ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV) is a typical gammaherpesvirus that establishes persistent lifelong infection in host cells. In order to establish successful infection, KSHV has evolved numerous immune evasion strategies to bypass or hijack the host immune system. However, host cells still produce immune cytokines abundantly during primary KSHV infection. Whether the immune effectors produced are able to inhibit viral infection and how KSHV successfully conquers these immune effectors remain largely unknown. The guanylate-binding protein 1 (GBP1) gene is an interferon-stimulated gene and exerts antiviral functions on several RNA viruses; however, its function in DNA virus infection is less well understood. In this study, we found that KSHV infection increases both the transcriptional and protein levels of GBP1 at the early stage of primary infection by activating the NF-κB pathway. The overexpression of GBP1 significantly inhibited KSHV infection, while the knockdown of GBP1 promoted KSHV infection. The GTPase activity and dimerization of GBP1 were demonstrated to be responsible for its anti-KSHV activity. Furthermore, we found that GBP1 inhibited the nuclear delivery of KSHV virions by disrupting the formation of actin filaments. Finally, we demonstrated that replication and transcription activator (RTA) promotes the degradation of GBP1 through a proteasome pathway. Taken together, these results provide a new understanding of the antiviral mechanism of GBP1, which possesses potent anti-KSHV activity, and suggest the critical role of RTA in the evasion of the innate immune response during primary infection by KSHV.
IMPORTANCE GBP1 can be induced by various cytokines and exerts antiviral activities against several RNA viruses. Our study demonstrated that GBP1 can exert anti-KSHV function by inhibiting the nuclear delivery of KSHV virions via the disruption of actin filaments. Moreover, we found that KSHV RTA can promote the degradation of GBP1 through a proteasome-mediated pathway. Taken together, our results elucidate a novel mechanism of GBP1 anti-KSHV activity and emphasize the critical role of RTA in KSHV evasion of the host immune system during primary infection.
KEYWORDS: KSHV, GBP1, GTPase activity, actin filaments, RTA, Kaposi's sarcoma-associated herpesvirus
INTRODUCTION
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) belongs to the Gammaherpesviridae subfamily. It is a DNA tumor virus that causes several malignancies such as KS, primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (1–3). KSHV displays two different phases in its life cycle: latent infection and lytic reactivation (4). To establish successful infection, KSHV needs to cross the plasma membrane and deliver its genome to the nucleus, in which viral gene expression and viral genome replication take place (5). It has been demonstrated that KSHV utilizes both microtubules (MTs) and microfilaments (MFs) to transport its virions to the nucleus. Disruption of either the microtubule or the microfilament system inhibits the nuclear delivery of KSHV virions (5–7). During KSHV infection, virion proteins and leaked nuclear acids can be detected by the host innate immune system. The Toll-like receptor (TLR) pathway or the cytosolic DNA-sensing pathway is activated to inhibit viral infection (8–10). Although KSHV has evolved numerous immune evasion strategies to bypass or hijack the host immune system (11), host cells still produce immune cytokines abundantly during primary KSHV infection (12). Whether the immune effectors produced are able to inhibit viral infection and how KSHV successfully conquers these immune effectors remain largely unknown.
Guanylate-binding protein 1 (GBP1) is an interferon (IFN)-inducible protein abundantly expressed during the innate immune response (13, 14). It is one of the large GTPases, with a relative molecular mass of 67 kDa, which can hydrolyze GTP to GDP and subsequently to GMP (15). GBP1 can also be induced by a large number of inflammatory cytokines such as tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β) (14). The cRel (NF-κB) motif and the interferon-stimulated response element (ISRE) can be found in the promoter of GBP1 (16). GBP1 is composed of an N-terminal globular GTP-binding domain and a C-terminal helical domain, which contains seven α-helices (15). It is a large self-activating GTPase, and its dimerization is necessary for sufficient GTP-hydrolyzing activity (17). It was reported previously that the GTPase activity of GBP1 is responsible for its antiviral activity (18). Previous studies have shown that GBP1 can act against various RNA viruses such as vesicular stomatitis virus (VSV), dengue virus, influenza A virus (IAV), classical swine fever virus, and hepatitis C virus (HCV) (19–24). In addition, new evidence shows that the homologous gene of GBP1 in mice exerts strong antibacterial function (13, 25). Whether GBP1 has an antiviral effect against DNA viruses, and its potential mechanism, is still unclear.
In this study, we found that GBP1 was highly upregulated at an early stage during de novo KSHV infection, at both the mRNA and protein levels. The overexpression of GBP1 significantly inhibited KSHV infection, while the knockdown of GBP1 promoted KSHV infection. The induced GBP1 exerted its antiviral function against KSHV by inhibiting the nuclear delivery of KSHV virions during infection. Moreover, we found that the GTPase activity and dimerization of GBP1 were responsible for antiviral activity in restricting KSHV infection. We also found that the KSHV replication and transcription activator (RTA) protein can target GBP1 for degradation, which may help the virus to antagonize the antiviral function of GBP1 during the early stages of de novo infection.
RESULTS
KSHV infection increases GBP1 expression at an early stage of primary infection by activating NF-κB signaling.
In order to establish successful infection, KSHV needs to overcome multiple defense reactions elicited by host cells (1, 8). To counteract these defense reactions, KSHV has evolved several strategies to evade and hijack antiviral responses and preexisting host signal pathways (11, 26–31). It was demonstrated previously that KSHV can activate NF-κB as early as 5 to 15 min postinfection and that moderate NF-κB activation is sustained during the whole process of primary infection (12). In addition, various inflammatory cytokines induced by primary KSHV infection may contribute to the inflammatory environment that is required for viral and host gene expression and for KSHV infection (12, 22, 32).
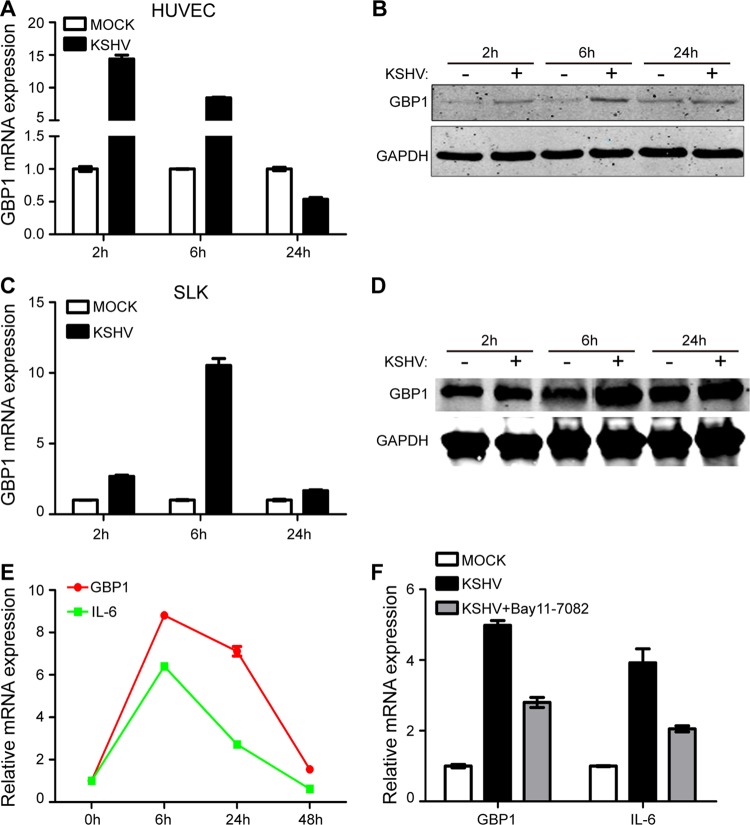
To study early events of KSHV infection, human umbilical vein endothelial cells (HUVECs) were de novo infected with purified KSHV virions, and the expression of inflammation-related genes was examined. We found that the RNA level of GBP1 in the KSHV-infected group was upregulated more than 10-fold compared with that in the mock group at 2 and 6 h postinfection (hpi); this fold change declined rapidly at 24 hpi (Fig. 1A). We also detected the protein level of GBP1 using Western blotting. As shown in Fig. 1B, the GBP1 level was increased at 2 and 6 hpi in the KSHV-infected group and had recovered at 24 hpi. To confirm this result, SLK cells were de novo infected with purified KSHV virions under similar conditions. As shown in Fig. 1C, a remarkable increase of the GBP1 RNA level was found in the KSHV-infected group at 6 hpi, whereas the RNA level was increased only slightly at 24 hpi. The protein level of GBP1 was significantly upregulated in the KSHV-infected group at 6 hpi and slightly upregulated at 24 hpi, similar to the change in RNA levels (Fig. 1D).
FIG 1.
KSHV infection increases GBP1 expression levels early during primary infection by activating NF-κB signaling. (A and B) HUVECs were infected with or without KSHV.BAC16.RGB (MOI, 10), and cells were collected at 2, 6, and 24 hpi. Transcriptional and protein expression levels of GBP1 were detected by qPCR (A) and Western blotting (B). (C and D) SLK cells were infected with or without KSHV.BAC16.RGB (MOI, 5), and cells were subjected to qPCR (C) and Western blotting (D) at the indicated time points. (E) SLK cells infected with KSHV.BAC16 (MOI, 5) were collected at the indicated time points and subjected to qPCR to measure the RNA levels of GBP1 and IL-6. (F) SLK cells were preincubated with or without 10 μM Bay11-7082 for 24 h before infection by KSHV. Cells were infected with KSHV.BAC16.RGB at an MOI of 5, the supernatant was removed at 2 hpi, and new culture medium with or without 10 μM Bay11-7082 was added to the cells. Cells were harvested for RNA extraction at 6 hpi.
Previous studies showed that GBP1 can be induced by the NF-κB signaling pathway, and NF-κB signaling can be strongly induced during the early stages of KSHV infection (12, 16). To explore whether the upregulation of GBP1 was due to NF-κB activation, we detected the RNA levels of GBP1 and IL-6, a reporter gene of NF-κB activation, during primary infection. As shown in Fig. 1E, the RNA levels of GBP1 and IL-6 displayed similar patterns, varying with time. Next, we used an inhibitor of the NF-κB signaling pathway, Bay11-7082, to detect the influence of NF-κB activation on the expression of GBP1. Indeed, the upregulated RNA level of GBP1 was attenuated when the inhibitor was added (Fig. 1F). The inhibition of the NF-κB signaling pathway by the inhibitor was determined by Western blotting of p65 and phospho-p65 (data not shown). We also detected the activation of the NF-κB signaling pathway during primary KSHV infection. The NF-κB signaling pathway was activated at 2 and 6 hpi in HUVECs and at 6 hpi in SLK cells (data not shown). Our results also showed that 10 μM Bay11-7082 did not influence the viability of SLK cells (data not shown). Taken together, these results suggested that GBP1 was upregulated in response to primary KSHV infection and that the increase in the GBP1 level was largely mediated by the NF-κB signaling pathway.
Overexpression of GBP1 inhibits KSHV infection and gene expression.
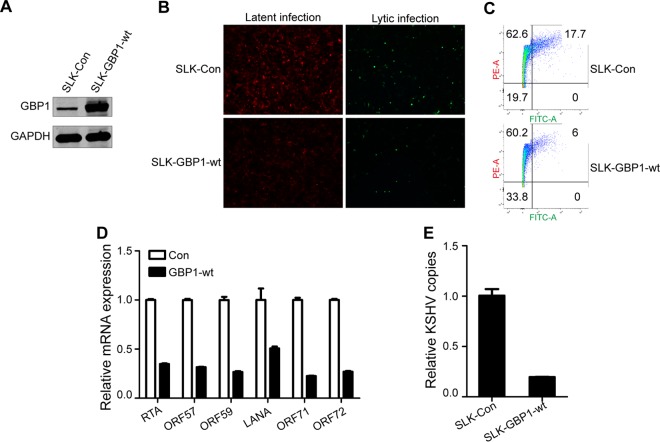
GBP1 is known as an interferon-stimulated gene that has antiviral activity against various viruses (13, 18); therefore, we investigated subsequently whether the upregulation of GBP1 would affect KSHV infection. For this purpose, we constructed two stably transfected SLK cell lines, SLK-Con and SLK-GBP1-wt. The results of Western blotting showed that GBP1 expression was significantly increased in the SLK-GBP1-wt cell line (Fig. 2A). The purified recombinant KSHV.BAC16.RGB virus (33) was used to infect the two stable cell lines at a multiplicity of infection (MOI) of 5. KSHV-infected cells are indicated by red fluorescence, and cells with lytic KSHV infection are indicated by green fluorescence. As shown in Fig. 2B, the intensity of red fluorescence was remarkably weakened, and the ratio of green fluorescence was decreased in the SLK-GBP1-wt group in which GBP1 was overexpressed, compared with the control.
FIG 2.
Overexpression of GBP1 inhibits KSHV infection. (A) SLK cells were stably transfected with lentiviruses containing a GBP1 overexpression plasmid or a vector plasmid and were named SLK-Con and SLK-GBP1-wt, respectively. The overexpression of GBP1 was detected by Western blotting. (B) SLK-Con and SLK-GBP1-wt cells were infected with KSHV.BAC16.RGB (MOI, 5), and fluorescence was visualized by using an inverted fluorescence microscope (catalog number DMI3000; Leica) at 48 hpi. KSHV-infected cells are indicated by red fluorescence, and cells with lytic KSHV infection are indicated by green fluorescence. (C) Cells infected as described above for panel B were fixed with 4% PFA and disaggregated by filtration at 48 hpi. The infected cells, indicated by red fluorescence (phycoerythrin [PE]), and cells with lytic KSHV infection, indicated with green fluorescence (fluorescein isothiocyanate [FITC]), were subsequently analyzed by flow cytometry. (D) RNA was extracted from infected cells at 48 hpi to investigate the expression levels of several KSHV ORFs: RTA, ORF57, ORF59, LANA, ORF71, and ORF72 (by qPCR). (E) The KSHV genome was extracted from infected cells at 48 hpi. KSHV genome copies are indicated by DNA copies of KSHV K9.
To validate these results further, infected cells were subjected to flow cytometry (FCM) analysis. Both red fluorescence and green fluorescence were decreased in GBP1-overexpressing cells (Fig. 2C). Using quantitative PCR (qPCR), we detected the expressions of six KSHV genes: three latent genes, open reading frame 71 (ORF71), ORF72, and LANA, and three lytic genes, ORF57, ORF59, and RTA. We found that the expression levels of both the latent and lytic genes of KSHV were decreased in the SLK-GBP1-wt cell line (Fig. 2D). We also detected copies of the KSHV genome in cells at 48 hpi. These results showed that there were fewer copies of the KSHV genome in SLK-GBP1-wt cells, which suggested that GBP1 can effectively restrict KSHV infection (Fig. 2E). Taken together, these results suggest that GBP1 possesses a potent antiviral function against KSHV infection.
Knockdown of GBP1 expression promotes KSHV infection and gene expression.
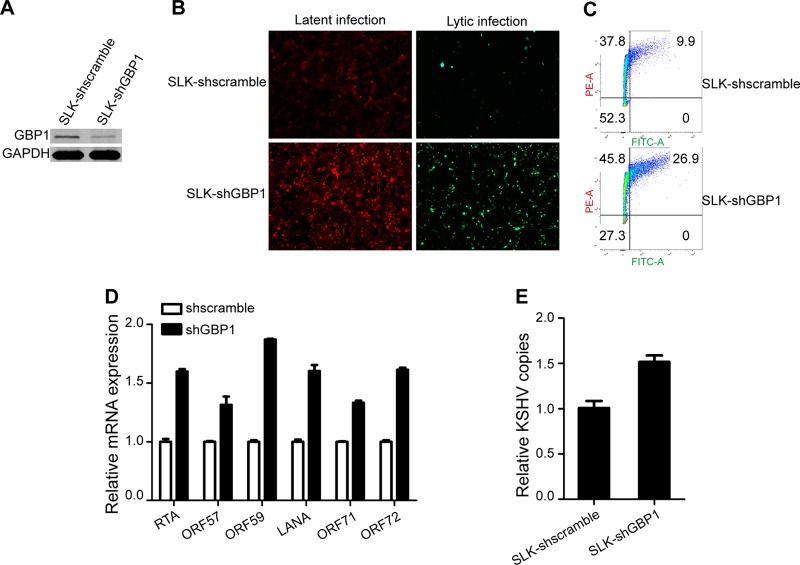
To further confirm the anti-KSHV activity of GBP1, we performed a loss-of-function experiment to determine the effect of GBP1 knockdown on KSHV infection. We constructed two stably transfected cell lines, named SLK-shscramble and SLK-shGBP1. The expression of GBP1 was significantly reduced by short hairpin RNA (shRNA) in SLK-shGBP1 cells (Fig. 3A). The two cell lines were infected with KSHV.BAC16.RGB at an MOI of 5. As shown in Fig. 3B, the knockdown of GBP1 promoted infection by KSHV. The FCM results showed that the frequencies of both red and green fluorescence-positive cells were increased for SLK-shGBP1 cells (Fig. 3C). Consistently, the expression levels of KSHV latent and lytic genes were increased in SLK-shGBP1 cells, as determined by qPCR (Fig. 3D). Similarly, the number of copies of the KSHV genome in SLK-shGBP1 cells was elevated compared with that in SLK-shscramble cells (Fig. 3E). Based on these results, we confirmed that GBP1 has effective antiviral activity against KSHV infection.
FIG 3.
Knockdown of GBP1 expression promotes KSHV infection. (A) SLK-shscramble and SLK-shGBP1 cells were established as described in the text. The knockdown efficiency was determined by Western blotting. (B) SLK-shscramble and SLK-shGBP1 cells were infected as described in the legend of Fig. 2B. Fluorescence was visualized and photographed at 48 hpi. (C) Infected cells and cells with reactivation of infection were analyzed by FCM as described in the legend of Fig. 2C. (D and E) The expression levels of KSHV latent and lytic genes (D) and intracellular genome copy numbers (E) were detected by qPCR at 48 hpi as described in the legend of Fig. 2C and D.
The GTPase activity and dimerization of GBP1 are required for the antiviral activity of GBP1 during KSHV infection.
As demonstrated by previous studies, GTPase activity is critical for the antiviral activity of GBP1 against many RNA viruses, and dimerization of GBP1 is necessary for its self-activating GTPase activity. There are three crucial GTP-binding motifs of GBP1 that mediate the hydrolysis of GTP to GDP and GMP (34, 35). Mutation of K51 in the (phosphate-binding) P loop of the N-terminal globular domain abrogates the GTP-binding and GTP hydrolytic activities of GBP1 (36). In addition, arginine residue R240 of GBP1 within the guanine cap is responsible for its dimerization and self-activation. Mutation of this site causes the accumulation of a nonactivated GBP1 monomer and thus decreases its GTPase activity (37). There is a CAAX motif within the C terminus of GBP1 that mediates the farnesylation and plasma membrane association of GBP1. Deletion of the CAAX motif abolishes the IFN-γ-induced Golgi localization of GBP1 (38, 39).
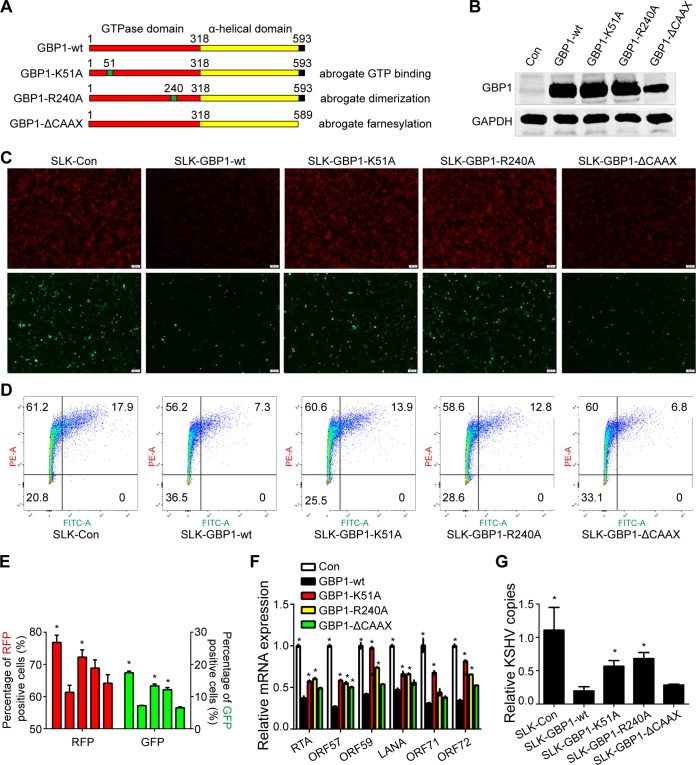
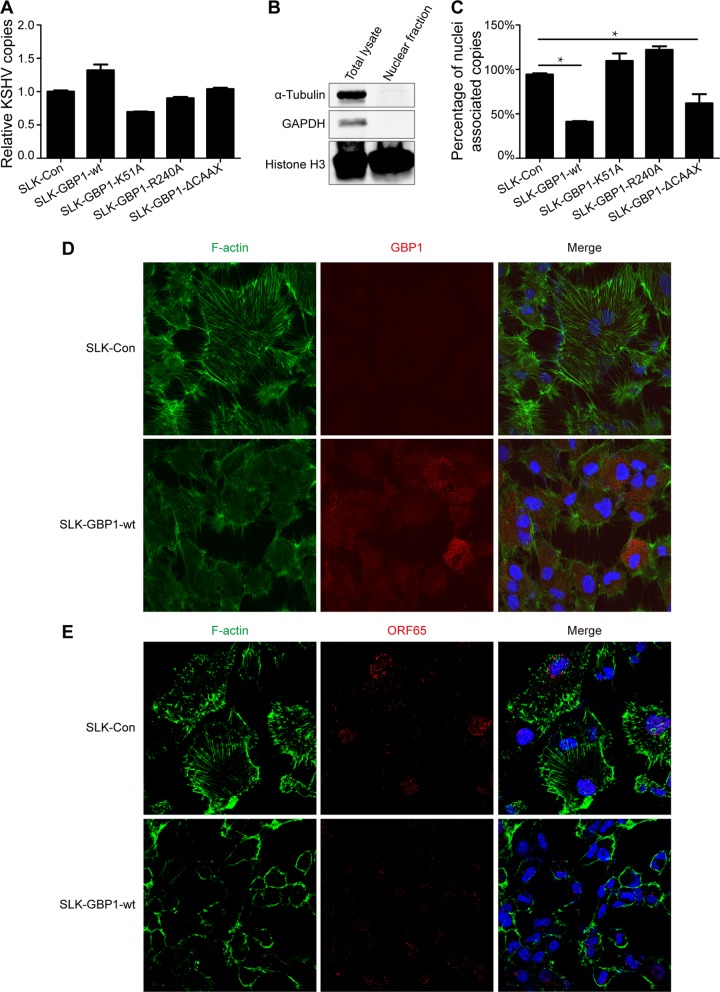
To determine whether GTPase activity and dimerization are responsible for the inhibition of KSHV infection, we constructed three GBP1 mutants, GBP1-K51A, GBP1-R240A, and GBP1-ΔCAAX, and three corresponding stably transfected SLK cell lines, SLK-GBP1-K51A, SLK-GBP1-R240A, and SLK-GBP1-ΔCAAX (Fig. 4A). The expression of these mutants in stable cell lines was verified by Western blotting (Fig. 4B). These three cell lines, along with SLK-Con and SLK-GBP1-wt, were infected by KSHV.BAC16.RGB at an MOI of 5, and the efficiency of infection was determined. As shown in Fig. 4C, mutations at K51and R240 restored the intensity of red fluorescence and the ratio of green fluorescence compared to the wild-type (WT) GBP1 and control groups, while the deletion of the CAAX motif had no influence on the intensity of red fluorescence and the ratio of green fluorescence (Fig. 4C, right). The FCM results showed a similar pattern (Fig. 4D and E). The expression of KSHV latent and lytic genes (Fig. 4F) and intracellular copies of the KSHV genome (Fig. 4G) were also partially recovered in SLK-GBP1-K51A and SLK-GBP1-R240A cells compared with SLK-GBP1-wt cells, while the deletion of the CAAX motif seemed to have no significant effect on the inhibition of the expression of KSHV genes and copies of the KSHV genome (Fig. 4F and G). Collectively, these results demonstrated that GTPase activity and dimerization of GBP1 are necessary for its anti-KSHV function, while the plasma membrane association is dispensable in this case.
FIG 4.
The GTPase activity and dimerization of GBP1 are critical for the anti-KSHV activity of GBP1. (A) Schematic representation of wild-type GBP1 and three site-specific mutants. (B) The three GBP1 mutants GBP1-K51A, GBP1-R240A, and GBP1-ΔCAAX were constructed by using a site-directed mutagenesis kit and subsequently stably transfected into SLK cells. The overexpression of these mutants in SLK cells was validated by Western blotting. (C) The five stably transfected SLK cell lines were infected with KSHV.BAC16.RGB (MOI, 5), and fluorescence was analyzed as described in the legend of Fig. 2B. (D) The fluorescence of the five stable cell lines infected with KSHV.BAC16.RGB was analyzed by FCM as described in the text. (E) Statistical analysis of the FCM results shown in panel D. RFP, red fluorescent protein; GFP, green fluorescent protein. (F and G) RNA or DNA was extracted from infected SLK cells. Expression levels of KSHV genes (F) and KSHV genome copy numbers (G) were detected by qPCR using the corresponding primers. Statistical analysis was conducted by using Student's t test. *, P < 0.05. The SLK-GBP1-wt group was used as a control.
GBP1 inhibits KSHV infection by disrupting the formation of actin filaments.
To explore the mechanism by which GBP1 inhibits KSHV infection, we first determined whether the inhibition of KSHV infection by GBP1 was due to the blockage of KSHV entry. We quantified the intracellular copies of the KSHV genome in infected cells at the very beginning of primary KSHV infection, using qPCR. The five stably transfected cell lines, SLK-Con, SLK-GBP1-wt, SLK-GBP1-K51A, SLK-GBP1-R240A, and SLK-GBP1-ΔCAAX, were incubated with purified KSHV.BAC16.RGB. After 6 h of incubation, the cells were subjected to thorough washing and collected for DNA extraction. As shown in Fig. 5A, no significant difference in the numbers of intracellular copies of the KSHV genome was observed across groups, which suggested that GBP1 did not affect the entry of KSHV into cells. However, we indeed found fewer copies of the KSHV genome in SLK-GBP1-wt cells at 48 hpi (Fig. 2D). This result indicated that GBP1 may not function at the very beginning of infection but may actually decrease the number of intracellular KSHV copies at the postentry stage.
FIG 5.
GBP1 disrupts the formation of actin filaments, thus inhibiting the nuclear delivery of KSHV virions. (A) The five stably transfected wild-type and mutant GBP1-overexpressing cell lines, grown in six-well plates, were incubated with KSHV.BAC16 for 6 h, washed twice with PBS, and then incubated with trypsin-EDTA for 5 min at 37°C. The cells were spun down and then washed once with PBS. Total DNA was isolated and subjected to qPCR. (B) Collected cells were resuspended in 500 μl hypotonic buffer and incubated on ice for 15 min, and 25 μl 10% NP-40 was added, followed by vortexing for 10s. The cell nuclei were spun down, and the pellet was collected. Histone H3 was used as the marker for the nucleus, while α-tubulin and GAPDH were used as the markers for the cytoplasm. (C) The five cell lines were infected with KSHV as described above for panel A. The nuclear fraction was isolated, and the number of nucleus-associated KSHV genome copies was estimated as described in Materials and Methods. (D) SLK-Con and SLK-GBP1-wt cells were fixed and stained with rat anti-GBP1 antibody, followed by incubation with goat anti-mouse IgG(H+L) Alexa Fluor 555 (red). The actin filaments were then stained with Cytopainter phalloidin-iFluor 488 (green) for 90 min. Slides were photographed by using a digital camera and software. (E) SLK-Con and SLK-GBP1-wt cells were incubated with KSHV for 6 h. Cells were fixed and stained with mouse anti-ORF65 antibody and then further stained with the secondary antibody goat anti-mouse IgG(H&L) Alexa Fluor 555 (red), followed by incubation with Cytopainter phalloidin-iFluor 488 (green) for 90 min.
Given that the overexpression of GBP1 did not influence KSHV entry, we wondered whether GBP1 inhibited the nuclear delivery of KSHV virions. To investigate the nucleus-associated KSHV copies that represent successful infection, we isolated the nuclear fraction from infected cells at the very beginning of primary KSHV infection. The purified nuclear fraction was positive for the nuclear marker histone H3 and negative for the cytosolic markers α-tubulin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. 5B, right), whereas the total lysate was positive for all these markers (Fig. 5B, left). The nuclear fraction was used for genome extraction and further quantification of the number of nucleus-associated KSHV genome copies by qPCR. As shown in Fig. 5C, the overexpression of GBP1 significantly reduced the number of copies of the nucleus-associated KSHV genome. Mutations at K51 and R240 abolished the ability to inhibit KSHV nuclear delivery of GBP1, while the deletion of the CAAX motif did not affect its function.
A recent study showed that GBP1 can disturb the natural cytoskeletal structure by disrupting the formation of actin filaments through a direct interaction with the actin protein (40). A natural and intact cytoskeletal structure is pivotal for infection by KSHV. Disorder of either microtubules or microfilaments would severely inhibit the entry and nuclear delivery of KSHV virions (6, 7). To determine whether GBP1 could efficiently disrupt the microfilament structure in order to block nuclear delivery in SLK-GBP1-wt cells, we investigated the formation of actin filaments and intracellular virions by an immunofluorescence assay (IFA). In line with previously reported findings, GBP1 was located predominantly in the cytoplasm, in a granular structure (Fig. 5D, red). The actin filaments were impaired in SLK-GBP1 cells, while there was a very distinct microfilament structure in SLK-Con cells (Fig. 5D, green). To visualize the intracellular KSHV virions, we used a monoclonal antibody against ORF65, one of the capsid proteins of KSHV, to detect the viral particles (41). SLK-Con and SLK-GBP1-wt cells were incubated with KSHV.BAC16.RGB for 6 h and then washed extensively before an immunofluorescence assay. The viral particles were stained with a red fluorescence-conjugated secondary antibody, and the actin filaments were stained with green fluorescence-conjugated phalloidin. There were more KSHV virions that accumulated in the perinuclear area in SLK-Con cells than in SLK-GBP1-wt cells, which may suggest an inhibition of nuclear delivery by GBP1 (Fig. 5E). Taken together, these results suggested that GBP1 inhibits the nuclear delivery of KSHV particles via disruption of the formation of actin filaments, which disturbs the natural cytoskeletal structure that is necessary for KSHV infection.
RTA reduces the protein level of GBP1 by proteasome-mediated degradation.
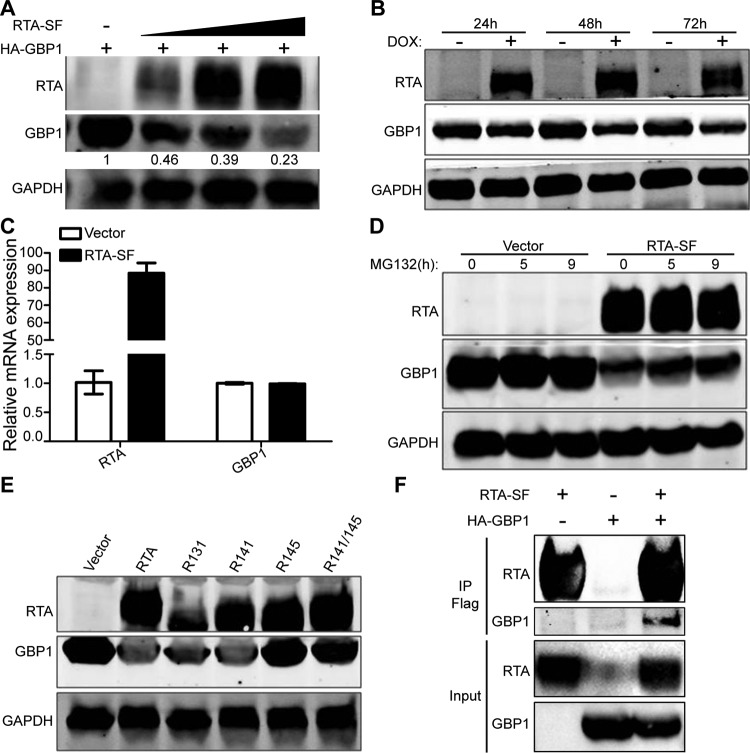
The above-described results strongly suggest that GBP1 is a potent anti-KSHV effector during de novo infection. Indeed, it is highly upregulated at an early stage of infection owing to the immune response from host cells. We speculated that there could be certain viral or host factors that can work against the antiviral effect mediated by GBP1 and facilitate the establishment of a successful latent infection. It has been well recognized that RTA is a critical gene for the regulation of the KSHV life cycle, and RTA is also known to be packaged into virions of KSHV and released into target cells to suppress the immune response elicited by KSHV infection (42, 43). Two previous studies, including ours, demonstrated that RTA can target the important immune adaptors interferon regulatory factor 7 (IRF7) and MyD88 for proteasome-mediated degradation (26, 28). Given that RTA is an important and powerful immune regulatory protein in the early stages of primary infection, we wondered whether RTA could also target GBP1 for degradation. HEK 293T cells were transfected with hemagglutinin (HA)-GBP1 and increasing amounts of RTA-Strep-Flag. Western blotting showed that RTA decreased the protein level of GBP1 in a dose-dependent manner (Fig. 6A). Moreover, when the iSLK-GBP1 cell line, which is a stably transfected cell line overexpressing GBP1, was induced with doxycycline (DOX) to express RTA, the protein level of GBP1 was also reduced (Fig. 6B). To determine whether RTA regulates the transcription of GBP1, HeLa cells were transfected with RTA-SF and collected for qPCR at 36 h posttransfection. The results showed that RTA did not affect the RNA level of GBP1 (Fig. 6C). As RTA did not regulate GBP1 at the transcriptional level, we wondered whether RTA also degraded GBP1 through a proteasome pathway. HEK 293T cells were transfected with GBP1, with or without an RTA-expressing plasmid, and MG132 was added to block protein degradation at the indicated time points. As shown in Fig. 6D, MG132 partially inhibited the degradation of GBP1 by RTA. To confirm that RTA degraded GBP1 through a proteasome pathway, HEK 293T cells were transfected with GBP1 along with wild-type RTA and four RTA mutants (Fig. 6E). Previous studies have shown that residues C131, C141, and H145 of RTA are important for the E3 ligase activity of RTA; mutations among these sites might abolish its E3 ligase activity (28). Our data showed that the H145L and C141S/H145L mutants failed to decrease the protein levels of GBP1, whereas wild-type RTA and the C131S and C141S mutants succeeded (Fig. 6E). To investigate whether RTA degraded GBP1 through physical protein interactions, a coimmunoprecipitation assay was conducted. HEK 293T cells were transfected with either RTA-SF, HA-GBP1, or both, and the cell lysates were immunoprecipitated by using anti-Flag M2 affinity gel. As shown in Fig. 6F, the anti-Flag M2 affinity gel immunoprecipitated GBP1 from cells with both RTA-SF and HA-GBP1 but not from cells with only RTA-SF or HA-GBP1, which meant that RTA could interact with GBP1. Taken together, these results suggested that RTA interacts with and degrades GBP1 by functioning as a ubiquitin (Ub) E3 ligase and that residue H145 of RTA is required for GBP1 degradation.
FIG 6.
RTA reduces the protein level of GBP1 through proteasome-mediated degradation. (A) HEK 293T cells were transfected with HA-GBP1 and increasing amounts of RTA-SF. After 36 h, cell lysates were subjected to immunoblotting. (B) iSLK-GBP1 cells were established as described in Materials and Methods and were treated with or without 5 μg/ml DOX for the induction of RTA. Cell lysates were harvested at the indicated time points and analyzed by Western blotting. (C) HeLa cells were transfected with RTA-SF or the vector control. After 36 h, RNA was isolated for qPCR. (D) HEK 293T cells were transfected with HA-GBP1 with or without RTA-SF and treated with 10 μM MG132 as indicated. Anti-HA and anti-Flag antibodies were used for Western blotting. (E) HEK 293T cells were cotransfected with HA-GBP1 and RTA mutants. After 36 h, cells were collected for subsequent Western blotting. (F) RTA-SF and HA-GBP1 were transfected into HEK 293T cells as indicated. After 48 h, cells were lysed and immunoprecipitated by using anti-Flag M2 affinity gel. The purified proteins, along with input samples, were detected by Western blotting.
DISCUSSION
It has been demonstrated that KSHV utilizes multiple strategies to facilitate infection. During KSHV infection, host cells recognize viral proteins and nucleic acid and elicit innate immune responses to eliminate the invading viruses, while the viruses encode many immunomodulatory proteins to evade innate immune responses (8, 10, 27). The early events of the interaction of the virus and host cells during primary KSHV infection have not been fully demonstrated. Here, we demonstrated that KSHV infection increased both the transcriptional and protein levels of GBP1 by activating NF-κB signaling. In addition, the overexpression of GBP1 significantly inhibited KSHV infection, while the knockdown of GBP1 expression increased it. Moreover, we found that GBP1 inhibited the nuclear delivery of KSHV virions by disrupting the formation of actin filaments. The GTPase activity and dimerization of GBP1 were indispensable for mediating its anti-KSHV function. Furthermore, our data showed that RTA could act as an E3 ligase and reduce the GBP1 protein level by proteasome-mediated degradation, which may serve as another evasion strategy used by KSHV.
Previous studies demonstrated that GBP1 retains moderate antiviral function and works mainly in the regulation of cell migration and cell-cell junctions (14). It was shown previously that GBP1 can be induced by infection with several RNA viruses and that it exerts an antiviral function against these viruses (19–21). In this study, we found that GBP1 expression was induced by KHSV, a DNA virus, and that GBP1 possessed potent antiviral activity against KSHV infection. Our results showed that GBP1 was upregulated by KSHV infection only at 2 and 6 hpi and had recovered at 24 hpi. The transient nature of the change indicated that the upregulation of GBP1 may be a stress response of host cells due to KSHV infection; thus, GBP1 may exert its anti-KSHV activity during the early phase of KSHV infection. As shown in Fig. 1, the changes in GBP1 at the transcriptional level are more dramatic than those at the protein level in both HUVECs and SLK cells. This apparent discrepancy means that KSHV may utilize some strategies, such as promoting the degradation of GBP1 by RTA, to control the excessive induction of GBP1 expression for more efficient infection.
Regarding the mechanism of the antiviral function of GBP1, previous studies demonstrated that GBP1 inhibits the replication of VSV, IAV, and HCV and that its antiviral activity requires GTPase activity (19, 20, 23). In contrast to the direct inhibition of the replication of RNA viruses, our findings provided the first evidence that GBP1 can inhibit KSHV infection by disrupting the nuclear delivery of KSHV virions. Similar to the scenario for RNA infection, the GTPase activity of GBP1 was indispensable for mediating its anti-KSHV function, which indicates that the GTPase activity of GBP1 is responsible for the inhibition of infection by both RNA and DNA viruses. A recently reported study showed that GBP1 can bind directly to actin and inhibit the formation of actin filaments; this was mediated by its GTPase activity (40). In addition, GBP1 can be localized at microtubules and interact with prosurvival kinases such as PIM1 to initiate downstream signaling (44). All these studies indicate that GBP1 is able to interact with the cytoskeleton, thus exerting its downstream function. In our study, we demonstrated that the ability of GBP1 to disrupt the intact actin network is critical for its function of inhibiting the nuclear delivery of KSHV virions. In Fig. 5A, we demonstrated that GBP1 did not affect the entry of KSHV virions, but the lower intensity of red fluorescence signals in SLK-GBP1-wt cells in Fig. 5E seems to be contrary to the results shown in Fig. 5A. The lower intensity of red fluorescence signals in SLK-GBP1-wt cells may result from the resolution limitation of IFA in that the diffused weak fluorescence signal may not be sensitive enough to be detected from the background. Two previous studies also reported similar results showing that different inhibitors of microtubule formation inhibited the nuclear delivery of KSHV virions, with no effect on KSHV entry, and the corresponding IFA results showed fewer viral particles in cells treated with these inhibitors (5, 7). Our results demonstrated a detailed mechanism by which GBP1 exerts its anti-KSHV function, which may provide some new insights for the functional study of the activity of GBP1 against other viral infections.
RTA is the switch of KSHV lytic reactivation and contributes to the evasion of host innate immune responses (11, 26, 28, 43, 45). RTA has been reported to downregulate the protein levels of TRIF, IRF7, and MyD88 by direct interactions and acts as an E3 ligase for these proteins (26, 28). Our study showed that RTA also interacts with and degrades GBP1 through a proteasome-mediated pathway and that the E3 ligase activity of RTA, especially H145, is indispensable for the degradation of GBP1. The less drastic effect of RTA on GBP1 degradation shown in Fig. 6B than in Fig. 6A was due to different experimental systems. The experiment shown in Fig. 6A was done by transient transfection, while the experiment shown in Fig. 6B utilized an RTA-inducible stable cell line that expressed smaller amounts of RTA than in the transient-transfection system. RTA is the pivotal gene for the KSHV life cycle and mediates the reactivation of latent KSHV infection. No infectious KSHV virions can be produced without RTA. Moreover, it is known that RTA can be packaged into virions and exerts its function immediately after de novo infection (42, 43). Therefore, it is impossible to generate KSHV virions with the RTA protein totally excluded. The physiological effect of RTA on GBP1 in the context of viral infection remains to be further studied in the future.
During de novo infection of KSHV, various cytokines, such as IL-1β and TNF-α, and the NF-κB signaling pathway are stimulated at an early stage of infection (12). All these cytokines contribute to the anti-KSHV function of host innate immune responses (46). However, activated NF-κB has been demonstrated to be essential for viral gene expression and the establishment of latent infection (12, 22, 32, 47). This paradox means that a complex mechanism is utilized by KSHV: on one hand, KSHV evades certain fatal immune responses and activates certain signaling pathways that are controllable and essential for viral survival, and on the other hand, KSHV fights against the immune effectors elicited by the activated signal pathways in a delicate way. Given that GBP1 can be induced by both interferon signaling and the NF-κB pathway, it is upregulated only during the early stages of primary KSHV infection. RTA was packaged into KSHV virions and abundantly expressed during the early stages of de novo infection. The fact that RTA promotes the degradation of GBP1 may indicate that the virions released and primary expressed RTA are critical for restricting the antiviral activity of GBP1 in the early phase of de novo infection. These results showed that KSHV encodes RTA to directly target GBP1, the downstream effector of the innate immune response, for the successful establishment of latent infection, while most of the other viral genes targeted signal transduction proteins.
In conclusion, our study describes a new mechanism by which GBP1 inhibits the nuclear delivery of KSHV virions by disrupting the formation of actin filaments. Moreover, we found that RTA targets induced GBP1 for degradation early during infection to antagonize the antiviral activity of GBP1, which provided new insights into the immune evasion strategies utilized by KSHV.
MATERIALS AND METHODS
Cell lines, plasmids, and antibodies.
Human primary endothelial cells (HUVECs) from Lonza were maintained in EGM-2 (Lonza, Basel, Switzerland). Human embryonic kidney HEK 293T, HeLa, SLK, iSLK, and iSLK.BAC16.RGB cells were cultured in Dulbecco's modified Eagle's medium (Biological Industries) supplemented with 10% fetal bovine serum (FBS) (Biological Industries) and antibiotics (penicillin and streptomycin). iSLK.BAC16.RGB cells, which harbored WT BAC16.RGB, were kind gifts from Jae Jung (University of Southern California, Los Angeles, CA, USA). SLK and iSLK cells were gifts from Fanxiu Zhu (Florida State University, FL, USA). The stable cell lines SLK-Con, SLK-GBP1-wt, SLK-GBP1-K51A, SLK-GBP1-R240A, SLK-GBP1-ΔCAAX, SLK-shscramble, SLK-shGBP1, and iSLK-GBP1 were established by infection with the indicated lentiviruses according to the manufacturer's instructions (System Bioscience, Palo Alto, CA, USA).
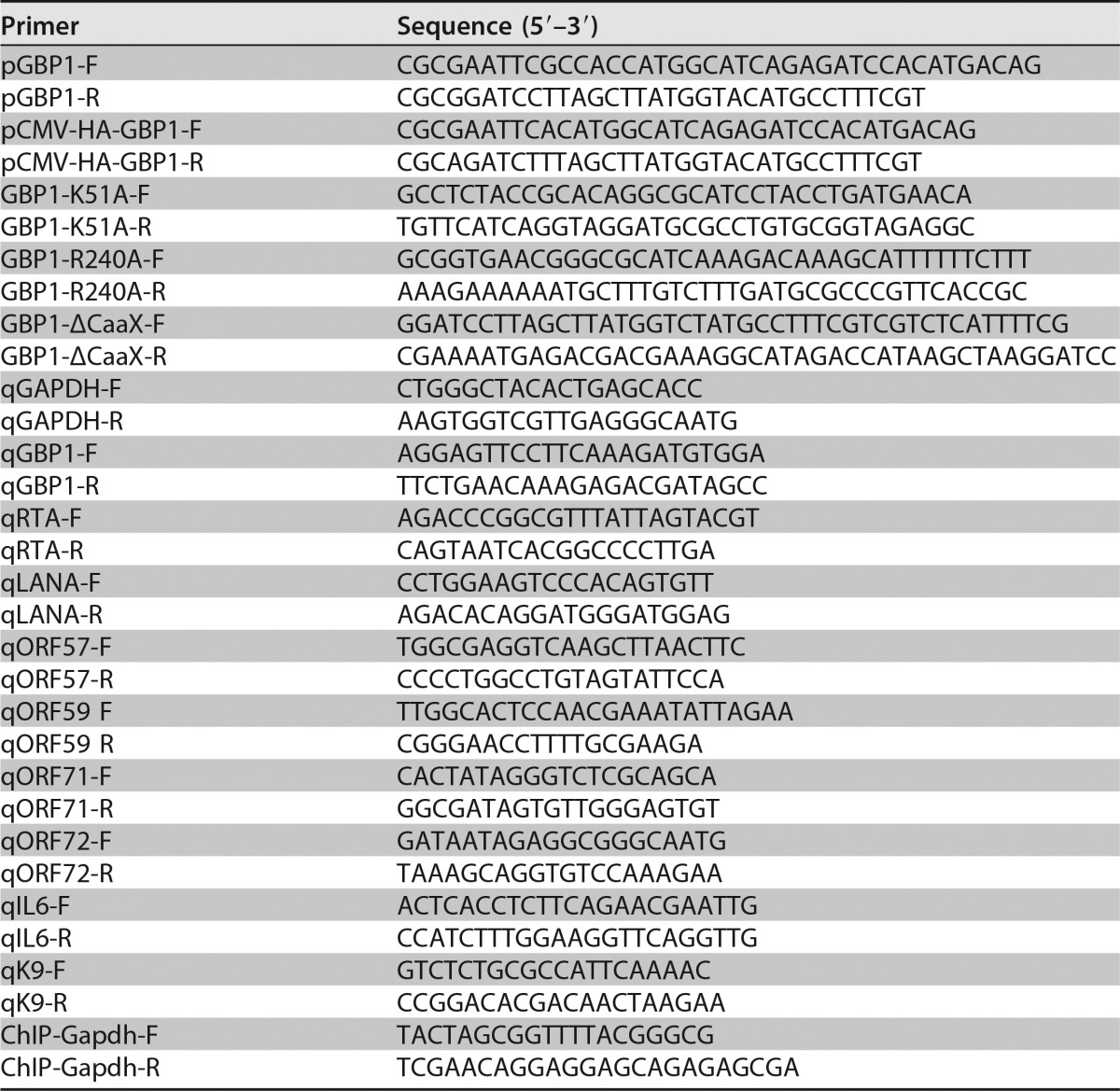
A GBP1 overexpression construct was amplified from the genomes of SLK cells and then inserted into pCDH-BLAST (System Bioscience); the plasmid was named GBP1-wt. Plasmid HA-GBP1 was subcloned from GBP1-wt and inserted into the pCMV-HA vector. shRNA targeting GBP1 (shGBP1) was converted into pLKO.1 by using the following primers: shGBP1-F (5′-CCGGCGGAAATTCTTCCCAAAGAAACTCGAGTTTCTTTGGGAAGAATTTCCGTTTTTG-3′) and shGBP1-R (5′-AATTCAAAAACGGAAATTCTTCCCAAAGAAACTCGAGTTTCTTTGGGAAGAATTTCCG-3′). RTA-SF, which encodes full-length RTA with tandem Strep-tag II and Flag epitopes at the C terminus, was described previously (26). One of the RTA E3 ligase mutants, H145L, was a kind gift from Yoshihiro Izumiya (UC Davis School of Medicine, Sacramento, CA, USA). The other three RTA mutants, C131S, C141S, and C141S/H145L, were described previously (26). The GBP1 mutants GBP1-K51A, GBP1-R240A, and GBP1-ΔCAAX were constructed by using a site-directed mutagenesis kit according to the manufacturer's protocol (QuikChange mutagenesis kit; Stratagene, Santa Clara, CA, USA). All the primers used for PCR amplification and qPCR are listed in Table 1.
TABLE 1.
Primers used in this study
Antibodies and reagents.
The antibodies used in this study were as follows: two antibodies used for the detection of endogenous GBP1, an antibody against GBP1 (1B1) (catalog number sc-53857; Santa Cruz, Dallas, TX, USA) and EPR8285 (catalog number ab131255; Abcam, Cambridge, MA, USA); anti-ORF65, which was a kind gift from Shoujiang Gao (University of Southern California, Los Angeles, CA, USA); anti-RTA (4D2; prepared in our laboratory); anti-GAPDH (catalog number E7EUT5; Abmart, Berkeley Heights, NJ, USA); anti-α-tubulin (catalog number T6199; Sigma, St. Louis, MO, USA); anti-histone H3 (catalog number BS1750; Bioworld, St. Louis, MO, USA); anti-p65 (catalog number 8242; Cell Signaling Technology, Danvers, MA, USA); anti-phospho-p65 (catalog number 3033; Cell Signaling Technology); anti-Flag (catalog number F1804; Sigma); and anti-HA (catalog number H6908; Sigma). The secondary antibodies used for Western blotting and the immunofluorescence assay were goat anti-mouse IRDye 800cw (catalog number 926-32210; Li-Cor, Lincoln, NE, USA), goat anti-rabbit IRDye 800cw (catalog number 926-32211; Li-Cor), goat anti-rat IRDye 800cw (catalog number 925-32219; Li-Cor), and goat anti-mouse IgG(H+L) Alexa Fluor 555 (catalog number A-21422; Invitrogen, Waltham, MA, USA). In addition, we used the NF-κB inhibitor Bay11-7082 (catalog number S1523; Beyotime, Haimen, China), a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) cell proliferation and cytotoxicity assay kit (catalog number C0009; Beyotime), anti-Flag M2 affinity gel (catalog number A2220; Sigma), MG132 (catalog number M7449; Sigma), Cytopainter phalloidin-iFluor 488 (catalog number ab176753; Abcam), and DAPI (4′,6-diamidino-2-phenylindole) (Sigma).
RNA extraction and qPCR.
To determine the RNA levels or genome copy numbers, qPCR was used. The cDNA was reverse transcribed with a genomic DNA remover reverse transcription (RT) kit (Toyobo, Osaka, Japan) from 0.5 μg of RNA extracted from cells harvested by using TRIzol reagent (Life Technologies, Grand Island, NY, USA). A SYBR green master mix kit (Toyobo) was used to perform qPCR. The qPCR program was conducted on the 7900HT sequence detection system (Life Technologies), and relative mRNA levels were normalized to GAPDH and calculated by the ΔΔCT method. All the primers used for qPCR are shown in Table 1. Quantification of intracellular KSHV genome copy numbers was performed as described previously (7).
Immunoprecipitation and Western blotting.
HEK 293T cells were seeded into 10-cm dishes and transfected with either RTA-SF or HA-GBP1 or cotransfected with RTA-SF and HA-GBP1; 48 h after transfection, the cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) for 1 h on ice, with brief vortexing every 20 min. Five percent of the cell lysate was taken as the input, while the remaining lysate was incubated with anti-Flag M2 affinity gel at 4°C for 3 h. The immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting.
KSHV infection and reactivation.
A KSHV stock was acquired by inducing iSLK.BAC16.RGB cells with DOX and valproate as described previously (33). The MOI of the concentrated viral stock was determined by infection of HEK 293T cells for 24 h. Infection of HEK 293T cells, SLK cells, and HUVECs was achieved by centrifugation at 1,250 × g at 37°C for 2 h; MOIs of 5 and 10 were used for infection of SLK cells and HUVECs, respectively. SLK cells were infected with KSHV at an MOI of 10, without centrifugation, for 6 h in the experiments investigating the mechanism by which GBP1 inhibits KSHV entry or nuclear delivery. iSLK cells were induced with 5 μg/ml DOX to stimulate the expression of RTA.
Inhibition of NF-κB induction.
Bay11-7082 was purchased from Beyotime and diluted in dimethyl sulfoxide (DMSO) for the stock. SLK cells were preincubated with or without 10 μM Bay11-7082 for 24 h before infection by KSHV. The cells were infected with KSHV.BAC16 at an MOI of 10 by centrifugation at 1,250 × g for 2 h, the supernatant was removed, and new culture medium with or without 10 μM Bay11-7082 was added to the cells. Cells were harvested by using TRIzol for RNA extraction, and the inhibition of the NF-κB pathway was indicated by a decrease in the IL-6 mRNA level.
Flow cytometry.
Stable cell lines were constructed as described above. Cells were infected with KHSV.BAC16.RGB at an MOI of approximately 5 by centrifugation at 1,250 × g for 2 h and collected at 48 hpi. The cells were then resuspended and fixed with 4% paraformaldehyde for 15 min, followed by washing with phosphate-buffered saline (PBS) three times. Cells were disaggregated by resuspension with PBS and filtration using cell strainer cap (Corning, Corning, NY, USA). Disaggregated cells were subsequently analyzed by using a fluorescence-activated cell sorter (BD Fortessa; BD, Sparks, MD, USA).
Isolation of KSHV genomes and nuclear extraction.
The kit used for the isolation of KSHV genomes from infected cells was purchased from Tiangen. SLK cells infected with KSHV at the indicated time points were washed twice with PBS to remove the unbound virus and then incubated for 5 min with trypsin-EDTA at 37°C to remove the viruses that were bound to the cell surface but not internalized. Cells were spun down and then washed once with PBS. To obtain the KSHV genomes within the cell nucleus, the cells acquired from the last step were resuspended with 500 μl hypotonic buffer (20 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2), incubated on ice for 15 min, and then added to 25 μl 10% NP-40, followed by vortexing for 10s. The cell nucleus was spun down by centrifugation at 956 × g at 4°C for 10 min.
Immunofluorescence assay.
SLK-Con and SLK-GBP1-wt cells that were uninfected or infected by KSHV were washed two or three times with PBS and then fixed with 4% paraformaldehyde (PFA) for 30 min. Next, cells were permeabilized by using 0.2% Triton X-100 for 30 min, followed by blocking in 10% goat serum (Life Technologies) for 30 min. The cells were incubated with anti-GBP1 rat monoclonal antibody or with anti-ORF65 mouse monoclonal antibody overnight at 4°C and then further stained with the secondary antibody goat anti-mouse IgG(H+L) Alexa Fluor 555 at a 1:1,000 dilution for 1 h. The actin filaments were stained with Cytopainter phalloidin-iFluor 488 for 90 min. Cell nuclei were stained with DAPI. Slides were photographed by using a digital camera and software (FV-1200; Olympus).
ACKNOWLEDGMENTS
This work was supported by grants from the Natural Science Foundation for Distinguished Young Scholar (81425017), the Ministry of Science and Technology of China (2016YFA0502100), the Key Project of the Natural Science Foundation of China (81230037), and the National Institutes of Health (1R01AI116442) to Ke Lan.
We thank Jae Jung, Shoujiang Gao (University of Southern California, Los Angeles, CA, USA), and Fanxiu Zhu (Florida State University, FL, USA) for cell lines and antibodies.
REFERENCES
- 1.Giffin L, Damania B. 2014. KSHV: pathways to tumorigenesis and persistent infection. Adv Virus Res 88:111–159. doi: 10.1016/B978-0-12-800098-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer 10:707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol Mech Dis 1:273–296. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- 4.Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J Clin Invest 120:939–949. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naranatt PP, Krishnan HH, Smith MS, Chandran B. 2005. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol 79:1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greene W, Gao SJ. 2009. Actin dynamics regulate multiple endosomal steps during Kaposi's sarcoma-associated herpesvirus entry and trafficking in endothelial cells. PLoS Pathog 5:e1000512. doi: 10.1371/journal.ppat.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raghu H, Sharma-Walia N, Veettil MV, Sadagopan S, Caballero A, Sivakumar R, Varga L, Bottero V, Chandran B. 2007. Lipid rafts of primary endothelial cells are essential for Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8-induced phosphatidylinositol 3-kinase and RhoA-GTPases critical for microtubule dynamics and nuclear delivery of viral DNA but dispensable for binding and entry. J Virol 81:7941–7959. doi: 10.1128/JVI.02848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brulois K, Jung JU. 2014. Interplay between Kaposi's sarcoma-associated herpesvirus and the innate immune system. Cytokine Growth Factor Rev 25:597–609. doi: 10.1016/j.cytogfr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bussey KA, Reimer E, Todt H, Denker B, Gallo A, Konrad A, Ottinger M, Adler H, Sturzl M, Brune W, Brinkmann MM. 2014. The gammaherpesviruses Kaposi's sarcoma-associated herpesvirus and murine gammaherpesvirus 68 modulate the Toll-like receptor-induced proinflammatory cytokine response. J Virol 88:9245–9259. doi: 10.1128/JVI.00841-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai MS, Li ML, Zheng CF. 2012. Herpesviral infection and Toll-like receptor 2. Protein Cell 3:590–601. doi: 10.1007/s13238-012-2059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HR, Lee S, Chaudhary PM, Gill P, Jung JU. 2010. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Future Microbiol 5:1349–1365. doi: 10.2217/fmb.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadagopan S, Sharma-Walia N, Veettil MV, Raghu H, Sivakumar R, Bottero V, Chandran B. 2007. Kaposi's sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J Virol 81:3949–3968. doi: 10.1128/JVI.02333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. 2012. IFN-inducible GTPases in host cell defense. Cell Host Microbe 12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens S, Howard J. 2006. The interferon-inducible GTPases. Annu Rev Cell Dev Biol 22:559–589. doi: 10.1146/annurev.cellbio.22.010305.104619. [DOI] [PubMed] [Google Scholar]
- 15.Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. 2000. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 16.Naschberger E, Werner T, Vicente AB, Guenzi E, Popolt K, Leubert R, Lubeseder-Martellato C, Nelson PJ, Sturzl M. 2004. Nuclear factor-κB motif and interferon-α-stimulated response element co-operate in the activation of guanylate-binding protein-1 expression by inflammatory cytokines in endothelial cells. Biochem J 379:409–420. doi: 10.1042/bj20031873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. 2010. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature 465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vestal DJ. 2005. The guanylate-binding proteins (GBPs): proinflammatory cytokine-induced members of the dynamin superfamily with unique GTPase activity. J Interferon Cytokine Res 25:435–443. doi: 10.1089/jir.2005.25.435. [DOI] [PubMed] [Google Scholar]
- 19.Itsui Y, Sakamoto N, Kakinuma S, Nakagawa M, Sekine-Osajima Y, Tasaka-Fujita M, Nishimura-Sakurai Y, Suda G, Karakama Y, Mishima K, Yamamoto M, Watanabe T, Ueyama M, Funaoka Y, Azuma S, Watanabe M. 2009. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50:1727–1737. doi: 10.1002/hep.23195. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z, Shi Z, Yan W, Wei J, Shao D, Deng X, Wang S, Li B, Tong G, Ma Z. 2013. Nonstructural protein 1 of influenza A virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity. PLoS One 8:e55920. doi: 10.1371/journal.pone.0055920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan W, Zuo X, Feng T, Shi X, Dai J. 2012. Guanylate-binding protein 1 participates in cellular antiviral response to dengue virus. Virol J 9:292. doi: 10.1186/1743-422X-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossmann C, Ganem D. 2008. Effects of NFkappaB activation on KSHV latency and lytic reactivation are complex and context-dependent. Virology 375:94–102. doi: 10.1016/j.virol.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson SL, Carton JM, Lou J, Xing L, Rubin BY. 1999. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256:8–14. doi: 10.1006/viro.1999.9614. [DOI] [PubMed] [Google Scholar]
- 24.Li LF, Yu J, Li Y, Wang J, Li S, Zhang L, Xia SL, Yang Q, Wang X, Yu S, Luo Y, Sun Y, Zhu Y, Munir M, Qiu HJ. 2016. Guanylate-binding protein 1, an interferon-induced GTPase, exerts an antiviral activity against classical swine fever virus depending on its GTPase activity. J Virol 90:4412–4426. doi: 10.1128/JVI.02718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. 2011. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science 332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Q, Liang D, Sun R, Jia B, Xia T, Xiao H, Lan K. 2015. Kaposi's sarcoma-associated herpesvirus-encoded replication and transcription activator impairs innate immunity via ubiquitin-mediated degradation of myeloid differentiation factor 88. J Virol 89:415–427. doi: 10.1128/JVI.02591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarviluoma A, Ojala PM. 2006. Cell signaling pathways engaged by KSHV. Biochim Biophys Acta 1766:140–158. [DOI] [PubMed] [Google Scholar]
- 28.Yu Y, Wang SE, Hayward GS. 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22:59–70. doi: 10.1016/j.immuni.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J Virol 74:5300–5309. doi: 10.1128/JVI.74.11.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coscoy L, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci U S A 97:8051–8056. doi: 10.1073/pnas.140129797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West J, Damania B. 2008. Upregulation of the TLR3 pathway by Kaposi's sarcoma-associated herpesvirus during primary infection. J Virol 82:5440–5449. doi: 10.1128/JVI.02590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matta H, Chaudhary PM. 2004. Activation of alternative NF-kappa B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1 beta-converting enzyme inhibitory protein (vFLIP). Proc Natl Acad Sci U S A 101:9399–9404. doi: 10.1073/pnas.0308016101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 174:12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerrit JK, Praefcke CH. 1999. Nucleotide-binding characteristics of human guanylate-binding protein 1 (hGBP1) and identification of third GTP-binding motif. J Mol Biol 292:321–332. [DOI] [PubMed] [Google Scholar]
- 35.Vetter IR, Wittinghofer A. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 36.Praefcke GJ, Kloep S, Benscheid U, Lilie H, Prakash B, Herrmann C. 2004. Identification of residues in the human guanylate-binding protein 1 critical for nucleotide binding and cooperative GTP hydrolysis. J Mol Biol 344:257–269. doi: 10.1016/j.jmb.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 37.Wehner M, Kunzelmann S, Herrmann C. 2012. The guanine cap of human guanylate-binding protein 1 is responsible for dimerization and self-activation of GTP hydrolysis. FEBS J 279:203–210. doi: 10.1111/j.1742-4658.2011.08415.x. [DOI] [PubMed] [Google Scholar]
- 38.Modiano N, Lu YE, Cresswell P. 2005. Golgi targeting of human guanylate-binding protein-1 requires nucleotide binding, isoprenylation, and an IFN-gamma-inducible cofactor. Proc Natl Acad Sci U S A 102:8680–8685. doi: 10.1073/pnas.0503227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Britzen-Laurent N, Bauer M, Berton V, Fischer N, Syguda A, Reipschlager S, Naschberger E, Herrmann C, Sturzl M. 2010. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One 5:e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostler N, Britzen-Laurent N, Liebl A, Naschberger E, Lochnit G, Ostler M, Forster F, Kunzelmann P, Ince S, Supper V, Praefcke GJ, Schubert DW, Stockinger H, Herrmann C, Sturzl M. 2014. Gamma interferon-induced guanylate binding protein 1 is a novel actin cytoskeleton remodeling factor. Mol Cell Biol 34:196–209. doi: 10.1128/MCB.00664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng F, He M, Jung JU, Lu C, Gao SJ. 2016. Suppression of Kaposi's sarcoma-associated herpesvirus infection and replication by 5′-AMP-activated protein kinase. J Virol 90:6515–6525. doi: 10.1128/JVI.00624-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bechtel JT, Winant RC, Ganem D. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J Virol 79:4952–4964. doi: 10.1128/JVI.79.8.4952-4964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lan K, Kuppers DA, Verma SC, Sharma N, Murakami M, Robertson ES. 2005. Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J Virol 79:7453–7465. doi: 10.1128/JVI.79.12.7453-7465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreoli M, Persico M, Kumar A, Orteca N, Kumar V, Pepe A, Mahalingam S, Alegria AE, Petrella L, Sevciunaite L, Camperchioli A, Mariani M, Di Dato A, Novellino E, Scambia G, Malhotra SV, Ferlini C, Fattorusso C. 2014. Identification of the first inhibitor of the GBP1:PIM1 interaction. Implications for the development of a new class of anticancer agents against paclitaxel resistant cancer cells. J Med Chem 57:7916–7932. doi: 10.1021/jm5009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan K, Kuppers DA, Verma SC, Robertson ES. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J Virol 78:6585–6594. doi: 10.1128/JVI.78.12.6585-6594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pozharskaya VP, Weakland LL, Offermann MK. 2004. Inhibition of infectious human herpesvirus 8 production by gamma interferon and alpha interferon in BCBL-1 cells. J Gen Virol 85:2779–2787. doi: 10.1099/vir.0.80214-0. [DOI] [PubMed] [Google Scholar]
- 47.Konrad A, Wies E, Thurau M, Marquardt G, Naschberger E, Hentschel S, Jochmann R, Schulz TF, Erfle H, Brors B, Lausen B, Neipel F, Sturzl M. 2009. A systems biology approach to identify the combination effects of human herpesvirus 8 genes on NF-kappaB activation. J Virol 83:2563–2574. doi: 10.1128/JVI.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]