ABSTRACT
Paramyxoviruses rely on the matrix (M) protein to orchestrate viral assembly and budding at the plasma membrane. Although the mechanistic details remain largely unknown, structural data suggested that M dimers and/or higher-order oligomers may facilitate membrane budding. To gain functional insights, we employed a structure-guided mutagenesis approach to investigate the role of canine distemper virus (CDV) M protein self-assembly in membrane-budding activity. Three six-alanine-block (6A-block) mutants with mutations located at strategic oligomeric positions were initially designed. While the first one includes residues potentially residing at the protomer-protomer interface, the other two display amino acids located within two distal surface-exposed α-helices proposed to be involved in dimer-dimer contacts. We further focused on the core of the dimeric interface by mutating asparagine 138 (N138) to several nonconservative amino acids. Cellular localization combined with dimerization and coimmunopurification assays, performed under various denaturing conditions, revealed that all 6A-block mutants were impaired in self-assembly and cell periphery accumulation. These phenotypes correlated with deficiencies in relocating CDV nucleocapsid proteins to the cell periphery and in virus-like particle (VLP) production. Conversely, all M-N138 mutants remained capable of self-assembly, though to various extents, which correlated with proper accumulation and redistribution of nucleocapsid proteins at the plasma membrane. However, membrane deformation and VLP assays indicated that the M-N138 variants exhibiting the most reduced dimerization propensity were also defective in triggering membrane remodeling and budding, despite proper plasma membrane accumulation. Overall, our data provide mechanistic evidence that the efficiency of CDV M dimerization/oligomerization governs both cell periphery localization and membrane-budding activity.
IMPORTANCE Despite the availability of effective vaccines, both measles virus (MeV) and canine distemper virus (CDV) still lead to significant human and animal mortality worldwide. It is assumed that postexposure prophylaxis with specific antiviral compounds may synergize with vaccination campaigns to better control ongoing epidemics. Targeting the matrix (M) protein of MeV/CDV is attractive, because M coordinates viral assembly and egress through interaction with multiple cellular and viral components. However, the lack of basic molecular knowledge of how M orchestrates these functions precludes the rational design of antivirals. Here we combined structure-guided mutagenesis with cellular, biochemical, and functional assays to investigate a potential correlation between CDV M self-assembly and virus-like particle (VLP) formation. Altogether, our findings provide evidence that stable M dimers at the cell periphery are required to productively trigger VLPs. Such stabilized M dimeric units may facilitate further assembly into robust higher-order oligomers necessary to promote plasma membrane-budding activity.
KEYWORDS: morbillivirus cell exit, matrix protein, dimerization efficiency, cell periphery accumulation, membrane deformation, VLP production
INTRODUCTION
The Morbillivirus genus, a group of enveloped, nonsegmented, single-stranded, negative-sense RNA viruses belonging to the Paramyxoviridae family, consists of several viruses, including measles virus (MeV), canine distemper virus (CDV), and rinderpest virus (1). The genomes of morbilliviruses consist of six tandemly linked transcription units encoding six structural and two accessory proteins: nucleocapsid (N), phosphoprotein (P), matrix (M), fusion (F), attachment (H), polymerase (L), and the V and C proteins (2). While N encapsidates the viral RNA genome, P and L (forming the viral RNA-dependent RNA polymerase complex) associate with the N-RNA complex, thereby constituting the minimal virus infectious unit, the ribonucleoprotein complex (RNP). Morbillivirus particles additionally carry two interacting surface glycoproteins (H and F) anchored in the lipidic envelope, which are strictly required for cell entry and spread. H recognizes specific entry receptors present on the host cell surface and initiates the membrane fusion process for cell entry and spread by activating the associated F protein at the right place and time (3–14). In turn, the activated F protein dramatically refolds in a process that is linked to the merging of the viral envelope with the host cell plasma membrane. Consequently, a fusion pore is formed, which enables injection of the RNP into the host cell cytoplasm (12, 15). C and V are involved in RNA synthesis and in counteracting the antiviral cell response (16).
M plays a major role in the assembly and budding of viral particles. M acts as an intermediate between the RNP and the surface glycoproteins by interaction of M with the C-terminal end of N and the cytoplasmic tails of H and F, thereby serving as the major viral component orchestrating the viral particle assembly process (17–24). Moreover, RNP accumulation at the cell periphery depends on stable expression of M and the ensuing proper N-M interaction, which likely results in relocating N (and thus the RNP) to viral budding platforms (25). Moreover, by interacting with the viral glycoproteins, M can contribute not only to the regulation of particle assembly but also to the process of membrane fusion. Indeed, M also controls viral lateral spread by downregulating the membrane fusion process, which is mediated by F, H, and a receptor expressed on neighboring contacting cells (20, 22, 26). The budding and assembly processes are additionally regulated by M interaction with cellular actin filaments, although through discrete molecular mechanisms depending on the viral strain studied (21, 27–29). Indeed, Wakimoto and colleagues recently demonstrated that actin molecules can specifically hamper M interaction with the cytoplasmic tail of H and that this regulation relies on a single residue of the MeV M protein (M-F50) (21).
Furthermore, the M proteins of MeV and several other members of the Paramyxoviridae and Pneumoviridae families are endowed with the ability to drive the formation of virus-like particles (VLPs) in the presence or absence of other viral components (25, 30–37). The inherent budding capacity relies on different properties: M can associate with cellular membranes (25, 38–41) and, more specifically for some paramyxoviruses, with detergent-resistant membrane domains that may act as virus budding platforms (40, 42–45). It has also been reported that M carries specific domains, the so-called L domains, that contribute to M's budding function by recruiting elements of the endosomal sorting complex required for transport (ESCRT) in order to efficiently complete virus particle release by promoting the membrane fission process (46). In addition, upon de novo synthesis in the cytoplasm, M accumulates at the cell periphery and is believed to trigger the formation of membrane protrusions, which may precede the final budding and cell exit processes (21, 25, 26, 28, 38, 40, 47–49). Remarkably, it was recently reported that the matrix protein of Nipah virus could additionally bind to the E3-ubiquitin ligase TRIM6, thereby contributing to the inhibition of the type I interferon antiviral response (50).
There is no clear evidence that the morbillivirus M protein can transiently traffic through the nucleus, as opposed to those of many other paramyxoviruses and pneumoviruses, including Nipah and Hendra viruses, respiratory syncytial virus (RSV), Sendai virus, and mumps virus (35, 51–57). In addition, the morbillivirus particle scission process seems to occur in an ESCRT-independent manner, in contrast to those of some other members of the Mononegavirales (e.g., Ebola virus and vesicular stomatitis virus) and other paramyxoviruses (e.g., parainfluenza virus type 5, Newcastle disease virus [NDV], and mumps virus) (37, 43, 58–60). Finally, inherent functions of M may rely on its capacity to self-assemble (19, 25, 42).
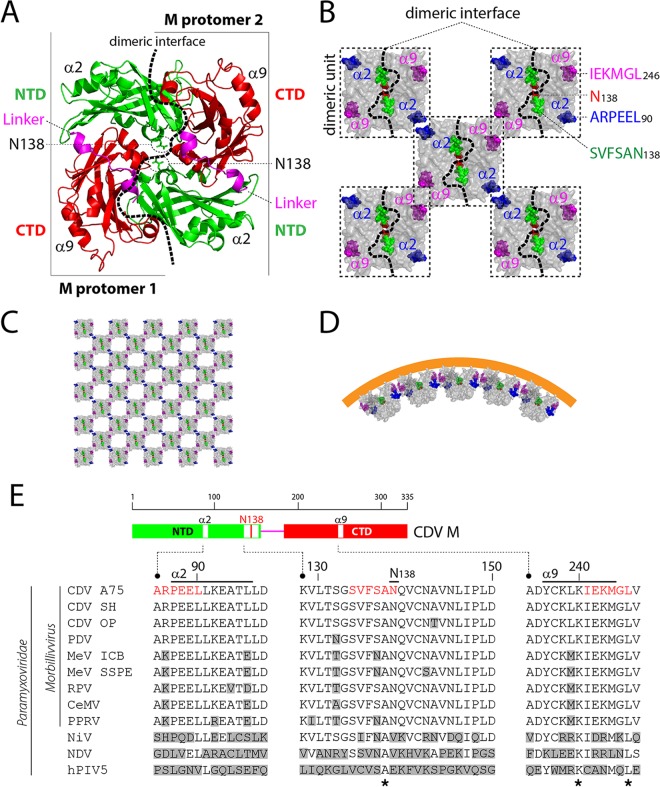
No structural data for any morbillivirus M protein are currently available. However, high-resolution atomic structures were determined for the M proteins of the related viruses NDV, human metapneumovirus, and RSV. Strikingly, these crystallographic studies illustrated high structural conservation: (i) the M protomer is composed of amino-terminal and carboxyl-terminal domains (NTD and CTD, respectively) that are linked by a putative flexible linker, and (ii) two protomers assemble into dimers in an antiparallel “head-to-tail” fashion through a large dimeric binding interface (Fig. 1A) (61–63). Additional electron microscopy (EM) analyses revealed that M can form curved higher-order oligomeric arrays upon free lipid treatment or upon membrane association within the viral particle (61, 62, 64). Both the NTD and the CTD were proposed to contribute to the formation of higher-order oligomers, putatively through two surface-exposed α-helices (namely, α2 in NTD and α9 in CTD) located on opposite sides of the dimeric binding interface (Fig. 1B and C). Interestingly, it has been hypothesized that the angle generated between two M dimers (at dimer-dimer interfaces) may contribute to the induction of the membrane curvature required for viral budding (Fig. 1D). Interestingly, EM data on MeV particles highlighted an alternative supramolecular organization in which M proteins formed helices coating the helical RNP rather than coating the inner leaflet of the membrane (65).
FIG 1.
Structural model of CDV M and derived putative oligomeric structures. (A) NDV M-based (PDB code 4G1G) structural homology model of a CDV M dimer. The three major domains of both protomers are colored green (NTD), pink (linker), and red (CTD). CDV M residue N138 is represented with green sticks, and surface-exposed helices suggested to be involved in dimer-dimer interactions are labeled. The protomer-protomer binding interface is depicted with a dashed black line. (B) Putative M dimer-dimer association. Surface-represented M dimeric units (dashed-line boxes) were manually assembled into putative higher-order oligomeric structures (61, 62). The three regions targeted for 6-alanine-block mutagenesis are represented by spheres and colored blue (ARPEEL), pink (IEKMGL), and green (SVFSAN). Asparagine 138 is shown in red, and the dimeric binding interface is represented by a dashed black line. (C) Manual assembly of M dimers in higher-order oligomers. (D) One-directional representation of M-mediated membrane curvature. M dimers were manually assembled with an angle at the dimer-dimer binding interface, forming curved oligomers (61, 63). (E) Sequence alignment of M proteins of various paramyxoviruses. At the top is a schematic representation of the M protein's main domains and encompassed regions targeted for mutagenesis (highlighted either in white or with a red bar). Accession numbers (GenBank or NCBI) for the virus sequences are as follows: CDV A75/17 (CDV A75), AAD49701; CDV Snyder Hill (CDV SH) (68), AFC40215; CDV Onderstepoort (CDV OP), NP_047204; phocine distemper virus (PDV), AGL33552; MeV IC-B (MeV ICB), BAM36454; MeV genotype B3, an SSPE strain (MeV SSPE), ALL29057; rinderpest virus (RPV), CAA53779; cetacean morbillivirus (CeMV), AEO51054; peste-des-petits-ruminants virus (PPRV), ACQ44669; Nipah virus (NiV), NP_112025; Newcastle disease virus (NDV), AEZ36128; and human parainfluenza virus 5 (hPIV5), ABC75775. The stars represent amino acids conserved throughout members of the subfamily. Amino acids differing from the reference CDV M sequence (strain A75/17) are shown in gray boxes.
In the present study, we aimed to gain mechanistic insights into the discrete inherent functions of CDV M that are required to achieve proper membrane-budding activity. Based on a homology model, we conducted a structure-guided mutagenesis approach which targeted residues located at critical putative oligomeric positions. The panel of derived M mutants was then subjected to a series of newly developed biochemical, cellular, and functional analyses. Our findings indicate that stable CDV M dimers are essential for triggering plasma membrane deformation and ensuing virus-like particle formation.
RESULTS
Cell periphery accumulation is preserved by single M-N138 mutants.
While several studies have demonstrated the key role of the morbillivirus M protein in regulating particle assembly, budding, and cell-cell fusion, our current knowledge on how M mechanistically proceeds to sustain these functions remains very limited. Here we aimed to shed the first light on the inherent mechanisms of M underlying viral budding and cell exit. A summary of the properties discovered in this study is provided in Table 1.
TABLE 1.
Summary of properties of M-wt and its derivative mutants
| Phenotype | Score or presence of phenotypea |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| M-wtb | ARPEEL90c | IEKMGL246c | SVFSAN138c | M-N138Fd | M-N138Ad | M-N138 Md | M-N138Rd | M-N138Ed | |
| Expression level | 4+ | 2+ | 3+ | 2+ | 2+ | 3+ | 2+ | 3+ | 3+ |
| Cell periphery accumulation | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes |
| Dimeric propensity | 4+ | NA | NA | NA | 3+ | 3+ | 3+ | 4+ | 3+ |
| M-M avidity of interaction | |||||||||
| coIP1 | 4+ | 1+ | 1+ | 1+ | 3+ | 3+ | 3+ | 2+ | 1+ |
| coIP2 | 4+ | 1+ | 1+ | 1+ | 3+ | 3+ | 3+ | 3+ | 2+ |
| coIP3 | 4+ | 1+ | 1+ | 1+ | 3+ | 4+ | 2+ | 4+ | 3+ |
| Cellular N relocalization | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes |
| Cell membrane deformation | |||||||||
| Branched filaments | Yes | No | No | No | No | Yes | No | Yes | No |
| Short filaments | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes |
| VLP formation efficiency | |||||||||
| 3 μg | 4+ | 1+ | 1+ | 1+ | 2+ | 3+ | 1+ | 5+ | 1+ |
| 1.75 μg | 4+ | 1+ | 1+ | 1+ | 1+ | 2+ | 1+ | 4+ | 1+ |
| 0.5 μg | 4+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 3+ | 1+ |
M activity scores: 5+, 133 to 165%; 4+, 100 to 132%; 3+, 67 to 99%; 2+, 34 to 66%; 1+, 1 to 33%; NA, not applicable. Yes, phenotype observed; no, phenotype not observed.
Wild-type M protein.
Six-alanine-block mutant.
Mutant with single mutated M residue.
To gain functional insights, we initially employed a comprehensive structure-guided mutagenesis approach to investigate putative correlations between the CDV M protein's self-assembly properties and membrane-budding activity. We thus generated a structural homology model based on the atomic coordinates of the related NDV M protein dimer. In the model, each CDV M protomer comprises an N- and a C-terminal domain (NTD and CTD, respectively) connected via a linker region. Two protomers interact in an antiparallel manner over a large dimeric interface (Fig. 1A). To potentially disrupt M dimers, we focused on the core of the dimeric interface by mutating the asparagine (N) at position 138 into several charged (glutamate [E] and arginine [R]), hydrophobic (methionine [M]), aromatic and hydrophobic (phenylalanine [F]), or small hydrophobic (alanine [A]) residues. Additionally, three mutants with strategically located six-alanine-block (6A-block) mutations were designed. The first one includes residues potentially residing at the protomer-protomer interface (SVFSAN138). The other two display amino acids potentially residing within or close to α2 (ARPEEL90) and α9 (IEKMGL246), the two α-helices proposed to be involved in dimer-dimer contacts (Fig. 1B to D). Interestingly, although residues analogous to those of the CDV M-N138 and 6A-block mutants are quite variable among other paramyxoviruses, they are very well conserved for all members of the Morbillivirus genus (Fig. 1E).
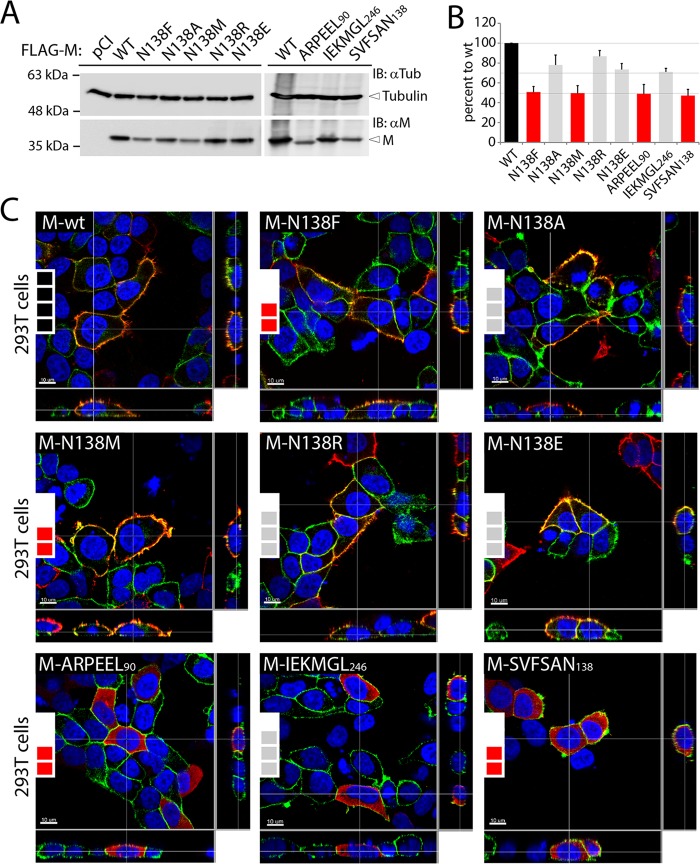
To investigate the impacts of the substitutions in regulating different M functions, each M mutant together with the parental wild type (M-wt; derived from the highly neurovirulent CDV strain A75/17) was initially expressed in 293T cells for 48 h. Western blot analyses of total protein extracts revealed that all mutants exhibited reduced expression profiles (Fig. 2A). While the N138A, N138R, N138E, and IEKMGL246 mutants were expressed at around 70 to 85% (Fig. 2B, gray bars) compared to the wt expression level (Fig. 2B, black bar), the N138F, N138M, ARPEEL90, and SVFSAN138 mutants exhibited a slightly more pronounced defect in intracellular expression levels (around 50% compared to the wt level) (Fig. 2B, red bars).
FIG 2.
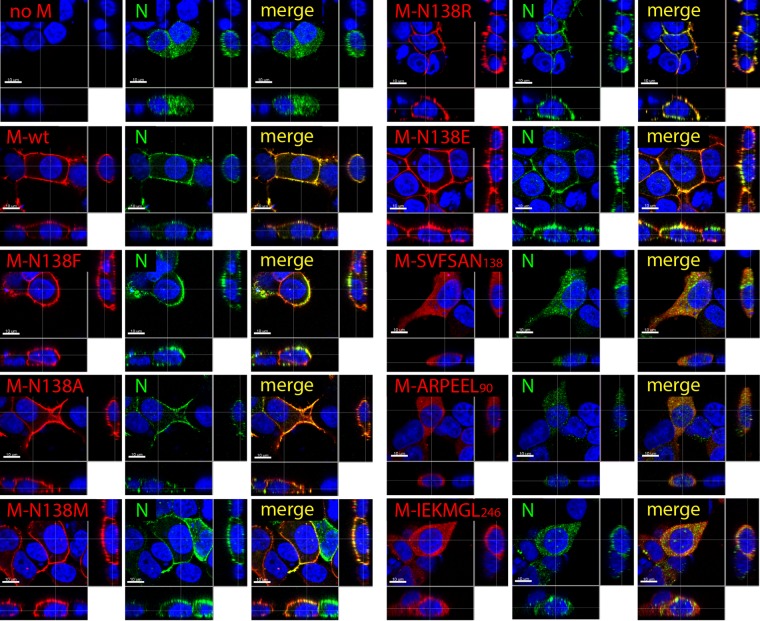
Cellular expression, accumulation, and localization of diverse CDV M mutants. (A) Assessment of expression levels of the various CDV M proteins by immunoblotting (IB) upon expression in HEK-293T cells. M antigenic materials were detected using a polyclonal anti-M antibody (αM). As a loading control, tubulin was revealed by employing a monoclonal anti-tubulin antibody (αTub). The different protein populations are labeled on the right. (B) Semiquantitative evaluation of M mutant expression levels. Expression values for each M protein were normalized to the tubulin score and the normalized CDV M-wt ratio. Means and standard errors of the means (SEM) for data from three independent experiments are shown. One-way ANOVA with Holm-Bonferroni correction and t tests rendered no significant difference. (C) FLAG-tagged M proteins were expressed in HEK-293T cells, and cellular localization was investigated by immunofluorescence analyses. Images were captured with a scanning confocal laser fluorescence microscope (Olympus). FLAG-tagged M proteins were stained red, and the plasma membranes and nuclei were counterstained in green and blue, respectively. The insets at the left of the panels represent the expression levels of the M protein mutants as acquired for panel B (4 black boxes, 100%; 3 gray boxes, ∼70 to 85%; and 2 red boxes, ∼50%).
Because a direct correlation between intracellular M expression and cell periphery accumulation has been proposed (25), we next determined the cellular localization of the various M mutants by immunofluorescence analyses. To highlight the cell periphery, plasma membranes were counterstained with an antibody targeting the plasma membrane-residing alpha 1 subunit of a sodium-potassium pump. Strikingly, independently of their steady-state levels, all mutated M-N138 proteins were efficiently transported to the cell periphery (Fig. 2C), whereas all three 6A-block mutants displayed a clear cytosolic localization phenotype (Fig. 2C). Thus, while impaired intracellular M expression may eventually lead to a plasma membrane targeting deficiency (25), our data provided evidence that the latter correlation is not a prerequisite. Taken together, while our results indicated that replacing the α2 and α9 helices and multiple residues located at the protomer-protomer binding interface strongly interfered with plasma membrane targeting, M mutants carrying single substitutions at position 138 efficiently preserved cell periphery targeting.
Cell periphery localization of M correlates with dimerization propensity.
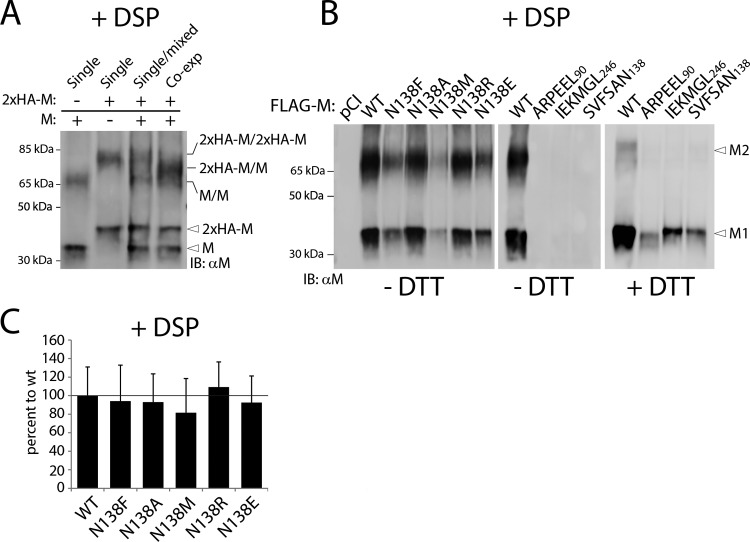
Structural and biochemical evidence suggested that dimerization of the matrix protein may represent an essential factor leading to cell periphery accumulation, cell membrane deformation, and virus-like particle formation (63). We therefore developed biochemical assays to investigate M dimerization. Unfortunately, several initial attempts failed to provide satisfactory data. For instance, native-PAGE gels returned a smeared population of M complexes, whereas SDS-PAGE gels under denaturing and nonreducing conditions proved not to be sensitive enough to properly reveal M dimer bands (data not shown). To overcome these technical issues, we treated M-expressing cells with the membrane-permeating and cleavable cross-linking agent dithiobis(succinimidyl propionate) (DSP) prior to protein extraction. Protein extracts were then subjected to denaturing and nonreducing conditions, heated, and loaded onto SDS-PAGE gels for fractionation.
Under these experimental conditions, two main populations of M-wt antigenic materials were detected. The higher population ran at a molecular mass of approximately 70 kDa, which was roughly twice the molecular mass of the second detected population (35 kDa) (Fig. 3A, left lane). To ensure that the slow-migrating M population represented M dimers, not M monomers cross-linked to a host cellular factor, two M proteins of different sizes (one without and one with an N-terminal 2×HA-linker tag) were expressed either separately or together in the same cells. As expected, mixing the total cell extracts of cells expressing the two M populations separately post-cell lysis led to band patterns representing the superimposition of the profiles obtained for “nonmixed” separate expressions (Fig. 3A, first three lanes). In sharp contrast, when both M variants were coexpressed in the same cells, a single major band of intermediate size was detected only at the level of the slow-migrating bands. Indeed, no difference was observed regarding the fast-migrating M population (Fig. 3A, right lane). Since such a population could be generated only by the assembly of M heterodimers (2×HA-linker-M/M), these experiments hence validated our cellular, DSP-based M dimerization assay.
FIG 3.
Cross-linking analyses reveal proper preservation of M dimeric populations. (A) FLAG-tagged and untagged M proteins (wt) were expressed alone (or in combination, as indicated in the text) in HEK-293T cells. Dimeric assembly was investigated by immunoblotting performed under nonreducing conditions. Prior to cell lysis, cells were treated with the membrane-permeating chemical cross-linker DSP (+DSP). M antigenic materials were detected using a polyclonal anti-M antibody. The different M populations are labeled on the right. (B) The dimeric propensity of the various M mutants was determined by immunoblotting under nonreducing conditions (−DTT), similar to that for panel A. For the last four lanes, however, wt and 6-alanine-block mutants were further treated with a reducing agent (+DTT). M1, M monomers; M2, M dimers. (C) Mean M dimeric propensity indices (calculated as indicated in Materials and Methods) and SEM for data from three independent experiments. One-way ANOVA rendered no significant difference.
The experiments were then repeated with the different M mutants. While all M-N138 variants efficiently displayed both monomeric and dimeric populations, surprisingly, neither M dimers nor M monomers were detected in the case of the three 6A-block mutants (Fig. 3B). Since misfolded proteins tend to form insoluble aggregates, the lysis products of cells expressing the 6A-block mutants were further treated with dithiothreitol (DTT) and subjected to Western blot analyses. This experimental setting returned a clear M monomeric population for all three 6A-block mutants, which in turn strongly suggested that these variants were trapped in insoluble fractions by DSP treatment (Fig. 3B, right panels). Finally, the M dimerization efficiency index was determined by calculating the dimer-to-monomer ratio, which confirmed the proper M-M interaction for all N138 mutants (Fig. 3C).
Overall, these findings provided evidence that the 6A-block M mutants had lost the ability to form wild-type-like dimers, a phenotype that very likely correlated with gross protein misfolding. Conversely, mutating residue 138, located at the core of the protomer-protomer binding interface, did not prevent dimerization.
Investigation of M-M interaction by coIP assays reveals modulation of self-assembly efficiency.
Since the DSP assay might have overestimated the efficiencies of M dimerization, we next detected self-assembly by performing coimmunopurification (coIP) assays.
In order to distinguish, in the same gel, IP and coIP M antigenic materials, 293T cells were cotransfected with plasmid DNAs encoding two M proteins of different sizes: FLAG-M and 2×HA-linker-M. Thus, upon total protein extraction under conditions with various levels of stringency (coIP1 to coIP3) (see Materials and Methods), 2×HA-linker-M proteins were purified from cell extracts by use of magnetic Dynabeads-based technology. Indeed, we noticed that CDV M proteins exhibited the strong tendency to sediment even in the absence of antibody (in the case of gravity force-dependent immunoprecipitation procedures). Purified M proteins were then boiled and fractionated in SDS-PAGE gels under reducing conditions. After transfer of the antigenic materials to nitrocellulose membranes, proteins were detected using a polyclonal anti-M antibody, which efficiently detected IP (2×HA-linker) and coIP (FLAG) M populations (Fig. 4B). Validation of the assay was confirmed with our controlled conditions (expression of FLAG-M or 2×HA-linker-M alone, or omission of both expression plasmids), which exhibited no detectable coIP of M (Fig. 4A, upper panel), whereas M proteins migrating with the expected molecular masses were visualized in total cell extracts (Fig. 4A, lower panel).
FIG 4.
Coimmunopurification experiments reveal critical differences in self-assembly efficiencies of diverse M mutants. (A to C) The inherent self-assembly efficiencies of modified and unmodified M proteins expressed in HEK-293T cells were determined by coimmunopurification (coIP) and immunoblot analyses. (A) Control experiments performed with FLAG- and HA-tagged M proteins. The specific antibodies used for the immunopurification (IP) and immunoblotting (IB) steps are indicated on the left. (B and C) coIP performed under experimental settings similar to those described for panel A, but with cell lysis buffers of various stringencies (coIP1 > coIP2 > coIP3 [as indicated in Materials and Methods]). TL, total cell lysates. The differently tagged protein populations and their mutations are labeled on the right and below the gels, respectively. (D) M-M avidities of interaction (calculated as indicated in Materials and Methods). Means and SEM for data from three independent experiments are shown. *, P < 0.05 by one-way ANOVA with Holm-Bonferroni correction and by t tests.
Regardless of the conditions used in the lysis buffer to extract total proteins, a coIP band was readily detected in cells expressing the M-N138F, -N138A, -N138M, and -N138R mutants (Fig. 4C, upper panels). In contrast, M proteins carrying the 6A-block mutations almost completely lost the ability to self-assemble (Fig. 4C, lower panels). Interestingly, M-N138E was efficiently coimmunopurified only when less stringent cell lysis conditions were applied (Fig. 4C, lower left panel). We next determined the coIP efficiency index (see Materials and Methods), and the data clearly confirmed that the 6A-block mutants were strongly deficient in M dimerization/oligomerization. Under the most stringent cell lysis conditions, all M-N138 variants featured self-assembly impairments, with the M-N138E and -N138R mutants (carrying charged amino acids) displaying the most pronounced deficiencies (Fig. 4D, black and dark gray bars). Remarkably, except for M-N138M and, to some extent, M-N138F (which did not exhibit drastic deficiencies under all cell lysis conditions), all other M-N138 variants recovered nearly wt-like M-M binding affinities when cells were lysed under less stringent conditions (Fig. 4D, light gray bars).
Taken together, while these findings confirmed the inability of the 6A-block M mutants to properly form oligomers, they additionally revealed that mutating M at N138 modulated the efficiency of self-assembly, though to different extents, depending on the nature of the substituted amino acid and the cell lysis conditions.
M-N138 mutants efficiently relocalize N to the cell periphery.
While a lack of M oligomerization of the 6A-block mutants likely resulted from gross protein misfolding, the data obtained in our coIP analyses nevertheless indicated discrepancies in M oligomerization efficiency when the protomer-protomer binding interface was disturbed by a substitution at position 138. To further probe whether such modulations were the result of major structural defects, we monitored the ability of M to relocate the viral nucleocapsid protein (N) to the cell periphery. Indeed, it was reported that while N is evenly distributed throughout the cytoplasm in the absence of M, in its presence, N is recruited to the cell periphery as a result of M-N interaction (25).
We thus initially expressed (hemagglutinin [HA] epitope-tagged) N alone or in combination with (FLAG-tagged) M-wt in 293T cells. At 24 h posttransfection, immunofluorescence analyses were performed with fixed and permeabilized cells that were subsequently dually decorated with anti-mouse FLAG and anti-rat HA monoclonal antibodies (MAbs). The cells were finally treated with red and green fluorescent dye-conjugated secondary antibodies to visualize the cellular localization of both proteins. As expected, expression of N alone returned evident cytoplasmic staining. Conversely, when N and M were coexpressed in the same cells, N not only was efficiently relocated to the cell periphery but also clearly colocalized with M, as revealed by confocal microscopy analyses (Fig. 5). Strikingly, except for the three 6A-block mutants, all M-N138 variants preserved the capacity to properly drag N to the cell periphery (Fig. 5).
FIG 5.
Mutating CDV M residue N138 does not alter N binding and subsequent relocation to the cell periphery. HA-tagged N proteins expressed alone (no M) or in combination with the indicated FLAG-tagged M proteins in HEK-293T cells were visualized by immunofluorescence analyses. Images were captured with a scanning confocal laser fluorescence microscope (Olympus). N proteins were stained in green, M proteins in red, and nuclei in blue.
Taken together, these findings strongly suggested that the overall structure of the M-N138 mutants, or at least the domain required to efficiently bind and relocate N to the cell periphery, remained properly folded. On the other hand, these data once again supported the misfolding phenotype of the 6A-block mutants.
M dimerization efficiency correlates with modulation of plasma membrane deformation and VLP formation.
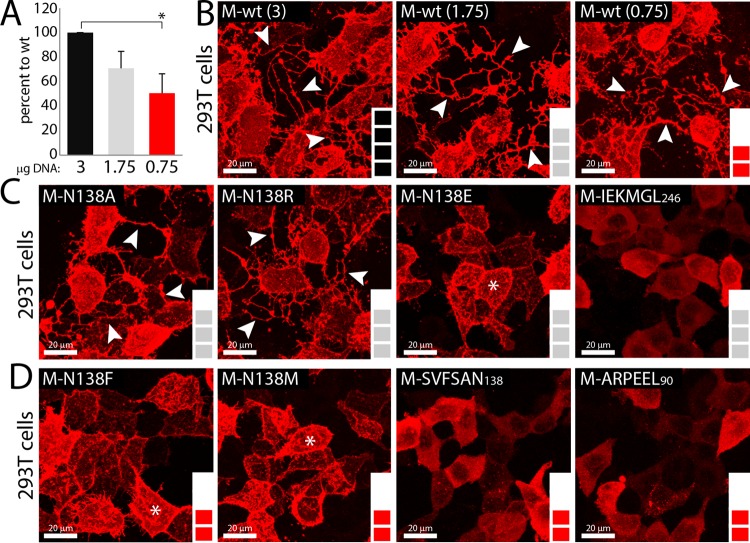
It was proposed that paramyxoviruses and pneumoviruses can induce membrane deformation and/or cell protrusions, which may serve as platforms for viral budding and ensuing cell exit. We thus expressed M-wt and the series of protein variants in 293T cells for 1 day and examined, by immunofluorescence analyses, the ability of each M protein to induce the deformation of the plasma membrane. To unambiguously highlight such membrane filamentous structures, we acquired 50 successive z-stacks of M-expressing cells by confocal microscopy and consequently merged them. Since differences in expression profiles might have influenced membrane deformation, we initially calibrated M-wt expression to match the two groups of expression levels displayed by the various M mutants (group 1, ∼70 to 85%; and group 2, ∼50%) (Fig. 6A and B). Regardless of the expression levels, M-wt was able to promote the generation of long cell-associated branched filaments that could interconnect with neighboring cells. Similarly, the M-N138A and M-N138R variants could also efficiently trigger the formation of these long cell membrane protrusions. In contrast, no sign of membrane deformation was observed in any of the three 6A-block M mutants (Fig. 6C and D). Interestingly, while long cell extensions were absent in cells expressing the N138E, N138F, and N138M mutants, short filamentous (filopodium-like) structures protruding from the cell surface were nevertheless highlighted (Fig. 6C and D). Furthermore, to unambiguously demonstrate that discrepancies in expression levels were not the main cause of the absence of filopodium-like and/or long cell process formation, the various M mutants were grouped by their overall expression profiles (∼70 to 85% [Fig. 6C] and ∼50% [Fig. 6D]), which clearly illustrated the absence of direct correlation.
FIG 6.
Differential triggering of membrane deformation by various M proteins. (A) Amounts of transfected M-wt DNA needed to match group 1 (∼70 to 85%) and 2 (∼50%) expression levels (color coding is as described in the legend to Fig. 2B and C). Expression was assessed by immunoblotting and semiquantitative evaluation, performed as described in the legend to Fig. 2B. *, P < 0.05 by one-way ANOVA with Holm-Bonferroni correction and by t tests. (B to D) Immunofluorescence analyses performed as described in the legend to Fig. 1C, without nuclear counterstaining. To gain an overall 3D view of cells, 50 z-stack images were acquired and digitally merged. (B) M-wt exhibiting 100, 70, and 50% expression levels, depending on the amount of plasmid DNA transfected. (C) M mutants exhibiting ∼70 to 85% expression levels. (D) M mutants exhibiting ∼50% expression levels. Images were captured with a scanning confocal laser fluorescence microscope (Olympus). White arrows indicate long cell protrusions, and white stars indicate short filopodium-like membrane structures.
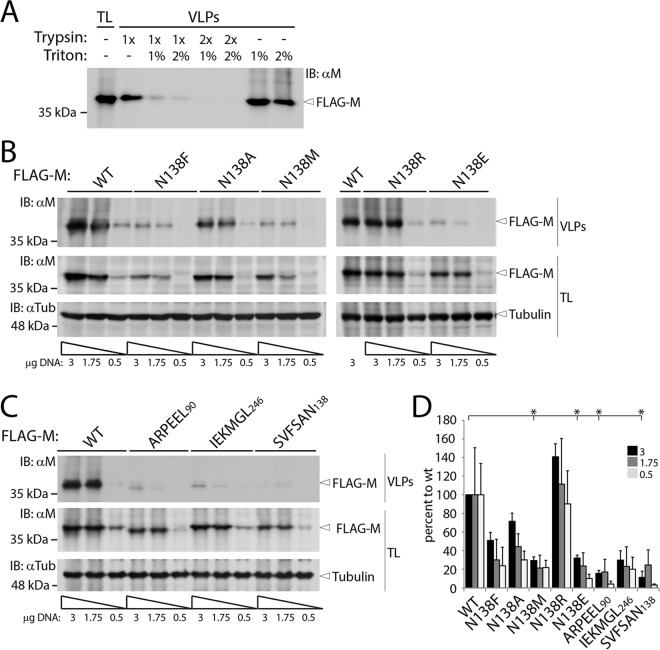
We next investigated the inherent capacity of CDV M-wt and its derivative mutants to release virus-like particles (VLPs). To this end, we expressed the M-wt protein and its derivative variants in 293T cells. At 48 h posttransfection, the supernatants were harvested, treated with trypsin, loaded onto a glycerol cushion, and subsequently centrifuged to purify VLPs. To validate the assay, the supernatants of M-wt-expressing cells were initially treated with trypsin (1× or 2×) and/or Triton X-100 (1% or 2%) prior to centrifugation. Clearly, pretreatment of supernatants with Triton X-100 and trypsin led to a drastic degradation of M, as indicated by Western blotting. In contrast, single treatment of supernatants did not result in substantial M degradation, thereby confirming that the assay was indeed focusing on M proteins protected by surrounding lipidic vesicles (Fig. 7A). As mentioned above, and presumably as a result of efficient spontaneous generation of large oligomers, M proteins did sediment even upon Triton X-100 treatment (Fig. 7A).
FIG 7.
Assessment of inherent membrane-budding ability of the panel of CDV M mutants. (A) Supernatants of HEK-293T cells expressing M-wt were loaded onto glycerol cushions and subjected to high-speed centrifugation. Cell supernatants were treated with trypsin and/or Triton X-100 prior to centrifugation, and the resulting pellets were subjected to immunoblotting. The left lane illustrates M expression in total cell lysates. (B and C) M proteins were expressed in HEK-293T cells, and the generated VLPs were assessed as described for panel A, but without Triton X-100 treatment. To control for M protein expression, decreasing amounts of plasmid DNA were transfected (as indicated below the blots). The different proteins are labeled on the right. VLPs, virus-like particles; TL, total cell lysates. (D) VLP formation efficiency indices (calculated as indicated in Materials and Methods). Means and SEM for data from three independent experiments are shown. *, P < 0.05 by one-way ANOVA with Holm-Bonferroni correction and by t tests.
Because of the reported variations in intracellular M expression depending on the substitution applied, we systematically decreased the amount of DNA transfected into cells in order to control total protein expression. Western blot analyses (Fig. 6A and 7B and C) indeed confirmed a direct correlation between decreasing amounts of plasmid DNA transfected and total amounts of M protein expressed. Importantly, and corroborating the data obtained in the cell protrusion formation assay, VLPs were efficiently produced in the case of M-wt and the N138A and N138R mutants (Fig. 7B), and this was confirmed by the VLP efficiency index (Fig. 7D) (see Materials and Methods). In contrast, the three 6A-block mutants as well as the N138M and N138E M mutants were strongly impaired in VLP formation (Fig. 7C and D). Finally, while M-N138A did preserve an intermediate ability to produce VLPs, the N138F mutant retained a residual activity only when large amounts of DNA were transfected into 293T cells (Fig. 7B and D).
In summary, these results indicated that the 6A-block M mutants, which exhibited severely impaired folding, oligomerization propensity, and cell periphery accumulation, were additionally defective in short and long cell protrusion triggering and, consequently, VLP formation. Interestingly, the M-N138E, -N138F, and -N138M mutants, which displayed proper cell periphery accumulation, exhibited defects in M-M interactions and in VLP production. However, in contrast to the 6A-block M mutants, they retained some ability to accumulate only in short filopodium-like membrane structures.
Coexpression of M-N138R with M-N138E restores dimerization and allows incorporation of M-N138E into VLPs.
The coIP assays, which were performed under various cell lysis conditions, revealed that mutation of asparagine 138 to charged amino acids resulted in the production of unstable M-M interactions (likely due to the engineering of electrostatic repulsion). Interestingly, the M-M avidity of interaction revealed that M-N138E was more affected than M-N138R, a phenotype that translated into impaired (N138E) versus efficient (N138R) VLP production. We thus decided to investigate whether coexpression of both mutants in the same cells could to some extent restore (i) the M-M interaction efficiency (switching repulsive to attractive electrostatics) and (ii) the incorporation of the N138E mutant into VLPs.
To this end, 2×HA-linker-M- and FLAG-M-expressing DNA plasmids were cotransfected alone or in various combinations into 293T cells. Remarkably, while the coIP assays (exclusively those performed under stringent conditions for cell lysis [coIP1]) confirmed that M-N138R and M-N138E expressed individually exhibited deficiencies in self-assembly, coexpressing both mutants in the same cells led to a clear restoration of M-M binding affinity (Fig. 8A and B).
FIG 8.
Dimerization and VLP formation efficiencies of heterodimeric M populations. (A and C) The self-assembly propensity and VLP formation efficiency of M-N138E- and M-N138R-coexpressing HEK-293T cells were determined as indicated in the legends to Fig. 4 and 7, respectively. (B and D) Dimerization and VLP formation indices (calculated as indicated in Materials and Methods). Means and SEM for data from three independent experiments are shown. *, P < 0.05; **, P < 0.01 by one-way ANOVA with Holm-Bonferroni correction and by t tests.
We next determined whether the mixed M dimeric population indeed correlated with the incorporation of M-N138E into VLPs. Remarkably, not only were VLPs successfully generated from 293T cells that expressed M heterodimers, but M-N138E was readily detected within the released VLPs (Fig. 8C). Interestingly, while the VLP efficiency index for mixed M dimer populations indicated an increase compared to that for VLPs induced by M-N138E expressed alone, VLP production was slightly more efficient when the FLAG-tagged M protein carried the “VLP-boosting” N138R substitution (Fig. 8D). We noticed in this set of experiments, however, that FLAG-tagged M proteins were slightly more expressed than the HA-tagged variants (Fig. 8C, lower panels). It was therefore not unexpected to purify more VLPs from the supernatant of cells coexpressing the FLAG-tagged M protein carrying the “VLP-boosting” N138R substitution.
Taken together, these findings clearly demonstrated the successful generation of a mixed dimeric M population composed of M-N138E- and M-N138R-carrying protomers. Importantly, M heterodimers not only exhibited proper stability but also could promote the release of VLPs into the supernatant, with efficient incorporation of M-N138E protomers. These phenotypes likely resulted from a switch from repulsive to attractive electrostatics, which provides strong evidence that residues 138 of two M protomers are indeed located in close spatial proximity within dimeric M units. In addition, these data further strengthen the notion that minimal M dimerization efficiency is required to mediate proper VLP production.
DISCUSSION
Although it has been reported that paramyxoviral and pneumoviral M proteins, by interacting with multiple cellular and viral components, play a key regulatory role in viral replication, assembly, egress, and membrane fusion (16, 46, 66), very little is known about the inherent ability of M to drive these discrete functions. In this study, we initiated a comprehensive structure-guided analysis to gain mechanistic insights into various M functionalities. We rationally targeted asparagine 138 of CDV M (M-N138) for substitution purposes because, in our structural homology model, this residue is located at the center of the M-M dimeric binding interface. Indeed, increasing evidence suggests that M dimers may act as an essential minimal unit to sustain the M protein's diverse activities (16, 25, 31, 61–63). Moreover, because it was proposed that M can also form higher-order oligomers, we focused our targeted mutational analyses on previously hypothesized critical M regions (α2 and α9 helices). Indeed, both helices may support two-dimensional dimer-dimer assembly, thereby putatively producing higher-order oligomeric networks (Fig. 1). Such M arrays may then drive the lipid curvature, which eventually may lead to the formation of cell protrusions and the ensuing platforms for viral budding (61, 62, 64).
Our data clearly indicated that dimerization/oligomerization is critical for proper M intracellular transport and cell periphery accumulation. Indeed, the three strategically designed 6A-block mutants (SVFSAN138, protomer-protomer interface; ARPEEL90, dimer-dimer interface; and IEKMGL246, dimer-dimer interface) failed to self-assemble into detectable dimers, as illustrated in our newly developed assays, i.e., the cellular DSP-based dimerization assay and a coIP assay based on multiple cell lysis conditions. Since several residues mapping to the SVFSAN peptide are involved in the protomer-protomer interface, a lack of dimerization was not unexpected. Strikingly, our data further demonstrated that when dimerization/oligomerization was severely impaired, cell periphery accumulation, recruitment of N to the plasma membrane, membrane deformation triggering, and VLP formation were additionally entirely defective. This strongly supported the notion that self-assembly must occur to promote multiple M functionalities. Interestingly, Runkler and colleagues described that mutating residue V101 of MeV M to alanine (M-V101A) also resulted in a complete loss of cell periphery accumulation, although in this case self-assembly seemed to be preserved to some extent (25). In our structural model, V101, which is well conserved among all members of the Morbillivirus genus, is located within the NTD and at the dimeric interface generated with the CTD of another protomer. It is noteworthy that the first serine residue of the 6A-block mutant SVFSAN138 (S133) folds at a position within the overall structure which is spatially very close to amino acid V101. Although it is only speculative at this stage, it is possible that mutation of both residues S133 and V101 to alanines may translate into a lack of (or inappropriate) M dimerization, thereby generating nonfunctional M proteins.
The fact that similar phenotypes were recorded with the 6A-block mutants carrying substitutions in the α2 and α9 helices was, however, more surprising. Indeed, based on our structural homology model, both helices are surface exposed and are located opposite the large protomer-protomer binding interface, which implies a role in higher-order oligomer organization but not in dimer assembly (Fig. 1). While it is possible that our model might not be entirely correct, we cannot exclude that mutating both helices might have translated into long-range effects indirectly affecting dimerization propensity. Alternatively, some residues in both helices might be involved in critical posttranslational modifications and/or functional interactions with a host cellular factor(s) required for proper dimerization. It is interesting that the lysine residue at position 240 of MeV M (M-K240; potentially located in α9 and N-terminal to the IEKMGL domain) undergoes posttranslational ubiquitination, although the functional role of such modification remains obscure (51). We can thus speculate that M ubiquitination is required for dimerization/oligomerization and that substitutions within the flanking IEKMGL peptide will influence K240 ubiquitination. Although additional work is needed to precisely understand the failure of self-assembly when the α2 and α9 helices are mutated, our data unambiguously highlight that dimerization is necessary for proper M transport to the cell periphery. Efficient M dimerization as a requirement for cell periphery accumulation may hence represent a basic feature shared by most (if not all) members of the Paramyxoviridae and Pneumoviridae families (63).
In sharp contrast to the phenotype reported with the 6A-block M variants, and regardless of the nature of the substitution performed, N138-mutated M proteins exhibited unaltered cell periphery accumulation. Preservation of M structural integrity was also assumed, since all mutants were still capable of (i) recruiting N proteins to the plasma membrane and (ii) self-assembly. However, although M-M physical interactions remained detectable, our newly developed differential coIP assay demonstrated substantial modulation of M self-assembly propensity depending on the side chain properties of the amino acid substitution at position 138. Indeed, under stringent cell lysis conditions (coIP1), the M-N138F and M-N138A mutants displayed slight reductions in self-interaction, whereas the M-N138M and M-N138R variants exhibited mild deficiencies in self-interaction and the N138E mutant showed severe deficiencies. These findings thus support the idea that when charged residues are inserted at the core position of the M-M binding interface, self-assembly is disturbed, likely as a consequence of introducing electrostatic repulsions. Conversely, and further upholding this hypothesis, the generation of heterodimers with opposite charges at position 138 led to a full restoration of dimerization efficiency, presumably as a result of engineered attractive electrostatics.
Importantly, when less stringent conditions were applied for cell lysis (coIP3), self-assembly was substantially restored for the M-N138A, M-N138R, and M-N138E mutants but not the M-N138M and M-N138F mutants. In fact, for the first three mutants, we speculate that specific to less stringent lysis conditions, coimmunopurified M proteins relied not only on dimeric protomer-protomer interactions but also on “dimer-dimer” contacts involved in higher-order M oligomer structures (Fig. 1B). In contrast, since the self-assembly efficiency of M-N138M and M-N138F did not increase substantially under less stringent cell lysis conditions, we suggest that both mutants associated into irregular dimers which failed to promote detectable dimer-dimer contacts. Combined, the data obtained in our coIP assays not only confirmed that N138 very likely is located at the core of the protomer-protomer binding interface but also that mutating this residue can interfere with dimer stability and/or an overall dimeric architecture compatible with higher-order oligomer assembly. Importantly, this also spotlights, for the first time, a biochemical approach potentially able to detect discrete M-M binding interfaces.
Although all N138 M variants were targeted to the cell periphery and capable of forming dimers, significant variations in their ability to trigger membrane deformation and VLP production were recorded. Therefore, while our findings highlighted the notion that triggering of membrane protrusions and VLP formation relies on M self-assembly, they additionally suggest that M dimers must preserve some structural stability. Indeed, M-wt, M-N138A, and M-N138R, which featured at least 50% dimerization interaction efficiencies, were able to promote the formation of long cell processes and to trigger the release of large amounts of VLPs into the supernatants. In contrast, M-N138E, which exhibited only around a 20% dimerization propensity, while still being able to induce short filopodium-like structures, failed to productively rearrange plasma membranes and generate VLPs.
Interestingly, while the N138F and N138M M mutants self-associated with substantial affinities (∼80% and ∼50%, respectively), they also triggered only small, filopodium-like membrane protrusions and were deficient in VLP formation. However, as suggested above, the size and hydrophobic nature of the phenylalanine and methionine residues may have translated into the assembly of irregular dimers unable to efficiently produce higher-order oligomers. Alternatively, M arrays may have formed, but with inappropriate angles at the dimer-dimer binding interface, in turn leading to nonproductive triggering of lipid curvatures. Thus, while a minimal dimeric stability is strictly required to promote membrane deformation and budding, our data further suggest that both phenotypes additionally rely on functional dimer-dimer contacts.
Remarkably, our VLP assays performed with mixed M dimers not only revealed restoration of the dimerization efficiency of inherently “self-assembly-defective” mutants (N138E and N138R) but also corroborated the substantial incorporation of M-N138E protomers into VLPs. Nevertheless, the amounts of VLPs released into the supernatants did not reach the levels induced by M-wt. Because both heterodimeric and homodimeric populations are generated in the context of cells coexpressing different M proteins, we hypothesize that the production of VLPs was affected by incorporation of unstable M-N138E–M-N138E homodimers into extending M arrays. Such a destabilized M mesh may in turn interfere with optimal lipid curvature and ensuing triggering of budding.
Altogether, our findings allowed us to propose a mechanistic model for how M may trigger membrane budding. M initially assembles into dimers in the cytosol, which in turn traffic intracellularly to finally reach the plasma membrane (Fig. 9A). Higher-order M oligomers subsequently assemble at the inner leaflet of the plasma membrane. Lipid curvature is induced as the result of two-dimensional dimer-dimer M assembly, with an angle possibly generated at both oligomeric binding interfaces (Fig. 9B). Plasma membrane deformation further proceeds in correlation with extension of the oligomerization process (Fig. 9C). This “array elongation” step depends on a minimal stability of M dimers to generate robust M oligomers able to sustain the physical forces required for the promotion of membrane curvature (Fig. 9C). Large and stable spherical M arrays ultimately contribute to the release of VLPs into the supernatant. Importantly, our model does not exclude the requirement for host cellular proteins and/or posttranslational modifications (which may also occur in the nucleus) in contributing to any stages of the proposed mechanism of M-induced membrane budding and scission.
FIG 9.
Model of M-mediated triggering of membrane budding. (A) Accumulation of M dimeric units at the inner leaflet of the plasma membrane (pm) upon intracellular trafficking. (B) Bidirectional dimer-dimer M assembly (dim 1 to dim 4) generates angles at the “oligomeric” binding interfaces (for simplicity, only one direction is shown), which directly correlate with the induction of lipid curvatures. (C) Oligomerization proceeds, and the derived assembling supramolecular complexes require stable dimeric units to build up robust M arrays. Ultimately, growing 2D M arrays generate spherical lipid curvatures, which are released from the host cells. The dimeric and oligomeric binding interfaces are highlighted in red and green, respectively.
In summary, our findings provide evidence that a critical amount of stable M dimers at the cell periphery is required to productively trigger VLPs. Such stabilized M dimeric units may facilitate further assembly into robust higher-order oligomers necessary to promote plasma membrane-budding activity.
MATERIALS AND METHODS
Cell cultures, plasmid construction, and transfections.
HEK-293T cells were grown in Dulbecco's modified Eagle medium (DMEM; Gibco, Life Technologies) supplemented with 10% fetal calf serum and 1% penicillin-streptomycin at 37°C in the presence of 5% CO2. Six-well cell culture plates (9.5 cm2) were used for seeding of HEK-293T cells for the diverse experiments performed in this study and were covered with glass coverslips specifically for indirect immunofluorescence and confocal microscopy assays.
The matrix protein gene was amplified with supplementary RsrII restriction sites, N-terminal tags, and a linker (i.e., HA-linker) from a plasmid coding for the full-length antigenome of the CDV A75/17 strain, using the following primers: 5′-ATATATCGGACCGATGTATCCATATGATGTTCCAGATTATGCTACCGGTGGTTCCGGTGGTTCCGGAACTGAGGTGTATGACTTCGATCAGTCTTCATGGGACACCAAAG-3′ (forward) and 5′-ATATATCGGTCCGTTAGAGAATTTTGAAAAGACCCTGATC-3′ (reverse) (65, 67). Amplicons were subcloned into the vector pCI by using the restriction endonuclease RsrII. All mutations to replace the HA tag with a FLAG tag, to elongate the HA tag or the linker, and to replace the amino acids at positions 138, 85 to 90, 133 to 138, and 241 were performed in the pCI-HA-linker-M construct by using a QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions. HA-tagged CDV N was generated following the same strategy. HEK-293T cells were transfected at 90% confluence with the various vector constructs by use of TransIT-LT1 (Mirus) following the supplier's instructions. Mutagenesis primer sequences will be communicated upon request.
Western blotting.
HEK-293T cells expressing different pCI constructs were harvested in phosphate-buffered saline (PBS) at 24 or 48 h posttransfection, cleared by centrifugation (10 min, 2,000 × g, 4°C), and frozen at −20°C. Cell pellets were resuspended in 0.2 M dithiothreitol (DTT)-1.6× Laemmli buffer (Bio-Rad) and then subjected to Western blot analysis using an in-house-generated rabbit polyclonal anti-CDV M antibody (1:1,000) (22) and a horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (1:9,000) (Dako) or HRP-conjugated mouse monoclonal anti-tubulin antibody (1:5,000) (AbCam). Signals were detected with Amersham ECL Prime Western blotting reagent (GE Healthcare Life Sciences) following the manufacturer's instructions. Anti-CDV M and anti-tubulin signal intensities from three independent experiments were quantified relative to the wild-type (wt) signals, which were set at 100%, using Aida image analyzer software, and total lysate (TL) expression level ratios (anti-CDV M/anti-tubulin) were calculated. For each mutant (mut), mean ratios are shown (Fig. 2B).
Indirect immunofluorescence analysis.
HEK-293T cells transfected with plasmids encoding M or derivative mutated M proteins were fixed with 4% paraformaldehyde at 24 h posttransfection and then permeabilized by use of 0.5% Triton X-100. Immunofluorescence analysis was performed by using a mouse anti-FLAG monoclonal antibody (1:1,000; Sigma-Aldrich) for FLAG-M constructs, a rabbit anti-sodium-potassium ATPase (anti-Na/K ATPase) monoclonal antibody (1:200; Abcam) for the plasma membrane, a rat anti-HA monoclonal antibody (1:1,000; Roche Diagnostics) for the HA-N construct, Alexa Fluor 555-conjugated goat anti-mouse IgG(H+L), Alexa Fluor 488-conjugated goat anti-rabbit IgG(H+L), and Alexa Fluor 488-conjugated goat anti-rat IgG(H+L) secondary antibodies (1:1,000; Life Technologies), and 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml; AppliChem). Pictures were acquired by laser scanning confocal microscopy using an Olympus IX81 confocal microscope with FluoView FV1000 software and processed into section pictures or three-dimensional (3D) reconstitutions with Imaris software.
Dimerization assay.
M-expressing cells were harvested in PBS 48 h after transfection and cleared by centrifugation (20 min, 400 × g, 4°C). Cells were incubated in PBS with 1 mM dithiobis(succinimidyl propionate) (DSP; an amine-reactive cross-linker) (Thermo Scientific) for 2 h with agitation at 4°C. After clearing by centrifugation (10 min, 2,000 × g, 4°C), cells were lysed in cold native sample buffer (NS buffer) (100 mM Tris-Cl, 10% glycerol, 0.0025% bromophenol blue [pH 8.6], 0.1% digitonin) supplemented with cOmplete EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics). The lysate was cleared by centrifugation as described above, and the supernatant was mixed with 4 mM urea Laemmli buffer (2.5% SDS, 20% glycerol, 120 mM Tris [pH 6.8], 0.02% bromophenol blue, 4 mM urea) at a 1:1 ratio. Samples were then heated at 50°C, loaded into 7% NuPAGE Tris-acetate gels (Novex; Life Technologies), and subjected to Western blot analysis with a rabbit polyclonal anti-CDV M primary antibody (1:1,000) (22) as described above. Dimer and monomer anti-CDV M signal intensities were measured simultaneously for each construct, using Aida software. Dimer-to-monomer ratios were normalized to that of the wild type. Mean ratios were determined for four independent experiments (Fig. 3C).
Coimmunopurification.
HEK-293T cells coexpressing FLAG-M and 2×HA-linker-M, both bearing the same mutations, were resuspended in PBS at 24 h posttransfection. Eighty percent of the total volume was used for coIP assay, and the rest was used for total lysate protein expression analysis. After centrifugation (10 min, 2,000 × g, 4°C), the PBS solution was removed, and cells were lysed in RIPA buffer (50 mM Tris, pH 7.2, 150 mM NaCl, 10 mM EDTA, 1% sodium deoxycholate, 1% Nonidet P-40, 0.05% SDS) or NS buffer. Lysates were incubated for 1 h with a mouse anti-HA monoclonal antibody (1:800; BioLegend) and overnight at 4°C with prewashed protein G Dynabeads (Novex; Life Technologies) at an adjusted antibody concentration of 1:1,000. After 3 washes with either RIPA buffer (after RIPA buffer lysis [coIP1]) or PBS (after RIPA buffer lysis [coIP2] or after NS buffer lysis [coIP3]), bead samples were resuspended in 33.3 mM glycine (pH 2.8), 0.1 M DTT, and 0.8× Laemmli buffer (Bio-Rad). Supernatants were then subjected to Western blot analysis with a rabbit polyclonal anti-CDV M primary antibody (1:1,000) (22) as described above. Anti-M band intensities from FLAG-M (coimmunopurified protein [coIP]) and 2×HA-linker-M (immunopurified protein [IP]) expression or coimmunopurification were determined simultaneously for each mutant, using Aida software. The coIP efficiency index was determined as the ratio of coIP to IP signals normalized to their respective total lysate expression levels. The mean index was normalized to the wild-type value to allow for direct comparison .
VLP assay.
HEK-293T cells were transfected with plasmids expressing different M constructs. Culture supernatants and cells were collected 48 h after transfection. Cellular debris was removed from supernatants by centrifugation (20 min, 500 × g, 4°C). Some of the clarified supernatants were adjusted to 1% or 2% Triton X-100, and then every supernatant was incubated for 30 min at 37°C with 100 μg or 200 μg of trypsin (trypsin-EDTA solution [10×]; Sigma-Aldrich). Subsequently, 25 μg or 50 μg, respectively, of soybean trypsin inhibitor (Roche Diagnostics) was added to neutralize the protease. The resulting mixture was loaded onto a 25% glycerol cushion and centrifuged at 16,100 × g for 1 h at 4°C. The pellet was resuspended in 0.2 M DTT–1.6× Laemmli buffer (Bio-Rad) and then subjected to Western blot analysis as described before, using a rabbit polyclonal anti-CDV M antibody (1:1,000) (22). As for total lysate expression levels, signal intensities were acquired relative to the 3-μg wild-type signals, which were set to 100%. Each mutant's efficiency index for VLP production was calculated relative to the wild-type index for the corresponding DNA amount transfected .
Statistical analysis.
One-way analysis of variance (ANOVA) and an unpaired two-tailed t test were used to analyze differences between the wild-type and mutant means. P values were adjusted by the Holm-Bonferroni sequential correction to control the type I error rate resulting from multiple comparisons (67). Differences were considered statistically significant if the resulting P values were <0.05 and are illustrated accordingly in the figures.
ACKNOWLEDGMENTS
We thank Marc Vandevelde for critically reviewing the manuscript.
This work was supported by the University of Bern and the Swiss National Science Foundation (grant 310030_173185 to P.P.).
REFERENCES
- 1.Amarasinghe GK, Bao Y, Basler CF, Bavari S, Beer M, Bejerman N, Blasdell KR, Bochnowski A, Briese T, Bukreyev A, Calisher CH, Chandran K, Collins PL, Dietzgen RG, Dolnik O, Durrwald R, Dye JM, Easton AJ, Ebihara H, Fang Q, Formenty P, Fouchier RA, Ghedin E, Harding RM, Hewson R, Higgins CM, Hong J, Horie M, James AP, Jiang D, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lee B, Leroy EM, Li M, Maisner A, Muhlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Pearson MN, Randall RE, Revill PA, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Smither SJ, Song Q, Stone DM, Takada A, Terregino C, Tesh RB, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Volchkov VE, Wahl-Jensen V, Walker PJ, Wang B, Wang D, Wang F, Wang LF, Werren JH, Whitfield AE, Yan Z, Ye G, Kuhn JH. 7 April 2017. Taxonomy of the order Mononegavirales: update 2017. Arch Virol doi: 10.1007/s00705-017-3311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T, Subbarao SM, Belsham GJ, Mahy BW. 1991. The molecular biology of the morbilliviruses, p 83–102. In Kingsbury DW. (ed), The paramyxoviruses. Plenum Press, New York, NY. [Google Scholar]
- 3.Tatsuo H, Ono N, Tanaka K, Yanagi Y. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 4.Baron MD. 2005. Wild-type rinderpest virus uses SLAM (CD150) as its receptor. J Gen Virol 86:1753–1757. doi: 10.1099/vir.0.80836-0. [DOI] [PubMed] [Google Scholar]
- 5.Tatsuo H, Ono N, Yanagi Y. 2001. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J Virol 75:5842–5850. doi: 10.1128/JVI.75.13.5842-5850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birch J, Juleff N, Heaton MP, Kalbfleisch T, Kijas J, Bailey D. 2013. Characterization of ovine Nectin-4, a novel peste des petits ruminants virus receptor. J Virol 87:4756–4761. doi: 10.1128/JVI.02792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noyce RS, Delpeut S, Richardson CD. 2013. Dog nectin-4 is an epithelial cell receptor for canine distemper virus that facilitates virus entry and syncytia formation. Virology 436:210–220. doi: 10.1016/j.virol.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Noyce RS, Richardson CD. 2012. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol 20:429–439. doi: 10.1016/j.tim.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Muhlebach MD, Mateo M, Sinn PL, Prufer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB Jr, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pratakpiriya W, Seki F, Otsuki N, Sakai K, Fukuhara H, Katamoto H, Hirai T, Maenaka K, Techangamsuwan S, Lan NT, Takeda M, Yamaguchi R. 2012. Nectin4 is an epithelial cell receptor for canine distemper virus and involved in neurovirulence. J Virol 86:10207–10210. doi: 10.1128/JVI.00824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ader-Ebert N, Khosravi M, Herren M, Avila M, Alves L, Bringolf F, Orvell C, Langedijk JP, Zurbriggen A, Plemper RK, Plattet P. 2015. Sequential conformational changes in the morbillivirus attachment protein initiate the membrane fusion process. PLoS Pathog 11:e1004880. doi: 10.1371/journal.ppat.1004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bose S, Jardetzky TS, Lamb RA. 2015. Timing is everything: fine-tuned molecular machines orchestrate paramyxovirus entry. Virology 479–480:518–531. doi: 10.1016/j.virol.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navaratnarajah CK, Rosemarie Q, Cattaneo R. 2015. A structurally unresolved head segment of defined length favors proper measles virus hemagglutinin tetramerization and efficient membrane fusion triggering. J Virol 90:68–75. doi: 10.1128/JVI.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brindley MA, Chaudhury S, Plemper RK. 2015. Measles virus glycoprotein complexes preassemble intracellularly and relax during transport to the cell surface in preparation for fusion. J Virol 89:1230–1241. doi: 10.1128/JVI.02754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plattet P, Plemper RK. 2013. Envelope protein dynamics in paramyxovirus entry. mBio 4:e00413-13. doi: 10.1128/mBio.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb RA, Parks GD. 2013. Paramyxoviridae: the viruses and their replication, p 957–995. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 17.Cathomen T, Mrkic B, Spehner D, Drillien R, Naef R, Pavlovic J, Aguzzi A, Billeter MA, Cattaneo R. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J 17:3899–3908. doi: 10.1093/emboj/17.14.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano A, Wang AH, Gombart AF, Wong TC. 1992. The matrix proteins of neurovirulent subacute sclerosing panencephalitis virus and its acute measles virus progenitor are functionally different. Proc Natl Acad Sci U S A 89:8745–8749. doi: 10.1073/pnas.89.18.8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki M, Takeda M, Shirogane Y, Nakatsu Y, Nakamura T, Yanagi Y. 2009. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J Virol 83:10374–10383. doi: 10.1128/JVI.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spielhofer P, Bachi T, Fehr T, Christiansen G, Cattaneo R, Kaelin K, Billeter MA, Naim HY. 1998. Chimeric measles viruses with a foreign envelope. J Virol 72:2150–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakimoto H, Shimodo M, Satoh Y, Kitagawa Y, Takeuchi K, Gotoh B, Itoh M. 2013. F-actin modulates measles virus cell-cell fusion and assembly by altering the interaction between the matrix protein and the cytoplasmic tail of hemagglutinin. J Virol 87:1974–1984. doi: 10.1128/JVI.02371-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiener D, Plattet P, Cherpillod P, Zipperle L, Doherr MG, Vandevelde M, Zurbriggen A. 2007. Synergistic inhibition in cell-cell fusion mediated by the matrix and nucleocapsid protein of canine distemper virus. Virus Res 129:145–154. doi: 10.1016/j.virusres.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Cathomen T, Naim HY, Cattaneo R. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol 72:1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahara M, Takeda M, Seki F, Hashiguchi T, Yanagi Y. 2007. Multiple amino acid substitutions in hemagglutinin are necessary for wild-type measles virus to acquire the ability to use receptor CD46 efficiently. J Virol 81:2564–2572. doi: 10.1128/JVI.02449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Runkler N, Pohl C, Schneider-Schaulies S, Klenk HD, Maisner A. 2007. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell Microbiol 9:1203–1214. doi: 10.1111/j.1462-5822.2006.00860.x. [DOI] [PubMed] [Google Scholar]
- 26.Mahapatra M, Parida S, Baron MD, Barrett T. 2006. Matrix protein and glycoproteins F and H of peste-des-petits-ruminants virus function better as a homologous complex. J Gen Virol 87:2021–2029. doi: 10.1099/vir.0.81721-0. [DOI] [PubMed] [Google Scholar]
- 27.Dietzel E, Kolesnikova L, Maisner A. 2013. Actin filaments disruption and stabilization affect measles virus maturation by different mechanisms. Virol J 10:249. doi: 10.1186/1743-422X-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klauschies F, Gutzkow T, Hinkelmann S, von Messling V, Vaske B, Herrler G, Haas L. 2010. Viral infectivity and intracellular distribution of matrix (M) protein of canine distemper virus are affected by actin filaments. Arch Virol 155:1503–1508. doi: 10.1007/s00705-010-0737-6. [DOI] [PubMed] [Google Scholar]
- 29.Katayama H, Hori M, Sato K, Kajita M, Ozaki H, Karaki H, Ohashi K, Kai C. 2004. Role of actin microfilaments in canine distemper virus replication in Vero cells. J Vet Med Sci 66:409–415. doi: 10.1292/jvms.66.409. [DOI] [PubMed] [Google Scholar]
- 30.Coronel EC, Murti KG, Takimoto T, Portner A. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J Virol 73:7035–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, Zhang S, Ding B, Yang X, Chen L, Yan Q, Jiang Y, Zhong Y, Chen M. 2014. A leucine residue in the C terminus of human parainfluenza virus type 3 matrix protein is essential for efficient virus-like particle and virion release. J Virol 88:13173–13188. doi: 10.1128/JVI.01485-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugahara F, Uchiyama T, Watanabe H, Shimazu Y, Kuwayama M, Fujii Y, Kiyotani K, Adachi A, Kohno N, Yoshida T, Sakaguchi T. 2004. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology 325:1–10. doi: 10.1016/j.virol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 33.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J Virol 75:11384–11391. doi: 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patch JR, Crameri G, Wang LF, Eaton BT, Broder CC. 2007. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol J 4:1. doi: 10.1186/1743-422X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciancanelli MJ, Basler CF. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol 80:12070–12078. doi: 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. 2006. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol 80:11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. 2009. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J Virol 83:7261–7272. doi: 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riedl P, Moll M, Klenk HD, Maisner A. 2002. Measles virus matrix protein is not cotransported with the viral glycoproteins but requires virus infection for efficient surface targeting. Virus Res 83:1–12. doi: 10.1016/S0168-1702(01)00379-3. [DOI] [PubMed] [Google Scholar]
- 39.Subhashri R, Shaila MS. 2007. Characterization of membrane association of rinderpest virus matrix protein. Biochem Biophys Res Commun 355:1096–1101. doi: 10.1016/j.bbrc.2007.02.088. [DOI] [PubMed] [Google Scholar]
- 40.Dietzel E, Anderson DE, Castan A, von Messling V, Maisner A. 2011. Canine distemper virus matrix protein influences particle infectivity, particle composition, and envelope distribution in polarized epithelial cells and modulates virulence. J Virol 85:7162–7168. doi: 10.1128/JVI.00051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bellini WJ, Englund G, Richardson CD, Rozenblatt S, Lazzarini RA. 1986. Matrix genes of measles virus and canine distemper virus: cloning, nucleotide sequences, and deduced amino acid sequences. J Virol 58:408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pohl C, Duprex WP, Krohne G, Rima BK, Schneider-Schaulies S. 2007. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J Gen Virol 88:1243–1250. doi: 10.1099/vir.0.82578-0. [DOI] [PubMed] [Google Scholar]
- 43.Salditt A, Koethe S, Pohl C, Harms H, Kolesnikova L, Becker S, Schneider-Schaulies S. 2010. Measles virus M protein-driven particle production does not involve the endosomal sorting complex required for transport (ESCRT) system. J Gen Virol 91:1464–1472. doi: 10.1099/vir.0.018523-0. [DOI] [PubMed] [Google Scholar]
- 44.Sanderson CM, Avalos R, Kundu A, Nayak DP. 1995. Interaction of Sendai viral F, HN, and M proteins with host cytoskeletal and lipid components in Sendai virus-infected BHK cells. Virology 209:701–707. doi: 10.1006/viro.1995.1308. [DOI] [PubMed] [Google Scholar]
- 45.Gosselin-Grenet AS, Marq JB, Abrami L, Garcin D, Roux L. 2007. Sendai virus budding in the course of an infection does not require Alix and VPS4A host factors. Virology 365:101–112. doi: 10.1016/j.virol.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 46.El Najjar F, Schmitt AP, Dutch RE. 2014. Paramyxovirus glycoprotein incorporation, assembly and budding: a three way dance for infectious particle production. Viruses 6:3019–3054. doi: 10.3390/v6083019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tahara M, Takeda M, Yanagi Y. 2007. Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion. J Virol 81:6827–6836. doi: 10.1128/JVI.00248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naim HY, Ehler E, Billeter MA. 2000. Measles virus matrix protein specifies apical virus release and glycoprotein sorting in epithelial cells. EMBO J 19:3576–3585. doi: 10.1093/emboj/19.14.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alldinger S, Baumgartner W, Orvell C. 1993. Restricted expression of viral surface proteins in canine distemper encephalitis. Acta Neuropathol 85:635–645. doi: 10.1007/BF00334674. [DOI] [PubMed] [Google Scholar]
- 50.Bharaj P, Wang YE, Dawes BE, Yun TE, Park A, Yen B, Basler CF, Freiberg AN, Lee B, Rajsbaum R. 2016. The matrix protein of Nipah virus targets the E3-ubiquitin ligase TRIM6 to inhibit the IKKepsilon kinase-mediated type-I IFN antiviral response. PLoS Pathog 12:e1005880. doi: 10.1371/journal.ppat.1005880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pentecost M, Vashisht AA, Lester T, Voros T, Beaty SM, Park A, Wang YE, Yun TE, Freiberg AN, Wohlschlegel JA, Lee B. 2015. Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins. PLoS Pathog 11:e1004739. doi: 10.1371/journal.ppat.1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang YE, Park A, Lake M, Pentecost M, Torres B, Yun TE, Wolf MC, Holbrook MR, Freiberg AN, Lee B. 2010. Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog 6:e1001186. doi: 10.1371/journal.ppat.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coleman NA, Peeples ME. 1993. The matrix protein of Newcastle disease virus localizes to the nucleus via a bipartite nuclear localization signal. Virology 195:596–607. doi: 10.1006/viro.1993.1411. [DOI] [PubMed] [Google Scholar]
- 54.Peeples ME, Wang C, Gupta KC, Coleman N. 1992. Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. J Virol 66:3263–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duan Z, Song Q, Wang Y, He L, Chen J, Zhu Y, Hu S, Liu X. 2013. Characterization of signal sequences determining the nuclear export of Newcastle disease virus matrix protein. Arch Virol 158:2589–2595. doi: 10.1007/s00705-013-1769-5. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida T, Nagai Y'Yoshii S, Maeno K, Matsumoto T. 1976. Membrane (M) protein of HVJ (Sendai virus): its role in virus assembly. Virology 71:143–161. doi: 10.1016/0042-6822(76)90101-X. [DOI] [PubMed] [Google Scholar]
- 57.Bauer A, Neumann S, Karger A, Henning AK, Maisner A, Lamp B, Dietzel E, Kwasnitschka L, Balkema-Buschmann A, Keil GM, Finke S. 2014. ANP32B is a nuclear target of henipavirus M proteins. PLoS One 9:e97233. doi: 10.1371/journal.pone.0097233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol 79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duan Z, Hu Z, Zhu J, Xu H, Chen J, Liu H, Hu S, Liu X. 2014. Mutations in the FPIV motif of Newcastle disease virus matrix protein attenuate virus replication and reduce virus budding. Arch Virol 159:1813–1819. doi: 10.1007/s00705-014-1998-2. [DOI] [PubMed] [Google Scholar]
- 61.Battisti AJ, Meng G, Winkler DC, McGinnes LW, Plevka P, Steven AC, Morrison TG, Rossmann MG. 2012. Structure and assembly of a paramyxovirus matrix protein. Proc Natl Acad Sci U S A 109:13996–14000. doi: 10.1073/pnas.1210275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leyrat C, Renner M, Harlos K, Huiskonen JT, Grimes JM. 2014. Structure and self-assembly of the calcium binding matrix protein of human metapneumovirus. Structure 22:136–148. doi: 10.1016/j.str.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forster A, Maertens GN, Farrell PJ, Bajorek M. 2015. Dimerization of matrix protein is required for budding of respiratory syncytial virus. J Virol 89:4624–4635. doi: 10.1128/JVI.03500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McPhee HK, Carlisle JL, Beeby A, Money VA, Watson SM, Yeo RP, Sanderson JM. 2011. Influence of lipids on the interfacial disposition of respiratory syncytial virus matrix protein. Langmuir 27:304–311. doi: 10.1021/la104041n. [DOI] [PubMed] [Google Scholar]
- 65.Liljeroos L, Huiskonen JT, Ora A, Susi P, Butcher SJ. 2011. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc Natl Acad Sci U S A 108:18085–18090. doi: 10.1073/pnas.1105770108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrison MS, Sakaguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol 42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaetano J. 2013. Holm-Bonferroni sequential correction: an EXCEL calculator (1.1). https://www.researchgate.net/publication/236969037_Holm-Bonferroni_Sequential_Correction_An_EXCEL_Calculator. Accessed May 2017.
- 68.Stettler M, Beck K, Wagner A, Vandevelde M, Zurbriggen A. 1997. Determinants of persistence in canine distemper viruses. Vet Microbiol 57:83–93. doi: 10.1016/S0378-1135(96)01281-3. [DOI] [PubMed] [Google Scholar]