ABSTRACT
Zika virus (ZIKV), a member of the Flaviviridae family, has recently emerged as an important human pathogen with increasing economic and health impact worldwide. Because of its teratogenic nature and association with the serious neurological condition Guillain-Barré syndrome, a tremendous amount of effort has focused on understanding ZIKV pathogenesis. To gain further insights into ZIKV interaction with host cells, we investigated how this pathogen affects stress response pathways. While ZIKV infection induces stress signaling that leads to phosphorylation of eIF2α and cellular translational arrest, stress granule (SG) formation was inhibited. Further analysis revealed that the viral proteins NS3 and NS4A are linked to translational repression, whereas expression of the capsid protein, NS3/NS2B-3, and NS4A interfered with SG formation. Some, but not all, flavivirus capsid proteins also blocked SG assembly, indicating differential interactions between flaviviruses and SG biogenesis pathways. Depletion of the SG components G3BP1, TIAR, and Caprin-1, but not TIA-1, reduced ZIKV replication. Both G3BP1 and Caprin-1 formed complexes with capsid, whereas viral genomic RNA stably interacted with G3BP1 during ZIKV infection. Taken together, these results are consistent with a scenario in which ZIKV uses multiple viral components to hijack key SG proteins to benefit viral replication.
IMPORTANCE There is a pressing need to understand ZIKV pathogenesis in order to advance the development of vaccines and therapeutics. The cellular stress response constitutes one of the first lines of defense against viral infection; therefore, understanding how ZIKV evades this antiviral system will provide key insights into ZIKV biology and potentially pathogenesis. Here, we show that ZIKV induces the stress response through activation of the UPR (unfolded protein response) and PKR (protein kinase R), leading to host translational arrest, a process likely mediated by the viral proteins NS3 and NS4A. Despite the activation of translational shutoff, formation of SG is strongly inhibited by the virus. Specifically, ZIKV hijacks the core SG proteins G3BP1, TIAR, and Caprin-1 to facilitate viral replication, resulting in impaired SG assembly. This process is potentially facilitated by the interactions of the viral RNA with G3BP1 as well as the viral capsid protein with G3BP1 and Caprin-1. Interestingly, expression of capsid proteins from several other flaviviruses also inhibited SG formation. Taken together, the present study provides novel insights into how ZIKV modulates cellular stress response pathways during replication.
KEYWORDS: Zika virus, unfolded protein response, translational arrest, stress granules, G3BP1, Caprin-1, capsid protein, NS3, NS4A
INTRODUCTION
Zika virus (ZIKV) belongs to the family Flaviviridae, a large group of RNA viruses that include the important human pathogens dengue virus (DENV), Japanese encephalitis virus (JEV), and West Nile virus (WNV), to name a few. It is transmitted mainly by the mosquito vectors Aedes spp., but sexual transmission of ZIKV has also been reported (1). Infection of humans is usually asymptomatic or limited to flu-like symptoms (2). However, the dramatic increase in the number of microcephaly cases during the recent ZIKV outbreak in Brazil (3–5) prompted intense investigation that eventually confirmed the virus as a teratogenic agent that can cause significant developmental defects in fetuses (6, 7). ZIKV infection in adults has also been linked to Guillain-Barré syndrome, a neurological condition that can lead to paralysis and, in some cases, death (8). Evidence from clinical and animal studies indicates that ZIKV is unique among flaviviruses in that it can persist in testes (9, 10) and fetal brain (11–13). Consistent with this idea, studies from our laboratory and others have revealed multiple mechanisms used by ZIKV to evade the host interferon system (14–16), an indispensable antiviral response that controls ZIKV infection and pathogenesis (17, 18). How the virus counteracts other cellular antiviral pathways is largely unknown.
Stress response pathways are among the first lines of defense that mammalian cells deploy against viruses (19). Their activation can lead to global translational arrest and formation of stress granules (SGs), which are dynamic cytoplasmic RNA granules composed of cellular mRNAs and stalled preinitiation complexes (reviewed in reference 20). SGs, whose formation can be induced by phosphorylation of the eukaryotic initiation factor 2α (eIF2α), maintain RNA homeostasis under stress conditions. EIF2α is a substrate for at least four kinases that are activated in response to different stress stimuli (20). These include HRI (heme-regulated inhibitor), which is activated by oxidative and heat shock stress; the endoplasmic reticulum (ER) membrane-resident PERK (protein kinase R [PKR]-like ER kinase), which detects unfolded protein stress in the ER lumen; GCN2 (general control nonderepressible-2) kinase, which recognizes uncharged tRNAs during nutrient starvation; and PKR (protein kinase R), which detects viral dsRNA (double-stranded RNA) during RNA virus infection. Phosphorylation of eIF2α by any of these kinases leads to inhibition of preinitiation complex formation, resulting in suppression of protein translation initiation (20). Binding of SG nucleating factors to the stalled polysomes then drives the formation of SGs through rapid condensation of RNA/protein aggregates. Several proteins with RNA-binding and self-aggregating properties have been identified as important SG nucleating factors, including Ras-GAP SH3 domain-binding protein (G3BP) (21), T-cell-restricted intracellular antigen 1 (TIA-1), and the TIA-1-related protein (TIAR) (22, 23). Assembly of SGs requires an intact microtubule network (24, 25) and can be affected by posttranslational modifications of assembly factors (26).
SGs can restrict viral access to translational machinery, and in this sense they are generally viewed as antiviral. Not surprisingly, some viruses have evolved ways to counteract SG assembly. For instance, the poliovirus 3Cpro protease cleaves G3BP, thereby inhibiting SG formation (27). Viruses can also block SG formation at earlier steps. For example, nonstructural protein 1 of influenza A virus sequesters viral dsRNA, preventing the activation of PKR and PKR-mediated phosphorylation of eIF2α (28, 29). In doing so, the virus circumvents translational arrest and SG formation (28, 29).
Some flaviviruses, including DENV and WNV, have been reported to exploit the SG proteins TIA-1 and TIAR to facilitate synthesis of viral genomes, a process that eventually leads to inhibition of SG formation (30). However, it is not clear how this occurs, and the viral proteins that facilitate this process have not been identified. Interestingly, the related hepatitis C virus (HCV) seems to induce assembly of SGs or SG-like foci in an oscillating fashion (31, 32). HCV also coopts G3BP1, TIAR, TIA-1, PABP, and USP10 to support different stages of the viral life cycle (32, 33). Finally, recent studies showed that ZIKV infection suppresses SG formation (34, 35) through a process that is not linked to eIF2α phosphorylation-mediated translational repression (34). However, the precise mechanism by which ZIKV prevents SG assembly remains to be elucidated.
Here, we endeavored to further understand the interplay between ZIKV and host stress pathways. We observed that despite inducing the unfolded protein response (UPR), PKR activation, eIF2α phosphorylation, and arrest of host cell protein synthesis, SG formation was strongly suppressed during ZIKV infection. Mapping studies revealed that multiple viral proteins block host cell translation and SG formation. Moreover, short interfering RNA (siRNA)-mediated depletion of the SG factors G3BP1, TIAR, and Caprin-1 had a negative impact on ZIKV replication. The ZIKV capsid protein, whose expression blocks SG formation, was shown to form stable complexes with G3BP1 and Caprin-1. Interestingly, expression of capsids from yellow fever virus (YFV) and Murray Valley encephalitis virus (MVEV), but not DENV or WNV, also negatively impacted SG biogenesis. Together, our data demonstrate that ZIKV manipulates the host stress response pathways to benefit viral replication.
RESULTS
ZIKV infection does not induce robust SG formation in A549 cells or primary HFAs.
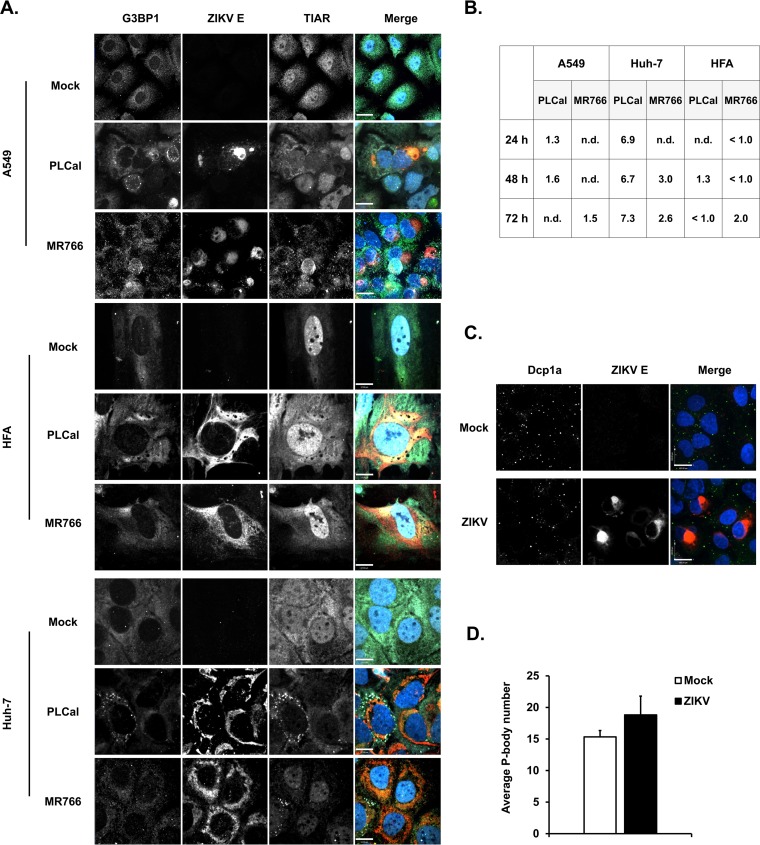
As a first step toward understanding how ZIKV infection affects cellular stress response pathways, we used confocal microscopy to monitor SG formation in A549 and Huh-7 cells infected with African-lineage (MR766) and Asian-lineage (PLCal) strains of ZIKV. SGs were identified by staining for G3BP1 and TIAR. ZIKV-infected cells were identified by positive staining for viral envelope (E) protein. As shown in Fig. 1A and B, very few G3BP1/TIAR-positive foci were observed in the cytoplasm of ZIKV PL-Cal- or MR766-infected A549 and Huh-7 cells at 24, 48, and 72 h postinfection (h.p.i.), indicating that unlike other RNA viruses, such as respiratory syncytial virus and HCV (31, 32, 36), ZIKV infection does not robustly induce SG formation. Similar results were obtained using ZIKV-infected primary human fetal astrocytes (HFAs) (Fig. 1A and B), a brain cell type that has recently been reported to support ZIKV replication (37, 38). As both ZIKV strains had similar effects on SG induction, all subsequent experiments were carried out using the PLCal strain.
FIG 1.
ZIKV infection does not induce robust SG formation in A549 cells or primary human fetal astrocytes (HFAs). A549 cells, primary HFAs, and Huh-7 cells were infected with ZIKV strain PLCal or MR766 (MOI of 3) for the indicated time periods and then processed for confocal microscopy analyses. Infected cells were detected using mouse anti-flavivirus E protein antibody; SGs were identified by staining for endogenous G3BP1 and TIAR using rabbit and goat polyclonal antibodies, respectively. (A) Representative images of mock- and ZIKV-infected cells at 48 h.p.i. are shown. (B) Quantification of SGs was performed using the software Volocity, and the average numbers of SGs per cell are shown in the table. A minimum of 30 cells was used for each sample. n.d., not detected. (C) A549 cells were infected with ZIKV (PLCal strain) for 48 h and then processed for confocal microscopy analyses. P-bodies were identified by staining Dcp1a with a rabbit polyclonal antibody. Primary antibodies were detected with donkey anti-mouse IgG conjugated to Alexa Fluor 546, donkey anti-rabbit IgG conjugated to Alexa Fluor 488, and chicken anti-goat IgG conjugated to Alex Fluor 647. Nuclei were stained with DAPI. Size bars are 17 μm (A) and 100 μm (C). Images were acquired on a spinning-disk confocal microscope. (D) Quantification of P-bodies was performed using Volocity software. A minimum of 30 cells were used for each sample. Values are expressed as the average number of P-bodies per cell ± standard errors from three independent experiments.
We next assessed whether ZIKV infection affects abundance of another cytoplasmic RNA granule, processing bodies (P-bodies), whose functions are closely linked to SGs (39). To visualize these structures, we used an antibody to the P-body resident decapping protein 1a (Dcp1a). As shown in Fig. 1C, the morphology and localization of P-bodies was not affected by ZIKV infection. Moreover, quantitation showed that there was no significant difference in the numbers of P-bodies between mock- and ZIKV-infected cells (Fig. 1D).
ZIKV inhibits SG formation induced by different stress stimuli.
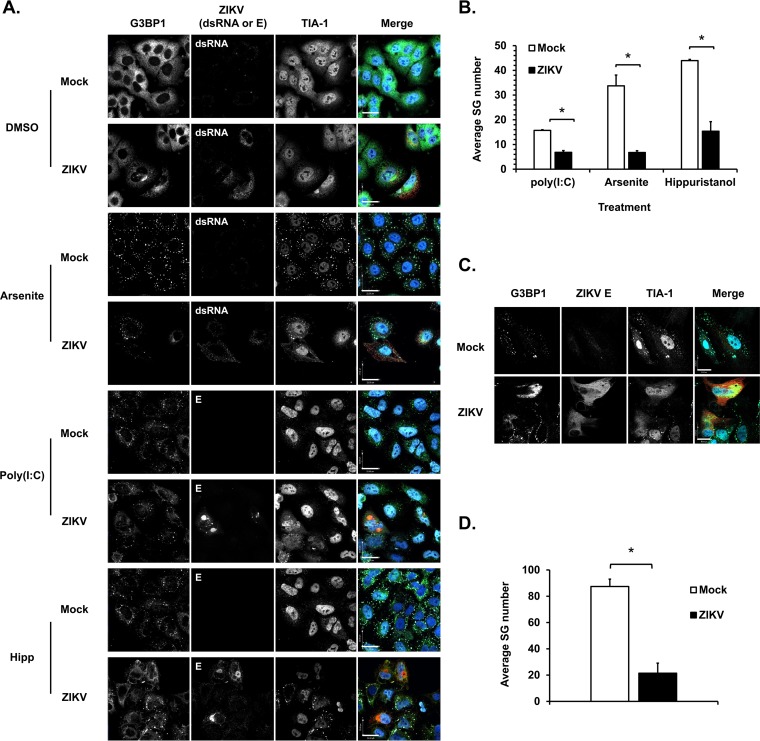
DENV and WNV have been shown to inhibit SG formation induced by oxidative stress (30, 34), indicating that flaviviruses employ active countermeasures to block formation of these structures. To investigate this further, we quantitated the numbers of SGs in ZIKV-infected cells following treatment with three different SG inducers [sodium arsenite, poly(I·C) and hippuristanol], all of which interfere with translation initiation. Specifically, sodium arsenite induces oxidative stress that leads to HRI-mediated phosphorylation of eIF2α (20), whereas poly(I·C) is a dsRNA analog that mimics viral replication intermediates. It induces PKR activation and subsequent eIF2α phosphorylation. In contrast, hippuristanol stimulates SG formation through an eIF2α-independent mechanism by interfering with the RNA-binding activity of the translation initiation factor eIF4A (40, 41).
The effect of poly(I·C), arsenite, and hippuristanol on SG formation (as assessed by quantifying G3BP1 and TIA-1 double-positive foci in the cytoplasm) varied significantly, with hippuristanol having the most robust effect (Fig. 2A and B). ZIKV-infected A549 cells contained 40 to 75% fewer SGs depending upon which inducer was employed (Fig. 2B). These data indicate that the virus actively blocks SG assembly mediated through HRI and PKR as well as through translational arrest mediated by inactivation of eIF4A. Similarly, in primary HFAs, the number of arsenite-induced SGs was significantly reduced by ZIKV infection (Fig. 2C and D).
FIG 2.
ZIKV inhibits SG formation induced by multiple stress stimuli. (A) A549 cells were infected with ZIKV (MOI of 3) for 24 h. Cells were then treated with 0.5 mM sodium arsenite (Arsenite) for 30 min, 1 μM hippuristanol (Hipp) for 25 min, or dimethyl sulfoxide (DMSO) (vehicle control) or were transfected with 0.5 μg of poly(I·C) for 10 h and then processed for confocal microscopy. Infected cells were detected with mouse antibodies to flavivirus E protein or dsRNA. SGs were identified using rabbit anti-G3BP1 and goat anti-TIA-1. Primary antibodies were detected with donkey anti-rabbit IgG conjugated to Alexa Fluor 488, donkey anti-mouse IgG conjugated to Alexa Fluor 546, and chicken anti-goat IgG conjugated to Alexa Fluor 647. Nuclei were stained with DAPI. Images were acquired using a spinning-disc confocal microscope. Size bar, 22 μm. (C) Primary human fetal astrocytes were infected with ZIKV (MOI of 5) for 48 h, left untreated or treated with sodium arsenite (0.5 mM) for 30 min, and processed for confocal microscopy. Infected cells were detected with mouse antibodies to flavivirus E protein. SGs were identified using rabbit anti-G3BP1 and goat anti-TIA-1. (B and D) Quantification of SGs was performed using Volocity software. A minimum of 30 cells was used for each sample. Values are the average number of SGs per cell ± standard errors from three independent experiments. P values were determined by Student's t test. *, P < 0.05 (significant).
ZIKV triggers activation of the stress response.
SG formation can occur when protein translation is blocked through phosphorylation of eIF2α (20). Since our data indicated that ZIKV can inhibit SG formation mediated through both HRI and PKR (Fig. 2A), we next determined whether ZIKV interferes with phosphorylation of the downstream target, eIF2α. Because other flaviviruses can trigger stress response pathways and phosphorylation of eIF2α through PKR and UPR (32, 42–44), we monitored activation of both pathways during ZIKV infection. Significant upregulation of phospho-PKR was observed at 24, 48, and 60 h.p.i. in A549 cells (Fig. 3A) and HFAs (Fig. 3B). Increased levels of phospho-eIF2α were observed at 24 and 48 h.p.i. in both cell types (Fig. 3A and B). In addition, ZIKV infection in A549 cells caused a dramatic increase in the level of spliced XBP1 transcripts (Fig. 3A, bottom), an indicator of UPR activation (45). This occurred as early as 12 h.p.i. and was sustained until at least 60 h.p.i. (Fig. 3A, bottom). These data indicate that ZIKV infection activates both PKR- and UPR-mediated signaling, as well as phosphorylation of the downstream target eIF2α, albeit with slightly different kinetics. To determine whether the delay in eIF2α phosphorylation at early time points (12 h.p.i.) was due to active suppression by the virus, ZIKV-infected A549 cells were treated 12 h.p.i. with poly(I·C), arsenite, or the ER stress inducer thapsigargin to induce eIF2α phosphorylation. Data shown in Fig. 3C to E revealed that irrespective of the stress-inducing agent, increased phosphorylation of eIF2α was observed in ZIKV-infected cells. In fact, levels of phospho-eIF2α were similar to those observed in mock-infected cells that were treated with stress inducers. Similarly, ZIKV infection did not affect activation of PKR or UPR, as indicated by comparable levels of phospho-PKR and spliced XBP1 between mock-infected and infected samples following poly(I·C) and thapsigargin treatment, respectively (Fig. 3C and E). Together, these data indicate that the block in SG formation during ZIKV infection is downstream of PKR- and UPR-induced phosphorylation of eIF2α.
FIG 3.
ZIKV infection does not interfere with early stress response signaling. (A) A549 cells were infected with ZIKV (MOI of 3) for the indicated time periods, after which total cell lysates were processed for immunoblotting or RT-PCR and agarose gel electrophoresis for detection of unspliced XBP1 (usXBP1) and spliced XBP1 (sXBP1) transcripts. (B) Primary human fetal astrocytes were infected with ZIKV (MOI of 5) for the indicated time periods, after which total cell lysates were processed for immunoblotting. (C and D) A549 cells were infected with ZIKV (MOI of 5) for 12 h and then transfected with 0.5 μg of poly(I·C) for 2 h (C) or treated with sodium arsenite (Ars; 0.5 mM) for 1 h before immunoblot analyses (D). (E) A549 cells were infected with ZIKV (MOI of 5) for 16 h and then treated with thapsigargin (Thap; 1 μM) for 1 h. Cell lysates were harvested and processed for immunoblotting (top) or RT-PCR analysis and agarose gel electrophoresis (bottom). Total PKR, phospho-PKR (P-PKR), total eIF2α, and phospho-eIF2α (P-eIF2α) were detected using appropriate antibodies. Levels of β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ZIKV envelope protein (ZIKV E) are shown as loading and infection controls, respectively. Quantification of P-PKR and P-eIF2α induction levels was performed using the software Image Studio Lite. Values are expressed as means from three independent experiments. Values that are underlined indicate statistical significance by Student's t test (P < 0.05).
ZIKV infection leads to translational arrest in host cells.
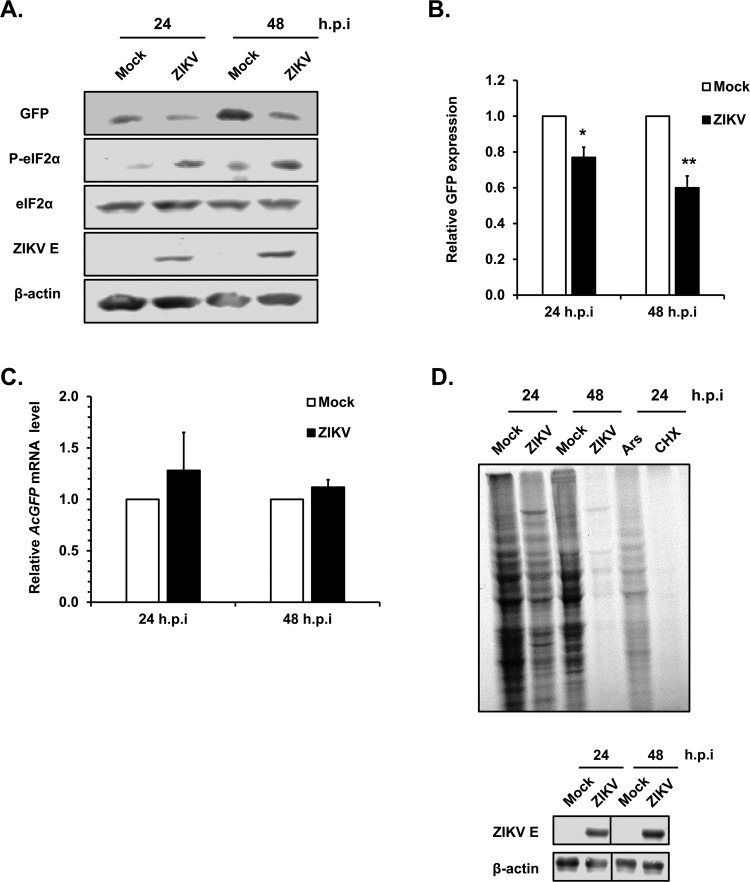
We next investigated whether ZIKV-induced eIF2α phosphorylation was associated with arrest of host cell protein synthesis. First, we measured the effect of viral infection on the expression of an inert ectopic protein (AcGFP). Consistent with the data shown in Fig. 3A, phospho-eIF2α was upregulated at 24 and 48 h.p.i. (Fig. 4A). Interestingly, while the level of viral E protein increased during this period, expression of AcGFP protein was significantly lower in ZIKV-infected cells than in mock-infected cells (Fig. 4B). This was not due to lower AcGFP mRNA levels in the presence of viral infection (Fig. 4C). We next monitored the level of nascent protein synthesis during ZIKV infection. Results from 35S labeling and autoradiography experiments indicate that ZIKV infection had a dramatic effect on global protein synthesis, similar to what was observed in cells treated with the translation inhibitors arsenite and cycloheximide (Fig. 4D, top). Infection of the cells by ZIKV was confirmed by immunoblotting for ZIKV E protein (Fig. 4D, bottom).
FIG 4.
ZIKV infection inhibits host cell protein synthesis. (A) A549 cells transduced with pTRIP-AcGFP lentivirus (MOI, 2) were infected 12 h later with ZIKV (MOI, 5). Cell lysates were harvested at 24 h.p.i. and 48 h.p.i. for immunoblot and qRT-PCR analyses. AcGFP was detected with antibodies to GFP. Levels of total eIF2α and phospho-eIF2α (P-eIF2α) are shown. Levels of β-actin and ZIKV envelope protein (ZIKV E) are shown as loading and infection controls, respectively. (B) The relative levels of AcGFP protein in mock- and ZIKV-infected cells were determined using Image Studio Lite software. Values are expressed as means ± standard errors from three independent experiments. P values were determined by Student's t test. *, P < 0.05 (significant); **, P < 0.01 (highly significant). (C) Total RNA was extracted from mock- and ZIKV-infected cells, and the relative mRNA levels for ACTB and AcGFP were determined by qRT-PCR. The values are expressed as means ± standard errors from three independent experiments. (D) A549 cells were infected with ZIKV (MOI, 5) for 24 and 48 h. Following depletion of amino acids for 1 h, cells were incubated with or without 110 μCi/ml of [35S]cysteine and methionine for 2 h. Cell lysates were harvested and processed for SDS-PAGE and autoradiography (top) or immunoblotting (bottom). Sodium arsenite (Ars; 0.5 mM) and cycloheximide (CHX; 50 μM) were added 30 min prior to 35S labeling as controls. For immunoblot analyses, levels of β-actin and ZIKV envelope protein (ZIKV E) are shown as loading and infection controls, respectively. Representative blots from three independent experiments are shown.
ZIKV NS3 and NS4A facilitate global translational arrest in host cells.
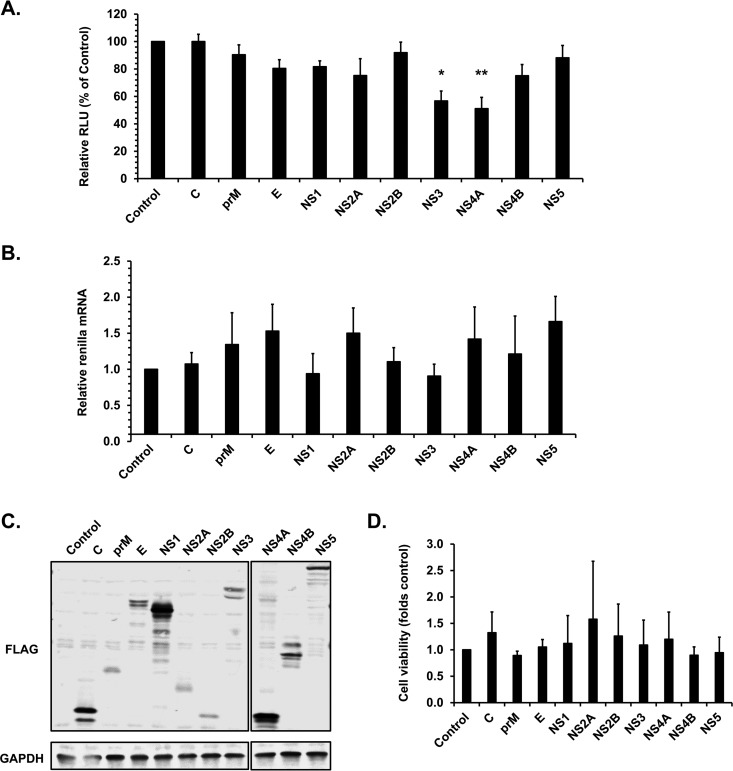
We next sought to determine which viral proteins inhibit translation in host cells. Individual ZIKV proteins were coexpressed with a renilla luciferase reporter in HEK293T cells, and the effect on reporter protein expression was assessed by luciferase assay. As shown in Fig. 5A, expression of NS3 or NS4A reduced luciferase expression in cotransfected cells by 40 to 45% but did not affect luciferase mRNA levels (Fig. 5B). The expression of individual viral proteins was confirmed by immunoblotting (Fig. 5C). The cell viability assay indicated expression of these proteins did not cause significant cytotoxic effects (Fig. 5D). Together, these data suggest that NS4A and/or NS3 plays a putative role in blocking host cell translation without affecting transcription.
FIG 5.
Expression of ZIKV NS3 and NS4A reduces cellular protein translation. HEK293T cells were cotransfected with plasmids encoding ZIKV proteins and renilla luciferase. Forty-eight hours posttransfection, cell lysates were subjected to luciferase assay, qRT-PCR analyses, immunoblotting, and cell viability assays. (A) Mean luciferase activities are expressed as relative luminescence units (RLU) ± standard errors from three independent experiments. P values were determined by Student's t test. *, P < 0.05 (significant); **, P < 0.01 (highly significant). (B) Relative levels of renilla luciferase mRNA (compared to ACTB) were determined by qRT-PCR. Values represent the means ± standard errors from three independent experiments. (C) Immunoblot analysis of FLAG-tagged ZIKV proteins as identified by an anti-FLAG antibody. GAPDH was used as the loading control. (D) Cell viability is expressed as the relative value ± standard errors from three independent experiments.
ZIKV does not alter expression of key SG proteins.
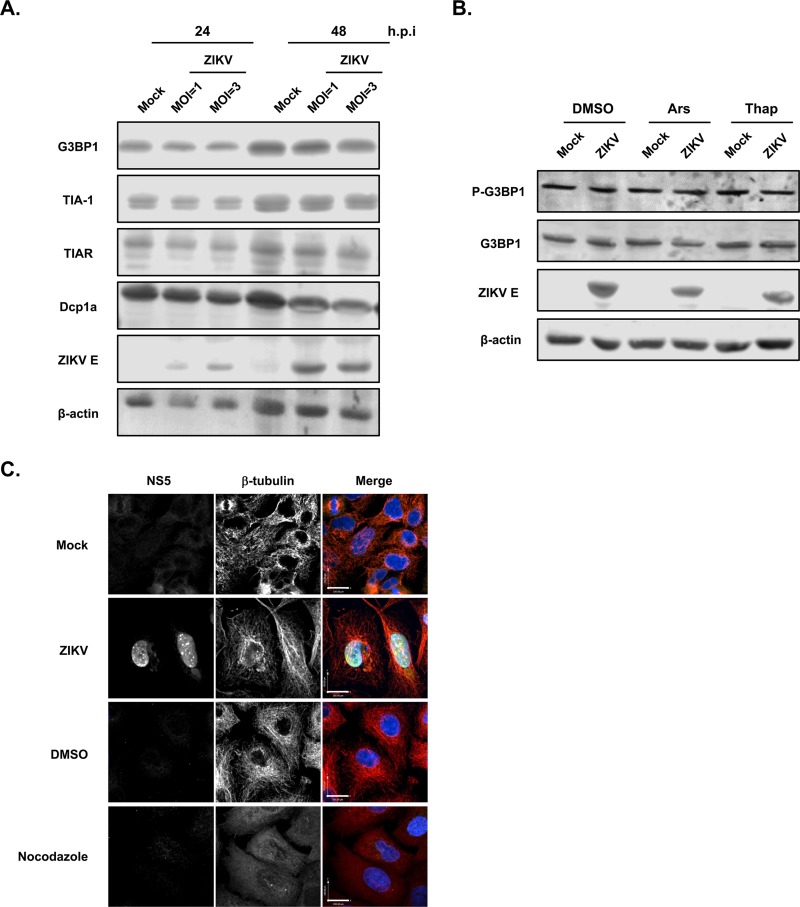
One of the strategies used by viruses to antagonize SG formation is cleavage of key SG factors. A classic example is the poliovirus protease 3Cpro, which cleaves G3BP and eIF4GI, thus preventing SG assembly (27, 46). To determine whether ZIKV inhibits SG assembly by cleaving or altering the expression of SG proteins, immunoblot analysis was performed to compare the steady-state levels of G3BP1, TIA-1, and TIAR in mock- and ZIKV-infected A549 cells. Data depicted in Fig. 6A show that relative levels of these three core SG proteins were not affected by ZIKV infection, and there was no evidence of intermediate cleavage products (data not shown).
FIG 6.
ZIKV infection does not affect steady-state levels of key SG-associated proteins or disrupt the microtubule network. (A) A549 cells infected with ZIKV (MOI, 1 or 3) were harvested at 24 or 48 h.p.i., and then levels of G3BP1, TIA-1, TIAR, and Dcpa1 were determined by immunoblotting. Levels of β-actin and ZIKV envelope protein (ZIKV E) are shown as loading and infection controls, respectively. (B) A549 cells infected with ZIKV (MOI, 5) for 16 h were left untreated or were treated with sodium arsenite (Ars; 0.5 mM) or thapsigargin (Thap; 1 μM) for 1 h. Levels of total G3BP1 and phospho-G3BP1 (P-G3BP1) were determined by immunoblotting. Levels of β-actin and ZIKV envelope protein (ZIKV E) are shown as loading and infection controls, respectively. (C) A549 cells were infected with ZIKV (MOI, 3) for 48 h and processed for confocal microscopy. As positive controls, cells were treated with 10 μM nocodazole for 8 h prior to fixing. Infected cells were detected using goat anti-NS5 and donkey anti-goat IgG conjugated to Alexa Fluor 488. Microtubules were stained with mouse anti-β-tubulin and donkey anti-mouse IgG conjugated to Alexa Fluor 546. Nuclei were stained for DAPI. Images were acquired on a spinning-disk confocal microscope. Size bars, 100 μm.
As well as altering levels of SG nucleating factors (21, 22), posttranslational modification of these proteins can influence SG formation. For example, dephosphorylation of G3BP1 at serine 149 or disruption of this regulatory site by mutagenesis abrogates SG formation (26). Accordingly, we investigated whether serine 149 phosphorylation of G3BP1 was affected by ZIKV infection. Results shown in Fig. 6B indicate that G3BP1 phosphorylation was not affected by ZIKV infection regardless of whether cells were treated with agents that stimulate SG formation. Because microtubule-mediated transport of stalled polysome complexes is also important for aggregation of SGs, we assessed the effect of ZIKV infection on microtubule morphology. As a positive control, A549 cells were treated with nocodazole, a chemical agent which interferes with microtubule polymerization (47). As shown in Fig. 6C, the β-tubulin-positive structures in nocodazole-treated cells lacked the characteristic filamentous organization of microtubules seen in mock-treated cells. Significant disruption of the microtubule network was not evident in ZIKV-infected cells (Fig. 6C).
Expression of ZIKV capsid, NS3, NS2B-3, or NS4A protein inhibits SG formation.
Based on the data described above, a potential mechanism by which ZIKV disrupts SG assembly is interfering with a signaling step downstream of translational arrest and ribonucleoprotein complex transport. To identify the viral component(s) responsible for blocking SG formation, we expressed individual FLAG-tagged ZIKV proteins in A549 cells and induced SG assembly with hippuristanol. With the exception of capsid, NS3, NS2B-3, and NS4A, hippuristanol robustly stimulated formation of G3BP1- and TIAR-positive SGs in cells expressing individual ZIKV proteins (Fig. 7A). However, in cells expressing capsid, NS3, NS2B-3, or NS4A, the average number of SGs was reduced by ∼70% (Fig. 7B). Moreover, in some cells expressing these viral proteins, there was a complete absence of G3BP1/TIAR-positive foci (Fig. 7A). Expression of prM had a modest but statistically significant effect on SG formation (Fig. 7B).
FIG 7.
Expression of capsid, NS3, NS2B-3, or NS4A proteins blocks SG formation. (A) A549 cells were transfected with the indicated ZIKV expression plasmids for 48 h and treated with hippuristanol (1 μM) for 25 min before fixation and processing for confocal microscopy. Samples were incubated with mouse anti-FLAG epitope, rabbit anti-G3BP1, and goat anti-TIAR, followed by donkey anti-rabbit IgG conjugated to Alexa Fluor 488, donkey anti-mouse IgG conjugated to Alexa Fluor 546, and chicken anti-goat IgG conjugated to Alexa Fluor 647. Nuclei were stained for DAPI. Images were acquired using a spinning-disk confocal microscope. Size bars, 100 μm. (B) SGs in transfected cells were quantified using Volocity software. A minimum of 15 cells were used for each sample. The average number of SGs per cell ± standard errors from three independent experiments are shown. P values were determined by Student's t test. *, P < 0.05 (significant); **, P < 0.01 (highly significant); ***, P < 0.001 (highly significant).
The SG proteins G3BP1, TIAR, and Caprin-1 are important for ZIKV replication.
Given the link between SG components and replication of DENV and WNV (30), we examined whether SG nucleation factors play a role in ZIKV replication. G3BP1, TIA-1, TIAR, and Caprin-1 were transiently depleted using siRNAs in A549 cells, after which cells were infected with ZIKV for 48 h. Compared to cells transfected with nonsilencing siRNAs (NS), knockdown of G3BP1, TIAR, or Caprin-1, but not TIA-1, resulted in an ∼70% reduction in viral titers (Fig. 8A), an effect that was not due to reduction in cell viability (Fig. 8B).
FIG 8.
SG proteins G3BP1, TIAR, and Caprin-1 are important for ZIKV production. (A) A549 cells were transfected with control nonsilencing siRNAs (NS) or siRNAs specific for G3BP1, TIAR, TIA-1, or Caprin-1 for 48 h. Cells were then infected with ZIKV (MOI, 0.5) for an additional 48 h, after which they were processed for immunoblotting to assess knockdown efficiency, plaque assays to determine viral titers, and cell viability assay. Average viral titers ± standard errors from four independent experiments are shown. P values were determined by Student's t test. *, P < 0.05 (significant). (B) Cell viability is expressed as relative value ± standard errors from three independent experiments. (C) HEK293T cells were transfected with a control plasmid (pcDNA3.1) or the indicated ZIKV protein expression constructs for 48 h. Cell lysates were harvested and processed for immunoprecipitation (IP) using mouse anti-FLAG or anti-myc antibody (negative control) followed by immunoblotting with anti-FLAG, anti-G3BP1, and anti-Caprin-1 antibodies. (D and E) A549 cells infected with ZIKV (MOI, 5) were harvested 48 h.p.i. and processed for RNA-IP. Anti-G3BP1, anti-TIAR, and anti-myc antibodies were used for IP. The bound RNAs were processed for qRT-PCR to quantify the relative ZIKV RNAs (D), and proteins were processed for immunoblotting (E). Values represent the relative amount of ZIKV RNA normalized to myc-IP samples ± standard errors from three independent experiments. P values were determined by Student's t test. **, P < 0.01 (highly significant). ZIKV replication was determined by qRT-PCR and is shown as relative viral RNA levels (normalized to mock infection).
Since capsid, NS3, and NS4A proteins block SG assembly (Fig. 7), we assessed whether these viral proteins interacted with G3BP1, TIAR, or Caprin-1. ZIKV NS1 protein, whose expression had no effect on SG assembly (Fig. 7B), was used as a negative control for these experiments. FLAG-tagged ZIKV proteins were transiently expressed in HEK293T cells for 48 h, after which immunoprecipitation (IP) was performed using an anti-FLAG antibody. Capsid protein, NS3, G3BP1, and TIAR are all RNA-binding proteins; therefore, to avoid RNA-dependent interactions, cell lysates were treated with a nuclease that degrades both DNA and RNA prior to IP. Stable interaction between capsid, G3BP1, and Caprin-1 was readily detected (Fig. 8C); however, we did not observe specific interaction between TIAR and capsid, NS3, or NS4A proteins (data not shown). We also conducted pulldown with FLAG-tagged prM, E, NS2A, NS2B, NS4B, and NS5 but did not detect stable interactions with G3BP1 or TIAR (data not shown).
As reported for WNV and DENV, the viral RNAs have been shown to interact with several SG components, such as TIA-1 and TIAR (30). To determine if G3BP1 and/or TIAR interact with ZIKV RNA during infection, RNA-IP assays were performed using antibodies against G3BP1 and TIAR in ZIKV-infected cells. Anti-myc antibody was used as a negative control for the IP. Quantitative reverse transcription-PCR (qRT-PCR) was used to quantify the relative amounts of viral RNA in the immunoprecipitates. Viral infection was confirmed by quantifying the relative viral RNA levels (Fig. 8D, right), and IP of G3BP1 and TIAR was confirmed by immunoblotting (Fig. 8E). Based on the data shown in Fig. 8D (left), ZIKV RNA was shown to bind G3BP1 but not TIAR. Together, these results support a scenario in which the proviral effects of SG proteins are linked in part to interactions between G3BP1 and Caprin-1 with the capsid protein as well as the interaction of G3BP1 with viral genomic RNA.
Capsid proteins of YFV and MVEV suppress SG formation.
JEV capsid was reported to interact with Caprin-1 and block SG formation (48). Based on the interaction of ZIKV capsid with G3BP1 and Caprin-1 and its negative impact on SG formation, we tested whether capsid proteins from other flaviviruses have a similar effect on SG assembly. Capsid proteins from JEV, DENV, MVEV, WNV, and YFV were transiently expressed in A549 cells, after which SG formation was induced with hippuristanol. Expression of the capsid proteins was confirmed by immunoblotting (Fig. 9C). Significant repression of SG formation was observed in JEV, MVEV, and YFV capsid-expressing cells, with the latter showing an ∼60% reduction in SG numbers compared to control cells (Fig. 9A and B). However, expression of DENV or WNV capsid did not significantly inhibit SG formation. These data suggest that capsid-mediated suppression of SG assembly is a strategy employed by some, but not all, flaviviruses to overcome host defenses during viral infection.
FIG 9.
Expression of flavivirus capsid proteins has differential effects on SG formation. (A) A549 cells were transfected with the indicated myc-tagged capsid expression plasmids for 48 h, and 25 min before fixation, cells were treated with hippuristanol (1 μM). Samples were incubated with mouse anti-myc, rabbit anti-G3BP1, and goat anti-TIAR antibodies, followed by donkey anti-rabbit IgG conjugated to Alexa Fluor 488, donkey anti-mouse IgG conjugated to Alexa Fluor 546, and chicken anti-goat IgG conjugated to Alexa Fluor 647. Nuclei were stained for DAPI. Images were acquired using a spinning-disk confocal microscope. Size bars, 100 μm. (B) SGs in transfected cells were quantified using Volocity software. A minimum of 15 cells were used for each sample. The average number of SGs per cell ± standard errors from three independent experiments are shown. P values were determined by Student's t test. *, P < 0.05 (significant); **, P < 0.01 (highly significant). (C) HEK293T cells were transfected with the indicated myc-tagged capsid expression constructs. After 48 h, cell lysates were harvested and processed for SDS-PAGE and immunoblotting. Anti-myc antibody was used to detect the capsid proteins, and β-actin was used as the loading control.
DISCUSSION
Stress responses are integral to cellular defense against viral infection (49). By inhibiting protein translation and inducing SG assembly, the host cell can redirect resources toward antiviral responses and limit viral access to key translation factors. During flavivirus infection, cellular stress pathways can be triggered by PKR-mediated detection of viral dsRNA as well as accumulation of unfolded proteins in the endoplasmic reticulum. This in turn can lead to phosphorylation of the translation initiation factor eIF2α, resulting in global arrest of protein synthesis and formation of SGs. Flaviviruses have evolved different strategies to counteract the formation of SGs in order to establish productive infections (30, 34, 48, 50). For example, DENV and WNV block SG formation by sequestering TIA-1 and TIAR and exploiting them for viral genome synthesis (30). Here, we report the molecular mechanisms deployed by ZIKV to antagonize SG biogenesis pathways (Fig. 10).
FIG 10.
Proposed model for ZIKV-mediated suppression of SG formation. During ZIKV infection, activation of PKR and UPR occurs, which leads to phosphorylation of eIF2α, resulting in formation of stalled translation initiation complexes and global translational shutoff. ZIKV RNA and capsid protein hijack key SG nucleating factors such as G3BP1 (and potentially others) for viral replication, leading to inhibition of SG assembly. In addition, the viral determinants, including capsid, NS3, and NS4A, further block the condensation of these ribonucleoprotein complexes, possibly through binding to other SG-associated proteins, such as Caprin-1. Together, these actions impair biogenesis of SGs.
Unlike a number of other RNA viruses (31, 32, 36), ZIKV infection poorly induces formation of SGs. While the majority of ZIKV-infected A549 cells and HFAs failed to produce any SGs, ∼25% of ZIKV-infected Huh-7 cells contained SGs (data not shown), which is in agreement with Roth et al. (34). The virus was shown to efficiently block the formation of SGs induced by viral RNA mimics (poly[I:C]), translational inhibitors (hippuristanol), and oxidative stressors (arsenite). The latter is consistent with recent reports showing that ZIKV blocks arsenite-induced SG formation (34, 35). Importantly, our study demonstrated that ZIKV-mediated inhibition of SGs occurs in physiologically relevant cell types, such as primary HFAs. The fact that ZIKV infection induces the UPR and phosphorylation of PKR and its downstream target, eIF2α, indicates that the block in SG formation occurs downstream of the early signaling step that stimulates SG biogenesis. Similar observations were reported for DENV and JEV, in which upregulation of eIF2α phosphorylation was detected during viral infections (51, 52). However, other studies showed that DENV and WNV do not induce phosphorylation of eIF2α (34, 35, 42, 43). The differences in cell types and stress-inducing protocols employed might explain the discrepancies observed among these studies.
ZIKV-induced phosphorylation of eIF2α was associated with global inhibition of cellular protein synthesis. While a number of earlier studies reported that WNV and DENV evade eIF2α-induced host translation shutoff (42, 43), our present findings are consistent with a more recent study showing that DENV, WNV, and ZIKV do in fact inhibit host cell translation (34). In ZIKV-infected cells, NS3 and NS4A appear to be important for this process. To our knowledge, this is the first study to implicate specific ZIKV proteins in blocking host cell translation. While previous studies have linked WNV NS4A and NS4B as well as DENV NS2B-3 with upregulated UPR signaling (43, 45), it is not clear if these proteins negatively regulate host cell translation. Global shutdown of protein synthesis serves to restrict viral protein production as part of the antiviral response; however, according to our observations, it appears that ZIKV and potentially other flaviviruses have evolved a way(s) to evade this block in protein translation and establish productive infection.
Unlike poliovirus or encephalomyocarditis virus infection, in which SG formation is blocked by cleavage of nucleating factors (46, 53), there was no significant change in steady-state levels of SG proteins G3BP1, TIA-1, and TIAR in ZIKV-infected cells. Moreover, we found no evidence to suggest lack of SG formation was due to changes in phosphorylation of G3BP1 or disruption of the microtubule network. Blocking SG formation would seem to be an important process for ZIKV, as we found that individual expression of three viral proteins, capsid, NS3, and NS4A, were suppressors of SG assembly. The capsid protein of JEV also inhibits (arsenite-induced) SG formation by interacting with the SG protein Caprin-1 (48). Similar to JEV and ZIKV capsids, we observed that expression of capsid proteins from two other flaviviruses, YFV and MVEV, also block SG formation. Interestingly, WNV and DENV capsids did not significantly repress SG assembly, suggesting these flaviviruses employ alternative strategies to counter the antiviral effect of SGs. While G3BP1, Caprin-1, and TIAR are important for ZIKV production, only G3BP1 and Caprin-1 formed stable complexes with capsid protein. Of note, TIAR and G3BP1, as well as TIA-1, are important for replication of HCV, particularly at early time points during infection (32, 33). Collectively, these findings suggest that exploitation of SG components is a common strategy used by flaviviruses to increase replication and/or blunt antiviral defenses.
As well as there being protein-protein interactions between flaviviruses and SGs, WNV and DENV genomic RNA has been shown to bind several SG assembly factors, including TIA-1, TIAR, G3BP1, G3BP2, DDX6, and Caprin-1 (30, 54–56). Based on the observation that ZIKV genomic RNA and G3BP1 form a stable complex and viral replication is impaired in cells depleted of SG factors, we propose that ZIKV RNA-mediated recruitment of G3BP1 constitutes an additional mechanism to promote genome replication and to inhibit SG formation (Fig. 10). However, as G3BP1 binds to ZIKV capsid, we cannot rule out the possibility that the interaction between G3BP1 and viral RNA is facilitated by capsid. Given that G3BP1 and Caprin-1 also bind noncoding subgenomic flaviviral RNAs (sfRNAs) during DENV infection (55), it will be of interest to determine if ZIKV-associated sfRNAs (57) play a role in sequestration of SG-associated proteins.
In summary, our data indicate that ZIKV blocks host cell translation and inhibits SG formation (Fig. 10). The interactions of the viral genomic RNA and capsid protein with G3BP1 support a scenario in which ZIKV hijacks SG components for viral replication and simultaneously blocks SG assembly. Abrogation of SG assembly mediated by viral capsids appears to be a conserved defense mechanism among several flaviviruses. Together with recent studies on other flaviviruses, our data suggest that interfering with biogenesis of SGs is a critical aspect of flavivirus biology.
MATERIALS AND METHODS
Cell culture and virus infection.
A549 cells, HEK293T cells, and Vero cells were purchased from the American Type Culture Collection (Manassas, VA) and were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 100 U/ml penicillin and streptomycin, 1 mM HEPES (Gibco), 2 mM glutamine (Gibco), 10% heat-inactivated fetal bovine serum (FBS; Gibco) at 37°C in 5% CO2. Primary human fetal astrocytes (HFAs) were prepared as previously described (58) from 15- to 19-week aborted fetuses with written consent approved under protocol 1420 by the University of Alberta Human Research Ethics Board (Biomedical) and cultured as described earlier (15). The Zika virus (strain PLCal) was kindly provided by David Safronetz at the Public Health Agency of Canada. Virus manipulations were performed according to level-2 containment procedures. Virus stocks were generated in C6/36 cells and titrated by plaque assay using Vero cells.
Plasmids and transfection.
The triple FLAG-tagged ZIKV C, prM, E, NS1, NS2A, NS2B, NS4A, and NS4B cDNAs were generated by PCR using constructs encoding the corresponding viral proteins (15) as templates. The resulting PCR products were then cloned into pcDNA 3.1(−) plasmid using the restriction sites NheI and XhoI. For NS3 and NS5 constructs, single FLAG-tagged NS3 and NS5 constructs (15) were used as templates for PCR. The resulting PCR products were cloned into the same single FLAG-tagged NS3 or NS5 plasmid using the restriction sites AfeI and XhoI (for NS3) as well as BsrG1 and XhoI (for NS5). All reverse primers used for the above-described PCRs encoded a double-FLAG epitope (Table 1). For indirect immunofluorescence analysis in A549 cells, transfection of the appropriate expression plasmids was performed using TransIT-LT1 (Mirus). For immunoblot and luciferase reporter analysis in HEK293T cells, plasmid transfection was performed using Lipofectamine 2000 (Invitrogen). Poly(I·C) (Sigma-Aldrich) was transfected into cells using Lipofectamine 2000 (Invitrogen).
TABLE 1.
PCR primer sequences used
| ZIKV PCR target(s) | Primer sequence (5′→3′) |
|---|---|
| Capsid | Forward: ATATATGCTAGCGTTTAAACGCCACCATGAAAAACCCAAAAAAGAAATCC |
| prM | Forward: ATATATGCTAGCGTTTAAACGCCACCATGGGCGCAGATACTAGTGTCGGAAT |
| Envelope | Forward: ATATATGCTAGCGTTTAAACGCCACCATGGGAAGCTCAACGAGCCAAAAAGTC |
| NS2A | Forward: ATATATGCTAGCGTTTAAACGCCACCATGGGAAGCTCAACGAGCCAAAAAGTCATATACTTGGTCATGATACTGCTGATTGCCCCGGCATACAGCATCAGGTGCATAGGAGTCAGTAACTTAGTAAGGTCAAT |
| NS2B | Forward: ATATATGCTAGCGTTTAAACGCCACCATGAGCTGGCCCCCTAGCGAAGTACTC |
| NS3 | Forward: ATGCCACCTTCACTTCACGTCTAC |
| NS2B-3 | Forward: ATATATGCTAGCGTTTAAACGCCACCATGAGCTGGCCCCCTAGCGAAGTACTC |
| NS4A | Forward: ATATATGCTAGCGTTTAAACGCCACCATGGGAGCGGCTTTTGGAGTGATGGAA |
| NS4B | Forward: ATATATGCTAGCGTTTAAACGCCACCATGTCTCCCCAGGACAACCAAATGGCA |
| NS5 | Forward: GAGGAGAGTGCCAGAGTTGTGTGT |
| All 3× FLAG targets | Reverse: GGCGGGAGCGGCGGGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGACTACAAGGATGACGATGACAAGTAGGGCGCGCCCTCGAGATATAT |
| myc-DENV capsid | Forward: ATTAGCGCTAGCATGGAACAAAAACTCATCTCAGAAGAGGATCTGAATGACCAACGGAAAAAGGC |
| Reverse: GTACGGATCCTTATCTGCGTCTCCTATTCAAGA | |
| myc-JEV capsid | Forward: ATTAGCGCTAGCATGGAACAAAAACTCATCTCAGAAGAGGATCTGACTAAAAAACCAGGAGGGC |
| Reverse: GTACGGATCCTTATCTTTTGTTTTGCTTTCTGCC | |
| myc-MVEV capsid | Forward: ATTAGCGCTAGCATGGAACAAAAACTCATCTCAGAAGAGGATCTGTCTAAAAAACCAGGAGGAC |
| Reverse: GTACGGATCCTTATCTTTTCTTTTGTTTTTTGCC | |
| myc-WNV capsid | Forward: ATTAGCGCTAGCATGGAACAAAAACTCATCTCAGAAGAGGATCTGTCTAAGAAACCAGGAGGGCC |
| Reverse: GATCGGATCCTTATCTTTTCTTTTGTTTTGAGC | |
| myc-YFV capsid | Forward: ATTAGCGCTAGCATGGAACAAAAACTCATCTCAGAAGAGGATCTGTCTGGTCGTAAAGCTCAGGG |
| Reverse: GTACGGATCCTTATTAACGGCGTTTCCTTGAG |
Antibodies and compounds.
The following antibodies were purchased from the indicated sources. Rabbit anti-G3BP1 (07-1801) and mouse anti-flavivirus E protein 4G2 (MAB10216) were from Millipore; rabbit anti-PKR (ab32052), rabbit anti-phospho-PKR (ab131447), goat anti-GFP (ab5450), and rabbit anti-Dcp1a (ab47811) were from Abcam; goat anti-TIA-1 (sc-1751) and goat anti-TIAR (sc-1749) were from Santa Cruz; rabbit anti-Caprin-1 (SAB1101135), mouse anti-FLAG (F3165), mouse anti-β-actin (a3853), rabbit anti-phospho-G3BP1 (S149) (G8046), and mouse anti-β-tubulin (T5201) were from Sigma-Aldrich; rabbit anti-eIF2α (9722) and rabbit anti-phospho-eIF2α (9721) were from Cell Signaling Technologies; mouse monoclonal anti-double-strand RNA J2 (10010200) was from Scicons, Hungary; mouse anti-ZIKV E protein (MBS5304716) was from MyBioSource; mouse anti-myc (9E10) (ATCC CRL-1729) was from the ATCC; and goat anti-ZIKV NS5 was generated in-house (see details below). Sodium arsenite, thapsigargin, and poly(I·C) were purchased from Sigma-Aldrich. Hippuristanol was kindly provided by Jerry Pelletier at McGill University, Canada. For 35S labeling, the EasyTag EXPRESS35S protein labeling mix was purchased from PerkinElmer.
ZIKV NS5 protein expression, purification, and antibody generation.
ZIKV strain Zika SPH2015:KU321639.1 (59) polyprotein sequence ALU33341.1 was used to generate the construct for the expression of ZIKV-NS5 (amino acids 2525-[GETL-…-YLGEE]-3416). The design was based on previously described protocols for the full-length DENV-NS5 construct (60). An N-terminal Strep-tag (61), followed by the tobacco etch virus (TEV) protease cleavage site, were engineered to facilitate purification of full-length NS5 that contains MTase, linker, and RdRp domains as well as flanking viral amino acids. The pFastBac-1 (Invitrogen) plasmid with the codon-optimized synthetic DNA sequence (GenScript) was used as a starting material for NS5 expression in insect cells (Sf9; Invitrogen). We employed the MultiBac (Geneva Biotech) system according to protocols provided by Bieniossek et al. and Berger et al. (62, 63). NS5 was purified using Strep-tag affinity chromatography according to the manufacturer's specifications (IBA), followed by cleavage of the Strep-tag with TEV protease (Sigma-Aldrich). The identity of the purified NS5 protein was confirmed by mass spectrometry analysis (Jack Moore, Alberta Proteomics and Mass Spectrometry). NS5 samples were buffer exchanged (PD10; GE Healthcare) in Tris-HCl, pH 8, buffer [50 mM Tris (BioShop), 100 mM NaCl (BioShop), 2 mM Tris(2-carboxyethyl)phosphine reductant (Sigma)] supplemented with 40% glycerol (BioShop) and concentrated with Amicon-Ultra (Millipore) centrifugal membranes. The concentration was determined using the extinction coefficient 232,820 M−1 cm−1, calculated by GPMAW-lite free software (Alphalyse). Purified ZIKV NS5 was used to generate goat polyclonal antibody (ProSci Incorporated). The specificity of the antibody was determined by Western blotting and immunofluorescence analysis.
Immunoprecipitation.
HEK293T cells were transfected with individual triple-FLAG-tagged ZIKV protein constructs using Lipofectamine 2000 for 48 h. The cells were pelleted and resuspended in IP buffer (137 mM NaCl, 50 mM Tris, pH 7.5, 1% NP-40, 1 mM NaF, 1 mM dithiothreitol [DTT], protease inhibitor cocktail [Roche], and Benzonase nuclease [Millipore]). The supernatants were clarified by centrifugation at 16,000 × g for 15 min. Lysates were then preincubated with protein G-Sepharose beads (Sigma-Aldrich) at 4°C for 1 h to remove proteins nonspecifically bound to the beads. Aliquots of the precleared cell lysates were incubated overnight with the appropriate primary antibody at 4°C, followed by incubation with protein G-Sepharose beads for 2 h. After washes with IP buffer (without Benzonase), SDS sample buffer was added to the beads and then boiled for 5 min to release bound proteins, which were then resolved by SDS-PAGE.
RNA immunoprecipitation.
A549 cells were infected with ZIKV (MOI of 5) for 48 h. Cells were then washed with ice-cold phosphate-buffered saline (PBS), trypsinized, and pelleted at 800 × g for 5 min at 4°C. Pellets were resuspended in RNA-IP buffer (137 mM NaCl, 50 mM Tris, pH 7.5, 1% NP-40, 1 mM NaF, 1 mM DTT, protease inhibitor cocktail [Roche], and the RNase inhibitor RNaseOUT [Invitrogen]). Protein G-Sepharose beads were incubated overnight at 4°C with anti-G3BP1, anti-TIAR, or anti-myc. Subsequently, cell lysates were incubated with the antibody-bead complexes for 2 h at 4°C, followed by five washes with RNA-IP buffer. An aliquot of the protein-antibody-bead complexes was boiled and processed for SDS-PAGE, while the rest of the complexes were processed for RNA extraction.
Immunoblotting.
A549 cells or HEK293T cells collected at designated time points postinfection, postsilencing, or posttransfection were washed three times with PBS before lysing with SDS sample buffer containing β-mercaptoethanol (2%) and 1 U of Benzonase per sample. The samples were incubated at 98°C for 10 min to denature proteins, which then were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes for immunoblotting as described previously (64). Lastly, blots were imaged and analyzed using the Odyssey CLx imaging system (LI-COR Biosciences).
Autoradiography.
A549 cells were infected with ZIKV (MOI of 5) for 24 and 48 h. Twenty-four hours postinfection, growth medium was replaced with DMEM depleted of cysteine and methionine (Gibco) and 10% dialyzed FBS (Gibco) for 1 h at 37°C. Subsequently, cells were incubated with 110 μCi/ml of [35S]cysteine and methionine for 2 h at 37°C before washing with cold PBS, lysis in SDS-protein loading buffer, and then SDS-PAGE. The gels were then stained with Coomassie blue (Bio-Rad) and processed for autoradiography.
Confocal microscopy.
A549 cells or HFAs grown on coverslips were fixed for 15 min at room temperature with 4% electron microscopy-grade paraformaldehyde (Electron Microscope Sciences) in PBS. Samples were then washed three times with PBS and incubated in blocking buffer (0.2% Triton X-100 [VWR Internationals] and 3% bovine serum albumin [BSA; Sigma-Aldrich] in PBS) at room temperature for 1.5 h. Incubations with primary antibodies in blocking buffer were carried out overnight at 4°C. Cells were then washed three times in wash buffer (0.3% BSA and 0.02% Triton X-100 in PBS). Subsequently, samples were incubated with the indicated secondary antibodies in blocking buffer for 1 h at room temperature, followed by three washes in wash buffer. The indicated secondary antibodies (Invitrogen) were used at 1:1,000 dilutions in blocking buffer. In the meantime, samples were also stained with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich). After three washes, coverslips were mounted on microscope slides using Prolong Gold antifade mounting reagent (Life Technologies). Images were acquired using an Olympus IX-81 spinning-disk confocal microscope equipped with a 40× and 60×, 1.42-numerical-aperture oil PlanApo N objective. Images were analyzed using Volocity 6.2.1 software (PerkinElmer).
qRT-PCR.
Total RNA from A549 cells was isolated using the RNA NucleoSpin kit (Macherey-Nagel) and reverse transcribed using random primers (Invitrogen) and the Improm-II reverse transcriptase system (Promega) at 42°C for 2 h. For RNA samples isolated from RNA-IP, the RNase inhibitor RNaseOUT was added to the reverse transcription reaction mixtures. The resulting cDNAs were mixed with the appropriate primers (Integrated DNA Technologies [IDT]) and the PerfecTa SYBR green SuperMix with low ROX (Quanta Biosciences) and amplified for 40 cycles (30 s at 94°C, 40 s at 55°C, and 20 s at 68°C) in a Stratagene Mx3005P qRT-PCR machine. The gene targets and primers used are listed in Table 2. The changes in threshold cycle (ΔCT) values were calculated using β-actin mRNA as the internal control. The ΔΔCT values were determined using control samples as the reference value. Relative levels of mRNAs were calculated using the 2−ΔΔCT formula. For RNA-IP analysis, the ZIKV RNA CT values were directly normalized to that of the myc-IP samples.
TABLE 2.
List of primer sequences used for qRT-PCR
| Target (reference) | Primer sequences (5′→3′) |
|---|---|
| Zika virus (65) | Forward: CCTTGGATTCTTGAACGAGGA |
| Reverse: AGAGCTTCATTCTCCAGATCAA | |
| ACTB | Forward: CCTGGCACCCAGCACAAT |
| Reverse: GCCGATCCACACGGAGTACT | |
| GFP | Forward: AAGCTGACCCTGAAGTTCATCTGC |
| Reverse: CTTGTAGTTGCCGTCGTCCTTGAA | |
| XBP1 (66) | Forward: TTACGAGAGAAAACTCATGGCC |
| Reverse: GGGTCCAAGTTGTCCAGAATGC | |
| Renilla luciferase (67) | Forward: GGAATTATAATGCTTATCTACGTGC |
| Reverse: CTTGCGAAAAATGAAGACCTTTTAC |
Quantitation of XBP1 mRNA.
Total RNA from A549 cells following infection and/or treatment with thapsigargin was isolated and reverse transcribed into cDNA using the Improm-II reverse transcriptase system. To amplify XBP1, PCR was done using the cDNAs, primers for human XBP1 (Table 2), and Platinum Taq DNA polymerase (Invitrogen) and amplified for 40 cycles (94°C for 30 s, 58°C for 30 s, and 68°C for 1 min). The PCR products were separated on 2.5% agarose gels containing ethidium bromide (Sigma-Aldrich). Images were acquired using a Molecular Imager GelDoc XR+ imaging system (Bio-Rad).
Luciferase reporter assay.
HEK293T cells were transfected with the renilla reporter plasmid pRL-TK (Promega), together with the indicated ZIKV protein constructs. Cells were harvested at the indicated time points by washing the cells once with PBS and then lysing in 250 μl of luciferase lysis buffer (0.1% [vol/vol] Triton X-100, 25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, and 1 mM DTT). Samples were frozen at −80°C and then thawed at room temperature for efficient lysis. An aliquot of lysates was processed for RNA isolation and another for SDS-PAGE/immunoblotting. For renilla luciferase measurements, samples were aliquoted in duplicates, and the substrate coelenterazine (Gold Biotechnology USA) was prepared at a final concentration of 1.4 μM in luciferase assay buffer (25 mM glycylglycine [pH 7.8], 15 mM K2PO4 [pH 7.8], 15 mM MgSO4, and 4 mM EGTA). Luciferase activity was measured using an Illuminator plate reader (BioTek).
siRNA experiments.
A549 cells were transfected with the indicated siRNAs using RNAiMAX (Invitrogen). Control siRNA (D-001810-10-05; Dharmacon), G3BP1 (L-012099-00-0005; Dharmacon), Caprin-1 (M-016057-01-0005; Dharmacon), control siRNA (sense, 5′rCrGrUrUrArArUrCrGrCrGrUrArUrArArUrArCrGrCrGrUAT3′; antisense, 5′rArUrArCrGrCrGrUrArUrUrArUrArCrGrCrGrArUrUrArArCrGrArC3′, where “r” indicates ribonucleotide; IDT), TIA-1 (sense, 5′rGrUrArCrUrArCrCrArUrUrGrArArGrGrUrCrArUrGrUrUGT3′; antisense, 5′rArCrArArCrArUrGrArCrCrUrUrCrArArUrGrGrUrArGrUrArCrCrA3′; IDT), and TIAR (sense, 5′rGrGrUrArArArGrArArUrCrUrCrCrUrGrArUrArUrGrArCTA3′; antisense, 5′rUrArGrUrCrArUrArUrCrArGrGrArGrArUrUrCrUrUrUrArCrCrCrC3′; IDT). Forty-eight hours later cells were mock infected or infected with ZIKV (MOI, 0.5) for 48 h. Total cell lysates were isolated and processed for immunoblotting. Titers of the virus supernatants were determined in Vero cells using plaque assay.
Cell viability assay.
The cells were harvested in PBS and lysed by freezing and thawing at −80°C. The lysates were processed for cell viability assays using the CellTiter-Glo luminescent cell viability assay system (Promega) according to the manufacturer's instructions.
Statistical analyses.
A paired/unpaired Student's t test was used for pairwise statistical comparison. The means ± standard errors of the means are shown in all bar graphs. All statistical analyses were performed using Microsoft Excel and GraphPad Prism software.
ACKNOWLEDGMENTS
We thank Daniel Limonta for constructive and helpful insights and advice on primary cell culture and Justin Pare and Joaquin Lopez-Orozco for sharing their expertise in RNA granule research. We also thank Eileen Reklow and Valeria Mancinelli for excellent technical support. We thank Jerry Pelletier (McGill University) for providing hippuristanol.
This work was funded by grants from the Canadian Institutes of Health Research (CIHR MOP-125903; PJT-148699; ZV1-149782), the Li Ka Shing Institute of Virology, and the Women & Children's Health Research Institute (WCHRI) to T.C.H. S.H. is the recipient of a WCHRI graduate studentship. A.K. is funded by a postdoctoral fellowship from the Alberta Innovates-Health Solutions. A.M.A. is the recipient of a doctoral scholarship from CIHR. M.G. is funded by CIHR MOP-126149. T.C.H. holds a Canada Research Chair award.
S.H., A.K., and T.C.H. conceived the research. S.H., A.K., and T.C.H. drafted the manuscript. S.H., A.K., Z.X., A.M.A., C.P.W., and I.S. carried out the experiments. M.G. and E.T. generated ZIKV NS5 protein. C.P. and W.B. isolated primary human fetal astrocytes. S.H. and A.K. created the figures. All authors discussed the results and contributed to the revision of the final manuscript.
We have no competing financial interests to declare.
REFERENCES
- 1.Moreira J, Peixoto TM, Machado de Siqueira A, Lamas CC. 2017. Sexually acquired Zika virus: a systematic review. Clin Microbiol Infect 23:296–305. [DOI] [PubMed] [Google Scholar]
- 2.Simpson DI. 1964. Zika virus infection in man. Trans R Soc Trop Med Hyg 58:335–338. doi: 10.1016/0035-9203(64)90200-7. [DOI] [PubMed] [Google Scholar]
- 3.Zanluca C, Melo VC, Mosimann AL, Santos GI, Santos CN, Luz K. 2015. First report of autochthonous transmission of Zika virus in Brazil. Mem Inst Oswaldo Cruz 110:569. doi: 10.1590/0074-02760150192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Araújo TV, Rodrigues LC, de Alencar Ximenes RA, de Barros Miranda-Filho D, Montarroyos UR, de Melo AP, Valongueiro S, de Albuquerque MF, Souza WV, Braga C, Filho SPB, Cordeiro MT, Vazquez E, Di Cavalcanti Souza Cruz D, Henriques CMP, Bezerra LCA, da Silva Castanha PM, Dhalia R, Marques-Júnior ETA, Martelli CMT. 2016. Association between Zika virus infection and microcephaly in Brazil, January to May, 2016: preliminary report of a case-control study. Lancet Infect Dis 16:1356–1363. doi: 10.1016/S1473-3099(16)30318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ventura CV, Maia M, Bravo-Filho V, Góis AL, Belfort R. 2016. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 6.Driggers RW, Ho C-Y, Korhonen EM, Kuivanen S, Jääskeläinen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, Timofeev J, Rodriguez FJ, Levanov L, Razak J, Iyengar P, Hennenfent A, Kennedy R, Lanciotti R, du Plessis A, Vapalahti O. 2016. Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N Engl J Med 374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- 7.Calvet G, Aguiar RS, Melo ASO, Sampaio SA, de Filippis I, Fabri A, Araujo ESM, de Sequeira PC, de Mendonça MCL, de Oliveira L, Tschoeke DA, Schrago CG, Thompson FL, Brasil P, Dos Santos FB, Nogueira RMR, Tanuri A, de Filippis AMB. 2016. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis 16:653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 8.Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial A-L, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra J-C, Despres P, Fournier E, Mallet H-P, Musso D, Fontanet A, Neil J, Ghawché F. 2016. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, Platt DJ, Gorman MJ, Richner JM, Caine EA, Salazar V, Moley KH, Diamond MS. 2016. Zika virus infection damages the testes in mice. Nature 540:438–442. doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansuy JM, Pasquier C, Daudin M, Chapuy-Regaud S, Moinard N, Chevreau C, Izopet J, Mengelle C, Bujan L. 2016. Zika virus in semen of a patient returning from a non-epidemic area. Lancet Infect Dis 16:894–895. doi: 10.1016/S1473-3099(16)30153-0. [DOI] [PubMed] [Google Scholar]
- 11.Adams Waldorf KM, Stencel-Baerenwald JE, Kapur RP, Studholme C, Boldenow E, Vornhagen J, Baldessari A, Dighe MK, Thiel J, Merillat S, Armistead B, Tisoncik-Go J, Green RR, Davis MA, Dewey EC, Fairgrieve MR, Gatenby JC, Richards T, Garden GA, Diamond MS, Juul SE, Grant RF, Kuller L, Shaw DWW, Ogle J, Gough GM, Lee W, English C, Hevner RF, Dobyns WB, Gale M Jr, Rajagopal L. 2016. Fetal brain lesions after subcutaneous inoculation of Zika virus in a pregnant nonhuman primate. Nat Med 22:1256–1259. doi: 10.1038/nm.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandão WN, Rossato C, Andrade DG, Faria D de P, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM de A, Peron JPS, Muotri AR, Beltrão-Braga PCB. 2016. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Saucedo-Cuevas L, Regla-Nava JA, Chai G, Sheets N, Tang W, Terskikh AV, Shresta S, Gleeson JG. 2016. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell 19:593–598. doi: 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, García-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. 2016. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep 17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meertens L, Labeau A, Dejarnac O, Cipriani S, Sinigaglia L, Bonnet-Madin L, Le Charpentier T, Hafirassou ML, Zamborlini A, Cao-Lormeau V-M, Coulpier M, Missé D, Jouvenet N, Tabibiazar R, Gressens P, Schwartz O, Amara A. 2017. Axl mediates ZIKA virus entry in human glial cells and modulates innate immune responses. Cell Rep 18:324–333. doi: 10.1016/j.celrep.2016.12.045. [DOI] [PubMed] [Google Scholar]
- 17.Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ETDA, Cherry S, Sadovsky Y, Coyne CB. 2016. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 19:705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. 2016. A mouse model of Zika virus pathogenesis. Cell Host Microbe 19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol 20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson P, Kedersha N. 2002. Stressful initiations. J Cell Sci 115:3227–3234. [DOI] [PubMed] [Google Scholar]
- 21.Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M. 2013. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 18:135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 22.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartoli KM, Bishop DL, Saunders WS. 2011. The role of molecular microtubule motors and the microtubule cytoskeleton in stress granule dynamics. Int J Cell Biol 2011:939848. doi: 10.1155/2011/939848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov PA, Chudinova EM, Nadezhdina ES. 2003. RNP stress-granule formation is inhibited by microtubule disruption. Cell Biol Int 27:207–208. doi: 10.1016/S1065-6995(02)00341-4. [DOI] [PubMed] [Google Scholar]
- 26.Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.White JP, Lloyd RE. 2011. Poliovirus unlinks TIA1 aggregation and mRNA stress granule formation. J Virol 85:12442–12454. doi: 10.1128/JVI.05888-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khaperskyy DA, Hatchette TF, McCormick C. 2012. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J 26:1629–1639. doi: 10.1096/fj.11-196915. [DOI] [PubMed] [Google Scholar]
- 29.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. 2014. Influenza A virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10:e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A 104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest J-P, Mazur J, Bankhead P, Hiet M-S, Kallis S, Alvisi G, Samuel CE, Lohmann V, Kaderali L, Rohr K, Frese M, Stoecklin G, Bartenschlager R. 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garaigorta U, Heim MH, Boyd B, Wieland S, Chisari FV. 2012. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J Virol 86:11043–11056. doi: 10.1128/JVI.07101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol 85:6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K, Lohmann V, Bartenschlager R, Fackler OT, Locker N, Stoecklin G, Ruggieri A. 2017. Flavivirus infection uncouples translation suppression from cellular stress responses. mBio 8:e02150-16. doi: 10.1128/mBio.02150-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu M, Courtney SC, Brinton MA. 2017. Arsenite-induced stress granule formation is inhibited by elevated levels of reduced glutathione in West Nile virus-infected cells. PLoS Pathog 13:e1006240. doi: 10.1371/journal.ppat.1006240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE. 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J Virol 84:12274–12284. doi: 10.1128/JVI.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Retallack H, Lullo ED, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Leon WRM, Krencik R, Ullian EM, Spatazza J, Pollen AA, Mandel-Brehm C, Nowakowski TJ, Kriegstein AR, DeRisi JL. 2016. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci U S A 113:14408–14413. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamel R, Ferraris P, Wichit S, Diop F, Talignani L, Pompon J, Garcia D, Liégeois F, Sall AA, Yssel H, Missé D. 2017. African and Asian Zika virus strains differentially induce early antiviral responses in primary human astrocytes. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis 49:134–137. doi: 10.1016/j.meegid.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Anderson P, Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 40.Cencic R, Pelletier J. 2016. Hippuristanol–a potent steroid inhibitor of eukaryotic initiation factor 4A. Translation 4:e1137381. doi: 10.1080/21690731.2015.1137381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bordeleau M-E, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J. 2005. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci U S A 102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peña J, Harris E. 2011. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem 286:14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J Virol 85:2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharyya S, Sen U, Vrati S. 2014. Regulated IRE1-dependent decay pathway is activated during Japanese encephalitis virus-induced unfolded protein response and benefits viral replication. J Gen Virol 95:71–79. doi: 10.1099/vir.0.057265-0. [DOI] [PubMed] [Google Scholar]
- 45.Yu C-Y, Hsu Y-W, Liao C-L, Lin Y-L. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol 80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Samson F, Donoso JA, Heller-Bettinger I, Watson D, Himes RH. 1979. Nocodazole action on tubulin assembly, axonal ultrastructure and fast axoplasmic transport. J Pharmacol Exp Ther 208:411–417. [PubMed] [Google Scholar]
- 48.Katoh H, Okamoto T, Fukuhara T, Kambara H, Morita E, Mori Y, Kamitani W, Matsuura Y. 2013. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with Caprin-1 and facilitates viral propagation. J Virol 87:489–502. doi: 10.1128/JVI.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lloyd RE. 2012. How do viruses interact with stress-associated RNA granules? PLoS Pathog 8:e1002741. doi: 10.1371/journal.ppat.1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Courtney SC, Scherbik SV, Stockman BM, Brinton MA. 2012. West Nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response. J Virol 86:3647–3657. doi: 10.1128/JVI.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tu Y-C, Yu C-Y, Liang J-J, Lin E, Liao C-L, Lin Y-L. 2012. Blocking double-stranded RNA-activated protein kinase PKR by Japanese encephalitis virus nonstructural protein 2A. J Virol 86:10347–10358. doi: 10.1128/JVI.00525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umareddy I, Pluquet O, Wang QY, Vasudevan SG, Chevet E, Gu F. 2007. Dengue virus serotype infection specifies the activation of the unfolded protein response. Virol J 4:91. doi: 10.1186/1743-422X-4-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng CS, Jogi M, Yoo J-S, Onomoto K, Koike S, Iwasaki T, Yoneyama M, Kato H, Fujita T. 2013. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol 87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J Virol 76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bidet K, Dadlani D, Garcia-Blanco MA. 2014. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a Dengue virus non-coding RNA. PLoS Pathog 10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward AM, Gunaratne J, Garcia-Blanco MA. 2014. Identification of dengue RNA binding proteins using RNA chromatography and quantitative mass spectrometry. Methods Mol Biol 1138:253–270. doi: 10.1007/978-1-4939-0348-1_16. [DOI] [PubMed] [Google Scholar]
- 57.Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y, Shi P-Y, Nix JC, Beckham JD, Kieft JS. 2016. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354:1148–1152. doi: 10.1126/science.aah3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vivithanaporn P, Asahchop EL, Acharjee S, Baker GB, Power C. 2016. HIV protease inhibitors disrupt astrocytic glutamate transporter function and neurobehavioral performance. AIDS 30:543–552. doi: 10.1097/QAD.0000000000000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cunha MS, Esposito DLA, Rocco IM, Maeda AY, Vasami FGS, Nogueira JS, Souza RP de, Suzuki A, Addas-Carvalho M, de Barjas-Castro ML, Resende MR, Stucchi RSB, de Boin IFSF, Katz G, Angerami RN, da Fonseca BAL. 2016. First complete genome sequence of Zika virus (Flaviviridae, Flavivirus) from an autochthonous transmission in Brazil. Genome Announc 4:e00032-16. doi: 10.1128/genomeA.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y, Soh TS, Zheng J, Chan KWK, Phoo WW, Lee CC, Tay MYF, Swaminathan K, Cornvik TC, Lim SP, Shi P-Y, Lescar J, Vasudevan SG, Luo D. 2015. A crystal structure of the Dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog 11:e1004682. doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt TG, Skerra A. 1994. One-step affinity purification of bacterially produced proteins by means of the “Strep tag” and immobilized recombinant core streptavidin. J Chromatogr A 676:337–345. doi: 10.1016/0021-9673(94)80434-6. [DOI] [PubMed] [Google Scholar]
- 62.Berger I, Fitzgerald DJ, Richmond TJ. 2004. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol 22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 63.Bieniossek C, Richmond TJ, Berger I. 2008. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr Protoc Protein Sci Chapter 5:Unit 5.20. [DOI] [PubMed] [Google Scholar]
- 64.You J, Hou S, Malik-Soni N, Xu Z, Kumar A, Rachubinski RA, Frappier L, Hobman TC. 2015. Flavivirus infection impairs peroxisome biogenesis and early antiviral signaling. J Virol 89:12349–12361. doi: 10.1128/JVI.01365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balm MND, Lee CK, Lee HK, Chiu L, Koay ESC, Tang JW. 2012. A diagnostic polymerase chain reaction assay for Zika virus. J Med Virol 84:1501–1505. doi: 10.1002/jmv.23241. [DOI] [PubMed] [Google Scholar]
- 66.Shang J. 2011. Chapter sixteen–quantitative measurement of events in the mammalian unfolded protein response, p 293–308. In Conn PM. (ed), Methods in Enzymology. Academic Press, Inc, New York, NY. [DOI] [PubMed] [Google Scholar]
- 67.Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, Willis AE, Bushell M. 2008. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci U S A 105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]