ABSTRACT
The human immunodeficiency virus type 1 (HIV-1)/simian immunodeficiency virus (SIV) envelope spike (Env) mediates viral entry into host cells. The V3 loop of the gp120 component of the Env trimer contributes to the coreceptor binding site and is a target for neutralizing antibodies. We used cryo-electron tomography to visualize the binding of CD4 and the V3 loop monoclonal antibody (MAb) 36D5 to gp120 of the SIV Env trimer. Our results show that 36D5 binds gp120 at the base of the V3 loop and suggest that the antibody exerts its neutralization effect by blocking the coreceptor binding site. The antibody does this without altering the dynamics of the spike motion between closed and open states when CD4 is bound. The interaction between 36D5 and SIV gp120 is similar to the interaction between some broadly neutralizing anti-V3 loop antibodies and HIV-1 gp120. Two conformations of gp120 bound with CD4 are revealed, suggesting an intrinsic dynamic nature of the liganded Env trimer. CD4 binding substantially increases the binding of 36D5 to gp120 in the intact Env trimer, consistent with CD4-induced changes in the conformation of gp120 and the antibody binding site. Binding by MAb 36D5 does not substantially alter the proportions of the two CD4-bound conformations. The position of MAb 36D5 at the V3 base changes little between conformations, indicating that the V3 base serves as a pivot point during the transition between these two states.
IMPORTANCE Glycoprotein spikes on the surfaces of SIV and HIV are the sole targets available to the immune system for antibody neutralization. Spikes evade the immune system by a combination of a thick layer of polysaccharide on the surface (the glycan shield) and movement between spike domains that masks the epitope conformation. Using SIV virions whose spikes were “decorated” with the primary cellular receptor (CD4) and an antibody (36D5) at part of the coreceptor binding site, we visualized multiple conformations trapped by the rapid freezing step, which were separated using statistical analysis. Our results show that the CD4-induced conformational dynamics of the spike enhances binding of the antibody.
KEYWORDS: cryo-electron tomography, image processing, electron microscopy, immunology, AIDS, HIV
INTRODUCTION
Viral surface human immunodeficiency virus type 1 (HIV-1)/simian immunodeficiency virus (SIV) envelope spikes (Env) each consist of three gp120 glycoprotein protomers noncovalently associated with three gp41 membrane-spanning glycoproteins. Env mediates entry of HIV/SIV into the host cell through a two-step process. After binding CD4, Env undergoes a conformational change that exposes chemokine coreceptor binding interfaces. The host cell surface CXCR4 or CCR5 chemokine coreceptors then bind gp120, inducing a further conformational change leading to gp41 activation to form a coiled-coil structure. Fusion of the virus membrane with the host cell membrane follows, leading to entry of the viral genome into the host cell (1, 2).
The gp120 protomer is made up of five constant regions (C1 to C5) and five variable regions (V1 to V5) (3, 4). Of those, the V3 variable loop is required for efficient chemokine receptor binding. Neutralizing antibodies are often directed against the V3 loop. Some of these antibodies block the interaction between gp120 and CD4, and others appear to exert their action by blocking the binding of CD4-activated gp120 to chemokine receptor-expressing cells (5, 6). Thus, the V3 loop plays a role in both receptor and coreceptor binding as well as serving as an important target for antibody neutralization. The structures of gp120 and gp41 alone and in complex with different ligands have been determined by X-ray crystallography (7–15). Structures of gp120 trimers in the native and CD4- and antibody-liganded states have been obtained by cryo-electron tomography (cryoET) (7, 13, 16–18). Through the combination of X-ray crystallography and cryoET, empirical atomic models of HIV-1 trimer spikes have been built to provide insights into the conformation of the envelope spike (7, 13, 16, 18, 19). However, determination of the atomic structure of the Env trimer, especially for the native state, has been difficult.
Recently, several atomic structures of a soluble, recombinant trimer, dubbed SOSIP, were obtained by X-ray crystallography and cryo-electron microscopy (cryoEM) (20–22). SOSIP trimers are engineered to covalently stabilize the interaction between gp120 and the truncated extramembrane portion of gp41 by incorporating a disulfide bond (SOS). An I-to-P substitution in gp41 further stabilizes the interactions between the three gp120-gp41 protomers (23, 24). This SOSIP trimer binds all broadly neutralizing monoclonal antibodies (MAbs), implying that gp120 is properly folded and can serve as a close mimic of the membrane-bound native trimer (25).
A single-molecule fluorescence resonance energy transfer (smFRET) study of the HIV-1 virion revealed that unliganded gp120 is highly dynamic (26). Native gp120 was shown to transit between three distinct conformations, corresponding to a closed ground state, a CD4-bound open state, and a coreceptor-bound state. The ground state is the most frequently occupied state. The binding of broadly neutralizing antibodies was shown to stabilize the ground state, whereas binding of CD4 and that of a coreceptor to gp120 were able to stabilize the other two states, respectively.
Primate SIV models of infection have commonly been used as surrogates for HIV-1 and to aid in the development of vaccines against HIV-1. Yet there are relatively few structural studies of antibody-bound SIV Env compared to those for HIV. This is due in part to the fact that very few broadly neutralizing MAbs targeting SIV have been characterized. Some structural studies of unliganded SIV Env and SIV Env with bound soluble CD4 (sCD4), performed by cryoET at nanometer resolution, have been reported (18, 27, 28), but an atomic model of SIV Env and structural details of its interaction with neutralizing antibodies are still lacking.
In this study, we used cryoET to study SIV Env bound with sCD4 and the anti-V3 loop neutralizing antibody 36D5 (29, 30). We found the liganded Env to be conformationally and compositionally variable. CryoET is well adapted to deciphering a complex mixture of states, because the three-dimensional (3D) image provides a complete view of the structure. The details of the interaction between SIV and 36D5 will help us to understand the unliganded SIV envelope trimer structure and a neutralization mechanism in the SIV-macaque model and also will help to inform the design of new proof-of-concept virus neutralization trials with SIV models in anticipation of future HIV-1 neutralization efficacy trials.
RESULTS AND DISCUSSION
MAb 36D5.
The MAb used in this study, 36D5, was generated using soluble SIVmacCP-MAC gp140 as the immunizing agent (29). SIVmacCP-MAC was derived from a clone of SIVmac251 (BK28) (31). MAb 36D5 was strongly neutralizing against SIVmacCP-MAC Env in a GHOST assay, did not affect sCD4 binding, and blocked CCR5 binding (29). Although SIVmacCP-MAC can infect cells in a CD4-independent manner, infection with SIVmac239 requires CD4 binding (32, 33). In a separate study, MAb 36D5 was effective at neutralizing SIVmac239 (30). The virus strain used in this study, SIVmac239/251 tail (unpublished data), was developed by James Hoxie (University of Pennsylvania) and is a recombinant variant of SIVmac239 truncated at position 724 (34). As such, it maintains an essential endocytosis signal at positions 721 to 724 (35). This virus strain has been used in numerous studies of Env in intact virions (27, 36, 37).
SIV gp120 trimer with and without bound sCD4/36D5.
An example of a central slice through one of the raw tomograms (Fig. 1) shows the Env spikes on the surface of the virion. The aldrithiol-2 (AT-2) treatment used to neutralize the virus may have disrupted the virion structure within the viral envelope, as reported previously, but this treatment does not affect the structure of Env (38, 39).
FIG 1.
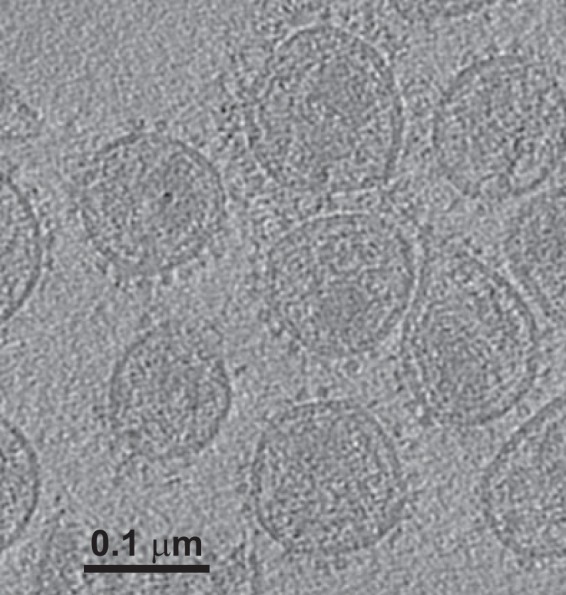
Tomogram of SIV. The image shows a single section from one tomogram of strain SIVmac239/251 tail/Supt-CCR5 CL.30, revealing envelope spikes on the virion surface. These virions were preincubated with sCD4 as well as MAb 36D5. Ligands are difficult to see unless they lie within the section plane.
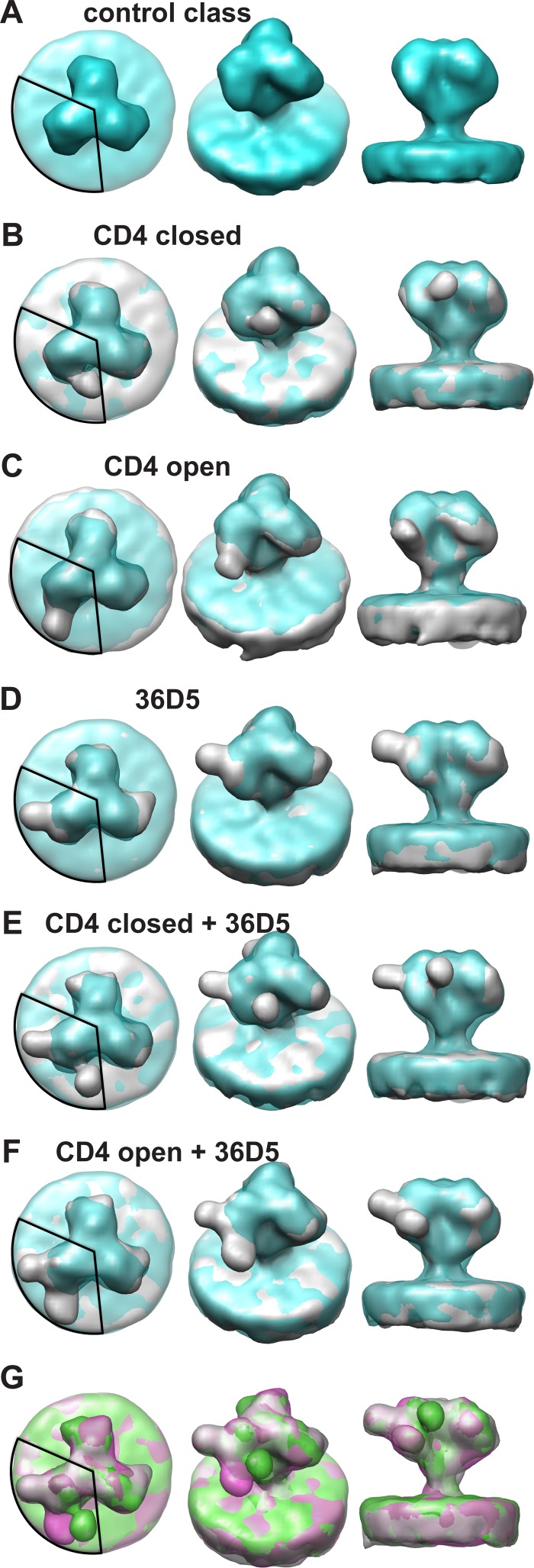
Single-arm classification (see Materials and Methods) of the aligned trimers revealed six classes (Fig. 2). The class that lacked apparent extra density attributable to bound ligands was chosen as the control class, and 3-fold symmetry was applied to generate the symmetrical “control” spike (Fig. 2A). Because it was derived from the same data set as that for the liganded spike averages, this unconventionally obtained control spike has the advantage of having qualities similar to those of the other class averages with respect to signal-to-noise (S/N) ratio, specimen quality, data collection parameters, and resolution. We consider this class a close approximation of an unliganded SIV envelope spike, and its structural features compare very favorably with those of previously published unliganded HIV/SIV envelope trimers (16, 18, 21).
FIG 2.
Characteristic class averages. The control class is shown in blue and the other classes in gray. A 120° circular segment is superimposed on each left panel to indicate the region of the density used for the classification. MDA was used to look only for density patterns within this region. However, averaging was done over entire spikes to retain the context. This means that the other two arms of gp120 usually do not resemble the classified arm. (A) Views of the control class, which lacks bound sCD4 or 36D5. Panels B to G are superimposed on the control class. (B) Views of the class liganded with sCD4 in position 1. (C) Views of the class liganded with sCD4 in position 2. (D) Views of the class with bound 36D5. (E) Views of the class liganded with 36D5 and with sCD4 in position 1. (F) Views of the class liganded with 36D5 and with sCD4 in position 2. (G) Overlap of the classes shown in panels D (gray), E (green), and F (magenta).
The other five classes, none of which were 3-fold symmetrized, have protruding densities bound at three different locations on the single gp120 arm upon which the classification was conducted (Fig. 2B to G). Note that classification was carried out on only a single gp120 arm, but averaging was done over complete Env spikes. Consequently, the pair of unclassified gp120 arms carried along in the process will usually show little or no pattern of bound ligands, unless ligand binding among gp120 arms is coupled, which we have not seen. The pairs of unclassified spike arms are averages over heterogeneous states, both liganded and unliganded and both open and closed. Because unliganded and closed conformations dominate open conformations 3:2 (Table 1), the averages for the remaining gp120 arms will tend to appear closed rather than open. The classes with protruding densities on the classification arm (Fig. 2B to F) also compare favorably with sCD4-bound Env spikes from strains SIVmneE11s, SIVmac239, and SIVmacCP-MAC, which show two CD4 binding conformations (18). Env spikes from strains SIVmneE11s and SIVmac239, which are in a closed conformation when ligand free, remain so even when liganded with sCD4. On the other hand, Env spikes from strain SIVmacCP-MAC, which are in an open conformation even without CD4 binding, remain in the open conformation after CD4 binding. The orientations of bound CD4 in the open and closed conformations are quite different. The CD4 binding conformation for SIVmneE11s and SIVmac239, which we call the “CD4 closed” state, is a state wherein the gp120 protomers meet at the spike apex. The CD4 binding conformation for SIVmacCP-MAC, which we call the “CD4 open” state, is a state in which the gp120 protomers are separated at the spike apex. Previous structural studies of HIV-1 Env spikes bound with sCD4 (17) show a structure in an open state similar to the SIVmacCP-MAC “CD4 open” state. In short, previous studies revealed that CD4 binding to HIV-1 Env induces a conformational change to the open state, whereas the conformation of SIV Env can be in the open or closed state after CD4 binding, depending on the strain. Unlike the strain-specific effects on Env conformation induced by CD4, we found that CD4 binding produced both open and closed spike conformations in a single strain of SIV (SIVmac239/251 tail/Supt-CCR5).
TABLE 1.
Relative proportions of spike arm structures
| Class | Conformation | Proportion (%) |
|---|---|---|
| 1 | Unliganded “control” | 33.40 |
| 2 | CD4 closed | 14.00 |
| 3 | CD4 open | 20.60 |
| 4 | 36D5 | 1.70 |
| 5 | 36D5 + CD4 closed | 12.00 |
| 6 | 36D5 + CD4 open | 18.20 |
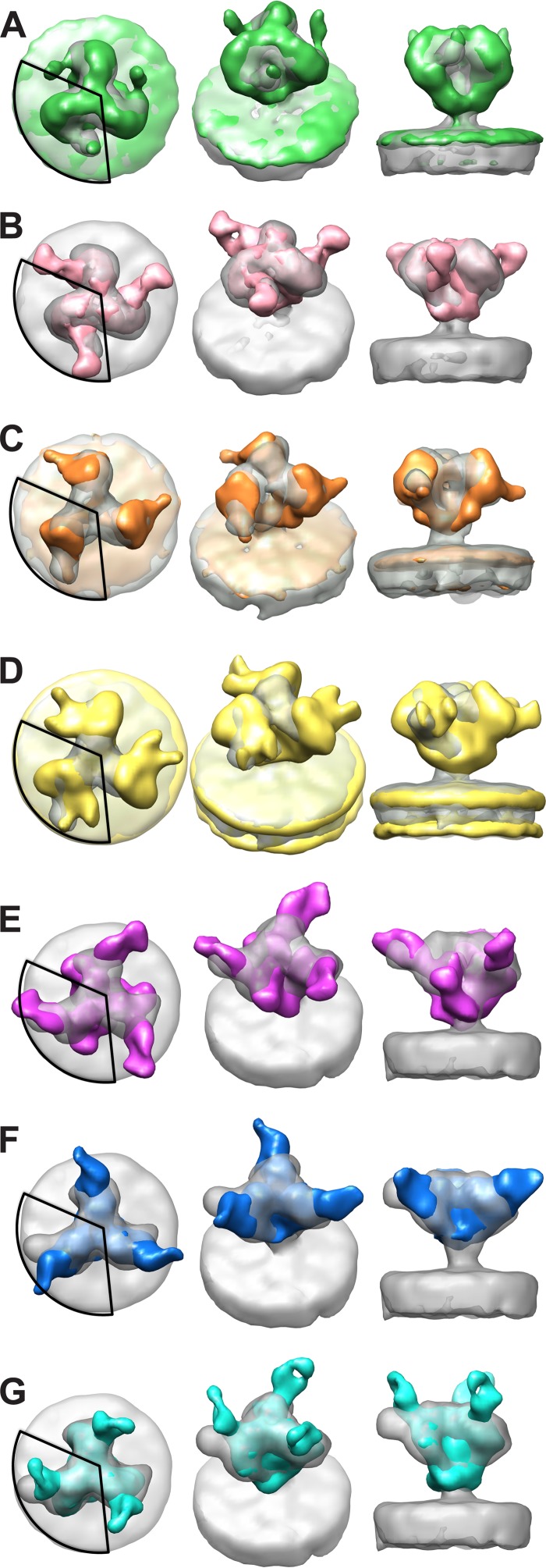
In the present study, the two classes with protruding density where CD4 binding is expected reveal two distinct conformations. One class (Fig. 2B) has a protruding density where CD4 is expected to bind in the “CD4 closed” position, similar to the structures of SIVmac239 with bound CD4 (Fig. 3A) and HIV-1 bound with the CD4 binding site MAb VRC03 (Fig. 3B) (18). The second class (Fig. 2C) shows a protruding density due to bound CD4 in the same CD4 binding site position and orientation as those in the “CD4 open” conformation of SIVmacCP-MAC (Fig. 3C) (18) and HIV-1 (Fig. 3D) (17).
FIG 3.
Comparison of current results with previously published structures. Structures obtained in this study are shown in gray. A 120° circular segment is superimposed on each left panel to indicate the region of the density used for the classification, which is where the comparison should be made. (A and B) Comparisons of the “CD4 closed” class (from Fig. 2B) with the SIVmac239-CD4 spike complex (green) (A) and the HIV-1 spike in complex with VRC03 (EMD-5458) (hot pink) (B). (C and D) Comparisons of the “CD4 open” class (from Fig. 2C) with the SIVmacCP-MAC spike in complex with CD4 (orange) (C) and the HIV-1 spike in complex with CD4 (EMD-5455) (yellow) (D). (E to G) Comparisons of the “36D5” class (from Fig. 2D) with the HIV-1 spike in complex with PGT128 Fab (EMD-1970) (purple) (E), PGT135 (EMD-2331) (blue) (F), and PGT122 (EMD-5624) (cyan) (G).
In addition to the apparent CD4 densities corresponding to both the closed and open conformations, a third protruding density was revealed at a position on the opposite side of the gp120 spike arm from where CD4 binds (Fig. 2D). We believe this density to be the Fab arm of MAb 36D5.
The interactions of HIV-1 V3 loop MAbs PGT128 (13), PGT135 (7), and PGT122 (40) with SOSIP envelope trimers show that these MAbs bind at the base of V3 but that each approaches the Env trimer from a different angle (Fig. 3E to G). This is not surprising, since these MAbs recognize epitopes that, in addition to V3 segments, incorporate elements of different neighboring variable loops and glycans. In contrast to those for HIV-1, structural studies on the interactions between V3 antibodies and the Env trimers of SIV are lacking. Comparisons of one class average from the present study (Fig. 2D) with HIV-1 bound with V3 loop antibodies PGT128 (Fig. 3E), PGT135 (Fig. 3F), and PGT122 (Fig. 3G) show that 36D5 binds gp120 roughly at the same region, suggesting that 36D5 binds SIV at the V3 loop base. We therefore named this class “the 36D5 state.”
Two other class averages have densities corresponding to both bound 36D5 and CD4. One of these shows CD4 binding gp120 in the closed state (Fig. 2E), and the other shows CD4 binding gp120 in the open state (Fig. 2F). Superposition of the three classes showing 36D5 density either alone or in combination with CD4 suggests that 36D5 binds in a single orientation, whereas CD4 has two binding orientations (Fig. 2G).
We determined the occupancy of each class from the class membership (Table 1). Unliganded gp120 protomers accounted for 33.4% of the total, and liganded gp120 protomers accounted for 66.6%. Spike arms with 36D5 binding alone occurred infrequently (<2%); spike arms with CD4 density in the open state, with or without 36D5, occurred more frequently (∼39%) than CD4-bound spike arms in the closed state, with or without 36D5 binding (∼26%).
The binding of human sCD4 to SIVmac239 gp120 appears to be somewhat weak in that only 2/3 of spike arms showed binding. The binding in the present case might suggest a cross-species effect. Biochemical studies suggest both multimeric (41) and monomeric (42, 43) binding of human sCD4 to SIV Env, and previous structural studies selected specifically for high binding (18). It has been shown that HIV poorly infects certain simian species (macaque) due to low affinity of the virus for simian CD4 (44). The comparatively poor saturation of SIVmac239 Env gp120 by human sCD4 might represent a cross-species effect.
Fitting the atomic models into the EM density.
Several atomic structures of unliganded and liganded HIV-1/SIV gp120 protomers have been published (9, 11, 12, 14, 15, 45–48), but the atomic structure of the complete Env trimer remains elusive. Compared to that of HIV-1, much less is known about the structure of SIV Env. There are published SIV gp120 atomic structures (PDB ID 2BF1 and 3FUS) (45). A comparison of SIV gp120 (PDB ID 3FUS) and HIV-1 gp120 (PDB ID 4NCO) models shows that the CD4 contact region and the V3 loop region overlap well. HIV-1 and SIV share good overall structural similarity, as revealed by 3D reconstruction of the structures of native HIV-1 and SIV (16, 19, 28). Also, the HIV-1 and SIV gp120s share relatively high sequence similarity (45). Good overall structural similarity, good sequence similarity, and good overlap of the atomic models at the CD4 and 36D5 regions between HIV-1 and SIV ensured that the trimer model of HIV-1 gp120 could be used to fit the EM densities of the SIV Env spikes in the current study, and the information about the CD4 binding site and the V3 loop site should be applicable to SIV.
The V3 loop of gp120 serves as a component of the coreceptor binding site and is an important target for neutralizing antibodies (49–52). V3-mediated neutralization sensitivity varies widely among HIV-1 strains (53, 54). An important mechanism for neutralization resistance derives from the strain-specific shielding of V3 by the V1/V2 loop (55–64). The interactions of several antibodies with the HIV-1 V3 loop have been studied at the structural level (7, 13, 17, 20, 40, 65, 66), but there are currently no structural studies of antibody interactions with the V3 loop of SIV gp120.
MAb 36D5 targets an epitope comprised of residues 321 to 340 (homologous to HIV-1 residues 310 to 328), located at the V3 base (29). Analysis of the interaction of HIV-1 gp120 with MAbs PGT128, PGT135, and PGT122 showed that these antibodies bind gp120 at the V3 base of the loop and are influenced by interactions with neighboring variable loops and glycans (7, 13, 17, 20). Comparisons of our 36D5 EM density and those for the HIV-1 V3 base-targeting MAbs show considerable overlap in the contact regions, even though 36D5 approaches from a different angle (Fig. 3E to G).
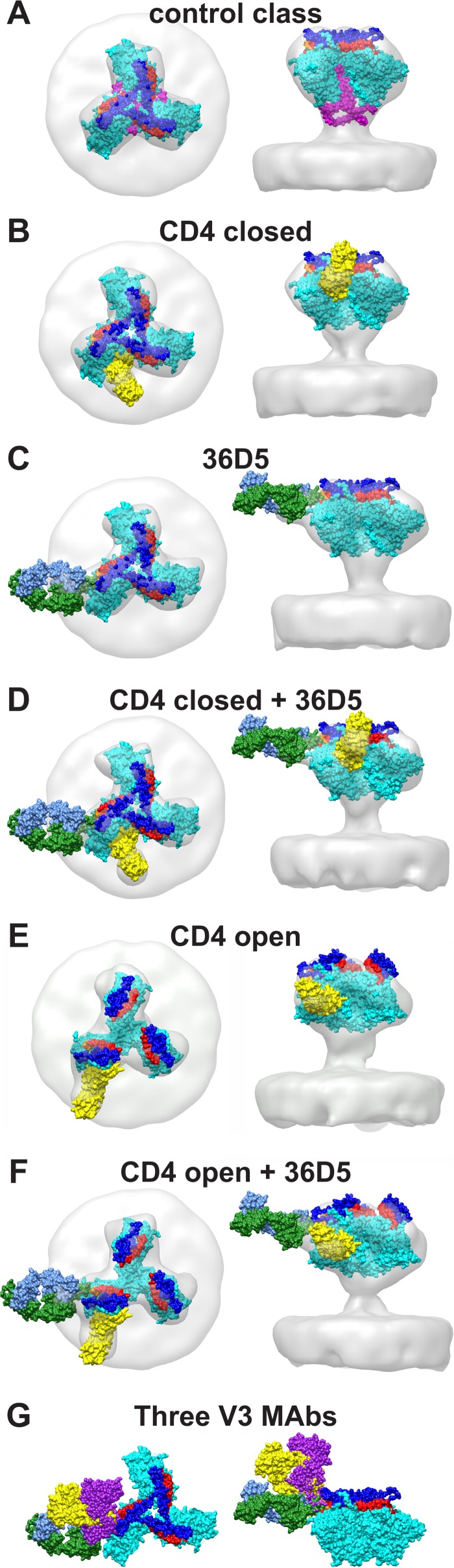
Due to the lack of an atomic structure for 36D5, the atomic structure of PGT122 was used as a mimic for 36D5 and was fitted into the EM density of 36D5 in the current work. The gp140 portion of PDB structure 4NCO was used as a starting model in the current study (20). The gp140 trimer in the 4NCO structure is in the closed state, and the structure is a mimic of the native unliganded Env trimer. Our control spike (Fig. 2A), though obtained unconventionally, is closest in structure to the native SIV envelope trimer. The gp140 trimer atomic model from the 4NCO structure was fitted into the EM density of the control spike (Fig. 2A) by rigid-body fitting using the “fit into map” function of UCSF Chimera (67) (Fig. 4A). The SOSIP module fits the electron density of our control SIV Env trimer well. The V1/V2 loop is located at the top of the spike, as predicted previously (18, 19), indicating that the SOSIP trimer 4NCO structure can serve as a mimic of the SIV spike trimer in the unliganded state.
FIG 4.
Class averages viewed with corresponding atomic models. Atomic models are displayed in space-filling mode. The corresponding reconstructions without the atomic models can be seen in Fig. 2A to F. The color scheme shows gp120 in cyan, V1/V2 in blue, V3 in red, CD4 in yellow, and 36D5 in forest green and cornflower blue. Residues close to 36D5 at the V3 base are shown in orange. (A) Atomic model for the control class. (B) Atomic model for the “CD4 closed” class. (C) Atomic model for the “36D5” class. (D) Atomic model for the “CD4 closed + 36D5” class. (E) Atomic model for the “CD4 open” class. (F) Atomic model for the “CD4 open + 36D5” class. (G) Comparison of the interaction between 36D5 and gp120 and those with PGT128 (yellow) and PGT122 (purple).
For model 1, the atomic structure of CD4 from PDB structure 2B4C was added to one of the arms of the gp120 trimer model fitted into the control class. The CD4 structure was added to gp120 as described previously (19), such that the contact region, relative location, and orientation between CD4 and gp120 were kept the same as in the 2B4C structure. The gp120 trimer model with added CD4 directly fits the EM density of the “CD4 closed” class well, without further modification (Fig. 4B). The trimer model in this fitting is the same as that for the control spike, except for the added CD4. It is worth noting that all the classes in the current study were generated from classification of ensembles of aligned Env spikes, and consequently, all the classes are in the same frame of reference as the control class. The excellent fitting of the gp120 trimer model with added CD4 into the “CD4 closed” class implies that gp120 remains in the closed state, consistent with the previous study (18).
Previous studies of HIV-1 gp120 bound with PGT128 and PGT122 (13, 20) indicated that the gp120 trimer remains in the closed state after binding V3 loop antibodies. In the current study, the model 1 gp120 trimer fit without alteration into the EM density of the “36D5” class (Fig. 4C). We used the atomic model of PGT122 as a mimic for 36D5 and fit it separately into the protruding density of 36D5 (Fig. 4C). The 36D5 Fab mimic contacts the V3 base at residues 323 to 327. The excellent fitting of the gp120 trimer model for the control class into the “36D5” class indicates that 36D5 binding does not open the gp120 trimer.
Atomic model 1 for the “CD4 closed” class (Fig. 4B) was also a good fit when 36D5 was present (“CD4 closed + 36D5” class) (Fig. 4D). The atomic model for the “36D5” class (Fig. 4C) also fits without modification into the “CD4 closed + 36D5” class (Fig. 4D), indicating that the gp120 trimer remains in the closed state with both ligands bound. When the 36D5 atomic model is compared to the published structures of V3 MAbs interacting with gp120 (Fig. 4G), the comparison shows that 36D5 approaches gp120 at a different angle but makes contact near the V3 base, similarly to PGT128 and PGT122. Comparison of the models for the control class and the 36D5 class (closed conformation) and those for the “CD4 closed” classes with and without 36D5 shows that the gp120 trimers are the same and remain in the closed conformation (Fig. 4A to D).
Rigid-body fittings of model 1 into the “CD4 open” class (Fig. 2C) and the “CD4 open + 36D5” class (Fig. 2F) were poor, necessitating building of model 2. A model 1 gp120 subunit with added CD4 was manually fit as a single rigid body into the EM densities of the “CD4 open” class (Fig. 2C), and a gp120 trimer model was generated by applying 3-fold symmetry (Fig. 4E). Model 2 also fits the “CD4 open + 36D5” class well, without further modification (Fig. 4F). When the atomic model of the 36D5 mimic is fitted into the protruding 36D5 density, its contact with gp120 occurs at the same region as in the “36D5” class, i.e., near gp120 residues 323 to 327 (Fig. 4F).
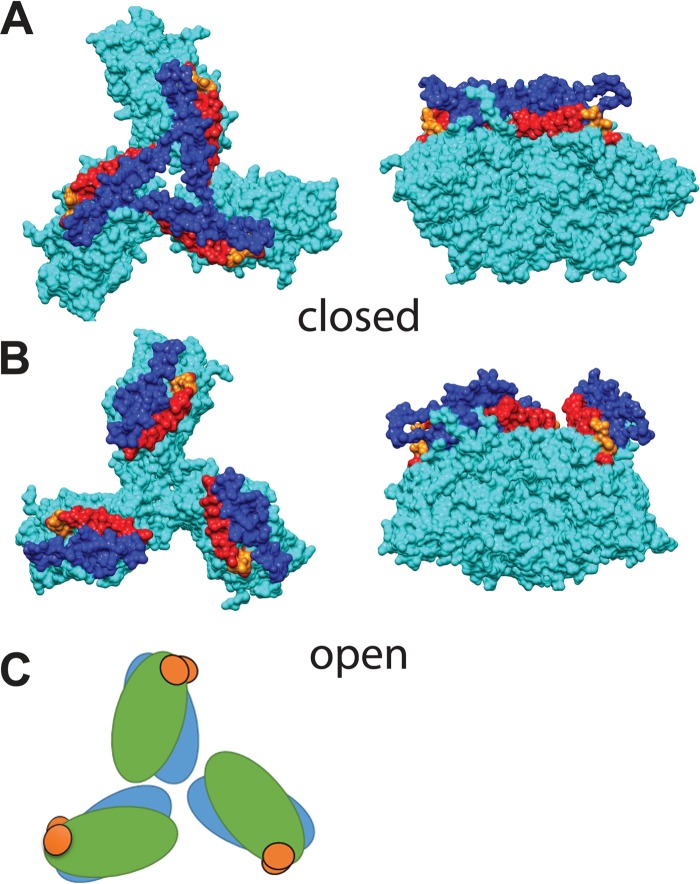
Our atomic model fitting suggests two conformations for the gp120 trimers. Comparison of gp120 models 1 and 2 (Fig. 5A and B) shows that even though the diameter of the gp120 trimer does not change dramatically, the apical part, consisting of the V1/V2 and V3 loops, moves to the periphery of the trimer and makes the apex of the trimer considerably more open. Moreover, comparison of our open and closed gp120 models shows little change in the position of the gp120 residues proximal to the 36D5 density, suggesting that the conformational change involves the gp120 monomer rotating around the V3 base, used as a pivot point (Fig. 5C), as previously suggested for HIV-1 (68). This observation is consistent with the two “CD4 open” SIV gp120 trimers, one with and one without 36D5 bound, being in the open state. The other four classes, i.e., the control class, the “36D5” class, and both “CD4 closed” classes (one with and one without 36D5 bound), are in the closed state. In the closed state, the V1/V2 loop is located directly above the V3 loop, thus preventing its interaction with V3 loop neutralizing antibodies. In the open state, V1/V2 moves to the periphery of the trimer and V3 moves more toward the surface becoming exposed, which makes the binding of coreceptor possible. This suggestion is similar to that proposed previously for the HIV-1 interaction with CD4, even though at the time there was no atomic structure of gp120 with complete V1/V2 and V3 loops available (16, 68). This observation indicates that HIV-1 and SIV share some similarities in Env activation and that discoveries from one can be applied to the other.
FIG 5.
Comparison of open and closed gp120 trimer models. The color scheme shows gp120 in cyan, V1/V2 in blue, V3 in red, and residues at the V3 base, close to the 36D5 binding site, in orange. (A) Top view (left) and side view (right) of the closed conformation. (B) Top view (left) and side view (right) of the open conformation. (C) Schematic illustration of the conformational change between the open (blue) and closed (green) states, with the V3 base (orange) as the pivot point.
Simultaneous binding of CD4 and 36D5.
Our results show two conformations, open and closed, of the Env trimer after binding with CD4. The interactions between CD4 and the Env trimers of SIV and HIV-1 have been studied, and similar open and closed conformations have been observed for both SIV and HIV-1 (12, 16–18, 69), but usually not simultaneously in the same strain under the same conditions of ligand binding. Some neutralizing antibodies have been shown to exert their effect by stabilizing the Env trimer spike in the closed state (16, 17, 21, 40).
Why does our analysis reveal open and closed conformations in a single strain of SIV for Env bound with sCD4, when previous results have shown only one? We think that this is due to very different approaches in the data analysis, with one optimized for the highest resolution and the other optimized for untangling heterogeneity. Highest-resolution imaging is driven by an approach which can be summarized as “find the largest number of homogeneous structures” to align and average. High-resolution imaging is driven to maximize ligand saturation in the specimen preparation as well as conformational homogeneity in the particles averaged. Consistent with that philosophy, outliers are discarded, sometimes as many as 50% of the total. With a large number of particles, this approach yields the desired result.
In the present case and one described previously (66), spike analysis by more conventional methods produced what can be described as “watered-down” versions of the class averages reported here, which indicated that incomplete ligand binding and/or conformational mobility was operating. Just how incomplete the ligand binding was can be seen in Fig. 2 by looking at the two spike arms on which classification was not performed but which were carried along to provide context. The potential conformers of gp120 protomers encompassed unliganded, 36D5-bound, CD4-bound, and open and closed forms, all of which were apparently independent and randomly distributed over the three protomers.
The solution we found was alignment of raw spikes through their class averages, thereby bringing all raw spikes to a common frame of reference. All raw spike protomers could then be superimposed on a single site, without a separate alignment step, by triplicating the spikes and using rotations about the symmetry axis predicted for the trimer stoichiometry. Subsequent classification based on a single gp120 protomer was a much simpler task because rather than requiring simultaneous combinations of ligands and states on separate spike protomers to define a pattern, the pattern recognition problem was reduced to the simultaneous occurrence of ligand densities on a single spike protomer. The fact that the classification produced a closed, unliganded gp120 protomer, as expected, very few 36D5-liganded protomers, consistent with the poor binding to a CD4-dependent strain of SIV, and CD4 and 36D5 binding to the expected location suggests that the classification performed as designed. The only surprise was the presence of both open and closed conformers in the presence of sCD4 in a strain where a closed conformation was expected in both the absence and presence of CD4.
The single largest difference between the present result and those that preceded it is the ligand saturation; the spike structures shown here are all partially liganded, whereas previous results selected for saturated spikes. Our analysis was necessitated by the partial ligand binding. If Env spikes display dynamics between open and closed conformations, then the analysis strategy must be designed to capture the different states.
The static images of the SIV and HIV-1 Env trimers in various conformations revealed here are consistent with a metastable nature of Env trimers in the prefusion state. Interaction of Env with CD4 is believed to cause a structural rearrangement in gp120 that exposes the coreceptor binding site, which is masked and unapproachable in the ground state (21, 40, 70, 71). The conformational change involves the movement of the V1/V2 loop from the apical location in the unliganded ground state to the trimer periphery in the “activated” state (16, 18, 19, 68). Subsequent interaction with the coreceptor induces conformational changes within gp41 and facilitates the fusion between the viral and host cell membranes.
In addition to various static images of HIV-1 Env obtained by cryoEM and crystallography studies, direct measurements of the dynamic features of HIV-1 Env trimers on the surfaces of native virions by single-molecule fluorescence resonance energy transfer (smFRET) revealed that unbound gp120 is intrinsically dynamic, transiting between three distinct prefusion conformations (26). The predominant gp120 state is the ground/closed state, with the other two states corresponding to the CD4-bound and coreceptor-bound open states. Binding of gp120 to CD4 and a coreceptor stabilizes the last two states, which are expressed only transiently in the absence of ligand binding. Various broadly neutralizing antibodies have been shown to capture and stabilize gp120 in one or another of these distinct conformations, thus preventing structural progression toward membrane fusion. SIV Env spikes might also have intrinsic dynamics similar to those of HIV-1, with binding of CD4 and antibodies capturing the different conformations. This likely explains why two conformations of CD4 binding were observed in the current study. CD4 binding to SIV is capable of occurring in two states: “CD4 closed” and “CD4 open.” The “CD4 open” class predominates (20.6% versus 14.0%) (Table 1). Whether the “CD4 closed” and “CD4 open” forms interconvert cannot be determined from our static images, but it seems likely. CD4 binding to HIV-1 Env trimers captures only the open state (corresponding here to “CD4 open”), whereas in SIV, apparently both the open and closed states are captured. There are several possible reasons for why CD4 binding is capable of capturing both open and closed conformations in SIV but can capture only the open conformation in HIV-1. One possibility is that the intrinsic dynamics of HIV-1 and SIV, e.g., the occupancy of and transit between each of the three states, may differ, thereby influencing CD4 binding behavior. Another possibility is that the interaction between CD4 and HIV-1 might be different from the interaction between CD4 and SIV. For example, a previous study showed that CD4 binding with SIV gp120 is strain dependent and, for some strains, nonobligatory (18).
We observed two classes of gp120 bound with CD4 together with 36D5 (Fig. 2E and F). The proportion of SIV Env spikes liganded with CD4 alone or with 36D5 shows a distinct bias against 36D5 binding alone (Table 1), consistent with the CD4 dependency of our virus strain. The CD4 dependency of 36D5 binding indicates that CD4 binding influences the accessibility or conformation of the 36D5 epitope; it appears that the epitope for 36D5 is poorly accessible in the native Env spike. Although the resolution of our structures is not high enough to exclude a small conformational effect, comparison of the two “CD4 closed” classes, one with and one without bound 36D5, indicates that no large change in conformation is caused by 36D5 binding (Fig. 2B, D, and E); most of the conformational change is induced or trapped by CD4 binding. Similarly, comparison of the two “CD4 open” classes, with and without bound 36D5, also indicates no large change in conformation due to 36D5 binding (Fig. 2C, D, and F). Thus, 36D5 neutralizes the virus by blocking coreceptor binding, with little additional conformational change beyond that caused by CD4 binding.
The classes which show density due to 36D5 binding have that additional density in the same general position, indicating that when the Env spike changes from the closed conformation to the open conformation, the gp120 protomer rotates, with the V3 base serving as its pivot point. Thus, MAb 36D5's neutralizing effect may involve partial or total blockage of coreceptor binding; it does not appear to block the closed-to-open conformational change. Recently, a similar observation of a broadly neutralizing MAb binding to open and closed Env conformations was made for a SOSIP trimer derived from HIV-1 (72). The MAb, 8ANC195, bound to the Env stalk, and its antigen was composed of segments from both gp120 and gp41. In that case, the MAb was proposed to block the conformational change needed for coreceptor binding and/or to prevent exposure of the fusion peptide. Thus, antibody neutralization need not require a block against the open-to-closed conformational change but can neutralize virus by blocking a subsequent step in virus-host membrane fusion.
Conclusions.
The V3 loop is a very important structural element of gp120 in that it functions as part of the coreceptor binding site and also as a neutralizing antibody target. Even though there are some studies on the interaction of HIV-1 gp120 with V3 loop antibodies, comparable studies on the interaction between SIV gp120 and V3 loop antibodies are lacking. The current cryoET study indicates that MAb 36D5 binds SIV gp120 at the base of the V3 loop and exerts its neutralization effect by blocking the coreceptor binding site. The results also reveal that the interaction between SIV gp120 and CD4 shares some similarity with its HIV-1 counterpart but also expresses some unique characteristics. The results reinforce the concept that the manner in which CD4 and gp120 interact is sequence/strain dependent and that unliganded SIV and HIV-1 envelope trimers might have different inherent dynamics.
MATERIALS AND METHODS
Viruses, antibodies, and ligands.
Strain SIVmac239/251 tail/Supt-CCR5 CL.30, lot P3974, was provided by the AIDS Vaccine Program (SAIC Frederick, NCI, Maryland, USA). Human recombinant sCD4 from Progenics Pharmaceuticals, obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, was provided by James Binley (San Diego Biomedical Research Institute). This construct contains only the first two domains. Full-length SIV MAb 36D5 was provided by James Hoxie (University of Pennsylvania). The frozen virus was thawed at room temperature for 1 h. sCD4 and MAb 36D5 were then added, mixed, and allowed to incubate at room temperature for an additional hour.
CryoET sample preparation.
CryoET sample preparation and microscopy were conducted at The University of Texas-Houston Medical School, Department of Pathology & Laboratory Medicine. For better tracking during tilt series data collection, 10-nm colloidal gold beads (BBI Solutions, Cardiff, United Kingdom) were mixed with the virus-sCD4-36D5 solution at a 1:10 ratio, and the combined solution was deposited on carbon-coated reticulated carbon grids (Quantifoil, Inc.), which were then rapidly vitrified by plunging into liquid ethane by use of a home-made cryoplunger.
CryoET.
The vitrified specimens were examined under low-dose conditions, using an FEI G2 Polara electron microscope equipped with a field emission gun and a TemCam-F415 4,096- by 4,096-pixel charge-coupled device camera (TVIPS, Gauting, Germany). The data were collected at 300 kV and a magnification of ×31,000, giving an effective pixel size of 0.57 nm after 2 × 2 binning. The FEI “batch tomography” capability of Xplor3D software (FEI) was used to collect the single-axis tilt series.
Defocus was set to a 4- to 5-μm underfocus, with a resulting cumulative dose of ∼100 e−/Å2. Each tilt series consisted of 130 images covering an angular range of −65° to 65°, with 1° fixed increments. A total of 24 tilt series were used for the structure determination.
Tilt series alignment.
Protomo (73) was used to align the tilt series, based on the intrinsic features of the individual projection images rather than the gold fiducials (74). Before the alignment of the tilt series, the colloidal gold particles were identified and replaced with a density corresponding to the mean value for the whole image. Weighted back-projection tomograms were computed after multicycle alignments.
Subvolume processing.
A total of 10,003 spikes were manually picked. The average number of spikes per virion was 27. After the raw spikes were picked, the initial Euler angles were assigned to each spike by assuming that the spike was oriented perpendicular to the viral surface and assigning the z axis of the spike subvolume to the vector between the center of the virus particle and the center of the extracted spike. Each raw spike was transformed according to the assigned initial Euler angle, and all spike volumes were averaged to generate a global average. Since the spikes were randomly oriented with respect to one another over the surfaces of the virions, this initial average had no regions that were data poor due to the missing wedge. This initial average was used as a reference to align the raw repeats for the first cycle, in which only translational alignment was applied.
The subsequent subvolume alignment and classification were done as described previously, using a procedure known as “alignment by classification” (19, 75). This procedure utilizes multivariate data analysis (MDA) and hierarchical ascendant classification to group the aligned repeats into 10 to 50 classes (73, 76). These class averages were aligned as a group in both translation and rotation by multireference alignment, using the class averages themselves as the multireferences. The class average with the best alignment cross-correlation coefficient (CCC) was selected, and the translation and rotation values for each class average obtained relative to this reference class average were then applied to the stored alignment parameters of the constituent members of each of the other classes. The resulting new set of parameters was stored for the next alignment cycle. Thus, classifications were performed on the aligned raw spike subvolumes, but alignments were performed using only the class averages, and the results for the alignment parameters of each class average were applied to the individual constituent raw spike subvolumes. Since the class averages have much higher signal-to-noise ratios than those for the raw spikes, this method greatly reduces the possibility of reference bias compared to that for the direct alignment of raw spikes. The appearance of the class averages no longer changed after the procedure was cycled 16 times. The mask used in the multireference alignment of class averages included the entire spike and a small portion (outer leaflet) of the membrane, because all class averages show the virus envelope.
Classification of aligned raw spikes, on the other hand, was conducted with a mask sufficiently large to include the entire Env head while excluding any contribution from the membrane, because membrane density in raw spike subvolumes varies depending on the latitude of the spike on the virion surface. The variation in membrane density is a missing wedge effect. The densities of sCD4, 36D5, and unliganded spike arms were revealed in class averages in the form of six different arrangements of densities on each Env arm.
Single-arm classification.
To reduce the complexity of the images and improve the signal-to-noise ratio, we devised the following method. Briefly, two copies of the aligned Env spikes were generated, with a +120° rotation about the 3-fold axis applied to one copy and a +240° rotation applied to the other. In this way, all three arms of each Env spike were brought to the site of a single Env arm. The method increased the number of subvolumes for classification 3-fold, from 10,003 to 30,009, reduced the size of the classification mask to 1/3 of the original mask, and also simplified the pattern complexity for MDA. Classification was then conducted with a 120° circular segment mask that included only one arm. The one-arm class averages represent density patterns derived from the combined averaging for all Env spike arms. The method revealed densities corresponding to sCD4 and 36D5 with better clarity than that by classification conducted over the whole trimer.
The classes having no apparent extra density due to bound ligands were combined, and the resulting average was masked by a 120° circular segment mask. The masked arm was then rotated 120° and 240°, and a symmetrical “control” spike was generated by summing up the density of the three arms, followed by a low-pass filtering step to smooth any seams generated during the process.
After the triplication procedure, we tested how many of the 30,009 spike arms from different spikes approached sufficiently close that they might contact and generate a spurious class average. By this process, we excluded 3% of the total number of spike arms. A further 12% of the spike arms contributed to class averages that were uninterpretable due to low average numbers of members or were simply poorly preserved spikes.
Atomic model building.
The crystal structure of a SOSIP gp140 HIV-1 envelope trimer in complex with Fab PGT122 (PDB ID 4NCO) served as the atomic model for fitting into the EM density and for the building of new models. Two models were used to accommodate the diversity of the EM densities revealed. Because there is no crystal structure for the 36D5 antibody, the crystal structure of another anti-V3 antibody, PGT122, was used as a mimic of 36D5. For both models 1 and 2, PGT122 was fitted into the EM density to illustrate the location of 36D5 relative to the whole gp120 trimer.
For model 1 (gp120 + CD4), the CD4 ligand from the PDB structure 2B4C was added to the 4NCO structure as described in our previous study (19). Basically, the gp120 portions of the 2B4C and 4NCO structures were aligned. Because of the close correspondence of the two aligned gp120 structures, correct positioning of the CD4 portion of the 2B4C structure with respect to the gp120 of the 4NCO structure was ensured. The combined trimer model of 4NCO gp120 with bound CD4 was then directly fitted into the EM densities of selected classes. For some classes, direct fitting of the model 1 trimer into the EM densities produced a poor fit. A second model (model 2) was therefore built to accommodate the complexity of the features observed. To build model 2, a gp120 subunit with bound CD4 from model 1 was fitted into the Env average by using the arm on which the classification had been conducted. The fit was conducted in a rigid-body manner, without modifying the atomic structure of gp120 or CD4 or the contact between them. Threefold symmetry was subsequently applied to gp120 to generate trimer model 2.
Accession number(s).
The six class averages described in this paper are available in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-6538, EMD-6539, EMD-6540, EMD-6541, EMD-6542, and EMD-6543. The atomic models used to interpret the class averages are available in the Protein Data Bank (www.pdb.org) under accession codes 3JCB and 3JCC.
ACKNOWLEDGMENTS
Virus samples were generously supplied and analyzed by the AIDS Vaccine Program (SAIC Frederick, NCI, Frederick, MD). We thank James Hoxie (University of Pennsylvania) for the gift of MAb 36D5.
This research was funded by NIH grant R01 AI055461 to K.H.R. and K.A.T.
REFERENCES
- 1.Wilen CB, Tilton JC, Doms RW. 2012. HIV: cell binding and entry. Cold Spring Harb Perspect Med 2:a006866. doi: 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumenthal R, Durell S, Viard M. 2012. HIV entry and envelope glycoprotein-mediated fusion. J Biol Chem 287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modrow S, Hahn BH, Shaw GM, Gallo RC, Wong-Staal F, Wolf H. 1987. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol 61:570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoxie JA. 2010. Toward an antibody-based HIV-1 vaccine. Annu Rev Med 61:135–152. doi: 10.1146/annurev.med.60.042507.164323. [DOI] [PubMed] [Google Scholar]
- 5.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 6.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 7.Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog 8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. 2011. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J Virol 85:12114–12123. doi: 10.1128/JVI.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu G, Liu J, Taylor KA, Roux KH. 2011. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J Virol 85:2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu P, Liu J, Bess J Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 28.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog 2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edinger AL, Ahuja M, Sung T, Baxter KC, Haggarty B, Doms RW, Hoxie JA. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J Virol 74:7922–7935. doi: 10.1128/JVI.74.17.7922-7935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yen PJ, Mefford ME, Hoxie JA, Williams KC, Desrosiers RC, Gabuzda D. 2014. Identification and characterization of a macrophage-tropic SIV envelope glycoprotein variant in blood from early infection in SIVmac251-infected macaques. Virology 458–459:53–68. doi: 10.1016/j.virol.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaBranche CC, Sauter MM, Haggarty BS, Vance PJ, Romano J, Hart TK, Bugelski PJ, Hoxie JA. 1994. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J Virol 68:5509–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori K, Ringler DJ, Kodama T, Desrosiers RC. 1992. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J Virol 66:2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edinger AL, Blanpain C, Kunstman KJ, Wolinsky SM, Parmentier M, Doms RW. 1999. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol 73:4062–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank I, Piatak M Jr, Stoessel H, Romani N, Bonnyay D, Lifson JD, Pope M. 2002. Infectious and whole inactivated simian immunodeficiency viruses interact similarly with primate dendritic cells (DCs): differential intracellular fate of virions in mature and immature DCs. J Virol 76:2936–2951. doi: 10.1128/JVI.76.6.2936-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fultz PN, Vance PJ, Endres MJ, Tao B, Dvorin JD, Davis IC, Lifson JD, Montefiori DC, Marsh M, Malim MH, Hoxie JA. 2001. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J Virol 75:278–291. doi: 10.1128/JVI.75.1.278-291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu P, Chertova E, Bess J Jr, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A 100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. 2007. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog 3:e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chertova E, Crise BJ, Morcock DR, Bess JW Jr, Henderson LE, Lifson JD. 2003. Sites, mechanism of action and lack of reversibility of primate lentivirus inactivation by preferential covalent modification of virion internal proteins. Curr Mol Med 3:265–272. doi: 10.2174/1566524033479889. [DOI] [PubMed] [Google Scholar]
- 39.Rossio JL, Esser MT, Suryanarayana K, Schneider DK, Bess JW Jr, Vasquez GM, Wiltrout TA, Chertova E, Grimes MK, Sattentau Q, Arthur LO, Henderson LE, Lifson JD. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J Virol 72:7992–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Earl PL, Doms RW, Moss B. 1992. Multimeric CD4 binding exhibited by human and simian immunodeficiency virus envelope protein dimers. J Virol 66:5610–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooks ET, Jiang P, Franti M, Wong S, Zwick MB, Hoxie JA, Robinson JE, Moore PL, Binley JM. 2008. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology 377:364–378. doi: 10.1016/j.virol.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M, Chen B, Hussey RE, Chishti Y, Montefiori D, Hoxie JA, Byron O, Campbell G, Harrison SC, Reinherz EL. 2001. The stoichiometry of trimeric SIV glycoprotein interaction with CD4 differs from that of anti-envelope antibody Fab fragments. J Biol Chem 276:42667–42676. doi: 10.1074/jbc.M104166200. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, Brocca-Cofano E, Deleage C, Hao X, Chuang GY, Gorman J, Gardner M, Lewis MG, Hatziioannou T, Santra S, Apetrei C, Pandrea I, Alam SM, Liao HX, Shen X, Tomaras GD, Farzan M, Chertova E, Keele BF, Estes JD, Lifson JD, Doms RW, Montefiori DC, Haynes BF, Sodroski JG, Kwong PD, Hahn BH, Shaw GM. 2016. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113:E3413–E3422. doi: 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Lu M, Poon BK, Wang Q, Ma J. 2009. Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement. Acta Crystallogr D Biol Crystallogr 65:339–347. doi: 10.1107/S0907444909003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaRosa GJ, Davide JP, Weinhold K, Waterbury JA, Profy AT, Lewis JA, Langlois AJ, Dreesman GR, Boswell RN, Shadduck P, Holley LH, Karplus M, Bolognesi DP, Matthews TJ, Emini EA, Putney SD. 1990. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science 249:932–935. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- 50.Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. 2004. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J Virol 78:2394–2404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, Self S, Williams C, Gorny MK, Zolla-Pazner S. 2010. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS One 5:e10254. doi: 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schweighardt B, Liu Y, Huang W, Chappey C, Lie YS, Petropoulos CJ, Wrin T. 2007. Development of an HIV-1 reference panel of subtype B envelope clones isolated from the plasma of recently infected individuals. J Acquir Immune Defic Syndr 46:1–11. [DOI] [PubMed] [Google Scholar]
- 53.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mascola JR, D'Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol 79:10103–10107. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol 71:9808–9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis KL, Bibollet-Ruche F, Li H, Decker JM, Kutsch O, Morris L, Salomon A, Pinter A, Hoxie JA, Hahn BH, Kwong PD, Shaw GM. 2009. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J Virol 83:1240–1259. doi: 10.1128/JVI.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, Montefiori DC, Graham BS, Keefer MC, Pinter A, Morris L, Hahn BH, Shaw GM. 2009. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387:414–426. doi: 10.1016/j.virol.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson WE, Morgan J, Reitter J, Puffer BA, Czajak S, Doms RW, Desrosiers RC. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J Virol 76:2075–2086. doi: 10.1128/jvi.76.5.2075-2086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol 75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol 79:780–790. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pantophlet R, Burton DR. 2006. gp120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol 24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 62.Saunders CJ, McCaffrey RA, Zharkikh I, Kraft Z, Malenbaum SE, Burke B, Cheng-Mayer C, Stamatatos L. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J Virol 79:9069–9080. doi: 10.1128/JVI.79.14.9069-9080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stamatatos L, Cheng-Mayer C. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J Virol 72:7840–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lusso P, Earl PL, Sironi F, Santoro F, Ripamonti C, Scarlatti G, Longhi R, Berger EA, Burastero SE. 2005. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J Virol 79:6957–6968. doi: 10.1128/JVI.79.11.6957-6968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Earl LA, Lifson JD, Subramaniam S. 2013. Catching HIV ‘in the act’ with 3D electron microscopy. Trends Microbiol 21:397–404. doi: 10.1016/j.tim.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dutta M, Liu J, Roux KH, Taylor KA. 2014. Visualization of retroviral envelope spikes in complex with the V3 loop antibody 447-52D on intact viruses by cryo-electron tomography. J Virol 88:12265–12275. doi: 10.1128/JVI.01596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Moscoso CG, Sun Y, Poon S, Xing L, Kan E, Martin L, Green D, Lin F, Vahlne AG, Barnett S, Srivastava I, Cheng RH. 2011. Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Proc Natl Acad Sci U S A 108:6091–6096. doi: 10.1073/pnas.1016113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci U S A 108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harrison SC. 2008. Viral membrane fusion. Nat Struct Mol Biol 15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 72.Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. 2015. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell 162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winkler H. 2007. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol 157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 74.Winkler H, Taylor KA. 2006. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy 106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 75.Winkler H, Zhu P, Liu J, Ye F, Roux KH, Taylor KA. 2009. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol 165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winkler H, Taylor KA. 1999. Multivariate statistical analysis of three-dimensional cross-bridge motifs in insect flight muscle. Ultramicroscopy 77:141–152. doi: 10.1016/S0304-3991(99)00035-2. [DOI] [Google Scholar]