβ in the mitochondria by either immunoblot analysis (lane 2) or immunofluorescence following transfection into HEK239T kidney cells (Fig. 1G). This suggested that the large GFP C-terminal tag may affect an MLS or MTH that is located at the C terminus (Fig. S1H). In the HEK293T cells, the SOD2 MLS Polβ-GFP was detected in the mitochondria after transfection as measured by both immunoblotting (Fig. 1F, lane 3) and immunofluorescence; however, this protein was still predominantly in the nucleus (Fig. 1F [compare lanes 7 and 3] and S1K). Removing the N-terminal 17 residues [(D17N) Polβ-GFP] caused a drastic change in the cellular distribution of Polβ. The protein was now exclusively cytoplasmic and mitochondrial, with only limited nuclear localization in HEK293T cells (Fig. 1F, lane 4). We remade the constructs with a FLAG rather than a GFP tag on the C terminus. The FLAG-tagged Polβ was also predominantly nuclear (Fig. 1G), unlike the cytoplasmic/nuclear antibody staining previously observed with endogenous Polβ (Fig. S1E). However, we did observe some mitochondrial localization with the smaller FLAG-tagged FL Polβ, consistent with the Western blot results (Fig. 1B). The aberrant expression pattern of the artificially expressed Polβ does suggest that some further posttranslational modification or protein-protein interaction is needed to transport and potentially stabilize mitochondrial Polβ.
The N-terminal domain of Polβ interacts with mtDNA maintenance and import proteins.
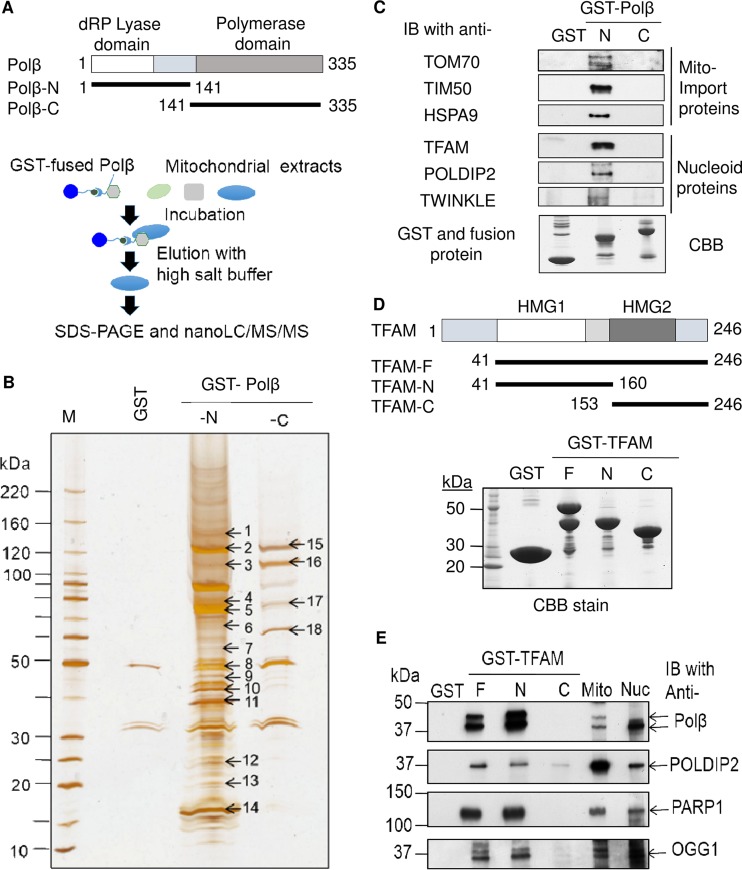
The deletion of the N-terminal 17 aa residues of Polβ does not hinder mitochondrial localization of the protein (see above). However, the N-terminal domain may be necessary for mitochondrial protein-protein interactions. We determined which mitochondrial proteins interact with Polβ and which region is most pertinent to the interactions. This was achieved by using glutathione S-transferase (GST)–Polβ fragments containing either the N terminus (aa 1 to 141) or the C terminus (aa 141 to 335) of Polβ (Fig. 2A). The fragments were expressed in Escherichia coli, purified, and bound to a glutathione-Sepharose column. This column was then exposed to proteinase-treated mitochondrial extracts derived from HEK293T cells overexpressing Polβ in the presence of DNase, RNase, and Benzonase to eliminate protein-DNA/RNA binding (see Fig. S2A to C in the supplemental material). The bound proteins were electrophoresed on a denaturing gel and stained (Fig. 2B), and the numbered bands were identified by mass spectrometry. The proteins that were identified fell into four broad categories (Fig. S2D). We had particular interest in proteins involved in mtDNA maintenance and import (summarized in Table 1). The DNA damage sensor PARP1 (ARTD1) and BER core protein and Polβ partner Lig3 were identified in both N- and C-terminal fragment pulldowns. The C-terminal construct bound proteins TDP1 and PNKP, which are associated with DNA end processing and have recently been reported in the mitochondria (14, 16). Of note, all the repair proteins bound by the C-terminal fragment had roles in both the nucleus and mitochondria. In contrast, we identified exclusively mitochondrial DNA maintenance proteins binding to the N-terminal construct, including TFAM and SSBP1. Further, proteins HSPA9 (also known as mtHSP70/Mortalin/GRP75) and HSPD1 (also known as HSP60), which are involved in correct protein folding after successful mitochondrial import, also interacted with only the N terminus of Polβ. Moreover, the matrix chaperone GRPEL1 may also be required for effective transport into the mitochondrial nucleoid.
FIG 2.
The N terminus of Polβ interacts with mitochondrial transport and mtDNA maintenance components. (A) HEK293T cells overexpressing Polβ (see Fig. S2A to C in the supplemental material) were incubated with GST-tagged N- or C-terminal Polβ recombinant fragments attached to glutathione-Sepharose beads. The bound proteins from the mitochondrial extracts were eluted after washing (see diagram). (B) Silver staining of gel used for subsequent mass spectrometry (also see Table 1 and Fig. S2D). (C) Immunoblotting (IB) of the GST pulldown used in mass spectrometry detected mitochondrial proteins HSPA9 and TFAM. Mitochondrial import proteins TOM70 and TIM50 and accessory mtDNA repair proteins TWINKLE and POLDIP2 were also detected. In all cases, mitochondrial proteins bound only to the N-terminal Polβ fragment. (D) GST-TFAM fragments were purified to establish which region of the TFAM protein bound to mitochondrial Polβ. (E) Identification of Polβ in the GST-TFAM-bound fraction using antibody detection. Polβ bound to the N-terminal fragment of TFAM containing the HMG1 domain. DNA maintenance proteins POLDIP2, PARP1, and OGG1 also bound to the HMG1-containing fragment.
TABLE 1.
Pertinent proteins identified using fragment pulldown and analysisa
| Macro function | Proteinsb |
|
|---|---|---|
| N terminus | C terminus | |
| DNA maintenance | PARP-1 (1, 2); Lig3, TOP1c (3); TFAM (12); SSBP1 (14) | PARP-1 (15), Lig3 (16), TDP-1 (17), PNKP (18) |
| Stress response/import | HSPA9 (5), HSPD1 (6), GRPEL1 (12) | |
The proteins identified show that the N terminus of Polβ interacts with exclusively mitochondrial proteins, including mtDNA repair components. In contrast, the C terminus interacted with proteins that functioned in both nuclear and mitochondrial DNA repair.
Numbers in parentheses correspond to the bands indicated in Fig. 2B.
Top1, DNA topoisomerase 1.
With the information gleaned from the mass spectrometry analysis, we developed insight into how Polβ might be transported into the mitochondria. This transport may require import initially through the outer mitochondrial membrane and then through the inner mitochondrial membrane using the TOM (translocase of outer membrane) and TIM (translocase of inner membrane) class of proteins. The TIM complex uses HSPA9 to facilitate ATP-dependent protein transfer (28, 29). We confirmed our mass spectrometry data using immunoblotting, which showed that HSPA9 does interact with the N terminus of Polβ (Fig. 2C). The N-terminal Polβ pulldown also bound the HSPA9-associated proteins Tom70 and Tim50. We also focused on potential interactions with principal mtDNA maintenance proteins, confirming the TFAM mass spectrometry result using immunoblotting (Fig. 2C). The identification of TFAM accessory proteins TWINKLE and POLDIP2 strongly suggests that Polβ participates in an mtDNA repair complex that has TFAM as a central component.
TFAM is essential for mitochondrial transcription and correct mtDNA packaging (30) and has also been directly implicated in mtDNA BER (31). TFAM was detected in the N-terminal Polβ pulldown and was confirmed to interact by immunoblotting. It was of particular interest to determine how Polβ interacts with TFAM and what other factors may also be involved in the TFAM-related DNA repair complex. Using a strategy similar to that used with GST-Polβ for Fig. 2A, GST constructs of TFAM that incorporated different functional regions of the protein were created (Fig. 2D and E). Polβ-overexpressing HEK293T mitochondrial extracts were again used as the input material. The full-length protein and the N terminus of TFAM were able to bind Polβ, but the C terminus of TFAM did not. These results indicate that it is the HMG1 domain of TFAM that binds Polβ. Importantly, the C-terminal region that comprises the HMG2 domain does not bind Polβ, suggesting a specific interaction. Further, the HMG1 domain appears to be central to the repair function of TFAM, binding the DNA damage sensor PARP1 and DNA repair proteins POLDIP2 and OGG1 (Fig. 2E). Thus, Polβ and TFAM interact through the N terminus of Polβ and the HMG1 domain of TFAM.
Polβ is required in mitochondria to provide enhanced mtDNA BER activity.
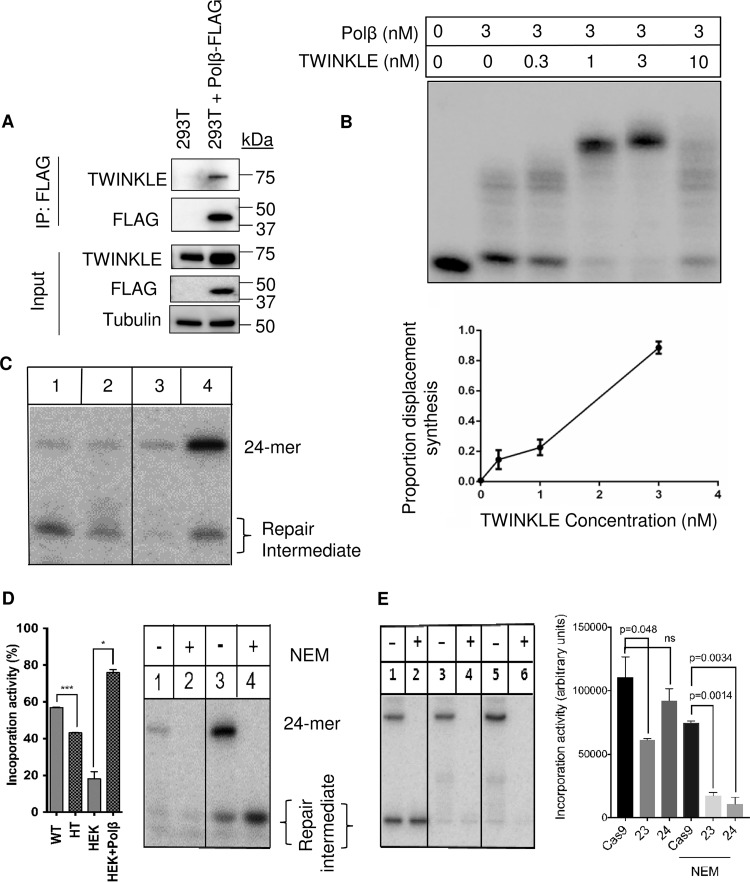
TWINKLE requires TFAM to maintain processivity, and the two proteins have been found in a complex together (32). To ascertain whether TWINKLE and Polβ interact in the mitochondria, we overexpressed FLAG-tagged Polβ in HEK293T cells and detected TWINKLE by immunoblotting in the FLAG pulldown (Fig. 3A). We then tested whether TWINKLE could stimulate Polβ-initiated strand displacement synthesis using purified proteins and a linear oligonucleotide substrate (Fig. 3B). TWINKLE can significantly increase Polβ strand displacement synthesis. This constitutes the first report of the activity of Polβ being stimulated by an mtDNA maintenance protein.
FIG 3.
Mitochondrial Polβ can modulate mtDNA repair through direct interaction with the mitochondrial DNA machinery. (A) To further validate the mass spectrometry results, HEK293T cells were transiently transfected with a Polβ construct with C-terminal FLAG. A protein interaction with TWINKLE protein was detected in the FLAG immunoprecipitation (IP). (B) Purified TWINKLE functionally interacts with purified Polβ to enhance the protein strand displacement synthesis activity. (C) To investigate the role of Polβ in mitochondrial repair, we measured incorporation and ligation activity on a circular substrate (see Fig. S3A in the supplemental material). Extracts derived from Polβ heterozygous mouse brain were deficient in nucleotide incorporation but not ligation activity (24-mer). Lane 1, WT mitochondrial extract (30 μg). Lane 2, HT mitochondrial extracts (30 μg). Overexpression of Polβ in the HEK293T cells resulted in an increase in incorporation and subsequent ligation of the substrate. Lane 3, HEK293T vector control mitochondrial extract (15 μg). Lane 4, HEK293T plus Polβ mitochondrial extracts (15 μg). (D) HEK293T mitochondrial extracts were exposed to the Polγ and ligase inhibitor NEM. With the addition of the inhibitor, the incorporation band (labeled Repair Intermediate) is still present in lanes 2 and 4. Ligation is also completely inhibited. Lane 1, HEK293T vector control mitochondria. Lane 2, HEK293T vector mitochondria plus 5 mM NEM. Lane 3, HEK293T plus Polβ FLAG mitochondria. Lane 4, HEK293T plus Polβ mitochondria plus 5 mM NEM. Mito-WT have significantly (***, P < 0.001) higher incorporation activity than HT samples. HEK293T-plus-Polβ mitochondrial extracts have a significant increase (*, P < 0.01) in incorporation activity compared to the vector control. The protein input was 15 μg. (E) In the absence of Polβ in the HEK293T mitochondria, there is a reduction of single-nucleotide incorporation activity. Lanes 1 and 2, HEK293T with cas9 and with (+) or without (−) NEM. Lanes 3 and 4, HEK293T clone 23 Polβ KO with (+) or without (−) NEM. Lanes 5 and 6, HEK293T clone 24 Polβ KO with (+) or without (−) NEM. Polβ KO HEK293T cells have only residual mitochondrial single-nucleotide insertion activity after NEM treatment, compared to the robust activity in the NEM-treated cas9 extracts. HEK293T Polβ KO assays were conducted in duplicate; all other assays were conducted in triplicate.
The accumulated evidence indicates that Polβ plays a DNA repair role in the mitochondria, analogous to its function in the nuclear compartment. Using Polβ KO MEF cells, we tested nucleotide incorporation activity in total cellular extracts using a circular DNA substrate and could find only limited differences in incorporation activity (not shown). Indeed, this and previous research have shown that there are alternate polymerases and repair pathways that are able to compensate for Polβ incorporation activity in the nucleus. We asked whether nucleotide incorporation rates would be affected in the mitochondrial extracts from cells with a reduction of Polβ. To increase mitochondrial relevance, we used a circular substrate with a synthetic analog of an apurinic/apyrimidinic (AP) site, 3-hydroxy-2-hydroxymethyltetrahydrofuran (THF) (see Fig. S3A in the supplemental material). For this experiment, we used brain tissue extracts from Polβ heterozygous (HT) mice. Polβ HT mice have accelerated age-related neurodegeneration, suggesting that the brain may be selectively affected by Polβ heterozygosity (25). WT mitochondrial samples (Fig. 3C, lane 1) had larger amounts of nucleotide incorporation than HT extracts (lane 2). However, there is only a limited difference in the amount of repair product (upper band), suggesting the rate-limiting step is not nucleotide incorporation. Previous research has reported that ligation activity is rate limiting in BER using this assay (33). The comparable ligation activity can be seen as indirect evidence that the samples have equal input protein concentration. Mitochondrial extracts from HEK293T cells overexpressing Polβ (Fig. 3C, lane 4) or vector alone (lane 3) were also assayed for incorporation. As observed with the mitochondrial brain extracts, nucleotide incorporation levels were heavily determined by Polβ protein levels in the HEK293T-derived mitochondria. However, compared to the brain-derived mitochondrial extracts (lanes 1 and 2), the HEK293T extracts also had higher levels of ligation, resulting in large increases in the completed repair product. This interesting side observation suggests that there are differences in which step in BER may be rate limiting, depending on the cell or tissue type.
To further ascertain whether Polβ was responsible for the previously described changes in incorporation activity, we used the potent polymerase inhibitor N-ethylmaleimide (NEM). NEM can completely inhibit Polγ incorporation at low millimolar (>2 mM) concentrations (34). The HEK293T vector- and Polβ-overexpressing extracts were retested in the presence of the NEM inhibitor (Fig. 3D, lane 4). As anticipated, there was reduced incorporation activity, suggesting that Polγ was inhibited in the reaction (graphed in Fig. S3B), and there was also complete inhibition of ligation activity. Despite the latter inhibition, there was still nucleotide incorporation visible both in the vector control and, more clearly, in the Polβ overexpression experiments, suggesting that Polβ may be responsible for a significant fraction of mitochondrial BER activity in this paradigm. To directly test this notion, we assayed mitochondrial extracts from the HEK293T/Polβ-KO cells and compared them to extracts from the HEK293T/cas9 parental cell line. We report that the Polβ KO extracts have reduced single-nucleotide incorporation activity but similar ligation activity (compare lanes 1 to 3 and 5 in Fig. 3E). With the addition of NEM, we report some reduction in nucleotide incorporation in the cas9 sample (Fig. 3E, lane 2), but this activity is nearly completely abolished in the Polβ KO samples after NEM treatment (lanes 4 and 6). These results suggest that Polβ in the mitochondria of HEK293T cells can act as a backup repair polymerase and can contribute a significant fraction of single-nucleotide incorporation activity.
To complete the analysis of mtBER, the 5′-dRP lyase activity was also investigated. Early in vivo experiments showed that it was not the nucleotide incorporation activity of Polβ but rather the 5′-dRP lyase activity that was cytoprotective (18). While Polγ has intrinsic 5′-dRP lyase capability, the rate of cleavage is extremely low compared to that of Polβ (35). We revisited these experiments in an effort to determine whether Polβ-deficient cells had lower 5′-dRP lyase activity, but we found no significant difference when comparing HT and WT mitochondrial brain extracts (Fig. S3C). This suggests that 50% of normal Polβ protein levels could be enough to rescue any deficiency in 5′-dRP lyase activity or, alternatively, that Polβ is not involved in this activity in the mitochondria.
Decline in Polβ causes changes in mtDNA damage and mutation level eventually resulting in mitochondrial dysfunction in animal and cellular models.
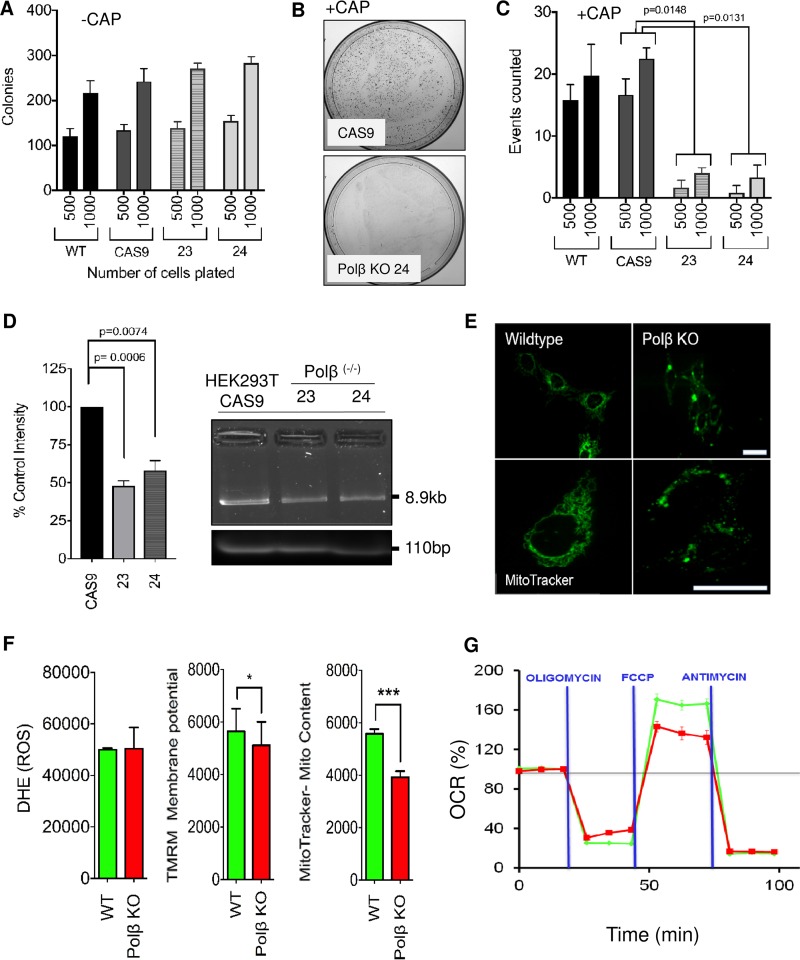
With the Polβ KO cells, we observed reduced levels of single-nucleotide incorporation activity in mitochondrial extracts. We asked whether this would have a direct impact on levels of mutagenesis and DNA damage in the HEK293T cells. The mutation potential of mtDNA was measured indirectly using the chloramphenicol (CAP) assay. Mitochondrial ribosomes differ from cytosolic ribosomes in that they are sensitive to bacterial ribosome inhibitors such as CAP. However, mitochondria can evolve resistance to CAP through single-nucleotide point mutations, altering the conformation of the mitochondrial ribosomes and inhibiting CAP binding. We initially confirmed that Polβ KO does not affect colony formation by plating a range of cell densities and leaving the cells to form colonies over 10 days (Fig. 4A; see Fig. S4A in the supplemental material). We did not measure a statistical difference in colony formation and also report that plating efficiency was comparable. Initial optimization experiments gave us a perspective of CAP survival levels in the HEK293T cells. We can confirm the previous literature that mutational events after CAP exposure are relatively rare. Despite this, the HEK293T/cas9 parental line had hundreds of mutational events when the plated amount of cells was high (104 to 105) (Fig. 4B). In contrast, very few mutational events conferring CAP resistance were observed with the two HEK293T/Polβ-KO cell lines. These preliminary observations were confirmed using a more quantitative approach (Fig. 4C). In this experiment, 500 or 1,000 cells were plated and then exposed to CAP. Again, we observed very few mutational events in the HEK293T/Polβ-KO cells. However, the number of mutational events did not correlate strongly with the number of cells plated in the parental lines. We believe that this is an artifact of the CAP treatment on the HEK293T cells, causing changes in cell morphology including loss of contact adhesion. Despite this, the data suggest that Polβ is potentially responsible for a large proportion of the mutational events in mtDNA.
FIG 4.
Reduction of Polβ causes elevated endogenous mtDNA damage with altered mutation rates. (A) The colony-forming ability of HEK293T cells is unaffected by Polβ KO. Five hundred and 1,000 cells were plated with similar plating efficiency across all groups (n = 6). Also see Fig. S4A in the supplemental material. (B) Cell were exposed to CAP, to which mutation of mitochondrial ribosomal genes can confer survival. In the optimization experiments 102 to 105 cells were plated to determine HEK293T CAP sensitivity. The example shows 104 cells plated, with hundreds of mutation events conferring survival counted in the cas9 cells; in comparison, the Polβ KO cells had relatively very few. (C) Reducing the plating numbers to 500 and 1,000 allowed for more accurate counting of events. Cells were exposed to CAP continuously for 10 days; compared to parental plus cas9, the Polβ KO clones had significantly lower survival rates (n = 6, assay conducted in duplicate). (D) mtDNA from parental+Cas9 cells had approximately 50% higher amplification than either of two HEK293T Polβ-deficient lines, suggesting that Polβ KO cells have higher levels of endogenous mtDNA damage. Assays were conducted in triplicate. Right, image of gel showing a single band with no smearing. Also shown is the small fragment PCR product as an input control. (E) MitoTracker signal shows that MEF Polβ KO cells have reduced mitochondrial content and a breakdown of the mitochondrial network. Upper scale bar = 60 μm (magnification, ×20); lower scale bar = 40 μm (magnification, ×40). (F) Measurements of mitochondrial parameters showed that Polβ KO MEFs had a decrease in mitochondrial content and membrane potential but not ROS levels (n = 3; *, P < 0.05; ***, P < 0.005). (G) Seahorse respiration analysis of MEF Polβ cells shows decreased reserve respiratory capacity after normalization to baseline (n = 5) in Polβ null MEF cells. Red line, Polβ KO; green line, WT.
To further substantiate the role of Polβ in mtDNA repair, we measured the level of unrepaired lesions in the mtDNA using long-amplification PCR (LA-PCR), as previously described (15). The basis of the assay is that a polymerase will be inhibited by certain lesions in the mtDNA, including but not limited to strand breaks and AP sites, lowering the overall amplification rate and resulting in less PCR product compared to that with an undamaged control. We normalized the amount of input mtDNA to mitochondrial copy number, which remained unchanged in the three cell lines (not shown), and quantitated the results using band densitometry after optimizing the assay for the HEK293T cells (Fig. S4B). The LA-PCR results clearly showed that mtDNA from the two HEK293T/Polβ-KO lines had significantly lower amplification than that from the HEK293T/cas9 parental line (Fig. 4D), evidence that these cells have higher levels of endogenous mtDNA damage. These data strongly link Polβ with mtDNA repair and mtDNA mutational load.
Polβ KO mice die just after birth. However, Polβ HT mice are viable but have altered life span characteristics (36) and elevated carcinogenic risk (37). We have previously extensively characterized the Polβ HT mouse and observed a range of differences, particularly in aged mice, that are reflective of an animal with mitochondrial dysfunction (25). Polβ HT mice have accelerated neurodegenerative decline and metabolic dysfunction, including an altered respiratory exchange rate (RER) (Fig. S4C). The elevated RER and higher food intake measured in these weight- and age-matched mice is indicative of an increase in energy demand. Higher endogenous levels of mtDNA damage can result in mitochondrial dysfunction and a shift toward glycolysis (38). Despite a shift toward glycolysis in the Polβ HT animals, there was little change in the expression of oxidative phosphorylation (OXPHOS) components, suggesting that 50% Polβ is enough in the absence of additional mtDNA damage to protect the animal from severe metabolic crisis (25). We used mouse embryos (5 WT and 5 Polβ KO), genotyped by RT-PCR and confirmed by immunoblotting (Fig. S4D), to test whether a complete KO of Polβ would accelerate metabolic dysfunction and cause aberrant expression of OXPHOS components. Microarray analysis showed a significant (greater than 2-fold change in Z-score) decrease in the expression of genes involved in all OXPHOS complexes except the exclusively nucleus-encoded complex 2 (Fig. S4E). Pathway analysis of the microarray revealed pathways associated with neurodegeneration, bioenergetics, and mitochondria to be particularly affected in the Polβ KO embryos (Table 2). Consequently, primary MEFs (also derived from the embryos) and transformed MEF Polβ KO cells were measured for a comprehensive range of mitochondrial parameters. Surprisingly, the primary KO MEFs did not exhibit mitochondrial dysfunction in any of the parameters tested (not shown). In contrast, the higher-passage (passage 75) simian virus 40 (SV40) large T-antigen-immortalized Polβ KO MEFs (Mβ19tsA) did have mitochondrial impairment (summarized in Table S1 in the supplemental material). This included a breakdown of the mitochondrial matrix visualized using MitoTracker (Fig. 4E). The mitochondrial content was significantly reduced (Fig. 4F, right), confirming the MitoTracker microscopy results. The mitochondrial membrane potential was also reduced in the Polβ KO cells compared to controls (Fig. 4F, middle). Despite the Polβ KO cells having only 60% of the mitochondrial content of the control cells, ROS measurement using dihydroethidium (DHE) showed that the cells had similar ROS levels (Fig. 4F, left). These results predicted that the Polβ KO MEFs would have altered OXPHOS function. We used the Seahorse XF24 bioanalyzer to measure oxidative phosphorylation (oxygen consumption rate [OCR]) and the extracellular acidification rate (ECAR) in the wild-type and KO cells. The reduced mitochondrial parameters culminated into a significant reduction in mitochondrial reserve capacity in the KO MEFs, though no change in basal respiration or extracellular acidification was measured (Fig. 4G and S4F).
TABLE 2.
Pathways heavily downregulated in Polβ MEF KO cellsa
| Affected pathway | Z-score |
|---|---|
| MOOTHA VoxPhos | −15.41821084 |
| Reactome diabetes pathways | −14.72676363 |
| Reactome electron transport chain | −14.44314437 |
| KEGG oxidative phosphorylation | −14.25314758 |
| KEGG Parkinson's disease | −13.13985838 |
| Wong mitochondrial gene module | −12.98398576 |
| KEGG Huntington's disease | −12.14384138 |
| KEGG Alzheimer's disease | −10.51044256 |
| MOOTHA mitochondria | −9.738233576 |
| Reactome glucose regulation of insulin secretion | −9.374372171 |
| MOOTHA human MitoDB | −9.149521789 |
Analysis of the MEF Polβ KO microarray showed that the most heavily affected pathways were those associated with mitochondrial respiration and neurodegenerative diseases (Z-scores range from −9 to −15).
DISCUSSION
We identified mitochondrial Polβ in multiple mammalian tissues and cell types. Polβ interacts with mitochondrion-specific DNA repair machinery, including the nucleoid proteins TWINKLE, SSBP1, and TFAM. The presence of Polβ in the mitochondria bolsters overall BER capacity, and KO of Polβ results in higher endogenous mtDNA damage and lower in vivo nucleotide incorporation activity. These results strongly suggest that Polβ is an important part of mtBER, at least in some tissues, with its N- and C-terminal ends having distinct roles in the protein's repair response. The N terminus of Polβ contains the 5′-dRP lyase domain (18) and also harbors single- and double-stranded DNA binding domains (39). However, this segment of the protein does not appear to contain a strong functional MLS, despite the indication otherwise by the MitoProt program. Conversely, deletion of the D17N region caused Polβ to be excluded from the nucleus, suggesting that this sequence contains a dominant NLS. A Polβ N-terminal nuclear localization sequence has been recently reported by another group, correlating with the results here (40). Indeed, even the addition of a confirmed MLS to the N terminus of FL Polβ could not completely disrupt the protein's transport to the nucleus. Only when the protein was not in the nucleus, as observed with the D17N construct, was there prominent Polβ mitochondrial localization. This observation can be interpreted a number of different ways. First, the N terminus of Polβ must be modified before the protein can localize to the mitochondria, potentially giving rise to an alternate mitochondrion-specific isoform. Indeed, there is an alternate in-frame start site 17 aa from the amino terminus of the protein. While not completely ruling out the possibility of an alternate isoform, the Polβ that was detected using Western analysis was larger, not smaller, than the FL Polβ protein, which is more consistent with a posttranslational modification. Alternatively, the N-terminal NLS may be cleaved or disrupted in the cytoplasm, resulting in mitochondrial localization, similar to what has been reported for the Polβ protein partner APE1 (41). The final possibility is that the addition of the large GFP tag at the C terminus artificially influences protein transport. Supporting this idea are the microscopy results confirming that cells overexpressing the smaller FLAG tag have a greater abundance of mitochondrial Polβ. The potential for a C-terminal GFP to influence mitochondrial transport raises the possibility that there is an MTH located at the C terminus. Indeed, the Polβ long-patch BER (LP-BER) partner DNA2 has an MTH domain that shares similarity to the segment in Polβ (42).
The C-terminal fragment of Polβ bound proteins that were highly relevant in single-strand break repair, but none of these proteins are exclusively mitochondrial. Previous research has highlighted the functionality of the C terminus of Polβ in nuclear BER. The region is required for binding the scaffold protein X-ray repair cross-complementing protein 1 (XRCC1), using residues 301 to 306 (43), with the interaction reported to protect and stabilize the protein in the nucleus (43). Further, the role of Polβ in nuclear LP-BER can be stimulated through a functional interaction with PCNA, and again, this interaction requires its C-terminal residues (44). Despite the strong evidence for the utility of the C terminus in nuclear DNA repair, a novel aspect of the present research is the finding that Polβ relies heavily on the N terminus for its mitochondrial protein interactions. The N-terminal fragment of Polβ bound proteins associated with mitochondrial transport, including the TOM70 import protein, implying that the TOM machinery is used to transport the protein from the outer mitochondrial membrane into the inner mitochondrial membrane. TOM complex transportation relies on the initial identification of mitochondrial preproteins by either TOM70, TOM22, or TOM20 complex subunits (45). Pertinently, TOM70 interacts with preproteins that have an internal MTH as opposed to an N-terminal MLS, additional indirect evidence that Polβ may have an MTH. After mitochondrial internalization, the protein is passed to the TIM complex, and TIM50 was identified as a likely Polβ interaction. Also identified in the pulldown were multiple proteins associated with assembly and stabilization of mitochondrial proteins once in the mitochondrial matrix, including GRPL-1, an essential part of the presequence translocase-associated motor (PAM) (46), and associated proteins HSPA9 (47) and HSPD1 (48). All mitochondrial transport components interacted with only the N-terminal fragment of Polβ. Indeed, the N terminus was important not only for transport but also for functional protein interactions in the mitochondrial nucleoid. The N terminus of Polβ bound to principal mtDNA maintenance proteins TFAM, TWINKLE, and SSBP1. Polβ and TFAM interacted using the N terminus of Polβ and the HMG1 domain of TFAM. The HMG1 domain directly binds mtDNA and was also required for the binding of Polβ BER protein partners PARP1 and OGG1.
We propose that Polβ is in the mitochondria to perform BER, and we support this notion using several lines of evidence. However, the lack of XRCC1 scaffold protein in mitochondria could argue against a role for Polβ in mtBER. Early reports suggested that the Polβ/XRCC1 interaction was indispensable for BER, stabilizing Polβ, facilitating recruitment, and enhancing processivity (49). Only very recently have two groups concluded that Polβ can function independently of XRCC1 in DNA repair (43, 50). It remains to be determined whether Polβ mtBER relies on an alternate scaffold protein in the mitochondria; however, TFAM would be the most obvious candidate. TFAM preferentially binds to damaged mtDNA and is required to maintain mitochondrial genomic stability. The interaction of Polβ with the TFAM-HMG1 motif in the mitochondria appears analogous to the interaction between Polβ and the high-mobility group box 1 (HMGB1) protein, which occurs in the nucleus (51). The nuclear HMGB1 protein binds to substrates containing 5′-dRP termini and directly interacts with and stimulates the activity of Polβ and associated BER proteins APE1 and flap endonuclease 1 (FEN1) (51). The same study found that the nuclear HMGB1-Polβ interaction inhibits 5′-dRP lyase activity of Polβ. Additional strong evidence for the role of Polβ in mtBER comes from its direct functional interaction with the TWINKLE helicase. This interaction was shown to facilitate Polβ strand displacement, suggesting that Polβ may also carry out LP-BER in the mitochondria. Nuclear LP-BER is divided into PCNA-dependent and -independent pathways. We did not detect PCNA in any of our mitochondrial fractions, indicating that Polβ participates in a PCNA-independent LP-BER pathway, potentially involving the associated proteins TWINKLE, FEN1, DNA2, and Lig3 (52, 53). Indeed, mitochondrial LP-BER has appeared in a number of related publications (54–56), and in vivo DNA repair work has shown that uracil and AP sites are resolved in mtDNA using both long- and short-patch mechanisms (34).
We have previously shown that Polβ may be particularly important in nuclear DNA repair in postmitotic systems due to the reduction of LP-BER factors closely associated with the replicative cycle, including FEN1, PCNA, and Polδ and -ε (52). In a postmitotic system, mitochondria contain the only DNA that is still replicating and hence is more susceptible to the long-term effects of DNA damage. Notably, we found high levels of Polβ in the mitochondrial extracts taken from brain tissue. However, other terminally differentiated tissue, including heart and muscle mitochondrial extracts, did not have detectable levels of Polβ, suggesting that terminal differentiation alone was not enough to confer mitochondrial Polβ localization. A previous report detected a Polβ-like enzyme in the bovine heart mitochondria using a much larger mitochondrial preparation. We cannot exclude the possibility that Polβ protein in the heart samples or other organs from mice was beyond the range of detection. We can speculate that the level of mitochondrial Polβ in the tissue may be correlative with regions that have particularly high levels of mtDNA damage that require the additional repair activity provided by mitochondrial Polβ. The mitochondrial role of Polβ does not appear to be related only to the 5′-dRP lyase activity. While it appears that Polγ has all the mtDNA repair activities required under endogenous conditions, this does not rule out a necessity for Polβ. The nucleus has multiple DNA polymerases, many that have overlapping substrates with Polβ, yet Polβ is still present in the nucleus and is required when the cell faces genotoxic stress. The nucleotide incorporation data presented here, in conjunction with the CAP mutational analysis and LA-PCR results, help us to build a more complete picture of the important role of Polβ in mitochondrial BER. Particularly interesting is the evidence that Polβ is responsible for a large proportion of mutational events that occur in mtDNA. This could place Polβ in a central causative role in progressive aging disorders and other diseases related to mitochondrial dysfunction, including cancer, a notion that requires further investigation.
In this research, we did not uncover any evidence that Polγ and Polβ work synergistically to repair mtDNA. The large spectrum of diseases associated with Polγ mutation is indirect evidence that Polβ is not able to compensate completely for Polγ (57–59). This is despite a homolog of Polβ being solely responsible for the repair and replication of mtDNA in the parasite Trypanosoma brucei (20, 60). However, in humans the two polymerases are vastly different, with Polγ dwarfing Polβ in size and complexity and encoding an efficient proofreading function. Polγ mutations attributed to serious disease cause loss of processivity of the protein, with the Polγ active site blocking the damage region and/or elevated misincorporation of nucleotides. The presence of Polβ in the mitochondria cannot compensate for these Polγ mutations, potentially due to Polβ not having proofreading activity or having the ability to bind to the blocked site. Some mutations in Polγ take years to manifest into disease in patients, and likewise, a lack of a mitochondrial phenotype in primary Polβ null MEFs is not entirely surprising. Despite not being able to completely compensate for Polγ, the discovery of Polβ in the mitochondria may address some of the peculiarities associated with Polγ mutations and disease outcome. Mutations in Polγ are attributed to a broad spectrum of disease phenotypes. Often the severity of the disease is not predicted by the amount of residual Polγ activity. Particularly perplexing is the Y955C mutation in POLGA, the most common and severe autosomal dominant mutation causing CPEO and Parkinsonism (61). The mutation causes Polγ incorporation levels to drop below 1% of control activity, yet the life span of these patients is measured not in days or weeks but rather in decades, and the individuals are often symptomless well into adulthood. This and other similar mutations in Polγ that cause nearly complete loss of protein activity with adult onset of symptoms (62) suggest the presence of a backup polymerase function in the absence of Polγ.
MATERIALS AND METHODS
Cell lines.
See the supplemental material for all information related to cell lines.
Animal protocols.
See the supplemental material and reference 25 for all information related to animal protocols. All mouse work was done in accordance with and with approval of the Animal Care and Use committee at the National Institute on Aging.
Large-scale mitochondrial extractions.
Mitochondrial extracts were prepared as described previously (34). Mitochondria were isolated from the cells collected from 15 cell culture dishes (150 mm) at ∼75 to 90% confluence. Cells were detached by trypsinization, collected by centrifugation at 400 × g, resuspended in hypotonic buffer (20 mM HEPES [pH 8.0], 5 mM KCl, and 1 mM dithiothreitol [DTT]), and incubated on ice until swollen; 2× MSH buffer (420 mM mannitol, 140 mM sucrose, 20 mM HEPES [pH 7.4], 4 mM EDTA, 2 mM EGTA, and 5 mM DTT) was then added (1:1, vol/vol), and the cells were broken with a Dounce homogenizer. The homogenate was centrifuged at 1,000 × g. This step was repeated until no nuclei were seen in the pellet (typically 2 or 3 times). The crude mitochondria were pelleted at 10,000 × g for 30 min, resuspended in 1× MSH–50% Percoll (GE Health Care Lifesciences, Pittsburgh, PA), loaded on top of a 1× MSH–50% Percoll mixture, and centrifuged at 50,000 × g for 75 min. The mitochondrial fraction was removed from the gradient and centrifuged in 1× MSH buffer at 3,000 × g for 10 min to remove Percoll. The mitochondrial pellet was washed once in 1× MSH buffer without EDTA and EGTA, resuspended in 0.15 ml of the same buffer, and then divided and treated with different concentrations of proteinase K (Qiagen, Valencia, CA) at 37°C for 20 min. Treatment of isolated mitochondria with ∼0.1 mg/ml proteinase K was sufficient to clear mitochondria of nuclear contamination. Phenylmethylsulfonyl fluoride (PMSF) was added at final concentration of 5 mM to inactivate proteinase K. Mitochondria were pelleted at 10,000 × g for 10 min and washed twice with 0.2 ml of a protease inhibitor mix (Complete proteinase inhibitor [Roche, New York, NY], 2× MSH buffer [1:1, vol/vol], and 5 mM PMSF). The mitochondria were suspended in buffer I (10 mM HEPES [pH 8.0] and 200 mM KCl), and then in an identical volume of buffer II (10 mM HEPES [pH 8.0], 2 mM DTT, 40% glycerol, 1% Nonidet P-40, 1% Triton X-100, Complete protease inhibitor, phosphatase inhibitor cocktails 1 and 2, 10 mM PMSF, and 200 mM KCl), incubated at 4°C for 90 min, and briefly sonicated. Mitochondrial debris was removed by centrifugation at 16,200 × g for 10 min, and the supernatant was collected and stored at −80°C. The protein concentration was measured using a bicinchoninic acid (BCA) assay using bovine serum albumin standards (Pierce, Thermo Fisher Scientific, Waltham, MA). Mitochondria derived from mouse organs were isolated from adult male C57BL/6 mice. Organs were cut into small pieces and homogenized in 5 ml of ice-cold 1× MSH buffer, pH 7.4. Tissue was disassociated using a Teflon Dounce homogenizer with lysate centrifuged at 600 × g for 10 min. The supernatant was removed and centrifuged at 7,000 × g for 10 min. The supernatant was carefully removed, and the pellet was suspended in 1× MSH–50% Percoll, loaded on top of a 1× MSH–50% Percoll mixture, and centrifuged at 50,000 × g for 75 min. The mitochondrial fraction was removed from the gradient and processed as described above.
DNA polymerase β protein quantitation by immunoblotting.
Protein lysate was separated on a mini-Protean TGX gradient (4 to 15%) or 12% gel (Bio-Rad, Hercules, CA) at 60 V for 120 min. Transfer was conducted overnight onto a Novex PVDF 0.2-μm membrane (Thermo Fisher Scientific) at 4°C and 40 V. Membranes were blocked in 4% milk protein for 60 min at room temperature and then exposed to primary antibody overnight at 4°C and to secondary antibody (1:10,000) (GE Life Sciences) for 120 min at room temperature. Molecular weight was predicted with broad-range molecular weight markers (Bio-Rad). Secondary horseradish peroxidase (HRP) conjugate was detected by chemiluminescence (Super Signal West Femto; Pierce) on an XRS+ ChemiDoc imager (Bio-Rad). Images were analyzed and quantitated using Image Lab (V3.0) (Bio-Rad).
Antibodies used in immunoblots were to Polβ (4445-MC-100, clone 61 [Trevigen, Rockville, MD], HB600-1026, clone 61 [Novus, Littleton, CO], and ab26343 [Abcam, Cambridge, MA], TFAM (SC-19050) and anti-laminin A/C (SC-206810) (Santa Cruz Biotechnology, Dallas, TX), VDAC (ab15895; Abcam), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ab181603; Abcam).
Polβ vector construction and validation.
Plasmid RG210765 (Origene, Rockville, MD) encoding human Polβ with a C-terminal GFP tag (Polβ-GFP) was PCR linearized with primers: Polβ-lin-F (5′-AGCAAACGGAAGGCGCCGCAG-3′) and pCMV6-lin-R (5′-GGCGATCGCGGCGGCAGATCT-3′). The purified PCR product was ligated with phosphorylated DNA duplex containing the mitochondrial targeting sequence of human superoxide dismutase 2 (5′-ATGTTGAGCCGGGCAGTGTGCGGCACCAGCAGGCAGCTGGCTCCGGCTTTGGGGTATCTGGGCTCCAGGCAG-3′), resulting in the MLS-Polβ-GFP plasmid. Similarly, plasmid D17N Polβ-GFP with the putative endogenous mitochondrial localization sequence of Polβ deleted was generated by PCR-mediated linearization (primers Polβ-lin2-F [5′-ATGCTCACAGAACTCGCAAACTTTG-3′] and pCMV6-lin-R [5′-GGCGATCGCGGCGGCAGATCT-3′]) followed by ligation of the PCR product. Immunoblot analysis was used to confirm the correct cellular localization of the vectors, conducted essentially as described for Western protein quantitation above. Small-scale mitochondrial extracts were produced using the magnetical activated cell sorting (MACS) mitochondrial isolation kit (Miltenyi Biotec, San Diego, CA) in accordance with the manufacturer's instructions. Whole-cell lysates were created using rapid freeze-thawing with sonication to disrupt nuclei. The antibodies used were to Polβ antibody (ab26343; Abcam), VDAC1 (SC-8828; Santa Cruz), and β-tubulin (SC-9935; Santa Cruz).
Transfection and microscopy.
HEK293T cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. At 24 h prior to transfection 6 × 105 cells were seeded in a 35-mm glass-bottom dish. Plasmids Polβ-GFP, SOD2 MLS-Polβ-GFP, and D17N Polβ-GFP were transfected using PolyJet (SignaGen, Rockville, MD) reagent according to the manufacturer's instructions. At 22 h posttransfection, the cells were stained for 15 min with 50 nM MitoTracker Deep Red FM (Thermo Fisher Scientific, Waltham, MA), washed with complete DMEM, and grown for an additional 2 h. Live cells were imaged using a Nikon Eclipse 2000E confocal microscope with a Yokogawa CSU 10 spinning-disk head (Improvision/PerkinElmer, Waltham, MA) equipped with an environmental chamber (InVivo Scientific, St. Louis, MO). Volocity software (Improvision, PerkinElmer) was employed for image acquisition and processing.
FLAG plasmid construction.
Origene plasmid RC210765 encoding human Pol β with a C-terminal Myc-FLAG tag (Pol β-FLAG) was PCR linearized with primers: PolB-lin-F (5′-AGCAAACGGAAGGCGCCGCAG-3′) and pCMV6-lin-R (5′-GGCGATCGCGGCGGCAGATCT-3′). The purified PCR product was ligated with phosphorylated DNA duplex containing the mitochondrial targeting sequence of human superoxide dismutase 2 (5′-ATGTTGAGCCGGGCAGTGTGCGGCACCAGCAGGCAGCTGGCTCCGGCTTTGGGGTATCTGGGCTCCAGGCAG-3′), resulting in the MLS-Pol β-FLAG plasmid. Similarly, plasmid d17N-Pol β-FLAG with the putative endogenous mitochondrial localization sequence of Pol β deleted was generated by PCR-mediated linearization (primers PolB-lin2-F [5′-ATGCTCACAGAACTCGCAAACTTTG-3′] and pCMV6-lin-R [5′-GGCGATCGCGGCGGCAGATCT-3′]) followed by ligation of the PCR product.
FLAG transfection and microscopy.
HEK293T cells were cultured in DMEM supplemented with 10% FBS and penicillin-streptomycin. Plasmids Pol β-FLAG, MLS-Pol β-FLAG, and d17N-Pol β-FLAG were transfected using PolyJet (SignaGen) reagent according to the manufacturer's instructions. At 22 h posttransfection, the cells were stained for 45 min with 500 nM MitoTracker Deep Red FM (Thermo Fisher), washed with complete DMEM, fixed, and permeabilized. Fixed cells were stained with anti-FLAG antibody (Sigma F7425), followed by anti-rabbit Alexa Fluor 488-labeled secondary antibody (A21206; Life Technologies), and mounted in Vectashield. Images were acquired using Nikon Eclipse 2000E confocal microscope with a Yokogawa CSU 10 spinning-disk head (Improvision/PerkinElmer) equipped with an environmental chamber (InVivo Scientific). Volocity software (Improvision) was employed for image acquisition and processing.
Affinity column screening using GST fusion protein and mass spectrometry.
To create the different Polβ and TFAM constructs used in the glutathione S-transferase (GST) affinity column, Polβ fragments Polβ-N (aa 1 to 141) and Polβ-C (aa 141 to 335) and TFAM fragments TFAM-F (aa 41 to 246), TFAM-N (aa 41 to 160), and TFAM-C (aa 153 to 246) regions were amplified by PCR and cloned into the pGEX vector (GE Healthcare). The constructed vectors were transfected into E. coli BL21 and cultured in 25 ml LB medium, followed by induction of expression with isopropyl-β-d-1-thiogalactopyranoside (IPTG) (1 mM). Cellular pellets were sonicated with 1.5 ml RP extraction buffer (50 mM Tris-HCl, 0.3 M NaCl, 0.1% NP-40, pH 7.5) and centrifuged at 1,700 × g for 10 min. The supernatants were incubated with 50 μl of glutathione-Sepharose at 4°C for 1 h, followed by washing twice with RP extraction buffer, twice with 1.2 M elution buffer (50 mM HEPES, 1.2 M NaCl, pH 7.5) and a final wash with cell extraction buffer (50 mM HEPES, 0.3 M NaCl, 0.1% NP-40, pH 7.5). The GST fusion protein on the beads was incubated with 1.5 ml of previously proteinase-treated mitochondrial extracts, DNase I (50 μg/ml; Wako, Richmond, VA), RNase A (50 μg/ml; Wako), and Benzonase (10 μg/ml; Novagen, Merck Bioscience, UK) at 4°C for 4 h and washed three times with 0.15 M washing buffer (50 mM HEPES, 0.15 M NaCl, 0.1% NP-40, pH 7.5) and two times with phosphate-buffered saline (PBS). The bead-bound proteins were eluted two times with 300 μl of 1.2 M elution buffer. The eluant (600 μl) had 1 ml of PBS added and was concentrated to 50 μl using Amicon Ultra-4 (Millipore, Billerica, MA). Bound proteins were separated by SDS-PAGE and stained by using a silver stain MS kit (Wako). Separated bands were analyzed by mass spectrometry (Japan Proteomics, Japan). Antibodies used in binding analysis were to OGG1 (SC-12076; Santa Cruz), PARP1 (SC-8007; Santa Cruz), Polβ (ab26343; Abcam), TFAM (7495S; Cell Signaling Technology, Danvers, MA), Tim50 (SC-393678; Santa Cruz), Tom70 (SC-26495; Santa Cruz), and TWINKLE (SC-155418; Santa Cruz).
Polβ strand displacement assay.
Polβ strand displacement synthesis was measured using a duplexed 34-mer oligonucleotide substrate with a single nucleotide gap at position 16. The 15-mer oligonucleotide was 5′ labeled with [γ-32P]ATP and subsequently hybridized to a 34-mer complementary strand and downstream 18-mer (63). Reactions (20-μl mixtures) were performed in 10 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 20 mM KCl, 2 mM DTT, 20 mM ATP, 0.5 mM deoxynucleoside triphosphate (dNTP) mix, and 0.1 mg/ml bovine serum albumin (BSA). Each reaction mixture contained 1 nM labeled substrate, either no Polβ or 3 nM Polβ, and increasing concentrations of TWINKLE, from 0 to 10 nM. Reaction mixtures were incubated at 37°C for 30 min. Subsequently, 10 μl of formamide stop buffer/loading dye was added to each reaction mixture prior to incubation at 95°C for 10 min. The samples were resolved in a denaturing 20% polyacrylamide gel containing 7 M urea. After electrophoresis, the gels were visualized using a Molecular Dynamics PhosphorImager (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). The images were analyzed using ImageQuant 5.2 software (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). The amount of displacement synthesis was quantitated by dividing the amount of radioactivity of the bands representing >1 nucleotide insertion by the total radioactivity of each lane.
THF BER assay.
An oligonucleotide containing a synthetic analog of an AP site, 3-hydroxy-2-hydroxymethyltetrahydrofuran (THF) (designated X), 5′-GATCCTCTAGAGTXGACCTGCA-3′, was annealed to single-stranded DNA (ssDNA) derived from plasmid pGEM-3Zf(+) containing guanine opposite the AP site. DNA synthesis was carried out with T4 gene 32 ssDNA-binding protein, T4 DNA polymerase, T4 DNA ligase, and dNTPs at 37°C for 120 min. The covalently closed double-strand circular DNA was purified from agarose gel. The repair reaction was carried out in 15 and 30 μg HEK293T and mouse mitochondrial extracts in 40 mM HEPES-KOH (pH 7.9), 1 mM DTT, 5 mM MgCl2, 120 mM KCl, 2 mM ATP, 0.36 mg/ml BSA, 20 μM dATP, 20 μM dTTP, 20 μM dGTP, 5 μM dCTP, 4.5 mM phosphocreatine, 50 ng/μl creatine kinase, 8 nM DNA substrate, and 80 nCi [α-32P]dCTP at 32°C for the indicated times in a volume of 25 μl. Where indicated, 5 mM N-ethylmaleimide (NEM) was added to the mixture before the addition of DNA substrate and the initiation of repair. The reaction was stopped by adding EDTA, and the mixture was further incubated with SDS and proteinase K at 42°C for 30 min. DNA was purified with phenol-chloroform extraction and salt precipitation, suspended in 10 mM Tris-HCl (pH 8.5), digested with XbaI and HindIII restriction enzymes, and separated in a 15% denaturing polyacrylamide gel at 300 V for 1 h. The repair experiments were carried out in duplicates.
dRP lyase assay.
The oligonucleotide sequences for the dRP lyase assay were 34U oligo (5′-CTGCAGCTGATGCGCUGTACGGATCCCCGGGTAC-3′) and 34templ oligo (5′-GTACCCGGGGATCCGTACGGCGCATCAGCTGCAG-3′). For substrate preparation, the 34U oligonucleotide was 3′ end labeled using terminal deoxynucleotide transferase (New England BioLabs) with 3′-[α-32P]dATP (5000 Ci/mmol; PerkinElmer) and annealed to the 34templ oligonucleotide by heating to 85°C and cooling at the rate of 1°C/min. The annealed duplex was purified with a Sephadex G-50 spin column equilibrated in Tris-EDTA (TE) and stored at −20°C. The dRP lyase assay was performed as previously published (64), with the following modifications. Uracil-containing 3′-32P-labeled DNA substrate (50 nM) was treated with 0.05 units of uracil DNA glycosylase (Trevigen; 0.1 U/μl) in 50 mM HEPES-NaOH (pH 7.5)–0.5 mM EDTA–0.2 mM DTT for 5 min at 37°C. Next, MgCl2 (5 mM final concentration) and AP endonuclease (10 nM final concentration) were added and incubated for additional 5 min at 37°C to generate AP site-containing substrate. The substrate was added (10 nM final concentration) to the dRP lyase reaction mixture (10-μl final volume; 50 mM HEPES-NaOH [pH 7.5], 20 mM KCl, 2 mM DTT) containing the indicated amount of mitochondrial extract or 20 nM Pol β and incubated for 30 min at 37°C. The reaction mixture was transferred to ice and reduced for 15 min by addition of 1 μl of 100 mM NaBH4. Finally, 0.5 μl of the reaction mixture was mixed with 10 μl loading dye (98% formamide, 20 mM EDTA, 0.02% bromphenol blue, 0.02% xylene cyanol), heated for 2 min at 75°C, and separated on 20% denaturing urea polyacrylamide gels. Gels were visualized using a Typhoon Trio+ phosphorimager (Molecular Dynamics) and quantified with Image Quant TL software.
Microarray analysis.
Gene expression analysis was conducted on 10 embryos, (WT, n = 5; Polβ KO, n = 5). Transcriptional profiling was determined using Illumina Sentrix BeadChips (Illumina, San Diego, CA). Total RNA was extracted using an RNA extraction kit (Qiagen) with further purification using a second column (RNA Clean and Concentrate; Zymo Research, Irvine, CA). Initial quantitation was conducted using a NanoDrop ND-1000 spectrophotometer. The quality of the RNA was inspected using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples below an RNA integrity number of 7.5 were discarded. The microarray analysis was performed by the Gene Expression and Genomics core facility (National Institute on Aging) and analyzed using DIANE 6.0 software (see http://www.grc.nia.nih.gov/branches/rrb/dna/diane_software.pdf for information). For further information regarding the analysis, see the supplemental material.
Flow cytometry.
Mitochondrial membrane potential, mitochondrial content, and both intracellular and mitochondrial ROS were measured by flow cytometry as previously described (65). Briefly, cells were plated in 6-well plate until semiconfluent for 24 h. Cells were harvested with trypsin, washed in PBS, and resuspended in PBS–10% FBS. Aliquots were separated for each experiment, followed by staining with specific dyes. Intracellular ROS were detected after treatment with dihydroethidium (DHE) (Thermo Fisher Scientific) at a concentration of 3 μM for 30 min. For mitochondrial membrane potential, cells were treated with 40 nM tetramethylrhodamine methyl ester perchlorate (TMRM) (Thermo Fisher Scientific) for 15 min, and for mitochondrial content, they were treated with 50 nM MitoTracker Green FM (Thermo Fisher Scientific) for 30 min. All the samples were incubated at 37°C, and the fluorescence signal detection was performed using an Accuri C6 flow cytometer (Accuri, San Jose, CA) and CFLOW Plus (version 1.0.227.4), counting 5 × 104 cells. FCS Express 4 (De Novo Software, Glendale, CA) was used to analyze data.
Cellular oxygen.
A Seahorse XF-24 Instrument (Seahorse Bioscience, Boston, MA) was used to measure cellular oxygen consumption rates and extracellular acidification. A two-step seeding process was used to seed samples in Seahorse 24-well cell culture plates. The number of cells for the experiments was defined according to previous tests for each cell line. At least three parallel experiments were done for each type of cell. After 24 h, the medium was changed to unbuffered XF assay medium (Seahorse Bioscience) supplemented with 25 mM glucose (Sigma-Aldrich, St. Louis, MO), 1 mM sodium pyruvate, and 1 mM GlutaMAX (Invitrogen). Cells were incubated for 1 h at 37°C in ambient concentrations of CO2 and O2. The cells were subjected to measurements of respiration in four blocks. First, the basal respiration rate was measured. The second measure was performed after addition of 1 μM oligomycin to inhibit mitochondrial complex V. Before the third block of measurements, 300 nM carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sigma-Aldrich) was added to uncouple respiration. Finally, the last measurements were performed after addition of 2 μM antimycin A (Sigma-Aldrich), which inhibited mitochondrial complex 3. After measurements, cells were trypsinized and counted using a Coulter Counter (Beckman Coulter, Brea, CA).
Statistics.
Comparison between two groups was done using the nonparametric Student t test with Welch's correction. Prism analysis software (PRISM 6; GraphPad, La Jolla, CA) was used throughout; the full statistical output is available on request.
Ethical approval of experiments.
All animal protocols were approved by the Animal Care and Use Committee (361-LMG-2014 and 361-LMG-2017) of the National Institute of Aging.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rachel Abbotts and Yujun Hou for critical reading of the manuscript. We also thank Qingming Fang for purified Polβ protein.
P.S., J.T., A.M., and T.K. carried out experiments for Fig. 1. S.K., J.T., and A.Y. carried out experiments for Fig. 2. A.M., B.A.B., and H.L. carried out experiments for Fig. 3. P.S., G.S.L., J.T., and K.A.B. carried out experiments for Fig. 4. P.S. and V.A.B. coordinated the study. P.S. wrote the article with input from D.L.C., D.M.W., R.W.S., and V.A.B. M.A. did experiments and helped write the paper.
Funding for this study was from the Intramural Research Program of the National Institutes of Health (NIH), National Institute on Aging (Z01-AG00735), and the Fundacao de Amparo a Pesquisa do Estado de Sao Paulo-FAPESP (2013/11052-1 to G.S.L.). Funding for open access was from the NIH Intramural Program. This work was supported by National Institutes of Health grant CA148629 (to R.W.S.) and the Abraham A. Mitchell Distinguished Investigator Fund (to R.W.S.).
R.W.S. is a scientific consultant for Trevigen, Inc. The authors have no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00237-17.
REFERENCES
- 1.Hoeijmakers JH. 2009. DNA damage, aging, and cancer. N Engl J Med 361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 2.Madabhushi R, Pan L, Tsai LH. 2014. DNA damage and its links to neurodegeneration. Neuron 83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. 2013. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harb Perspect Biol 5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agostino A, Valletta L, Chinnery PF, Ferrari G, Carrara F, Taylor RW, Schaefer AM, Turnbull DM, Tiranti V, Zeviani M. 2003. Mutations of ANT1, Twinkle, and POLG1 in sporadic progressive external ophthalmoplegia (PEO). Neurology 60:1354–1356. doi: 10.1212/01.WNL.0000056088.09408.3C. [DOI] [PubMed] [Google Scholar]
- 5.Qian Y, Kachroo AH, Yellman CM, Marcotte EM, Johnson KA. 2014. Yeast cells expressing the human mitochondrial DNA polymerase reveal correlations between polymerase fidelity and human disease progression. J Biol Chem 289:5970–5985. doi: 10.1074/jbc.M113.526418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian Y, Ziehr JL, Johnson KA. 2015. Alpers disease mutations in human DNA polymerase gamma cause catalytic defects in mitochondrial DNA replication by distinct mechanisms. Front Genet 6:135. doi: 10.3389/fgene.2015.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly Y, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 8.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, van, Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 9.Harman D. 1992. Free radical theory of aging. Mutat Res 275:257–266. doi: 10.1016/0921-8734(92)90030-S. [DOI] [PubMed] [Google Scholar]
- 10.Bohr VA, Stevnsner T, de Souza-Pinto NC. 2002. Mitochondrial DNA repair of oxidative damage in mammalian cells. Gene 286:127–134. doi: 10.1016/S0378-1119(01)00813-7. [DOI] [PubMed] [Google Scholar]
- 11.Prakash A, Doublie S. 2015. Base excision repair in the mitochondria. J Cell Biochem 116:1490–1499. doi: 10.1002/jcb.25103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilliam TA, Jozwiakowski SK, Ehlinger A, Barnes RP, Rudd SG, Bailey LJ, Skehel JM, Eckert KA, Chazin WJ, Doherty AJ. 2015. Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic Acids Res 43:1056–1068. doi: 10.1093/nar/gku1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh B, Li X, Owens KM, Vanniarajan A, Liang P, Singh KK. 2015. Human REV3 DNA polymerase zeta localizes to mitochondria and protects the mitochondrial genome. PLoS One 10:e0140409. doi: 10.1371/journal.pone.0140409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das BB, Dexheimer TS, Maddali K, Pommier Y. 2010. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci U S A 107:19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sykora P, Croteau DL, Bohr VA, Wilson DM III. 2011. Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc Natl Acad Sci U S A 108:7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahbaz N, Subedi S, Weinfeld M. 2012. Role of polynucleotide kinase/phosphatase in mitochondrial DNA repair. Nucleic Acids Res 40:3484–3495. doi: 10.1093/nar/gkr1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. 2006. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 18.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. 2000. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature 405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 19.Pinz KG, Bogenhagen DF. 2000. Characterization of a catalytically slow AP lyase activity in DNA polymerase gamma and other family A DNA polymerases. J Biol Chem 275:12509–12514. doi: 10.1074/jbc.275.17.12509. [DOI] [PubMed] [Google Scholar]
- 20.Fuenmayor J, Zhang J, Ruyechan W, Williams N. 1998. Identification and characterization of two DNA polymerase activities present in Trypanosoma brucei mitochondria. J Eukaryot Microbiol 45:404–410. doi: 10.1111/j.1550-7408.1998.tb05091.x. [DOI] [PubMed] [Google Scholar]
- 21.Torri AF, Kunkel TA, Englund PT. 1994. A beta-like DNA polymerase from the mitochondrion of the trypanosomatid Crithidia fasciculata. J Biol Chem 269:8165–8171. [PubMed] [Google Scholar]
- 22.Torri AF, Englund PT. 1995. A DNA polymerase beta in the mitochondrion of the trypanosomatid Crithidia fasciculata. J Biol Chem 270:3495–3497. doi: 10.1074/jbc.270.8.3495. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen-Preiss SM, Low RL. 2000. Identification of a beta-like DNA polymerase activity in bovine heart mitochondria. Arch Biochem Biophys 374:229–240. doi: 10.1006/abbi.1999.1590. [DOI] [PubMed] [Google Scholar]
- 24.Hansen AB, Griner NB, Anderson JP, Kujoth GC, Prolla TA, Loeb LA, Glick E. 2006. Mitochondrial DNA integrity is not dependent on DNA polymerase-beta activity. DNA Repair (Amst) 5:71–79. doi: 10.1016/j.dnarep.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Sykora P, Misiak M, Wang Y, Ghosh S, Leandro GS, Liu D, Tian J, Baptiste BA, Cong WN, Brenerman BM, Fang E, Becker KG, Hamilton RJ, Chigurupati S, Zhang Y, Egan JM, Croteau DL, Wilson DM III, Mattson MP, Bohr VA. 2015. DNA polymerase beta deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res 43:943–959. doi: 10.1093/nar/gku1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. 1996. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature 379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 27.Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 28.Moro F, Okamoto K, Donzeau M, Neupert W, Brunner M. 2002. Mitochondrial protein import: molecular basis of the ATP-dependent interaction of MtHsp70 with Tim44. J Biol Chem 277:6874–6880. doi: 10.1074/jbc.M107935200. [DOI] [PubMed] [Google Scholar]
- 29.Voos W, von Ahsen O, Muller H, Guiard B, Rassow J, Pfanner N. 1996. Differential requirement for the mitochondrial Hsp70-Tim44 complex in unfolding and translocation of preproteins. EMBO J 15:2668–2677. [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo HB, Lovely GA, Phillips R, Chan DC. 2014. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Commun 5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. 2010. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 9:1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell 14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akbari M, Keijzers G, Maynard S, Scheibye-Knudsen M, Desler C, Hickson ID, Bohr VA. 2014. Overexpression of DNA ligase III in mitochondria protects cells against oxidative stress and improves mitochondrial DNA base excision repair. DNA Repair (Amst) 16:44–53. doi: 10.1016/j.dnarep.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akbari M, Visnes T, Krokan HE, Otterlei M. 2008. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. 1998. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci U S A 95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabelof DC, Ikeno Y, Nyska A, Busuttil RA, Anyangwe N, Vijg J, Matherly LH, Tucker JD, Wilson SH, Richardson A, Heydari AR. 2006. Haploinsufficiency in DNA polymerase beta increases cancer risk with age and alters mortality rate. Cancer Res 66:7460–7465. doi: 10.1158/0008-5472.CAN-06-1177. [DOI] [PubMed] [Google Scholar]
- 37.Cabelof DC, Guo Z, Raffoul JJ, Sobol RW, Wilson SH, Richardson A, Heydari AR. 2003. Base excision repair deficiency caused by polymerase beta haploinsufficiency: accelerated DNA damage and increased mutational response to carcinogens. Cancer Res 63:5799–5807. [PubMed] [Google Scholar]
- 38.Ishikawa K, Hashizume O, Koshikawa N, Fukuda S, Nakada K, Takenaga K, Hayashi J. 2008. Enhanced glycolysis induced by mtDNA mutations does not regulate metastasis. FEBS Lett 582:3525–3530. doi: 10.1016/j.febslet.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 39.Husain I, Morton BS, Beard WA, Singhal RK, Prasad R, Wilson SH, Besterman JM. 1995. Specific inhibition of DNA polymerase beta by its 14 kDa domain: role of single- and double-stranded DNA binding and 5′-phosphate recognition. Nucleic Acids Res 23:1597–1603. doi: 10.1093/nar/23.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirby TW, Gassman NR, Smith CE, Zhao ML, Horton JK, Wilson SH, London RE. 2017. DNA polymerase beta contains a functional nuclear localization signal at its N-terminus. Nucleic Acids Res 45:1958–1970. doi: 10.1093/nar/gkw1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. 2006. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res 34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. 2008. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell 32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang Q, Inanc B, Schamus S, Wang XH, Wei L, Brown AR, Svilar D, Sugrue KF, Goellner EM, Zeng X, Yates NA, Lan L, Vens C, Sobol RW. 2014. HSP90 regulates DNA repair via the interaction between XRCC1 and DNA polymerase beta. Nat Commun 5:5513-. doi: 10.1038/ncomms6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kedar PS, Kim SJ, Robertson A, Hou E, Prasad R, Horton JK, Wilson SH. 2002. Direct interaction between mammalian DNA polymerase beta and proliferating cell nuclear antigen. J Biol Chem 277:31115–31123. doi: 10.1074/jbc.M201497200. [DOI] [PubMed] [Google Scholar]
- 45.Brix J, Dietmeier K, Pfanner N. 1997. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem 272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 46.Hohfeld J, Jentsch S. 1997. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J 16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amick J, Schlanger SE, Wachnowsky C, Moseng MA, Emerson CC, Dare M, Luo WI, Ithychanda SS, Nix JC, Cowan JA, Page RC, Misra S. 2014. Crystal structure of the nucleotide-binding domain of mortalin, the mitochondrial Hsp70 chaperone. Protein Sci 23:833–842. doi: 10.1002/pro.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostermann J, Horwich AL, Neupert W, Hartl FU. 1989. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature 341:125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- 49.Dianova II, Sleeth KM, Allinson SL, Parsons JL, Breslin C, Caldecott KW, Dianov GL. 2004. XRCC1-DNA polymerase beta interaction is required for efficient base excision repair. Nucleic Acids Res 32:2550–2555. doi: 10.1093/nar/gkh567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horton JK, Stefanick DF, Prasad R, Gassman NR, Kedar PS, Wilson SH. 2014. Base excision repair defects invoke hypersensitivity to PARP inhibition. Mol Cancer Res 12:1128–1139. doi: 10.1158/1541-7786.MCR-13-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno S, Asagoshi K, Hou EW, Khodyreva SN, Lavrik OI, Tomer KB, Yasui A, Wilson SH. 2007. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell 27:829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sykora P, Yang JL, Ferrarelli LK, Tian J, Tadokoro T, Kulkarni A, Weissman L, Keijzers G, Wilson DM III, Mattson MP, Bohr VA. 2013. Modulation of DNA base excision repair during neuronal differentiation. Neurobiol Aging 34:1717–1727. doi: 10.1016/j.neurobiolaging.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei W, Englander EW. 2008. DNA polymerase beta-catalyzed-PCNA independent long patch base excision repair synthesis: a mechanism for repair of oxidatively damaged DNA ends in post-mitotic brain. J Neurochem 107:734–744. doi: 10.1111/j.1471-4159.2008.05644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. 2008. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem 283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM III, Shen B, Demple B. 2008. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol 28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. 2009. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair (Amst) 8:1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blok MJ, van den Bosch BJ, Jongen E, Hendrickx A, de Die-Smulders CE, Hoogendijk JE, Brusse E, de Visser M, Poll-The BT, Bierau J, de Coo IF, Smeets HJ. 2009. The unfolding clinical spectrum of POLG mutations. J Med Genet 46:776–785. doi: 10.1136/jmg.2009.067686. [DOI] [PubMed] [Google Scholar]
- 58.Cohen BH, Naviaux RK. 2010. The clinical diagnosis of POLG disease and other mitochondrial DNA depletion disorders. Methods 51:364–373. doi: 10.1016/j.ymeth.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 59.Hudson G, Chinnery PF. 2006. Mitochondrial DNA polymerase-gamma and human disease. Hum Mol Genet 15(Spec No 2):R244–R252. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- 60.Saxowsky TT, Choudhary G, Klingbeil MM, Englund PT. 2003. Trypanosoma brucei has two distinct mitochondrial DNA polymerase beta enzymes. J Biol Chem 278:49095–49101. doi: 10.1074/jbc.M308565200. [DOI] [PubMed] [Google Scholar]
- 61.Estep PA, Johnson KA. 2011. Effect of the Y955C mutation on mitochondrial DNA polymerase nucleotide incorporation efficiency and fidelity. Biochemistry 50:6376–6386. doi: 10.1021/bi200280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graziewicz MA, Bienstock RJ, Copeland WC. 2007. The DNA polymerase gamma Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2′-deoxyguanosine. Hum Mol Genet 16:2729–2739. doi: 10.1093/hmg/ddm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. 2007. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Res 35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prasad R, Beard WA, Strauss PR, Wilson SH. 1998. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J Biol Chem 273:15263–15270. [DOI] [PubMed] [Google Scholar]
- 65.Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, Sengupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. 2014. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.