ABSTRACT
Microtubules (MTs) form a rapidly adaptable network of filaments that radiate throughout the cell. These dynamic arrays facilitate a wide range of cellular processes, including the capture, transport, and spatial organization of cargos and organelles, as well as changes in cell shape, polarity, and motility. Nucleating from MT-organizing centers, including but by no means limited to the centrosome, MTs undergo rapid transitions through phases of growth, pause, and catastrophe, continuously exploring and adapting to the intracellular environment. Subsets of MTs can become stabilized in response to environmental cues, acquiring distinguishing posttranslational modifications and performing discrete functions as specialized tracks for cargo trafficking. The dynamic behavior and organization of the MT array is regulated by MT-associated proteins (MAPs), which include a subset of highly specialized plus-end-tracking proteins (+TIPs) that respond to signaling cues to alter MT behavior. As pathogenic cargos, viruses require MTs to transport to and from their intracellular sites of replication. While interactions with and functions for MT motor proteins are well characterized and extensively reviewed for many viruses, this review focuses on MT filaments themselves. Changes in the spatial organization and dynamics of the MT array, mediated by virus- or host-induced changes to MT regulatory proteins, not only play a central role in the intracellular transport of virus particles but also regulate a wider range of processes critical to the outcome of infection.
KEYWORDS: virus, cytoskeleton, microtubules, nucleation, microtubule-associated proteins, plus-end-tracking proteins
INTRODUCTION
The dense cytosolic environment poses a major barrier to the free movement of macromolecules, making microtubule (MT)-based transport a critical aspect of virus replication. Indeed, almost as soon as these filaments were identified and characterized, virus particles were observed proximal to “microtubuli,” with evidence that MT-associated proteins (MAPs) might mediate this association. Chemicals that disrupt MT networks, still commonly used today, were reported to suppress virus infection. Moreover, either infection or expression of viral proteins was observed to alter the organization of these networks, suggesting that MTs not only facilitated infection but were also likely to be actively manipulated by viruses. In subsequent decades an enormous body of work helped unravel how virus particles exploit MT motors to traffic within infected cells, and we direct readers to a recent review of this subject (1). Here, we discuss recent advances in our understanding of how MTs themselves are regulated and function during infection, limiting this minireview to some illustrative examples in each case.
TWO ENDS TO THE STORY: THE BASICS OF MT POLARITY AND ORGANIZATION
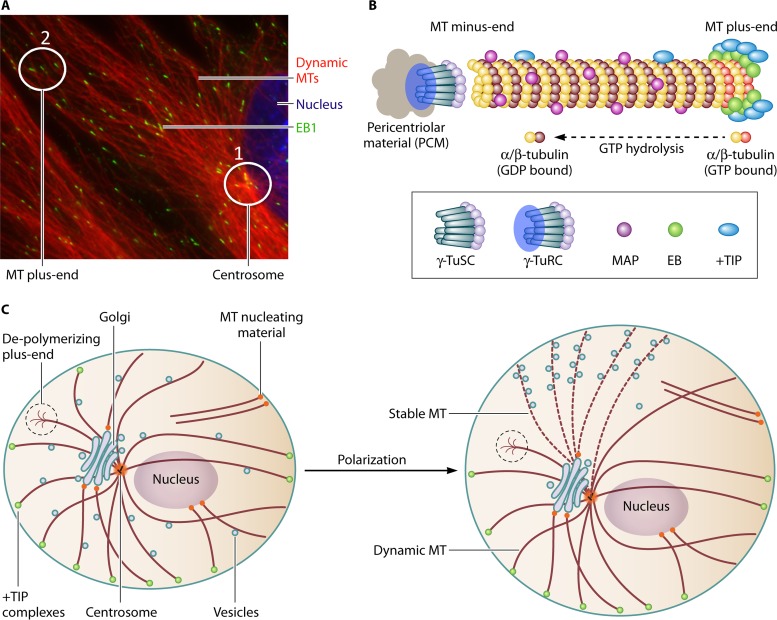
MTs form through the polymerization of α/β-tubulin heterodimers into polarized filaments that have minus and plus ends (Fig. 1A and B). MTs nucleate through minus-end seeding at MT-organizing centers (MTOCs), such as centrosomes (2). The centrosome core contains six γ-tubulin complex proteins (GCPs 1 to 6). GCPs 1 to 3 form the γ-tubulin small complex (γ-TuSC), which binds α-tubulin, and GCPs 4 to 6 assemble individual γ-TuSCs into a γ-tubulin ring complex (γ-TuRC), which acts as a template for MT formation. Surrounding the γ-TuRC is the pericentriolar matrix (PCM), an amorphous layer of proteins that regulate centrosome structure, positioning, and function. Many of us (with virologists particularly guilty!) casually refer to the centrosome as “the MTOC.” However, despite being the most prominent organizing center in many cell types, it is now well established that cells can contain several noncentrosomal nucleation sites, which include kinetochores, the Golgi apparatus, the nuclear envelope, regions of the plasma membrane, and even MTs themselves (3). Finally, MTs can also be severed from MTOCs to either depolymerize or attach at other sites, providing for a complex organization to the MT array and a diverse range of MTOCs with the potential to play important roles in infection.
FIG 1.
MT organization and regulation. (A) A primary fibroblast fixed and stained for EB1 and dynamic (tyrosinated) MTs. The nucleus is stained with Hoechst stain. 1, focal point of MTs emanating from the centrosome. 2, EB1 forms a “comet-like” staining pattern as it tracks growing MT plus ends. (B) Schematic overview of core features of MT nucleation at the centrosome. γ-TuSCs are organized into γ-TuRCs surrounded by pericentriolar material. γ-Tubulin binds α-tubulin at the minus end, while plus ends grow through the addition of new α/β-tubulin dimers loaded with GTP, which is rapidly hydrolyzed as the filament polymerizes. MAPs associate with the MT lattice and regulate stability. +TIPs can bind MTs but accumulate at MT plus ends through interactions with EB family members (EB1 to -3) that recognize GTP-tubulin. (C) Illustration showing how cells contain a mix of growing and depolymerizing MTs that can nucleate from multiple MTOCs, including the centrosome, Golgi apparatus, plasma membrane, and nucleus. Polarization of the MT array involves the stabilization of MT subsets captured at the plasma membrane in response to localized environmental signals. Stable MTs serve as specialized tracks that sort organelles and vesicles to specific regions of the cell.
After nucleating, MTs radiate from MTOCs, alternating rapidly between phases of growth, pause, or catastrophe largely through the addition or loss of α/β-tubulin at their more dynamic plus ends. This dynamic behavior allows MTs to continuously explore the intracellular environment and engage targets, such as organelles or the cell periphery, through a process of “search and capture.” The behavior and stability of MTs is regulated by MT-associated proteins (MAPs), a broad category of proteins that include motors and MT regulators. While many nonmotor MAPs associate along the length of the MT lattice and promote MT stabilization, a specialized subset of MAPs called end-binding (EB) proteins autonomously “tip-track” by specifically recognizing GTP-tubulin at growing MT plus ends before hydrolysis to GDP-tubulin (Fig. 1B) (4). EB proteins, comprising three family members, contain N-terminal regions sufficient to track MT plus ends and promote MT polymerization, while C-terminal domains mediate EB dimerization and binding to other proteins. EB-interacting proteins include other plus-end-tracking proteins (+TIPs), many of which can independently bind tubulin and MTs. However, their specific accumulation at growing MT plus ends requires binding to EBs. Different EB-interacting +TIPs fulfill different functions, such as linking MTs to targets or controlling MT growth and collapse (4). As such, EBs act as master regulators of MT plus-end behavior and function. +TIPs can respond to environmental cues to mediate changes in MT dynamics, including signaling through Rac1, glycogen synthase kinase 3β (GSK3β), or RhoA-diaphanous (Rho-Dia) pathways. Some of these signals trigger the stabilization of MT subsets, often to form a polarized array within cells that facilitates vesicular compartmentalization or the organization of cell surface proteins (5) (Fig. 1C). Stabilized MTs acquire distinguishing posttranslational modifications, including detyrosination or α-tubulin acetylation (5). These modifications do not impart stability but rather mark stable MTs, and they can be recognized by specific kinesin motors to serve as specialized tracks for vesicle trafficking. It is worth noting that the inherent polarity of both MT filaments and the cell itself is an important factor for infection in vivo, yet this polarity is often not recapitulated in cell culture systems.
FIRST ENCOUNTERS: CELL SURFACE INTERACTIONS AND CELL SIGNALING
Although they are intracellular, MTs influence infection from the outset. Viruses often attach to cells and enter at specific sites due to the spatial organization of surface receptors and entry factors, controlled in part by polarization of the MT array. For example, herpes simplex virus 1 (HSV-1) infects polarized epithelial cells basolaterally but not apically, unless cell-cell contacts are disrupted through depletion of extracellular calcium (6). Infection by HSV-1 in both cases requires MTs. MTs also function in human immunodeficiency virus type 1 (HIV-1) transcytosis, a form of particle “handoff” that underlies selective spread of chemokine receptor 5 (CCR5)-tropic HIV-1 in the upper gastrointestinal tract. This occurs because intestinal jejunal cells express CRR5 and glactosylceramide, an alternate HIV-1 receptor, which allows R5 HIV-1 to be endocytosed and transferred to CCR5-positive target cells in an MT-dependent manner (7). Viral attachment to cells through specific receptors also has the potential to trigger signal pathways that alter MT dynamics. Engagement of integrin-dependent focal adhesions by the glycoprotein B of Kaposi's sarcoma-associated herpesvirus (KSHV), a large DNA virus, induces rapid cytoskeletal remodeling that includes MT stabilization (8, 9). Notably, the cellular protein ezrin has been implicated as one of a number of effector targets for these early KSHV-induced cytoskeletal rearrangements. Ezrin-radixin-moesin (ERM) family members regulate actin-MT cross talk and MT stability. Similar to the case for KSHV, early stages of infection by HIV-1 requires focal adhesion proteins and ERM family members, at least in part through their ability to promote stable MT formation (10, 11). In contrast, hepatitis C virus (HCV) infection is negatively regulated by ERMs (12), in line with suggestions that HCV does not exploit stable MTs during early infection (13). As such, not only do MTs function in the organization of surface receptors used for cell attachment, but receptor engagement by viruses in turn influences MT dynamics to facilitate the very earliest stages of infection.
VIRUS ENTRY AND INTERNALIZATION
Viruses utilize diverse entry processes or combinations thereof. While some undergo membrane fusion that deposits capsids directly into the cytosol, others delay cytosolic exposure by trafficking within compartments such as endosomes. Many of the latter viruses initially exploit host sorting pathways that themselves require MTs. Intriguingly, the poliovirus (PV) receptor (PVR) (Necl-5/CD155) not only stimulates MT growth toward the plasma membrane, even outside the context of infection, but also functions in an MT-dependent manner during postattachment virus internalization. After entry by endocytosis, the cytoplasmic domain of PVR interacts with the dynein light chain, Tctex-1, to regulate retrograde transport on MTs (14, 15). MT depolymerization not only disrupts endosome trafficking, but can affect endosomal escape of virus particles and virus trafficking from early endosomes to lysosomes, as is the case for viruses such as Semliki Forest virus and adeno-associated virus type 2 (16, 17). Influenza virus translocates toward the nuclear periphery in a dynein-dependent manner, where initial acidification to exit from late endosomes occurs at the centrosome (18, 19). In this case, histone deacetylases control tubulin acetylation, and histone deacetylase 8 (HDAC8) inhibition decreases centrosome-associated MTs, suppresses centripetal endosome transport, and inhibits influenza virus infection (19).
Other routes of virus entry involve specific plasma membrane structures. Rhinoviruses, for example, rapidly activate MT-dependent transport of intracellular sphingomyelinase to the plasma membrane to facilitate their entry through cholesterol-rich lipid rafts (20). Interestingly, lipid rafts of endothelial cells are thought to facilitate changes in MT dynamics induced by KSHV (21), which, as discussed above, also involves integrin engagement. This highlights the complexity of underlying strategies used even by individual viruses to influence MT dynamics over the course of virus attachment and entry. Although not essential, unusual invaginations of the plasma membrane, called caveolae, can also function in the entry of the nonenveloped polyomavirus simian virus 40 (SV40). SV40 has been found to actively stimulate MT-dependent motility of caveolae to promote infection (22, 23).
After escaping endosomes to penetrate the cytosol, viruses can modulate microtubule dynamics to promote transport to the nucleus. For example, adenovirus (Ad) transiently activates protein kinase A (PKA) and p38 mitogen-activated protein kinase (p38MAPK) signaling to enhance dynein-mediated transport (24). Ad also activates Rac1 signaling that increases MT growth toward the cell periphery. This may play a role in promoting the formation of stable MT networks, which often form through initial growth and subsequent capture at the plasma membrane, that might be exploited for Ad transport (25). However, this increase in MT growth has also been proposed to enhance the ability of MTs to capture incoming Ad particles (26). A more detailed plus-end capture mechanism involving specific +TIPs has recently been shown for HSV-1. Intriguingly, HSV-1 particles directly bind the inward- and outward-directed motors, dynein and kinesin, as well as dynactin 1 (DCTN1) (27). Although DCTN1 binding has been proposed to stimulate dynein processivity to enhance infection (28), virus association with DCTN1 also plays a key role in the capture of HSV-1 particles by dynamic MT plus ends (29). In this capture process, EB1 bridges DCTN1 to MT plus ends through the +TIP cytoplasmic linker protein 170 (CLIP170). Depletion of EB1, CLIP170, or DCTN1 results in a near-complete block to HSV-1 transport after entry (29). These findings suggest that, for at least some viruses, particles do not randomly engage MTs but are captured by specific MT plus-end-associated complexes to initiate retrograde transport. Foot-and-mouth disease virus (FMDV) nonstructural protein 3A (NSP3A) binds and colocalizes with DCTN3 early in infection. This interaction with DCTN3 is required for virulence in cattle or cultured primary kidney cells but not in transformed kidney cell lines (30). Intriguingly, during serial passage, NSP3A mutants that reacquired virulence had mutated to reacquire DCTN3 binding (30). This underscores the importance of DCTN3 to FMDV infection in vivo and in normal cells and highlights potential dangers of studying transformed cell lines.
While viruses such as HSV-1 exploit +TIPs for capture by dynamic MTs, other viruses target +TIPs in a different manner during early infection. Soon after entry, the HIV-1 capsid (CA) protein binds the EB1-associated +TIP Kif4 to induce MT stabilization, and this is critical for particle translocation to the nucleus (31). Indeed, that study provided the first direct evidence that EB1 and +TIPs function in virus infection. HIV-1 CA also interacts with MAP1A and MAP1S, host factors that also promote early infection (32). While it was proposed that these MAPs either facilitate MT stabilization or mediate virus trafficking by tethering particles to MTs, these MAPs do not have motor activity. Most likely, MAP1A/1S and EB1-mediated MT stabilization provides more long-lived and potentially selective networks for HIV-1 to cross the cytosol. Notably, HIV-1 does not appear to bind MT motors directly but instead uses a kinesin adaptor to regulate its bidirectional movement (33). Although it is normally an outward motor, kinesin-1 depletion blocks HIV-1 transport to the nucleus and early infection (33, 34). Kinesins may facilitate retrograde HIV-1 transport on stable MTs, as kinesins can exhibit selectivity toward acetylated or detyrosinated filaments. HIV-1 undergoes a complex series of events during early infection. Reverse transcription of the viral genome and disassembly of the capsid shell, a poorly understood process termed “uncoating,” are intricately linked to bidirectional virus transport on MTs and may be aided by the forces generated by opposing motors (34, 35).
Many viruses mentioned so far, including retroviruses, adenoviruses, and herpesviruses, have been observed to accumulate at the centrosome as they journey to the nucleus. Intriguingly, HIV-1 has been reported to establish a latent state at the centrosome in quiescent T cells that can last several weeks, and it can return to productive infection after T cell stimulation (36). Even at the centrosome, viruses such as HSV-1 appear to exploit +TIPs such as dystonin to make the final transition from the perinuclear centrosome to the nuclear membrane (37). In effect, this is a brief anterograde move from the MT nucleation site outward to the nucleus. Nearing the end of their journey to the nucleus, adenoviruses appear to exploit the nuclear export factor CRM1/XPO1 (chromosome region maintenance-1/exportin-1) to regulate a switch from fast MT-dependent transport toward the nucleus to slower MT-independent motion for nuclear entry (38). Interestingly, during its nuclear entry process, human papillomavirus 16 (HPV16) associates with MTs at the trans-Golgi network (TGN) to enter at the onset of mitosis, where it resides within unusual membranous vesicles until the nuclear envelope reforms (39). As such, after entry, viruses use a diverse array of strategies to modulate their movement through the cytoplasm to reach their subcellular sites of replication or, in some cases, sites of transient latency.
CYTOPLASMIC REPLICATION: SPECIALIZED COMPARTMENTS NEED MTs TOO
While some RNA viruses and most mammalian DNA viruses must reach the nucleus in order to replicate, for others replication occurs completely in the cytoplasm. Cytoplasmic replication might predict that these viruses would have a more limited dependence on MT arrays, but this is not the case.
Poxviruses such as vaccinia virus (VacV), and African swine fever virus (ASFV), the sole asfivirus family member, are unusually self-sufficient DNA viruses that replicate within cytoplasmic compartments called viral “factories.” Upon entry, poxvirus cores exude mRNAs produced by their own transcription system. These mRNAs are organized into ribosome-dense punctate structures aligned with MTs and depend on MTs for their formation (40). Small early factories move in an MT-dependent manner toward the nuclear periphery to form larger factories, driven by MT-mediated changes in cell contractility and motility as opposed to the activity of dynein (41). Indeed, early in infection MTs retract, and cell rounding gathers organelles proximal to the nucleus (42). Once infection enters later stages, cells that had rounded earlier expand once again coincident with the regrowth of MTs. VacV encodes at least three proteins that modulate MT behavior at these later stages. The F11 protein stimulates MT growth by inhibiting RhoA-Dia signaling that stabilizes MTs (43). Two other proteins, A10 and L4, have MAP-like activities and generate stable MTs (44). Given how stable MTs form, through growth and capture at sites such as the plasma membrane, these three proteins may function together to generate stable MT arrays or may regulate distinct MT subsets that serve different purposes in the viral replication cycle. Interestingly, actin rearrangements observed in VacV-infected cells are controlled by changes in MT dynamics, highlighting the complexity of broader cytoskeletal regulation during infection (42). Three forms of infectious poxvirus progeny are produced, two of which are transported away from factories to the cell surface on MTs (45–49). ASFV also exploits MTs to exit replication sites and reach the plasma membrane (50). Finally, distinct from factories, some orthopoxviruses also form large structures called A-type inclusions (ATIs) that embed virions and protect infectivity after cell lysis. MTs not only mediate ATI formation through motility and coalescence of their core components but also mediate the transport and embedding of virus particles within these structures (51).
Many RNA viruses also utilize diverse cytoplasmic replication structures that rely on MTs to form and function. Noroviruses, which cause gastroenteritis, form membrane-derived replication complexes adjacent to “the MTOC” (52). This process involves the activity of at least two virus-encoded proteins. The viral nonstructural NS3 protein can induce formation of motile lipid-derived structures, while reorganization of acetylated MTs that may facilitate replication compartment formation appears to involve the structural viral protein VP1 (52, 53). Rotavirus, a double-stranded RNA (dsRNA) virus, forms “viroplasms,” which begin as many small structures that coalesce into larger ones in perinuclear regions through interactions between two viral nonstructural proteins, NSP2 and NSP5, with MTs (54, 55). Nocodazole blocks the movement and fusion of structures to form viroplasms, which, like norovirus replication complexes, appear to embed in acetylated MTs to maintain structural integrity (55, 56). Reoviruses replicate in filamentous inclusions formed at perinuclear sites. Interestingly, different strains of reovirus produce multifunctional mu2 proteins that either do or do not possess MAP-like activity, and this correlates with the formation of either MT-colinear filamentous or globular inclusion bodies, respectively (57). Thus, mu2 regulation of stable MTs may be a key determinant in the organization of cytoplasmic reovirus replication and assembly sites. Flaviviruses also form replication compartments. For example, Zika virus forms endoplasmic reticulum (ER)-membrane invaginations similar to those of its relative, dengue virus, reorganizing MTs into cages around replication compartments (58). HCV replication compartments consist of both large “membranous webs” that exhibit limited motility and smaller compartments that are motile. MTs maintain these two discrete compartment types, both of which are required for HCV replication (59–61). The association of replication compartments with MTs involves the viral nonstructural proteins NS3 and NS5A (59–61). HCV progeny are assembled on lipid droplets that are brought proximal to replication compartments. This is controlled, at least in part, by the HCV core protein, which is sufficient to coat lipid droplets and control their motility, causing MT-dependent aggregation at “the MTOC” (62, 63). HCV also exploits septin 9, a multifunctional GTP- and membrane-binding protein that interacts with MTs to promote lipid droplet formation (64). Collectively, these effects likely enhance interactions between replication compartments and lipid droplets at a perinuclear region. As part of the virus assembly process, the viral NS2 and NS3-4A proteins are thought to promote core trafficking from large, immotile lipid droplets during assembly (65). Once assembled, progeny HCV is then transported to the cell surface along MTs in endosomes (66, 67). As such, HCV represents an excellent example of the enormous complexity of MT functions in the replication of viruses that do not even need to reach the nucleus.
The complex role played by MTs in the cytoplasmic replication of viruses becomes even more apparent when one considers that the positive-strand RNA genome of viruses such as HCV or PV serves as a template for both negative-strand RNA synthesis and translation. These are two incompatible processes that cannot occur simultaneously on the same RNA molecule, and MTs facilitate the spatial separation of these processes. Upon infection, newly made PV protein and negative-sense RNA transport to perinuclear regions, followed by the production and translation of more positive-sense RNA strands at the ER. As this occurs, ER-proximal vesicles containing negative-sense RNA form and are transported to perinuclear regions (68). This active transport between the ER and perinuclear regions then establishes discrete sites of translation and replication, respectively. Demonstrating this, transcriptional inactivation followed by reversal results in two distinct vesicles types: those trapped at the ER/Golgi apparatus, which are engaged exclusively in translation, cannot resume replication, while those trapped at the nuclear periphery can resume replication by transporting positive-sense RNA back to the ER for translation. For different reasons, MT-dependent transport and compartmentalization of RNA also occur during infection by vesicular stomatitis virus (VSV). VSV has a nonsegmented negative-sense RNA genome that is delivered in a transcription-competent ribonucleoprotein core. Initial synthesis of VSV mRNAs occurs throughout the cytoplasm. However, synthesis of viral proteins induces the formation of inclusions, where viral mRNA synthesis becomes segregated. MTs play a key role in transporting viral RNAs away from inclusions for translation (69). Interestingly, it is unclear whether this segregation of negative-sense RNA transcription represents an optimal condition established by the virus or a host response to infection that must be overcome through MT-dependent transport of viral mRNAs back to regions where they can be translated.
VIRUS REPLICATION AND EGRESS: TIME TO INTRODUCE MORE STABILITY AND A TOUCH OF POLARITY?
Virus assembly and egress are complex and diverse processes that occur at different locations for different viruses. Herpesviruses, for example, replicate and package their DNA genomes into capsids within the nucleus. Capsids then bud from the nucleus and accumulate a layer of proteins (tegument) before acquiring their outer membranes, studded with viral glycoproteins, at the TGN. Although retroviruses have a nuclear stage, virus assembly occurs in the cytoplasm at centrosomes or the plasma membrane. In many instances, however, viruses appear to induce the formation of acetylated MTs later in infection, the functional significance of which may relate to their preferential affinity for outward kinesin motors to enable egress.
Once HSV-1 particles reach the nucleus and establish productive infection, punctate pericentrin staining is disrupted and MTs lose centrosomal focus (70, 71). While disrupting centrosomal function, HSV-1 stabilizes MTs that nucleate at the TGN through the activity of Us3, a viral Akt mimic that regulates broad cytoskeletal rearrangements during infection (70, 72). In particular, formation of stable MT arrays occurs through Us3-mediated activation of cellular proteins called cytoplasmic linker-associated proteins (CLASPs) (70). CLASPs are specialized +TIPs that function in both MT nucleation at the Golgi apparatus and MT capture at the cell periphery. Formation of acetylated MT arrays may be further aided by the HSV-1 protein VP22 (73) and host HDAC6 (74). Another +TIP, dystonin, also regulates HSV-1 transport during egress (75), but the underlying mechanism is currently unknown. However, it is tempting to speculate that dystonin contributes to MT nucleation or stabilization, as it is a +TIP with no known motor function. As outward kinesin motors exhibit a preference for acetylated or detyrosinated MTs, stabilizing MTs that originate from the site of envelope acquisition would ensure movement of new virus particles toward the cell surface (70). Moreover, stable MTs often form through capture at specific regions of the plasma membrane, and herpesvirus egress has been reported to occur at regions enriched for LL55β, an established CLASP-interacting protein (76, 77).
Among RNA viruses that have a nuclear replication stage, many retroviruses transit through the centrosome en route to the cell surface. This plays a complex role in the biology of their assembly, as reviewed by Afonso and colleagues (78). By way of limited examples, foamy virus, mouse mammary tumor virus, and Mason-Pfizer monkey virus assemble at “the MTOC,” where some then acquire their envelope at the TGN (79–81). In contrast, other retroviruses (discussed below) assemble only after their structural proteins and RNA genomes have transited through the centrosome en route to assembly sites. Progeny influenza virus vRNPs exit the nucleus and accumulate at “the MTOC” before being transported in endosomes by MTs and actin (82–84). Interestingly, vRNPs have been proposed to be directed to MTs by YB-1, a host transcriptional and translational regulator, although the underlying mechanism remains unclear (85). Although MT disruption has only small effects on infection, it disperses the normal polarized organization and reduces the motility of vRNPs. Influenza virus also induces MT acetylation, and the degree of acetylation correlates with the degree of influenza virus release, which involves delivery of viral components to the apical plasma membrane (86, 87). As such, polarity of acetylated MT arrays plays a role in the proper release of DNA and RNA viruses from specific regions at the cell surface.
While some viruses assemble prior to transport to the cell surface, others, including paramyxovirus and some retrovirus family members, assemble at specific plasma membrane regions. Many of these have also been found to actively polarize cells for their own purposes. Mutant Sendai virus strains suggest that the viral M protein controls MT organization and polarized budding (88). After exiting the nucleus, HIV-1 genomic RNA accumulates at “the MTOC,” mediated by MTs and the trafficking factor heterogenous ribonucleoprotein A2 (hnRNPA2) (89). HIV-1 assembly occurs at the plasma membrane after both the viral RNA and Gag polyprotein are transported to the cell surface (90, 91). At this stage of infection, the host protein suppressor of cytokine signaling 1 (SOCS1) colocalizes with the viral Gag polyprotein on MTs and promotes both MT stabilization and virus production, effects that are blocked by disrupting MTs with nocodazole or expression of stathmin, a host MT-depolymerizing factor (92). Notably, both unassembled and assembled forms of Gag are transported along MTs to the plasma membrane (91). Intriguingly, in macrophages, a natural target cell type for HIV-1 infection in vivo, assembly occurs in virus-containing compartments whose spatial organization is controlled by MTs. HIV-1 release from macrophages requires the kinesin Kif3A, which is dispensable in T cells, where the virus does not employ virus-containing compartments (93). This suggests cell-specific replication strategies for HIV-1 and distinct roles for MTs in each case. In lymphocytes, polarization of the MT array may facilitate HIV-1 cell-cell spread though virological synapses and polysynapses (94, 95). During infection by another retrovirus, human T cell leukemia virus type 1 (HTLV-1), intercellular contact polarizes MTs in the infected cell to the cell-cell junction, with the viral genome and Gag localizing to this contact site (96). Infected-cell polarization involves reorientation of “the MTOC” toward the site of contact with the uninfected cell and is mediated by the combined action of the viral protein Tax and intracellular adhesion molecule 1 (ICAM-1) signaling (97). These examples illustrate how polarization and stabilization of MT arrays contribute to the assembly and egress of virus particles at specific regions of the plasma membrane, as well as the formation of intercellular connections or synapses that facilitate virus spread.
THE CENTROSOME: TRANSIT AND ORGANIZATION CENTER OR DANGER ZONE?
In discussing different viral replication strategies, we have touched several times on viruses that accumulate at the centrosome as they enter or exit the cell. The centrosome is a primary focal point of MTs in many cell types and plays diverse roles in various stages of virus replication that go beyond simply serving as a transit station for virus particles.
As mentioned above, initial acidification and endosome escape for viruses such as influenza virus occur at the centrosome, and centrosome “splitting” suppresses influenza virus infection (18, 19). While some viruses, such as retroviruses (discussed above), exploit the centrosome as an assembly site or reorient it during cell polarization, others actively alter its organization for other purposes. Herpesviruses, poxviruses, and ASFV reduce the punctate localization of centrosomal proteins (44, 70, 71, 98). While often broadly described as centrosome damage or disruption, it is most likely that regulatory or nucleation factors are displaced rather than that the entire structure is destroyed. Indeed, the FMDV nonstructural protease NSP3Cpro reduces MT tethering in a manner associated with a specific loss of γ-tubulin but not pericentrin at centrosomes (99). Disrupting centrosome function may serve multiple purposes, such as switching MT nucleation to sites where the virus is replicating or acquiring its envelope prior to egress, as is the case for HSV-1 (70). Alternatively, targeting the centrosome may counter antiviral responses. It has been suggested that the cell may view viruses as misfolded proteins, which are routed to the centrosome destined for degradation. Indeed, clusters of Ad5 observed at the centrosome and Golgi apparatus do not appear to represent the infectious pathway (100). In line with an antiviral defense, once allowed to accumulate at the centrosome as part of their critical journey from the cell periphery toward the nucleus, transduction by adeno-associated virus vectors can be enhanced by then disrupting MTs and the centrosome, where virus particles become trapped (101). The centrosome is also a site of antigen processing (102), so disrupting its organization may dampen host antiviral responses.
The centrosome also serves as a site for the accumulation of aggresomes, which are involved in protein turnover and autophagy. Aggresomes not only function in host defense but may also be exploited by viruses as sites of replication (103). Late in Ad infection, cellular DNA repair enzymes can join viral genomes into concatemers and inhibit formation of infectious particles. To prevent this, Ad5-encoded E4orf3 protein inactivates core components of host double-strand break repair complexes by targeting them to centrosome-resident aggresomes in an MT- and dynein-dependent manner (104, 105). Influenza virus exploits aggresomes to promote infection by mimicking misfolded proteins to recruit HDAC6 and other aggresome factors to viral fusion sites (106), although HDAC6 has also been suggested to negatively impact influenza virus release due to effects on acetylated MTs that likely mediate egress (107). ASFV replication sites form at the centrosome and resemble aggresomes, potentially using the aggresome pathway for virus assembly (108). Finally, PV induces autophagosome-like structures that tether to MTs and influence the rate at which PV can be released from cells through a process termed autophagosome-mediated exit without lysis (AWOL) (109). As such, although aggresomes may seem “dangerous to play with,” several viruses have co-opted their functions in an MT-dependent manner to benefit virus replication in a variety of ways.
MODULATION OF HOST BEHAVIOR AND ANTIVIRAL RESPONSES
Manipulation of MTs by viruses not only is important for their intracellular trafficking and formation of replication compartments but also enables them to control broader aspects of infected-cell behavior. Virus-encoded factors such as the VacV F11 protein enhances MT dynamics to increase cell motility, thereby promoting virus spread (43, 110). Viruses also exploit MTs to control organelle structure and localization. Both DNA and RNA viruses have been found to cause MT-dependent changes in the spatial organization of mitochondria (42, 111, 112), including the recruitment of mitochondria to ASFV replication compartments (113). This potentially redirects energy supplies to sites of virus replication or away from the host cell. PV disrupts the Golgi apparatus in an MT-dependent manner (114). Another picornavirus, FMDV, encodes a protease called 3C that inhibits MT growth and also causes Golgi fragmentation (115). Strategies aimed at fragmenting the Golgi apparatus impair secretory pathways and likely play a role in suppressing antigen presentation.
MTs also mediate host responses to infection. Cells can encode specialized antiviral proteins or restriction factors. MTs and dynein facilitate the antiretroviral activity of tripartite motif-containing proteins (TRIMs), a family of proteins that protect cells against retroviral infection, as the efficacy of TRIMs is reduced by MT depolymerization (116, 117). Only low levels of another antiviral, called Fv1 (Friend virus susceptibility gene 1), are required for protection of mice against murine leukemia virus (MLV), but localization to MT-associated TGN tubules is critical for its activity (118). The guanine nucleotide exchange factor GEF-H1 is regulated by MT binding and couples MT and Rho-GTPase activity. MTs sequester GEF-H1 but release it to enable intracellular nucleic acid sensing by retinoic acid-inducible gene 1 (RIG-I)-like receptors, a key pathway for detecting intracellular pathogens (119). MAP4 controls the perinuclear redistribution of butyrophilin subfamily 3 member A1 (BTN3A1), a host protein involved in major histocompatibility complex (MHC)-associated antigen presentation. This redistribution activates interferon (IFN) responses to double-stranded RNA that is produced during the replication of many viruses (120). Given the roles played by these MT-associated proteins in antiviral responses, viruses have evolved countermeasures in order to successfully replicate. For example, to block IFN activation, rabies virus P protein switches the host antiviral signal transducer and Activator of transcription 1 (STAT1) from an MT-facilitated to an MT-inhibited process of nuclear import, blocking it from entering the nucleus and inducing interferon-stimulated gene (ISG) expression (121). HIV-1 encodes several proteins that counter antiviral or immune responses through modulating MT function. In infected macrophages, HIV-1 Vpr binds and interferes with the plus-end localization of EB1-DCTN1 complexes, suppressing centripetal movement and maturation of phagosomes critical to pathogen clearance (122). The HIV-1 trans-activating regulatory protein (Tat) binds the cytoplasmic tail of the interleukin 7 (IL-7) receptor CD127, causing MT-dependent internalization (123). Indeed, this represents just one example of viral strategies that target surface receptors that mediate immune cell responses to infection, and many others exert wider effects on natural killer cell activation and formation of cytolytic granules through their effects on MTs.
Besides blocking host antiviral responses, some viruses can also cause cellular transformation by dysregulating cell division. While these are too numerous to include in this minireview, SV40 represents a good example of the underling complexity of the process and potential roles for MTs. The SV40 large T antigen (TAg) binds MTs and affects p53 signaling (124). Moreover, by interacting with the MT stabilizing factor transforming acid coiled-coil-containing protein 2 (TACC2), TAg leads to disorganized mitotic spindles and chromosome missegregation (125, 126). SV40 TAg may also affect the nuclear cytoskeleton through interactions with MAP1 (127). SV40 also encodes another protein, small T antigen, that affects MT stability and induces chromosome missegregation through interactions with protein phosphatase 2A (PP2A) (128, 129). Other oncogenic viruses, such as Merkel cell polyomavirus, hepatitis B virus, Epstein-Barr virus, and HTLV-1, encode proteins that also cause centrosome and MT abnormalities that appear to underlie at least some of their effects on cell division and transformation.
MT FUNCTIONS DURING INFECTION OF PLANTS, INSECTS, AND BACTERIA
MT organization in plants is strikingly different from that in other kingdoms, including the transient formation of unusual preprophase bands during cell division and the use of specialized MAPs (130, 131). A variety of plant viruses encode what are termed “movement proteins” (MPs), which are involved in the transport of viral proteins, RNAs, replication compartments, and virions (130, 132). MPs have been found to bind plant MAPs, which negatively or positively regulate infection, as well as to mimic host MAP functions to stabilize plant MTs (133–136). Plant MPs have also been found to interact with +TIPs. Tobacco mosaic virus (TMV) MP, for example, interacts with Arabidopsis thaliana EB1a, and dynamic MTs promote TMV spread (137, 138). Other commonalities with mammalian viruses include MPs that affect centrosome function (135). Perhaps most striking about plant MTs, however, is their role in vector transmission. Cauliflower mosaic virus (CaMV) is spread by aphids. CaMV forms intracellular replication compartments as well as a specialized transmission body in its plant host. CaMV aphid transmission factor (ATF) forms paracrystalline structures that bind and stabilize MTs (139). Remarkably, transmission bodies react within seconds to the presence of aphids on the plant by rapid disruption and redistribution of the core transmission body helper protein, P2, onto MTs, driving the release of virions from the transmission body and factories onto MTs. Within minutes of the aphid leaving, virions return to factories and the transmission body, which rapidly reforms in a form of vector “perceptive behavior” through the plant host (140–142). Horizontal transmission of viruses between insects may also involve MT-regulated structures called occlusion bodies. The baculovirus Autographa californica nucleopolyhedrovirus (AcMNPV) encodes an auxiliary protein, P10, that associates with and restructures its insect host's MTs. P10 forms tubular structures that depend upon MTs for their assembly and may regulate occlusion body release during virus transmission (143). Finally, bacterial FtsZ proteins, tubulin-like homologs, form short cytomotive protofilaments that function in cell division, with structures very different from those of eukaryotic MTs. However, some bacteriophages encode tubulin-like PhuZ, which forms a filamentous, spindle-like array that positions phage DNA at the bacterial cell center for optimal virus replication (144–146). These examples serve to illustrate the broader importance of MTs or MT-like filaments in infection by diverse viruses across kingdoms and species.
CONCLUDING REMARKS
There are many more examples of viruses that alter MT behavior and function, which are unfortunately too numerous to include within the space limitations of this minireview. However, the examples cited serve to illustrate the incredible diversity by which different viruses exploit MT arrays to facilitate their replication. Indeed, given the widespread viral exploitation of these networks, it is surprising to see numerous reports directly question the importance of MTs to infection. Without citing specific cases, most if not all of these studies are based on the use of “poisons” that artificially stabilize or depolymerize MTs, in particular nocodazole. However, there are a number of important caveats to these approaches that often go unconsidered. Stable MTs, which clearly are used by many viruses, are relatively resistant to nocodazole. Surprisingly, some studies treat with drugs for just 2 h before washout for several days to allow for reporter gene expression as the means of measuring infection levels, not accounting for the fact that the effects of nocodazole are reversed within minutes. Moreover, readouts based on viral gene expression without visualizing virus particle behavior may not account for aberrant events involving a small number of particles within a virus population. For example, a single virus entering at a site proximal to the nucleus or Brownian motion of just a few particles may be sufficient to achieve high levels of reporter gene expression, particularly if endpoint assays are performed after several hours or days and not within a reasonable time frame that reflects the period of MT-based movement. Without accounting for significant caveats, results obtained using nocodazole often lead to the suggestion that free diffusion mediates virus transport, not MTs. Given that MTs radiate throughout the cell and often tether organelles, they likely contribute to the crowded environment within the cell. As such, nocodazole may also remove impediments and artificially generate an environment more favorable to diffusion. Caution is needed when using and interpreting findings with MT drugs if they are the sole means of assessing the contribution of MTs to infection, a concern also raised for plant systems (147).
In addition, for reports of MT disruption by viruses it is important to understand the biological context. For example, although Zika virus uses MTs to replicate, it disrupts MT networks, but this occurs only when replication is complete and the cell is dying (58). As such, observations of MT disruption or reorganization in infected cells should be interpreted carefully and in context. Another point is the study of viral proteins outside the context of infection. For example, for a small virus, HIV-1 encodes a remarkable numbers of proteins, too many to cite here, that are reported to promote MT polymerization, stabilization, or disruption. It may seem odd that a virus that actively stabilizes and uses MTs for infection would encode destabilizing factors. Here, again, context is critical. The presence of viral proteins that depolymerize MTs does not mean that the virus does not need MTs, as these factors may work in conjunction with others that promote MT formation as part of a broader strategy to remodel the host MT network during infection. Alternatively, MT disruption may not be a natural function of factors studied outside the context of infection, or disruption may serve other purposes unrelated to virus replication. HIV-1 again provides an ideal example. The HIV-1 Tat protein is reported to promote MT polymerization, stabilization, or disruption in various contexts. Complicating matters, zinc binding changes whether Tat induces MT stabilization or tubulin aggregates (148). Adding yet more layers of complexity, Tat is secretory, and many of its effects are likely to be involved in immune modulation and broader cellular toxicity that contributes to AIDS progression. As such, it is unclear if Tat contributes to the regulation of MTs within the infected cell where replication is occurring and utilizes MTs. Finally, some viruses exploit MAPs or encode tubulin-interacting factors for purposes unrelated to MT regulation. For example, MAP4 depletion suppresses HIV-1 reverse transcription but not nuclear translocation (149), while tubulin is coopted by Sendai virus to control transcription (150). While reports of viral proteins that interact with MT regulators or alter MTs when expressed in cells demonstrate their ability to do so, it is important to know if the observed effects also occur in infected cells and during periods of virus replication versus late-stage cell death to truly understand their role in virus replication.
We have amassed an incredible amount of information about how viruses exploit MT motors, and our understanding of viral manipulation of MTs themselves is rapidly growing. Future studies, in particular of host signaling pathways and downstream effector MAPs, will undoubtedly provide important new insights into the underlying mechanisms by which MTs function during viral infection.
ACKNOWLEDGMENTS
We apologize to all colleagues whose work was not cited due to space limitations or any oversights on our part.
This work was funded by grants from the National Institutes of Health to M.H.N. (R01GM101975 and P01GM105536) and D.W. (P01GM105536).
We declare no competing financial interests.
REFERENCES
- 1.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J 30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kollman JM, Merdes A, Mourey L, Agard DA. 2011. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol 12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petry S, Vale RD. 2015. Microtubule nucleation at the centrosome and beyond. Nat Cell Biol 17:1089–1093. doi: 10.1038/ncb3220. [DOI] [PubMed] [Google Scholar]
- 4.Akhmanova A, Steinmetz MO. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol 16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- 5.Janke C, Bulinski JC. 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 6.Marozin S, Prank U, Sodeik B. 2004. Herpes simplex virus type 1 infection of polarized epithelial cells requires microtubules and access to receptors present at cell-cell contact sites. J Gen Virol 85:775–786. doi: 10.1099/vir.0.19530-0. [DOI] [PubMed] [Google Scholar]
- 7.Meng G, Wei X, Wu X, Sellers MT, Decker JM, Moldoveanu Z, Orenstein JM, Graham MF, Kappes JC, Mestecky J, Shaw GM, Smith PD. 2002. Primary intestinal epithelial cells selectively transfer R5 HIV-1 to CCR5+ cells. Nat Med 8:150–156. doi: 10.1038/nm0202-150. [DOI] [PubMed] [Google Scholar]
- 8.Sharma-Walia N, Naranatt PP, Krishnan HH, Zeng L, Chandran B. 2004. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 envelope glycoprotein gB induces the integrin-dependent focal adhesion kinase-Src-phosphatidylinositol 3-kinase-rho GTPase signal pathways and cytoskeletal rearrangements. J Virol 78:4207–4223. doi: 10.1128/JVI.78.8.4207-4223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naranatt PP, Krishnan HH, Smith MS, Chandran B. 2005. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol 79:1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, Wen Y, Mott C, Gundersen GG, Goff SP. 2007. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. EMBO J 26:41–52. doi: 10.1038/sj.emboj.7601475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown C, Morham SG, Walsh D, Naghavi MH. 2011. Focal adhesion proteins talin-1 and vinculin negatively affect paxillin phosphorylation and limit retroviral infection. J Mol Biol 410:761–777. doi: 10.1016/j.jmb.2011.03.076. [DOI] [PubMed] [Google Scholar]
- 12.Bukong TN, Kodys K, Szabo G. 2013. Human ezrin-moesin-radixin proteins modulate hepatitis C virus infection. Hepatology 58:1569–1579. doi: 10.1002/hep.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roohvand F, Maillard P, Lavergne JP, Boulant S, Walic M, Andreo U, Goueslain L, Helle F, Mallet A, McLauchlan J, Budkowska A. 2009. Initiation of hepatitis C virus infection requires the dynamic microtubule network: role of the viral nucleocapsid protein. J Biol Chem 284:13778–13791. doi: 10.1074/jbc.M807873200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohka S, Matsuda N, Tohyama K, Oda T, Morikawa M, Kuge S, Nomoto A. 2004. Receptor (CD155)-dependent endocytosis of poliovirus and retrograde axonal transport of the endosome. J Virol 78:7186–7198. doi: 10.1128/JVI.78.13.7186-7198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller S, Cao X, Welker R, Wimmer E. 2002. Interaction of the poliovirus receptor CD155 with the dynein light chain Tctex-1 and its implication for poliovirus pathogenesis. J Biol Chem 277:7897–7904. doi: 10.1074/jbc.M111937200. [DOI] [PubMed] [Google Scholar]
- 16.Vonderheit A, Helenius A. 2005. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol 3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao PJ, Samulski RJ. 2012. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J Virol 86:10462–10473. doi: 10.1128/JVI.00935-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu SL, Zhang LJ, Wang ZG, Zhang ZL, Wu QM, Sun EZ, Shi YB, Pang DW. 2014. Globally visualizing the microtubule-dependent transport behaviors of influenza virus in live cells. Anal Chem 86:3902–3908. doi: 10.1021/ac500640u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamauchi Y, Boukari H, Banerjee I, Sbalzarini IF, Horvath P, Helenius A. 2011. Histone deacetylase 8 is required for centrosome cohesion and influenza A virus entry. PLoS Pathog 7:e1002316. doi: 10.1371/journal.ppat.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grassme H, Riehle A, Wilker B, Gulbins E. 2005. Rhinoviruses infect human epithelial cells via ceramide-enriched membrane platforms. J Biol Chem 280:26256–26262. doi: 10.1074/jbc.M500835200. [DOI] [PubMed] [Google Scholar]
- 21.Raghu H, Sharma-Walia N, Veettil MV, Sadagopan S, Caballero A, Sivakumar R, Varga L, Bottero V, Chandran B. 2007. Lipid rafts of primary endothelial cells are essential for Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8-induced phosphatidylinositol 3-kinase and RhoA-GTPases critical for microtubule dynamics and nuclear delivery of viral DNA but dispensable for binding and entry. J Virol 81:7941–7959. doi: 10.1128/JVI.02848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagawa A, Mezzacasa A, Hayer A, Longatti A, Pelkmans L, Helenius A. 2005. Assembly and trafficking of caveolar domains in the cell: caveolae as stable, cargo-triggered, vesicular transporters. J Cell Biol 170:769–779. doi: 10.1083/jcb.200506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert J, Benjamin T. 2004. Uptake pathway of polyomavirus via ganglioside GD1a. J Virol 78:12259–12267. doi: 10.1128/JVI.78.22.12259-12267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suomalainen M, Nakano MY, Boucke K, Keller S, Greber UF. 2001. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J 20:1310–1319. doi: 10.1093/emboj/20.6.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren JC, Rutkowski A, Cassimeris L. 2006. Infection with replication-deficient adenovirus induces changes in the dynamic instability of host cell microtubules. Mol Biol Cell 17:3557–3568. doi: 10.1091/mbc.E05-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren JC, Cassimeris L. 2007. The contributions of microtubule stability and dynamic instability to adenovirus nuclear localization efficiency. Cell Motil Cytoskeleton 64:675–689. doi: 10.1002/cm.20215. [DOI] [PubMed] [Google Scholar]
- 27.Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog 6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dohner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R, Sodeik B. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell 13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jovasevic V, Naghavi MH, Walsh D. 2015. Microtubule plus end-associated CLIP-170 initiates HSV-1 retrograde transport in primary human cells. J Cell Biol 211:323–337. doi: 10.1083/jcb.201505123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gladue DP, O'Donnell V, Baker-Bransetter R, Pacheco JM, Holinka LG, Arzt J, Pauszek S, Fernandez-Sainz I, Fletcher P, Brocchi E, Lu Z, Rodriguez LL, Borca MV. 2014. Interaction of foot-and-mouth disease virus nonstructural protein 3A with host protein DCTN3 is important for viral virulence in cattle. J Virol 88:2737–2747. doi: 10.1128/JVI.03059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabo Y, Walsh D, Barry DS, Tinaztepe S, de Los Santos K, Goff SP, Gundersen GG, Naghavi MH. 2013. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez J, Portilho DM, Danckaert A, Munier S, Becker A, Roux P, Zambo A, Shorte S, Jacob Y, Vidalain PO, Charneau P, Clavel F, Arhel NJ. 2015. Microtubule-associated proteins 1 (MAP1) promote human immunodeficiency virus type I (HIV-1) intracytoplasmic routing to the nucleus. J Biol Chem 290:4631–4646. doi: 10.1074/jbc.M114.613133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malikov V, da Silva ES, Jovasevic V, Bennett G, de Souza Aranha Vieira DA, Schulte B, Diaz-Griffero F, Walsh D, Naghavi MH. 2015. HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nat Commun 6:6660. doi: 10.1038/ncomms7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukic Z, Dharan A, Fricke T, Diaz-Griffero F, Campbell EM. 2014. HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol 88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawlica P, Berthoux L. 2014. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses 6:4195–4211. doi: 10.3390/v6114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamborlini A, Lehmann-Che J, Clave E, Giron ML, Tobaly-Tapiero J, Roingeard P, Emiliani S, Toubert A, de The H, Saib A. 2007. Centrosomal pre-integration latency of HIV-1 in quiescent cells. Retrovirology 4:63. doi: 10.1186/1742-4690-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McElwee M, Beilstein F, Labetoulle M, Rixon FJ, Pasdeloup D. 2013. Dystonin/BPAG1 promotes plus-end-directed transport of herpes simplex virus 1 capsids on microtubules during entry. J Virol 87:11008–11018. doi: 10.1128/JVI.01633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang IH, Burckhardt CJ, Yakimovich A, Morf MK, Greber UF. 2017. The nuclear export factor CRM1 controls juxta-nuclear microtubule-dependent virus transport. J Cell Sci doi: 10.1242/jcs.203794. [DOI] [PubMed] [Google Scholar]
- 39.DiGiuseppe S, Luszczek W, Keiffer TR, Bienkowska-Haba M, Guion LG, Sapp MJ. 2016. Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis. Proc Natl Acad Sci U S A 113:6289–6294. doi: 10.1073/pnas.1600638113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallardo M, Schleich S, Krijnse Locker J. 2001. Microtubule-dependent organization of vaccinia virus core-derived early mRNAs into distinct cytoplasmic structures. Mol Biol Cell 12:3875–3891. doi: 10.1091/mbc.12.12.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schramm B, de Haan CA, Young J, Doglio L, Schleich S, Reese C, Popov AV, Steffen W, Schroer T, Locker JK. 2006. Vaccinia-virus-induced cellular contractility facilitates the subcellular localization of the viral replication sites. Traffic 7:1352–1367. doi: 10.1111/j.1600-0854.2006.00470.x. [DOI] [PubMed] [Google Scholar]
- 42.Schepis A, Schramm B, de Haan CA, Locker JK. 2006. Vaccinia virus-induced microtubule-dependent cellular rearrangements. Traffic 7:308–323. doi: 10.1111/j.1600-0854.2005.00381.x. [DOI] [PubMed] [Google Scholar]
- 43.Arakawa Y, Cordeiro JV, Way M. 2007. F11L-mediated inhibition of RhoA-mDia signaling stimulates microtubule dynamics during vaccinia virus infection. Cell Host Microbe 1:213–226. doi: 10.1016/j.chom.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Ploubidou A, Moreau V, Ashman K, Reckmann I, Gonzalez C, Way M. 2000. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J 19:3932–3944. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rietdorf J, Ploubidou A, Reckmann I, Holmstrom A, Frischknecht F, Zettl M, Zimmermann T, Way M. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat Cell Biol 3:992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- 46.Sanderson CM, Hollinshead M, Smith GL. 2000. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J Gen Virol 81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- 47.Ward BM, Moss B. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J Virol 75:11651–11663. doi: 10.1128/JVI.75.23.11651-11663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hollinshead M, Rodger G, Van Eijl H, Law M, Hollinshead R, Vaux DJ, Smith GL. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J Cell Biol 154:389–402. doi: 10.1083/jcb.200104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geada MM, Galindo I, Lorenzo MM, Perdiguero B, Blasco R. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J Gen Virol 82:2747–2760. doi: 10.1099/0022-1317-82-11-2747. [DOI] [PubMed] [Google Scholar]
- 50.Jouvenet N, Monaghan P, Way M, Wileman T. 2004. Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J Virol 78:7990–8001. doi: 10.1128/JVI.78.15.7990-8001.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howard AR, Moss B. 2012. Formation of orthopoxvirus cytoplasmic A-type inclusion bodies and embedding of virions are dynamic processes requiring microtubules. J Virol 86:5905–5914. doi: 10.1128/JVI.06997-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hyde JL, Gillespie LK, Mackenzie JM. 2012. Mouse norovirus 1 utilizes the cytoskeleton network to establish localization of the replication complex proximal to the microtubule organizing center. J Virol 86:4110–4122. doi: 10.1128/JVI.05784-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cotton BT, Hyde JL, Sarvestani ST, Sosnovtsev SV, Green KY, White PA, Mackenzie JM. 18 January 2017. The norovirus NS3 protein is a dynamic lipid- and microtubule-associated protein involved in viral RNA replication. J Virol doi: 10.1128/JVI.02138-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cabral-Romero C, Padilla-Noriega L. 2006. Association of rotavirus viroplasms with microtubules through NSP2 and NSP5. Mem Inst Oswaldo Cruz 101:603–611. doi: 10.1590/S0074-02762006000600006. [DOI] [PubMed] [Google Scholar]
- 55.Eichwald C, Arnoldi F, Laimbacher AS, Schraner EM, Fraefel C, Wild P, Burrone OR, Ackermann M. 2012. Rotavirus viroplasm fusion and perinuclear localization are dynamic processes requiring stabilized microtubules. PLoS One 7:e47947. doi: 10.1371/journal.pone.0047947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Criglar JM, Hu L, Crawford SE, Hyser JM, Broughman JR, Prasad BV, Estes MK. 2014. A novel form of rotavirus NSP2 and phosphorylation-dependent NSP2-NSP5 interactions are associated with viroplasm assembly. J Virol 88:786–798. doi: 10.1128/JVI.03022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parker JS, Broering TJ, Kim J, Higgins DE, Nibert ML. 2002. Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J Virol 76:4483–4496. doi: 10.1128/JVI.76.9.4483-4496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P, Schorb M, Pruunsild P, Schwab Y, Chatel-Chaix L, Ruggieri A, Bartenschlager R. 2017. Ultrastructural characterization of Zika virus replication factories. Cell Rep 18:2113–2123. doi: 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolk B, Buchele B, Moradpour D, Rice CM. 2008. A dynamic view of hepatitis C virus replication complexes. J Virol 82:10519–10531. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai CK, Jeng KS, Machida K, Lai MM. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J Virol 82:8838–8848. doi: 10.1128/JVI.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eyre NS, Fiches GN, Aloia AL, Helbig KJ, McCartney EM, McErlean CS, Li K, Aggarwal A, Turville SG, Beard MR. 2014. Dynamic imaging of the hepatitis C virus NS5A protein during a productive infection. J Virol 88:3636–3652. doi: 10.1128/JVI.02490-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9:1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 63.Lyn RK, Hope G, Sherratt AR, McLauchlan J, Pezacki JP. 2013. Bidirectional lipid droplet velocities are controlled by differential binding strengths of HCV core DII protein. PLoS One 8:e78065. doi: 10.1371/journal.pone.0078065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akil A, Peng J, Omrane M, Gondeau C, Desterke C, Marin M, Tronchere H, Taveneau C, Sar S, Briolotti P, Benjelloun S, Benjouad A, Maurel P, Thiers V, Bressanelli S, Samuel D, Brechot C, Gassama-Diagne A. 2016. Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV. Nat Commun 7:12203. doi: 10.1038/ncomms12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Counihan NA, Rawlinson SM, Lindenbach BD. 2011. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog 7:e1002302. doi: 10.1371/journal.ppat.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. 2012. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog 8:e1002466. doi: 10.1371/journal.ppat.1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai CK, Saxena V, Tseng CH, Jeng KS, Kohara M, Lai MM. 2014. Nonstructural protein 5A is incorporated into hepatitis C virus low-density particle through interaction with core protein and microtubules during intracellular transport. PLoS One 9:e99022. doi: 10.1371/journal.pone.0099022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Egger D, Bienz K. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J Gen Virol 86:707–718. doi: 10.1099/vir.0.80442-0. [DOI] [PubMed] [Google Scholar]
- 69.Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. 2010. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog 6:e1000958. doi: 10.1371/journal.ppat.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naghavi MH, Gundersen GG, Walsh D. 2013. Plus-end tracking proteins, CLASPs, and a viral Akt mimic regulate herpesvirus-induced stable microtubule formation and virus spread. Proc Natl Acad Sci U S A 110:18268–18273. doi: 10.1073/pnas.1310760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasdeloup D, Labetoulle M, Rixon FJ. 2013. Differing effects of herpes simplex virus 1 and pseudorabies virus infections on centrosomal function. J Virol 87:7102–7112. doi: 10.1128/JVI.00764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Favoreel HW, Van Minnebruggen G, Adriaensen D, Nauwynck HJ. 2005. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an alphaherpesvirus are associated with enhanced spread. Proc Natl Acad Sci U S A 102:8990–8995. doi: 10.1073/pnas.0409099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elliott G, O'Hare P. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol 72:6448–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakashima H, Kaufmann JK, Wang PY, Nguyen T, Speranza MC, Kasai K, Okemoto K, Otsuki A, Nakano I, Fernandez S, Goins WF, Grandi P, Glorioso JC, Lawler S, Cripe TP, Chiocca EA. 2015. Histone deacetylase 6 inhibition enhances oncolytic viral replication in glioma. J Clin Invest 125:4269–4280. doi: 10.1172/JCI80713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. 2013. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J Virol 87:2857–2867. doi: 10.1128/JVI.02676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hogue IB, Bosse JB, Hu JR, Thiberge SY, Enquist LW. 2014. Cellular mechanisms of alpha herpesvirus egress: live cell fluorescence microscopy of pseudorabies virus exocytosis. PLoS Pathog 10:e1004535. doi: 10.1371/journal.ppat.1004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mingo RM, Han J, Newcomb WW, Brown JC. 2012. Replication of herpes simplex virus: egress of progeny virus at specialized cell membrane sites. J Virol 86:7084–7097. doi: 10.1128/JVI.00463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Afonso PV, Zamborlini A, Saib A, Mahieux R. 2007. Centrosome and retroviruses: the dangerous liaisons. Retrovirology 4:27. doi: 10.1186/1742-4690-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang G, Sharon D, Jovel J, Liu L, Wine E, Tahbaz N, Indik S, Mason A. 2015. Pericentriolar targeting of the mouse mammary tumor virus GAG protein. PLoS One 10:e0131515. doi: 10.1371/journal.pone.0131515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pereira LE, Clark J, Grznarova P, Wen X, LaCasse R, Ruml T, Spearman P, Hunter E. 2014. Direct evidence for intracellular anterograde co-transport of M-PMV Gag and Env on microtubules. Virology 449:109–119. doi: 10.1016/j.virol.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu SF, Eastman SW, Linial ML. 2006. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in Gag. Traffic 7:966–977. doi: 10.1111/j.1600-0854.2006.00448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Momose F, Kikuchi Y, Komase K, Morikawa Y. 2007. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes Infect 9:1422–1433. doi: 10.1016/j.micinf.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 83.Avilov SV, Moisy D, Munier S, Schraidt O, Naffakh N, Cusack S. 2012. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J Virol 86:1433–1448. doi: 10.1128/JVI.05820-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amorim MJ, Bruce EA, Read EK, Foeglein A, Mahen R, Stuart AD, Digard P. 2011. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J Virol 85:4143–4156. doi: 10.1128/JVI.02606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawaguchi A, Matsumoto K, Nagata K. 2012. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. J Virol 86:11086–11095. doi: 10.1128/JVI.00453-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Husain M, Harrod KS. 2011. Enhanced acetylation of alpha-tubulin in influenza A virus infected epithelial cells. FEBS Lett 585:128–132. doi: 10.1016/j.febslet.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 87.Rindler MJ, Ivanov IE, Sabatini DD. 1987. Microtubule-acting drugs lead to the nonpolarized delivery of the influenza hemagglutinin to the cell surface of polarized Madin-Darby canine kidney cells. J Cell Biol 104:231–241. doi: 10.1083/jcb.104.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tashiro M, Seto JT, Klenk HD, Rott R. 1993. Possible involvement of microtubule disruption in bipolar budding of a Sendai virus mutant, F1-R, in epithelial MDCK cells. J Virol 67:5902–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Levesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, DesGroseillers L, Gatignol A, Mouland AJ. 2006. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic 7:1177–1193. doi: 10.1111/j.1600-0854.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- 90.Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol 4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leblanc JJ, Perez O, Hope TJ. 2008. Probing the structural states of human immunodeficiency virus type 1 pr55gag by using monoclonal antibodies. J Virol 82:2570–2574. doi: 10.1128/JVI.01717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nishi M, Ryo A, Tsurutani N, Ohba K, Sawasaki T, Morishita R, Perrem K, Aoki I, Morikawa Y, Yamamoto N. 2009. Requirement for microtubule integrity in the SOCS1-mediated intracellular dynamics of HIV-1 Gag. FEBS Lett 583:1243–1250. doi: 10.1016/j.febslet.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 93.Gaudin R, de Alencar BC, Jouve M, Berre S, Le Bouder E, Schindler M, Varthaman A, Gobert FX, Benaroch P. 2012. Critical role for the kinesin KIF3A in the HIV life cycle in primary human macrophages. J Cell Biol 199:467–479. doi: 10.1083/jcb.201201144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sol-Foulon N, Sourisseau M, Porrot F, Thoulouze MI, Trouillet C, Nobile C, Blanchet F, di Bartolo V, Noraz N, Taylor N, Alcover A, Hivroz C, Schwartz O. 2007. ZAP-70 kinase regulates HIV cell-to-cell spread and virological synapse formation. EMBO J 26:516–526. doi: 10.1038/sj.emboj.7601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rudnicka D, Feldmann J, Porrot F, Wietgrefe S, Guadagnini S, Prevost MC, Estaquier J, Haase AT, Sol-Foulon N, Schwartz O. 2009. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J Virol 83:6234–6246. doi: 10.1128/JVI.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Igakura T, Stinchcombe JC, Goon PK, Taylor GP, Weber JN, Griffiths GM, Tanaka Y, Osame M, Bangham CR. 2003. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science 299:1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 97.Nejmeddine M, Negi VS, Mukherjee S, Tanaka Y, Orth K, Taylor GP, Bangham CR. 2009. HTLV-1-Tax and ICAM-1 act on T-cell signal pathways to polarize the microtubule-organizing center at the virological synapse. Blood 114:1016–1025. doi: 10.1182/blood-2008-03-136770. [DOI] [PubMed] [Google Scholar]
- 98.Jouvenet N, Wileman T. 2005. African swine fever virus infection disrupts centrosome assembly and function. J Gen Virol 86:589–594. doi: 10.1099/vir.0.80623-0. [DOI] [PubMed] [Google Scholar]
- 99.Armer H, Moffat K, Wileman T, Belsham GJ, Jackson T, Duprex WP, Ryan M, Monaghan P. 2008. Foot-and-mouth disease virus, but not bovine enterovirus, targets the host cell cytoskeleton via the nonstructural protein 3Cpro. J Virol 82:10556–10566. doi: 10.1128/JVI.00907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yea C, Dembowy J, Pacione L, Brown M. 2007. Microtubule-mediated and microtubule-independent transport of adenovirus type 5 in HEK293 cells. J Virol 81:6899–6908. doi: 10.1128/JVI.02330-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xiao PJ, Mitchell AM, Huang L, Li C, Samulski RJ. 2016. Disruption of microtubules post-virus entry enhances adeno-associated virus vector transduction. Hum Gene Ther 27:309–324. doi: 10.1089/hum.2016.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anton LC, Schubert U, Bacik I, Princiotta MF, Wearsch PA, Gibbs J, Day PM, Realini C, Rechsteiner MC, Bennink JR, Yewdell JW. 1999. Intracellular localization of proteasomal degradation of a viral antigen. J Cell Biol 146:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wileman T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu Rev Microbiol 61:149–167. doi: 10.1146/annurev.micro.57.030502.090836. [DOI] [PubMed] [Google Scholar]
- 104.Araujo FD, Stracker TH, Carson CT, Lee DV, Weitzman MD. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J Virol 79:11382–11391. doi: 10.1128/JVI.79.17.11382-11391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Y, Shevchenko A, Shevchenko A, Berk AJ. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J Virol 79:14004–14016. doi: 10.1128/JVI.79.22.14004-14016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Banerjee I, Miyake Y, Nobs SP, Schneider C, Horvath P, Kopf M, Matthias P, Helenius A, Yamauchi Y. 2014. Influenza A virus uses the aggresome processing machinery for host cell entry. Science 346:473–477. doi: 10.1126/science.1257037. [DOI] [PubMed] [Google Scholar]
- 107.Husain M, Cheung CY. 2014. Histone deacetylase 6 inhibits influenza A virus release by downregulating the trafficking of viral components to the plasma membrane via its substrate, acetylated microtubules. J Virol 88:11229–11239. doi: 10.1128/JVI.00727-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heath CM, Windsor M, Wileman T. 2001. Aggresomes resemble sites specialized for virus assembly. J Cell Biol 153:449–455. doi: 10.1083/jcb.153.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taylor MP, Burgon TB, Kirkegaard K, Jackson WT. 2009. Role of microtubules in extracellular release of poliovirus. J Virol 83:6599–6609. doi: 10.1128/JVI.01819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morales I, Carbajal MA, Bohn S, Holzer D, Kato SE, Greco FA, Moussatche N, Krijnse Locker J. 2008. The vaccinia virus F11L gene product facilitates cell detachment and promotes migration. Traffic 9:1283–1298. doi: 10.1111/j.1600-0854.2008.00762.x. [DOI] [PubMed] [Google Scholar]
- 111.Claus C, Chey S, Heinrich S, Reins M, Richardt B, Pinkert S, Fechner H, Gaunitz F, Schafer I, Seibel P, Liebert UG. 2011. Involvement of p32 and microtubules in alteration of mitochondrial functions by rubella virus. J Virol 85:3881–3892. doi: 10.1128/JVI.02492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim S, Kim HY, Lee S, Kim SW, Sohn S, Kim K, Cho H. 2007. Hepatitis B virus x protein induces perinuclear mitochondrial clustering in microtubule- and dynein-dependent manners. J Virol 81:1714–1726. doi: 10.1128/JVI.01863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rojo G, Chamorro M, Salas ML, Vinuela E, Cuezva JM, Salas J. 1998. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol 72:7583–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Beske O, Reichelt M, Taylor MP, Kirkegaard K, Andino R. 2007. Poliovirus infection blocks ERGIC-to-Golgi trafficking and induces microtubule-dependent disruption of the Golgi complex. J Cell Sci 120:3207–3218. doi: 10.1242/jcs.03483. [DOI] [PubMed] [Google Scholar]
- 115.Zhou Z, Mogensen MM, Powell PP, Curry S, Wileman T. 2013. Foot-and-mouth disease virus 3C protease induces fragmentation of the Golgi compartment and blocks intra-Golgi transport. J Virol 87:11721–11729. doi: 10.1128/JVI.01355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Elis E, Ehrlich M, Bacharach E. 2015. Dynamics and restriction of murine leukemia virus cores in mitotic and interphase cells. Retrovirology 12:95. doi: 10.1186/s12977-015-0220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pawlica P, Dufour C, Berthoux L. 2015. Inhibition of microtubules and dynein rescues human immunodeficiency virus type 1 from owl monkey TRIMCyp-mediated restriction in a cellular context-specific fashion. J Gen Virol 96:874–886. doi: 10.1099/jgv.0.000018. [DOI] [PubMed] [Google Scholar]
- 118.Yap MW, Stoye JP. 2003. Intracellular localisation of Fv1. Virology 307:76–89. doi: 10.1016/S0042-6822(02)00053-3. [DOI] [PubMed] [Google Scholar]
- 119.Chiang HS, Zhao Y, Song JH, Liu S, Wang N, Terhorst C, Sharpe AH, Basavappa M, Jeffrey KL, Reinecker HC. 2014. GEF-H1 controls microtubule-dependent sensing of nucleic acids for antiviral host defenses. Nat Immunol 15:63–71. doi: 10.1038/ni.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seo M, Lee SO, Kim JH, Hong Y, Kim S, Kim Y, Min DH, Kong YY, Shin J, Ahn K. 2016. MAP4-regulated dynein-dependent trafficking of BTN3A1 controls the TBK1-IRF3 signaling axis. Proc Natl Acad Sci U S A 113:14390–14395. doi: 10.1073/pnas.1615287113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moseley GW, Lahaye X, Roth DM, Oksayan S, Filmer RP, Rowe CL, Blondel D, Jans DA. 2009. Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. J Cell Sci 122:3652–3662. doi: 10.1242/jcs.045542. [DOI] [PubMed] [Google Scholar]
- 122.Dumas A, Le-Bury G, Marie-Anais F, Herit F, Mazzolini J, Guilbert T, Bourdoncle P, Russell DG, Benichou S, Zahraoui A, Niedergang F. 2015. The HIV-1 protein Vpr impairs phagosome maturation by controlling microtubule-dependent trafficking. J Cell Biol 211:359–372. doi: 10.1083/jcb.201503124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Faller EM, Sugden SM, McVey MJ, Kakal JA, MacPherson PA. 2010. Soluble HIV Tat protein removes the IL-7 receptor alpha-chain from the surface of resting CD8 T cells and targets it for degradation. J Immunol 185:2854–2866. doi: 10.4049/jimmunol.0902207. [DOI] [PubMed] [Google Scholar]
- 124.Maxwell SA, Ames SK, Sawai ET, Decker GL, Cook RG, Butel JS. 1991. Simian virus 40 large T antigen and p53 are microtubule-associated proteins in transformed cells. Cell Growth Differ 2:115–127. [PubMed] [Google Scholar]
- 125.Tei S, Saitoh N, Funahara T, Iida S, Nakatsu Y, Kinoshita K, Kinoshita Y, Saya H, Nakao M. 2009. Simian virus 40 large T antigen targets the microtubule-stabilizing protein TACC2. J Cell Sci 122:3190–3198. doi: 10.1242/jcs.049627. [DOI] [PubMed] [Google Scholar]
- 126.Hu L, Filippakis H, Huang H, Yen TJ, Gjoerup OV. 2013. Replication stress and mitotic dysfunction in cells expressing simian virus 40 large T antigen. J Virol 87:13179–13192. doi: 10.1128/JVI.02224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sato C, Nishizawa K, Yamaguchi N. 1984. Co-localization of SV40 T antigen and p53 with immunological analogues of microtubule-associated protein-1 on the nuclear skeleton. Cell Struct Funct 9:305–309. doi: 10.1247/csf.9.305. [DOI] [PubMed] [Google Scholar]
- 128.Kotadia S, Kao LR, Comerford SA, Jones RT, Hammer RE, Megraw TL. 2008. PP2A-dependent disruption of centrosome replication and cytoskeleton organization in Drosophila by SV40 small tumor antigen. Oncogene 27:6334–6346. doi: 10.1038/onc.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gaillard S, Fahrbach KM, Parkati R, Rundell K. 2001. Overexpression of simian virus 40 small-T antigen blocks centrosome function and mitotic progression in human fibroblasts. J Virol 75:9799–9807. doi: 10.1128/JVI.75.20.9799-9807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Niehl A, Pena EJ, Amari K, Heinlein M. 2013. Microtubules in viral replication and transport. Plant J 75:290–308. doi: 10.1111/tpj.12134. [DOI] [PubMed] [Google Scholar]
- 131.Wasteneys GO. 2002. Microtubule organization in the green kingdom: chaos or self-order? J Cell Sci 115:1345–1354. [DOI] [PubMed] [Google Scholar]
- 132.Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M. 2000. Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol 2:826–832. doi: 10.1038/35041072. [DOI] [PubMed] [Google Scholar]
- 133.Ruggenthaler P, Fichtenbauer D, Krasensky J, Jonak C, Waigmann E. 2009. Microtubule-associated protein AtMPB2C plays a role in organization of cortical microtubules, stomata patterning, and tobamovirus infectivity. Plant Physiol 149:1354–1365. doi: 10.1104/pp.108.130450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Harries PA, Palanichelvam K, Yu W, Schoelz JE, Nelson RS. 2009. The cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol 149:1005–1016. doi: 10.1104/pp.108.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ferralli J, Ashby J, Fasler M, Boyko V, Heinlein M. 2006. Disruption of microtubule organization and centrosome function by expression of tobacco mosaic virus movement protein. J Virol 80:5807–5821. doi: 10.1128/JVI.00254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ashby J, Boutant E, Seemanpillai M, Groner A, Sambade A, Ritzenthaler C, Heinlein M. 2006. Tobacco mosaic virus movement protein functions as a structural microtubule-associated protein. J Virol 80:8329–8344. doi: 10.1128/JVI.00540-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ouko MO, Sambade A, Brandner K, Niehl A, Pena E, Ahad A, Heinlein M, Nick P. 2010. Tobacco mutants with reduced microtubule dynamics are less susceptible to TMV. Plant J 62:829–839. doi: 10.1111/j.1365-313X.2010.04195.x. [DOI] [PubMed] [Google Scholar]
- 138.Brandner K, Sambade A, Boutant E, Didier P, Mely Y, Ritzenthaler C, Heinlein M. 2008. Tobacco mosaic virus movement protein interacts with green fluorescent protein-tagged microtubule end-binding protein 1. Plant Physiol 147:611–623. doi: 10.1104/pp.108.117481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Blanc S, Schmidt I, Vantard M, Scholthof HB, Kuhl G, Esperandieu P, Cerutti M, Louis C. 1996. The aphid transmission factor of cauliflower mosaic virus forms a stable complex with microtubules in both insect and plant cells. Proc Natl Acad Sci U S A 93:15158–15163. doi: 10.1073/pnas.93.26.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martiniere A, Bak A, Macia JL, Lautredou N, Gargani D, Doumayrou J, Garzo E, Moreno A, Fereres A, Blanc S, Drucker M. 2013. A virus responds instantly to the presence of the vector on the host and forms transmission morphs. eLife 2:e00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bak A, Gargani D, Macia JL, Malouvet E, Vernerey MS, Blanc S, Drucker M. 2013. Virus factories of cauliflower mosaic virus are virion reservoirs that engage actively in vector transmission. J Virol 87:12207–12215. doi: 10.1128/JVI.01883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Martiniere A, Gargani D, Uzest M, Lautredou N, Blanc S, Drucker M. 2009. A role for plant microtubules in the formation of transmission-specific inclusion bodies of Cauliflower mosaic virus. Plant J 58:135–146. doi: 10.1111/j.1365-313X.2008.03768.x. [DOI] [PubMed] [Google Scholar]
- 143.Carpentier DC, Griffiths CM, King LA. 2008. The baculovirus P10 protein of Autographa californica nucleopolyhedrovirus forms two distinct cytoskeletal-like structures and associates with polyhedral occlusion bodies during infection. Virology 371:278–291. doi: 10.1016/j.virol.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 144.Kraemer JA, Erb ML, Waddling CA, Montabana EA, Zehr EA, Wang H, Nguyen K, Pham DS, Agard DA, Pogliano J. 2012. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell 149:1488–1499. doi: 10.1016/j.cell.2012.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Oliva MA, Martin-Galiano AJ, Sakaguchi Y, Andreu JM. 2012. Tubulin homolog TubZ in a phage-encoded partition system. Proc Natl Acad Sci U S A 109:7711–7716. doi: 10.1073/pnas.1121546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Erb ML, Kraemer JA, Coker JK, Chaikeeratisak V, Nonejuie P, Agard DA, Pogliano J. 2014. A bacteriophage tubulin harnesses dynamic instability to center DNA in infected cells. eLife doi: 10.7554/eLife.03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seemanpillai M, Elamawi R, Ritzenthaler C, Heinlein M. 2006. Challenging the role of microtubules in Tobacco mosaic virus movement by drug treatments is disputable. J Virol 80:6712–6715. doi: 10.1128/JVI.00453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen D, Wang M, Zhou S, Zhou Q. 2002. HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J 21:6801–6810. doi: 10.1093/emboj/cdf683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gallo DE, Hope TJ. 2012. Knockdown of MAP4 and DNAL1 produces a post-fusion and pre-nuclear translocation impairment in HIV-1 replication. Virology 422:13–21. doi: 10.1016/j.virol.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ogino T, Iwama M, Ohsawa Y, Mizumoto K. 2003. Interaction of cellular tubulin with Sendai virus M protein regulates transcription of viral genome. Biochem Biophys Res Commun 311:283–293. doi: 10.1016/j.bbrc.2003.09.205. [DOI] [PubMed] [Google Scholar]