ABSTRACT
Infectious bursal disease virus (IBDV) is a double-stranded RNA (dsRNA) virus. Segment A contains two overlapping open reading frames (ORFs), which encode viral proteins VP2, VP3, VP4, and VP5. Segment B contains one ORF and encodes the viral RNA-dependent RNA polymerase, VP1. IBDV ribonucleoprotein complexes are composed of VP1, VP3, and dsRNA and play a critical role in mediating viral replication and transcription during the virus life cycle. In the present study, we identified a cellular factor, VDAC1, which was upregulated during IBDV infection and found to mediate IBDV polymerase activity. VDAC1 senses IBDV infection by interacting with viral proteins VP1 and VP3. This association is caused by RNA bridging, and all three proteins colocalize in the cytoplasm. Furthermore, small interfering RNA (siRNA)-mediated downregulation of VDAC1 resulted in a reduction in viral polymerase activity and a subsequent decrease in viral yield. Moreover, overexpression of VDAC1 enhanced IBDV polymerase activity. We also found that the viral protein VP3 can replace segment A to execute polymerase activity. A previous study showed that mutations in the C terminus of VP3 directly influence the formation of VP1-VP3 complexes. Our immunoprecipitation experiments demonstrated that protein-protein interactions between VDAC1 and VP3 and between VDAC1 and VP1 play a role in stabilizing the interaction between VP3 and VP1, further promoting IBDV polymerase activity.
IMPORTANCE The cellular factor VDAC1 controls the entry and exit of mitochondrial metabolites and plays a pivotal role during intrinsic apoptosis by mediating the release of many apoptogenic molecules. Here we identify a novel role of VDAC1, showing that VDAC1 interacts with IBDV ribonucleoproteins (RNPs) and facilitates IBDV replication by enhancing IBDV polymerase activity through its ability to stabilize interactions in RNP complexes. To our knowledge, this is the first report that VDAC1 is specifically involved in regulating IBDV RNA polymerase activity, providing novel insight into virus-host interactions.
KEYWORDS: IBDV, polymerase activity, virus replication
INTRODUCTION
Infectious bursal disease (IBD) is a serious viral disease of young chickens that causes severe immunosuppression and damages the lymphoid cells in the bursa of Fabricius. The virus responsible for IBD, infectious bursal disease virus (IBDV), is a nonenveloped, bisegmented, double-stranded RNA (dsRNA) virus belonging to the Avibirnavirus genus within the Birnaviridae family (1). The genome of the virus includes two segments of dsRNA: segments A and B (2). In segment A, there are two overlapping open reading frames (ORFs); the small ORF encodes the viral protein VP5, and the large one encodes a polyprotein that is self-cleaved by the viral protease VP4 to form pVP2, VP4, and VP3 (3, 4). Segment B contains a single ORF that encodes the viral RNA-dependent RNA polymerase (RdRp), VP1 (5). The VP3 protein is known to be a scaffolding protein with multiple roles which can interact with multiple proteins, including both host cell proteins and viral proteins. A study showed that interaction between VP3 and host cellular ribosomal protein L4 (RPL4) can effectively promote the replication of IBDV (6). In the viral life cycle, in addition to being a self-interacting protein (7), VP3 also interacts with pVP2 during particle morphogenesis (8), with VP1 as a transcriptional activator (9), and with VP1 and the dsRNA genome to compose ribonucleoprotein (RNP) complexes (10).
IBDV RNP complexes act as capsid-independent functional units during the IBDV replication process (11), and they are fully competent for initiating the IBDV replication process. The three components of IBDV RNPs colocalize to the same structure and are involved directly in RNA synthesis (12, 13). Although researchers have performed functional analyses to characterize the IBDV RNP complexes (12), the mechanism by which RNPs are involved in IBDV replication and whether cellular factors participate in the activity of RNPs require further investigation. In this study, we identify a host protein that interacts with VP1 and VP3 and enhances IBDV polymerase activity.
In mammals, voltage-dependent anion channel protein 1 (VDAC1) is the most abundant isoform of VDAC and is therefore the most extensively studied of the isoforms (14). VDAC1 is located in the outer mitochondrial membrane (OMM) of all eukaryotes (15) and serves as a gatekeeper for the entry and exit of mitochondrial metabolites, thus controlling cross talk between mitochondria and the cytosol (16). In recent years, studies on VDAC1 have focused on the bioenergetics of metabolism (17) and apoptosis (18). Data show that VDAC1 contributes to metabolism by mediating the ATP/ADP exchange across the OMM as well as the binding and channeling of mitochondrial ATP directly to hexokinase (HK) (19); the VDAC1-HK association protects cancer cells from cell death. During apoptosis, VDAC1 mediates the release of many apoptogenic molecules, such as cytochrome c, from mitochondria to the cytosol via OMM permeabilization (19). A previous study suggested that VDAC1 upregulates cytochrome c and decreases the release of cytoplasmic Ca2+ in a nasopharyngeal carcinoma cell line during Epstein-Barr virus (EBV) infection (20). However, the role of VDAC1 in the progression of IBDV infection remains to be elucidated.
To understand the host response to IBDV infection and the interaction between virus and host, we performed labeling of differentially abundant proteins in IBDV-infected cells and mock-infected cells by use of isobaric tags for relative and absolute quantification (iTRAQ), and the results indicated that VDAC1 is upregulated during IBDV infection. We then studied the role of VDAC1 in the progression of IBDV infection, and the results showed that VDAC1 knockdown by treatment of DF-1 cells with specific small interfering RNAs (siRNAs) impaired IBDV growth. Furthermore, we demonstrated that the effects of VDAC1 on IBDV replication are dependent on its involvement in mediating IBDV polymerase activity. We found that VDAC1 regulates IBDV polymerase activity by stabilizing the interaction between VP1 and VP3. Our results provide new insight into the IBDV replication strategy.
RESULTS
Endogenous VDAC1 expression is upregulated by IBDV infection.
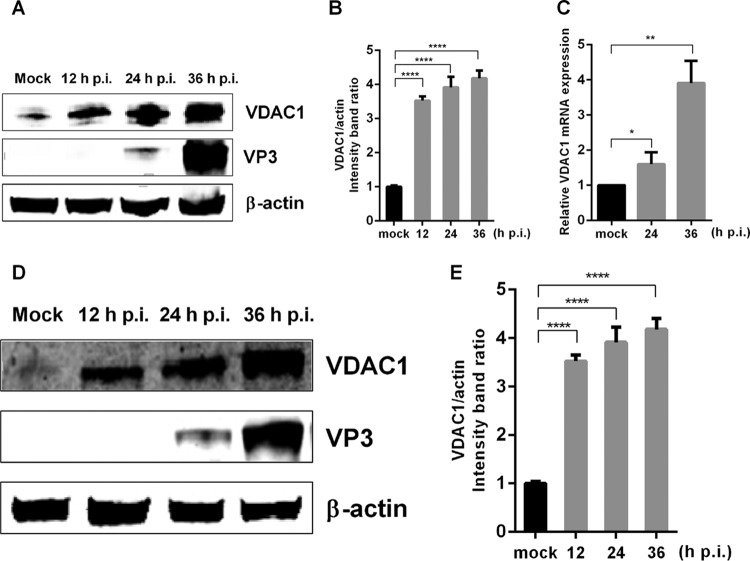
To understand the host response to IBDV infection and the interaction between the virus and its host, we performed iTRAQ labeling of differentially abundant proteins in IBDV-infected and mock-infected cells, and the results showed that viral infection alters the abundance of VDAC1 (data not shown), consistent with the results of previous studies (21, 22). To further verify the relationship between VDAC1 and IBDV infection, we infected DF-1 and 293T cells with IBDV at a multiplicity of infection (MOI) of 1. Then, at 12, 24, or 36 h postinfection, we collected cell cultures to detect VDAC1 protein expression by Western blotting and mRNA expression by quantitative real-time PCR (qRT-PCR). The results showed that IBDV infection increased the protein expression of VDAC1, resulting in levels up to 4-fold higher than those in mock-infected DF-1 cells (Fig. 1A and B). IBDV infection also resulted in increased VDAC1 levels in 293T cells (Fig. 1D and E), indicating that the observed upregulation of endogenous VDAC1 was not cell type specific. In IBDV-infected DF-1 cells, the expression of VDAC1 mRNA was also upregulated, reflecting changes similar to those observed at the protein level (Fig. 1C). These results indicate that IBDV infection induces upregulation of endogenous VDAC1 expression.
FIG 1.
IBDV infection upregulates the expression of VDAC1. (A) IBDV infection causes upregulation of VDAC1 expression in DF-1 cells. DF-1 cells were mock infected or infected with IBDV at an MOI of 1. Cells were harvested at 12, 24, and 36 h postinfection (p.i.) and then analyzed by Western blotting with anti-VDAC1 and -VP3 antibodies. (B) Relative intensities of VDAC1 in DF-1 cells. β-Actin was used as a protein loading control. Representative results are shown, with bars representing the intensities of VDAC1/actin normalized to the control condition. (C) IBDV infection causes upregulation of VDAC1 mRNA expression in DF-1 cells. DF-1 cells were mock infected or infected with IBDV at an MOI of 1. Cells were harvested at 24 and 36 h postinfection and then analyzed by qRT-PCR. (D) IBDV infection causes upregulation of VDAC1 expression in 293T cells. 293T cells were infected and analyzed as described for panel A. (E) Relative intensities of VDAC1 in 293T cells. β-Actin was used as a protein loading control. Representative results are shown, with bars representing the intensities of VDAC1/actin normalized to the control condition. Data are means and SD for three independent experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 versus control (unpaired t test).
RNAi-mediated knockdown of VDAC1 impairs IBDV replication.
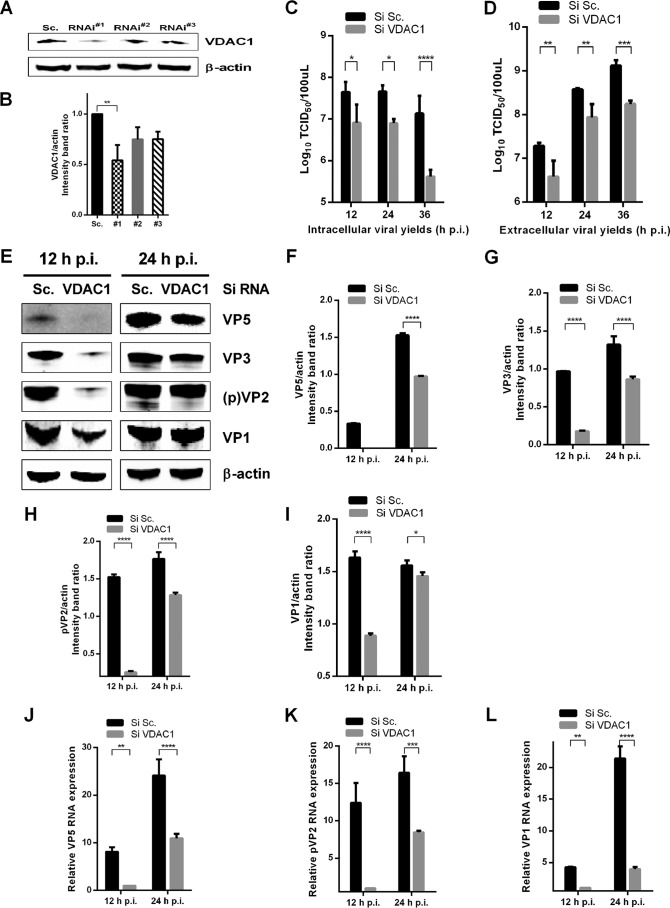
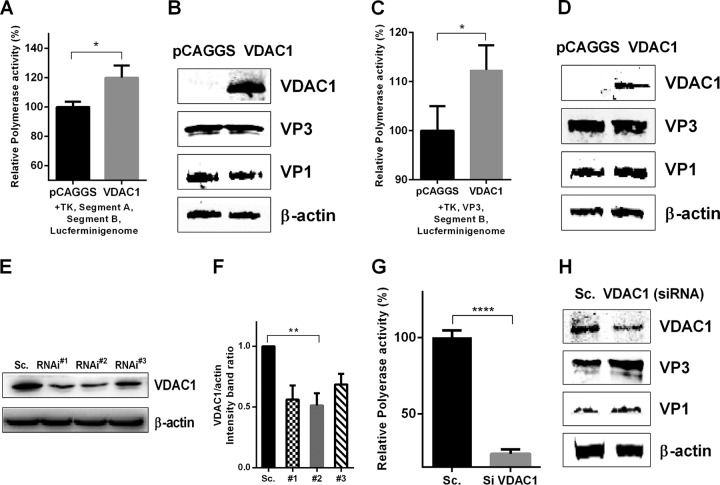
To further study the role of VDAC1 in the replication of IBDV, we used three VDAC1-specific siRNAs and found that one significantly reduced cellular levels of VDAC1 (Fig. 2A and B). We then knocked down VDAC1 in DF-1 cells by use of this competent siRNA and infected the cells with the IBDV Gt strain (MOI = 1). At different time points postinfection, we examined the viral loads in the supernatants and cell cultures of IBDV-infected cells by performing a 50% tissue culture infective dose (TCID50) assay. The results showed that knockdown of VDAC1 resulted in 5-fold, 6-fold, and 32-fold decreases in intracellular viral yields at 12, 24, and 36 h postinfection, respectively (Fig. 2C). Corresponding decreases in extracellular viral yields were observed at the same time points, with decreases of 5-fold, 4-fold, and 7-fold, respectively (Fig. 2D). At the same time, expression levels of viral proteins VP5, VP3, pVP2, and VP1 were detected by Western blotting, and downregulation of all viral proteins was observed in VDAC1 knockdown cells (Fig. 2E to I). Furthermore, we determined the relative expression of viral RNA by qRT-PCR and found that the viral RNA abundances of VP5, pVP2, and VP1 were downregulated in VDAC1 knockdown cells compared to the levels in scrambled siRNA-treated control cells (Fig. 2J to L). In conclusion, these data indicate that RNA interference (RNAi)-mediated knockdown of VDAC1 impairs IBDV replication.
FIG 2.
Knockdown of VDAC1 inhibits replication of IBDV. (A) Effects of VDAC1 RNAi on the expression of endogenous VDAC1. DF-1 cells were transfected with siRNA (RNAi#1, RNAi#2, or RNAi#3) or scrambled siRNA (Sc.). Cell lysates were harvested 48 h after the second transfection and examined by Western blotting with anti-VDAC1 antibody. Endogenous β-actin expression was used as an internal control. (B) Relative intensities of VDAC1 in knockdown cells. β-Actin was used as a protein loading control. Representative results are shown, with bars representing the intensities of VDAC1/actin normalized to the control condition. (C and D) Scrambled RNAi cells and VDAC1 RNAi cells were infected with IBDV at an MOI of 1. At different time points (12, 24, and 36 h) after IBDV infection, the infectious viral loads in the cell cultures (C) or supernatants (D) were determined by TCID50 analysis using 96-well plates. (E) Scrambled RNAi cells and VDAC1 RNAi cells were infected as described for panels C and D. Cells were harvested 12 and 24 h after infection and examined by Western blotting with antibodies against VP1, pVP2, VP3, and VP5. Endogenous β-actin expression was used as an internal control. (F to I) Relative intensities of viral proteins VP5 (F), VP3 (G), pVP2 (H), and VP1 (I) in VDAC1 siRNA-treated cells. (J to L) Control RNAi cells and VDAC1 RNAi cells were infected as described for panels C and D. Cells were harvested 12 and 24 h after infection, and RNA levels of VP5 (J), pVP2 (K), and VP1 (L) were determined by qRT-PCR. Data are means and SD for three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 versus control (two-way ANOVA).
VDAC1 interacts with both VP1 and VP3 of IBDV RNP complexes.
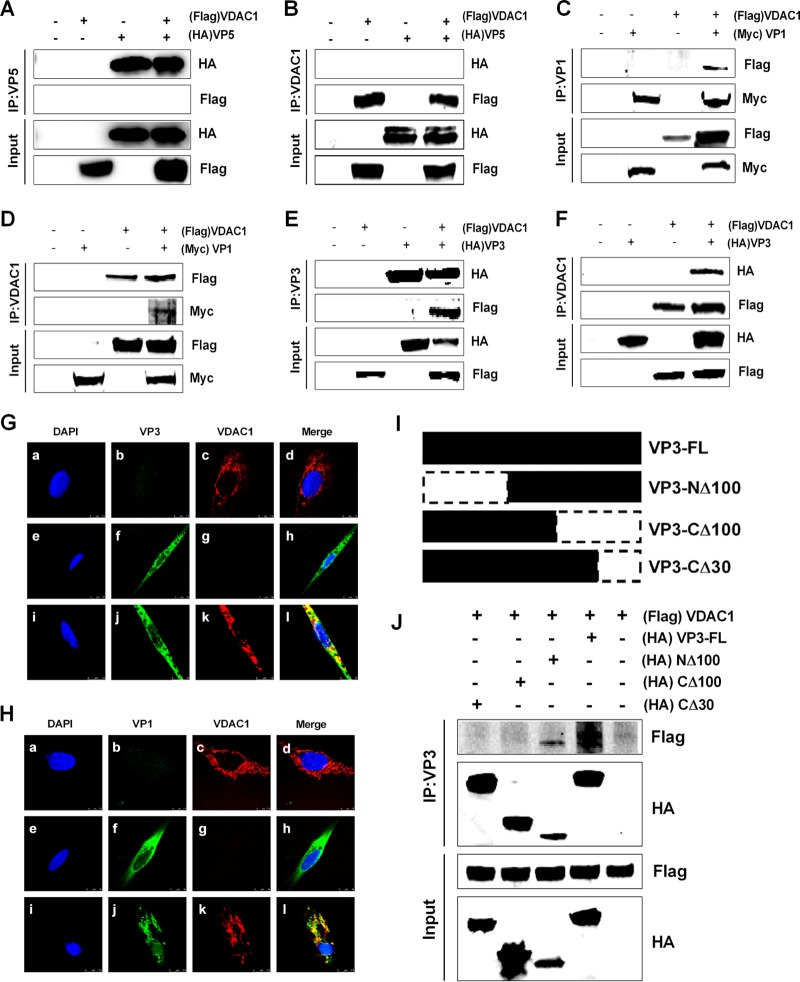
We decided to further analyze the possible mechanisms underlying the influence of VDAC1 on the progression of IBDV replication by searching for viral proteins that interact with VDAC1. Thus, we cotransfected 293T cells with FLAG-VDAC1 and MYC-GtVP1, HA-GtVP2, HA-GtVP3, HA-GtVP4, HA-GtVP5, or HA-GtpVP2. We then performed coimmunoprecipitation 48 h after transfection. When lysates of cells expressing both a viral protein and FLAG-VDAC1 were immunoprecipitated with a hemagglutinin (HA) or Myc antibody, FLAG-VDAC1 was detected in the precipitate, indicating that a particular viral protein interacted with VDAC1. The results showed that the viral protein VP5, which interacts with VDAC2 (23), displayed no interaction with VDAC1, while viral proteins VP1 and VP3 interacted with VDAC1 (Fig. 3A, C, and E). To confirm that the interactions occur in both directions, we performed an immunoprecipitation assay with a VDAC1 antibody and then performed Western blotting with an HA or Myc antibody. As shown in Fig. 3B, D, and F, VDAC1 could be associated specifically with VP1 and VP3 but not with VP5. These results demonstrate that VDAC1 interacts with VP1 and VP3 but not with VP5.
FIG 3.
VDAC1 interacts with viral proteins VP1 and VP3 but not with VP5. (A) 293T cells were transfected with VP5 and VDAC1 expression plasmids. Cell lysates were prepared 48 h after transfection, immunoprecipitated (IP) with anti-HA antibody, and immunoblotted with anti-FLAG or anti-HA antibody. (B) 293T cells were transfected as described for panel A. Cell lysates were prepared 48 h after transfection, immunoprecipitated with anti-VDAC1 antibody, and immunoblotted with anti-FLAG or anti-HA antibody. (C) 293T cells were transfected with VP1 and VDAC1 expression plasmids. Cell lysates were prepared 48 h after transfection, immunoprecipitated with anti-Myc antibody, and immunoblotted with anti-FLAG or anti-Myc antibody. (D) 293T cells were transfected as described for panel C. Cell lysates were prepared 48 h after transfection, immunoprecipitated with anti-VDAC1 antibody, and immunoblotted with anti-FLAG or anti-Myc antibody. (E) 293T cells were transfected with VP3 and VDAC1 expression plasmids. Cell lysates were prepared 48 h after transfection, immunoprecipitated with anti-HA antibody, and immunoblotted with anti-FLAG or anti-HA antibody. (F) 293T cells were transfected as described for panel E. Cell lysates were prepared 48 h after transfection, immunoprecipitated with anti-VDAC1 antibody, and immunoblotted with anti-FLAG or anti-HA antibody. (G) DF-1 cells were transfected with VP3 and/or VDAC1 for 24 h and then fixed and processed for dual labeling. Cell nuclei were counterstained with DAPI (blue). VP3 (green) and VDAC1 (red) proteins were visualized by immunostaining with monoclonal anti-HA and polyclonal anti-VDAC1 antibodies, respectively, and were analyzed using confocal microscopy. Bars, 10 μm. (a to d) Location of VDAC1; (e to h) location of VP3; (i to l) location of VP3 and VDAC1. VP3 staining is shown in green, and VDAC1 staining is shown in red; areas of colocalization in merged images are shown in yellow. (H) DF-1 cells were transfected with VP1 and/or VDAC1 for 24 h and then fixed and processed for dual labeling. Cell nuclei were counterstained with DAPI (blue). VP1 (green) and VDAC1 (red) proteins were visualized by immunostaining with monoclonal anti-Myc and polyclonal anti-VDAC1 antibodies, respectively, and were analyzed using confocal microscopy. Bars, 10 μm. (a to d) Location of VDAC1; (e to h) location of VP1; (i to l) location of VP1 and VDAC1. VP1 staining is shown in green, and VDAC1 staining is shown in red; areas of colocalization in merged images are shown in yellow. (I) Schematic representing the genes encoding the full-length VP3 protein and truncated VP3 molecules (CΔ30, CΔ100, and NΔ100). The numbers after “Δ” indicate the numbers of amino acids deleted from the C-terminal or N-terminal end of VP3. (J) Interaction between VDAC1 and truncated VP3 proteins. We transfected 293T cells with full-length HA-VP3 or truncated HA-VP3 molecules. Cell lysates were prepared 48 h after transfection, immunoprecipitated with anti-HA antibody, and immunoblotted with anti-FLAG or anti-HA antibody.
Furthermore, to verify whether VP1 and VP3 accumulate in locations where VDAC1 is present, colocalizations were observed by confocal microscopy. Briefly, DF-1 cells were transfected with plasmids expressing HA-VP3 and/or FLAG-VDAC1. The cells were fixed at 24 h posttransfection and incubated with DAPI (4′,6-diamidino-2-phenylindole), mouse anti-HA antibody, and/or rabbit anti-VDAC1 antibody. As shown in Fig. 3G, VDAC1 was found in the cytoplasm (a to d), and VP3 accumulated at the same positions as those of VDAC1 (e to h), suggesting that VP3 colocalized with VDAC1 in the cytoplasm (i to l). Next, we tested for colocalization of VDAC1 with VP1 by using confocal microscopy; we observed that VP1 was distributed in the cytoplasm and also exhibited colocalization with VDAC1 in the cytoplasm (Fig. 3H).
A previous study showed that the VP1 binding domain of VP3 maps to 16 residues at the C-terminal end of VP3 (24). To determine which region of VP3 is responsible for interacting with VDAC1, three different deletion mutants fused to HA tags were constructed: the VP3-CΔ30 and VP3-CΔ100 mutants lack 30 and 100 amino acids from the C-terminal end, respectively, and the VP3-NΔ100 mutant lacks 100 amino acids from the N-terminal end of VP3. Each VP3 mutant was expressed with VDAC1 in 293T cells, and the ability of each to interact with VDAC1 was examined by immunoprecipitation. The results showed that mutant VP3-NΔ100 was able to interact with VDAC1; however, mutants VP3-CΔ30 and VP3-CΔ100 exhibited no interactions with VDAC1, suggesting that the VDAC1 interaction domain of VP3 is located within the last 30 amino acids of the carboxyl terminus of VP3 (Fig. 3I and J). In conclusion, the results obtained from immunoprecipitation and confocal microscopy experiments show that VDAC1 interacts with IBDV VP1-VP3 complexes.
RNA bridges interactions between VDAC1 and IBDV VP1-VP3 complexes.
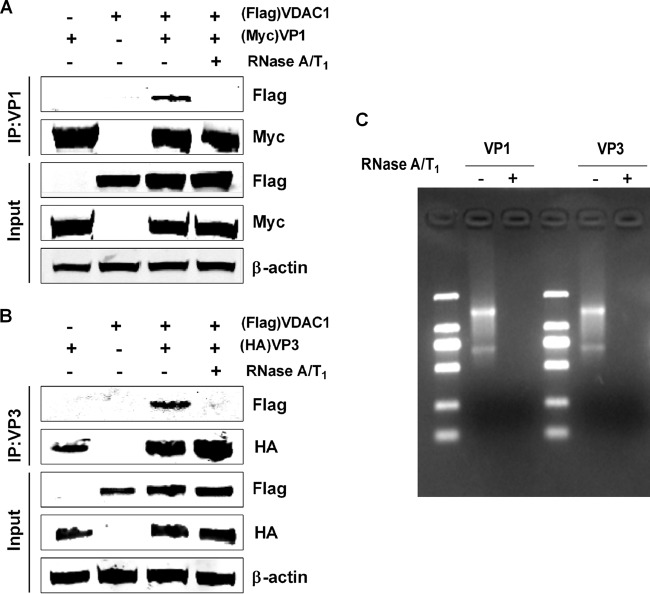
VP3 is an RNA binding protein capable of binding both single-stranded RNA and dsRNA (25). Hence, we sought to determine whether the VDAC1-VP1 or VDAC1-VP3 association was caused by protein interactions or RNA bridging. We treated cells transfected with VDAC1 plus VP1 or VDAC1 plus VP3 with RNase T1 and RNase A. As shown in Fig. 4C, RNase treatment led to complete digestion of internal RNA. We then performed immunoprecipitation assays with HA or Myc antibody, followed by Western blotting with the VDAC1 antibody. We found that before RNase digestion, VDAC1 was detected in the precipitate for cells expressing VP1 plus VDAC1 or VP3 plus VDAC1; however, VDAC1 was not detected in either precipitate following digestion with RNase (Fig. 4A and B). These findings suggest that the VDAC1-VP1 and VDAC1-VP3 associations are caused by viral and/or cellular RNA bridging.
FIG 4.
RNA bridges the interaction between VDAC1 and IBDV VP1-VP3 complexes. (A) 293T cells were transfected with VP1 and/or VDAC1 expression plasmids. Cell lysates were prepared 48 h after transfection, and then lysates with VDAC1-VP1 association were subjected to RNA digestion with RNase A and RNase T1 for 1 h at room temperature. Untreated cells were used as controls. All lysates were used for immunoprecipitation (IP). IP was performed with anti-Myc antibody, and lysates were immunoblotted with anti-FLAG or anti-Myc antibody. (B) 293T cells were transfected with VP3 and/or VDAC1 expression plasmids, and lysates of the VDAC1-VP3 cotransfection mixture were subjected to RNA digestion with RNase A and RNase T1 for 1 h at room temperature. Untreated cells were used as controls. All lysates were used for immunoprecipitation (IP). IP was performed with anti-HA antibody, and lysates were immunoblotted with anti-FLAG or anti-HA antibody. (C) A portion of each sample was analyzed for RNA content by electrophoresis on a 1% agarose gel. Lanes: −, samples without RNA digestion; +, samples with RNA digestion.
Overexpression of VDAC1 enhances IBDV polymerase activity, and knockdown of VDAC1 suppresses IBDV polymerase activity.
VP1-VP3 complexes have been shown to be involved in polymerase activation, during which VP3 binding promotes a conformational change that removes the steric blockade of the VP1 active site (9). To determine the role of VDAC1 in RNP complexes, we measured IBDV polymerase activity. In a previous study, researchers established a luciferase assay for detecting IBDV polymerase activity in DF-1 cells (26); here we used a similar method to assess polymerase activity in 293T cells exhibiting overexpression or knockdown of VDAC1. These cells were then transfected with IBDV-dependent luciferase reporter plasmids. The results showed that overexpression of VDAC1 enhances IBDV polymerase activity (Fig. 5A and B). In addition, VP3 was used to replace segment A in the polymerase activity detection system, and the results showed that VP3 was able to replace segment A to execute polymerase activity (Fig. 5C and D). Moreover, we used three VDAC1-specific siRNAs and found that one significantly reduced cellular levels of VDAC1 (Fig. 5E and F). Knockdown of VDAC1 led to a 4-fold decrease in IBDV polymerase activity (Fig. 5G and H).
FIG 5.
VDAC1 enhances IBDV polymerase activity in vitro. (A and B) Polymerase activity assays were performed with 293T cells expressing an IBDV segment B-driven luciferase minigenome system including segment A, segment B, TK, and VDAC1 or empty vector. VDAC1, VP1, and VP3 protein expression was analyzed by Western blotting. (C and D) Polymerase activity assays were performed as described for panels A and B, but with VP3 instead of segment A. VDAC1, VP1, and VP3 protein expression was analyzed by Western blotting. Endogenous β-actin expression was used as an internal control. (E) Effects of VDAC1 RNAi on the expression of endogenous VDAC1. 293T cells were transfected with specific siRNA (RNAi#1, RNAi#2, or RNAi#3) or a control. Cell lysates were harvested 48 h after the second transfection and examined by Western blotting with anti-VDAC1 antibody. Endogenous β-actin expression was used as an internal control. (F) Relative intensities of VDAC1 in VDAC1 siRNA-treated cells. β-Actin was used as a protein loading control. Representative results are shown, with bars representing the intensities of VDAC1/actin bands normalized to the control condition. (G and H) 293T cells were transfected with scrambled control or VDAC1 siRNA. Polymerase activity assays were performed 36 h after transfection as described for panels C and D. VDAC1, VP1, and VP3 protein expression was analyzed by Western blotting. Endogenous β-actin expression was used as an internal control.
VDAC1 enhances IBDV polymerase activity through its ability to stabilize interactions in RNP complexes.
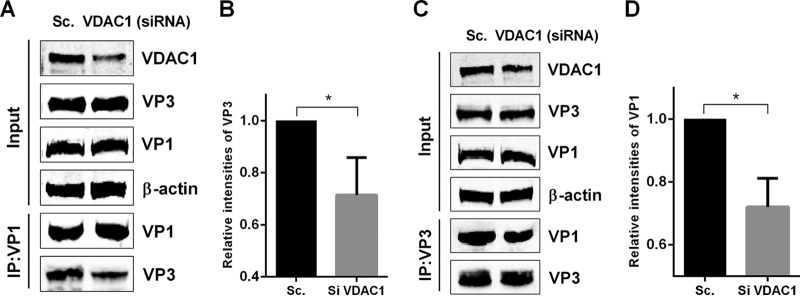
IBDV RNPs are composed of viral proteins VP1 and VP3 as well as dsRNA. The stability of the RNP components is considered to be a factor in mediating IBDV polymerase activity. As mentioned above, the VP1 binding domain of VP3 is located in the 16 residues at the C-terminal end of VP3 (24), and we found that the 30 amino acids at the C-terminal end of VP3 are responsible for interacting with VDAC1. To test the possibility that an increase in polymerase activity was due to a stabilizing effect of VDAC1 on IBDV RNPs, we transfected cells with a VDAC1-specific siRNA or a scrambled siRNA control, and 36 h later, we transfected cells with the plasmids HA-VP3 and MYC-VP1. We then performed an immunoprecipitation assay to detect an interaction between VP1 and VP3. When lysates of cells expressing both VP1 and VP3 were immunoprecipitated with the Myc antibody, VP3 was detected in the precipitates, indicating that VP1 interacts with VP3. Levels of VP3 detected in the precipitates of VDAC1 knockdown cells were lower than those in the precipitates of scrambled siRNA-treated cells (Fig. 6A and B). Moreover, when lysates of cells were immunoprecipitated with HA antibody, VP1 was detected in the precipitates, and levels of VP1 detected in the precipitates of VDAC1 knockdown cells were lower than those in the precipitates of scrambled siRNA-treated cells (Fig. 6C and D). These findings demonstrate that knockdown of VDAC1 weakens the interaction between VP1 and VP3 in both directions. This indicates that the observed increase in viral polymerase activity may be due to the stabilizing effect of VDAC1 on IBDV RNP complexes.
FIG 6.
VDAC1 expression enhances IBDV polymerase function through its ability to stabilize the interaction between VP1 and VP3. (A) 293T cells were transfected with scrambled control or VDAC1 siRNA. Cells were transfected with VP1 and VP3 36 h later. Cell lysates were prepared 36 h after transfection, immunoprecipitated with anti-Myc antibody, and immunoblotted with anti-Myc, anti-VDAC1, or anti-HA antibody. (B) Relative intensities of VP3 in VDAC1 siRNA-treated cells. β-Actin was used as a protein loading control. Representative results are shown, with bars representing the intensities of VP3/actin bands normalized to the control condition. (C) 293T cells were transfected as described for panel A. Cell lysates were prepared, immunoprecipitated with anti-HA antibody, and immunoblotted with anti-Myc, anti-VDAC1, or anti-HA antibody. (D) Relative intensities of VP1 in VDAC1 siRNA-treated cells. β-Actin was used as a protein loading control. Representative results are shown, with bars representing the intensities of VP1/actin bands normalized to the control condition. Data are means and SD for three independent experiments. *, P < 0.05; ****, P < 0.0001 versus control (unpaired t test).
DISCUSSION
Virus-encoded RdRp is a significant component in the life cycle of RNA viruses, being responsible for both genome replication and transcription. IBDV ribonucleoprotein (RNP) complexes are formed by the RdRp, the genome segments, and the dsRNA-binding VP3 polypeptide, and the RNP complexes execute viral dsRNA replication and transcription (11). The C-terminal end of VP3 is necessary for mediating VP1 and dsRNA binding and for modulating the activity of polymerase. Until now, rarely have cellular factors been identified as being involved in IBDV replication through mediating polymerase activity (27). In this study, we provide further evidence for the involvement of a cellular factor, VDAC1, which plays a role in enhancing IBDV polymerase activity during IBDV replication through its ability to stabilize interactions in RNP complexes.
VDAC1 is a pore-forming protein found in the OMM of all eukaryotes (15), and its expression is associated with many diseases (19, 28). Studies have shown that the expression of VDAC1 is much higher in tumor cells than in normal cells (29–31), and the increased expression of VDAC1 serves as an important factor in the process of mitochondrion-mediated apoptosis (19, 32–35). Viruses are known to modulate apoptosis at the mitochondrial level via multiple strategies (36–40). Previously, VDAC2 (a member of the VDAC family) was reported to be involved in regulating IBDV replication through modulating VP5-induced apoptosis (23, 41). In this study, we demonstrated that the expression of VDAC1 is upregulated following IBDV infection. In order to reveal the role of VDAC1 in the progress of IBDV replication, we first examined whether VDAC1 is involved in virus growth. Our results showed that knockdown of VDAC1 impairs IBDV replication, as viral loads, viral RNA abundances, and expression of viral proteins were much lower in VDAC1 knockdown cells than in scrambled siRNA-treated control cells. We therefore sought to elucidate the mechanisms of action of VDAC1. Although we found that knockdown of VDAC1 reduced IBDV-induced apoptosis and inhibited the activities of caspase-9 and caspase-3 (data not shown), we could not distinguish an effect on virus replication from a direct effect on apoptosis, because a viral replication reduction means that the cell receives less of the stimulatory signal to induce apoptosis. To illustrate this point, we identified the interactions between viral proteins and VDAC1. The viral proteins VP2 and VP5, which are associated with apoptosis, displayed no interaction with VDAC1, indicating that the mechanisms of action of VDAC1 in the process of IBDV replication are not directly related to apoptosis (Fig. 3A and B and data not shown). Interestingly, we found that both VP1 and VP3, the components of IBDV RNPs, interacted with VDAC1, and we also found that RNA bridges the interaction between VDAC1 and IBDV VP1-VP3 complexes. Both of these results indicate that VDAC1 may play a vital role in modulating polymerase activity.
To support this hypothesis, a minigenome assay was performed by overexpressing and knocking down VDAC1. The results indicate that overexpression of VDAC1 enhances polymerase activity, and in contrast, knockdown of VDAC1 reduces polymerase activity. How does the VDAC1 protein regulate IBDV polymerase activity? RNA and the VP1 and VP3 proteins can bind to VDAC1, and the three components of IBDV RNP complexes display a stable structure in which the dsRNA genome is wrapped with VP3 molecules and both ends of the dsRNA molecules are bound covalently with VP1 (12, 42); moreover, VP1 interacts with VP3 at the C-terminal tail of VP3. Considering the structure of RNPs and the fact that the VDAC1 binding domain of VP3 is located in the last 30 amino acids at the C-terminal end of the protein, together with our observation that the presence of VDAC1 can increase the polymerase activity of IBDV in vitro, we speculate that VDAC1 likely plays a role in modulating the formation of RNP complexes. A complementary immunoprecipitation approach led us to the conclusion that VDAC1 is necessary and sufficient for stabilizing the interaction between VP1 and VP3. Our findings in this study further provide novel insight into virus-host interactions that are specifically involved in stimulating viral RNA polymerase activities.
In conclusion, we have newly identified a member of the VDAC family, VDAC1, as an enhancer of IBDV replication. Our detailed molecular analysis revealed that VDAC1 interacts with IBDV RNP complexes and enhances viral RNA polymerase activity through its ability to stabilize the interaction between VP3 and VP1. These results not only increase our knowledge of host factors involved in regulating the IBDV RNA polymerase but also provide novel insight into our functional understanding of VDAC1 during viral infection.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
DF-1 (immortal chicken embryo fibroblast) cells and 293T human embryonic kidney cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, USA) supplemented with 10% fetal bovine serum (FBS), 1% glutamine (Gibco), and 1% penicillin-streptomycin (Gibco) in a 5% CO2 incubator. Primary chicken embryo fibroblast (CEF) cells were prepared from 9-day-old specific-pathogen-free chicken embryos. The animal experiment was approved by the Committee of the Ethics of Animal Experiments at the Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences. The Gt strain of IBDV was identified and preserved in our laboratory. Antibodies used in the study included mouse anti-FLAG M2 (F1804; Sigma, USA), rabbit anti-FLAG (F2555; Sigma), rabbit anti-VDAC1/Porn (ab15895; Abcam, United Kingdom), rabbit anti-HA (H6908; Sigma), mouse anti-HA (H9658; Sigma), mouse anti-β-actin monoclonal antibody (MAb) (A1978; Sigma), mouse anti-Myc (M4439; Sigma), rabbit anti-Myc (SAB4300319; Sigma), goat anti-mouse IgG(H+L)–Alexa Fluor 488 (A-11008; Invitrogen, USA), goat anti-rabbit IgG(H+L)–Alexa Fluor 633 (A-21070; Invitrogen), IRDye 800CW–goat anti-mouse IgG(H+L) (926-32210; LiCor Bio-Sciences, USA), and IRDye 800CW–goat anti-rabbit IgG(H+L) (926-32211; LiCor Bio-Sciences).
IBDV infection and titration.
For viral infection, DF-1 cells were grown to approximately 90% confluence and washed once with phosphate-buffered saline (PBS); the cells were then incubated in 1 ml DMEM with 2% FBS, and dilute viruses were incubated with cells for 1.5 h. Subsequently, the viral inoculum was removed, and cells were maintained with DMEM containing 10% FBS at 37°C until collection. The primary CEF cultures were used to titrate infectious progeny virus after various treatments. Infected cell cultures and supernatants were harvested at different time points (12, 24, and 36 h) after infection, and the titers of infectious viral progeny present in the cultures and supernatants were determined in terms of TCID50/100 μl, using the Reed-Muench formula (43). All experiments were repeated three times, and the means and standard deviations were calculated.
Construction of plasmids.
IBDV VP2, VP3, VP4, VP5, and pVP2 were cloned by inserting the viral genes of the IBDV Gt strain into the pCAGGS plasmid, with an HA tag inserted at the N terminus of the expressed proteins. IBDV VP1 was constructed with a Myc tag at the C terminus. VDAC1 was cloned from the cDNA of DF-1 cells by use of specific primers and was cloned into the pCAGGS plasmid with a FLAG tag. Truncated expression plasmids for VP3 (VP3-CΔ30, amino acids [aa] 1 to 227; VP3-CΔ100, aa 1 to 157; and VP3-NΔ100, aa 101 to 257) were cloned by inserting the corresponding genes into the pCAGGS plasmid with an HA tag at the C terminus. For luciferase assays, the expression plasmids were generated as part of a previously established RNA polymerase II reverse genetics system (44).
Transfection and siRNA knockdown.
DF-1 cells were transfected at 80% confluence by use of X-tremeGENE HP DNA transfection reagent (Sigma) according to the manufacturer's instructions. Three siRNAs specifically targeting the VDAC1 mRNA of Gallus or Homo sapiens were designed by the Genechem Company (China) to study viral replication and polymerase function. The siRNAs for knockdown of VDAC1 in DF-1 cells included RNAi#1 (sense, 5′-GUAGCUUGGAAACCAAAUATT-3′), RNAi#2 (sense, 5′-AUAUGGGCUGAUGUUCACGTT-3′), RNAi#3 (sense, 5′-CGGAAUAGCAGCCAAGUAUTT-3′), and a negative control (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′). The siRNAs for knockdown of VDAC1 in 293T cells included RNAi#1 (sense, 5′-CUGACGUUUACAGAGAAAUTT-3′), RNAi#2 (sense, 5′-GCGACAUGGAUUUCGACAUTT-3′), and RNAi#3 (sense, 5′-GCGGCUCCAUUUACCAGAATT-3′). siRNA transfections were performed using RNAiMAX (Invitrogen) according to the manufacturer's instructions. Double transfections were performed at 24-h intervals. Twenty-four hours after the second transfection, cells were harvested for further analysis. The siRNA with the highest knockdown efficiency was chosen for evaluation of the influence of VDAC1 on IBDV replication and polymerase function. For the replication study, siRNA-transfected cells were infected with the IBDV Gt strain at a multiplicity of infection (MOI) of 1 and cultured for an addition 12, 24, or 36 h. The supernatants and cell cultures were then collected to detect viral titers of IBDV. For the polymerase function study, Gt-Luc reporter plasmids were transfected into siRNA-transfected cells for another 36 h.
Coimmunoprecipitation and Western blot analysis.
First, 293T cells were seeded into 6-well plates and cultured for at least 12 h before being transfected with pCAGGS-HA-VP5, pCAGGS-HA-VP3, pCAGGS-MYC-VP1, and/or pCAGGS-FLAG-VDAC1 by use of X-tremeGENE HP DNA transfection reagent (Sigma). Next, 48 h after transfection, transfected cells were lysed in Pierce IP buffer (Thermo Fisher Scientific, USA) containing protease inhibitor cocktail (Roche, Switzerland). Supernatants were obtained by centrifugation and were incubated with 2 μg anti-HA or anti-Myc mouse MAb or anti-VDAC1 rabbit polyclonal antibody at 4°C for 6 to 8 h. After incubation with antibody, 25 μl of protein G Sepharose beads (Roche) was added, and the samples were incubated at 4°C overnight. Beads were washed five times with PBS containing protease inhibitor cocktail and then boiled with 2× SDS loading buffer for 10 min. Subsequently, the samples were fractionated by electrophoresis on 12% SDS-polyacrylamide gels, and resolved proteins were transferred onto nitrocellulose membranes. After blocking with 5% skim milk, the membranes were incubated with rabbit anti-Myc, rabbit anti-HA, and mouse anti-FLAG antibodies, followed by IRDye 800CW–goat anti-mouse IgG secondary antibody and IRDye 800CW–goat anti-rabbit IgG secondary antibody. The membrane blots were scanned using an Odyssey infrared imaging system.
Confocal microscopy assay.
To verify the interaction between VP3 and VDAC1, DF-1 cells were transfected with pCAGGS-FLAG-VDAC1 and/or pCAGGS-HA-VP3 for 24 h. At the same time, pCAGGS-FLAG-VDAC1 and/or pCAGGS-MYC-VP1 was also transfected into DF-1 cells for 24 h to determine the interaction between VDAC1 and VP1. The cells were washed with PBS three times and fixed with 4% formaldehyde for 30 min, followed by permeabilization with 0.1% Triton X-100 in PBS for 15 min. Samples were rinsed with PBS and blocked with 5% skim milk in PBS at 37°C for 1 h before being incubated with anti-VP3 (1:500) and anti-VDAC1 (1:100) antibodies or anti-Myc (1:200) and anti-VDAC1 (1:100) antibodies diluted in PBS for 1 h at 37°C. The cells were washed three times with PBS and then incubated with the Alexa 488–anti-mouse and Alexa 633–anti-rabbit (1:1,000) secondary antibodies. Finally, cells were stained with DAPI (Beyotime, China) at 25°C for 20 min. After staining, the cells were washed several times and examined using a Leica SP2 confocal microscope system (Leica Microsystems, Germany).
qRT-PCR.
Total RNA was extracted using an RNeasy minikit (Qiagen, Germany), and 2 μg of RNA was reverse transcribed into cDNA by use of a SuperScript III reverse transcription system (Invitrogen) in a 20-μl reaction mixture. cDNA was analyzed by qRT-PCR using Thunderbird probe qPCR mix (Toyobo, Japan). Specific primers and TaqMan probes for chicken actin as well as for IBDV VP1, pVP2, and VP5 were synthesized by Invitrogen (China). TaqMan probes for chicken VDAC1 were designed by Life Technologies, USA. qRT-PCR was performed under the following cycling conditions: 95°C for 1 min for initial denaturation, followed by 40 cycles of 95°C for 15 s for denaturation, 60°C for 1 min, and collection of PCR product signals. All controls and infected samples were examined in triplicate on the same plate. cDNA quantities were normalized to actin cDNA quantities measured from the same samples.
IBDV minigenome system for detection of polymerase activity.
Human 293T cells were seeded into 12-well plates. After 24 h, cells were transfected with an expression plasmid (1 μg of pCAGGS-FLAG-VDAC1 or empty vector), the vector pRL-TK (200 ng), 1 μg each of RNA polymerase plasmids pCAGGS-GtB, pCAGGS-GtA, and/or pCAGGS-GtVP3, and the luciferase reporter genome plasmid pCAGGS-Luc-Gt by use of X-tremeGENE HP DNA transfection reagent (Sigma). The pRL-TK vector was used as an internal control to correct for differences in both transfection and harvest efficiencies. Thirty-six hours after transfection, cells were lysed and analyzed by use of a dual-luciferase reporter assay kit according to the manufacturer's instructions (Promega, USA).
RNAi#2, targeting the VDAC1 mRNA of H. sapiens, and the negative siRNA control were transfected into 293T cells by use of RNAiMAX according to the manufacturer's instructions. Double transfections were performed at 24-h intervals; 24 h after the second transfection, cells were transfected with the pRL-TK vector (200 ng), 1 μg each of RNA polymerase plasmids pCAGGS-GtB and pCAGGS-GtVP3, and the luciferase reporter genome plasmid pCAGGS-Luc-Gt. Finally, 36 h after transfection, cells were harvested for analysis of polymerase activity.
Statistical analysis.
Data are presented as means ± standard deviations (SD). For some experiments, the significance of the variability between different groups was determined by two-way analysis of variance (ANOVA), using GraphPad Prism software (version 6.0). P values of <0.05 were considered statistically significant and are marked with asterisks in figures. For other experiments, the statistical analyses were done with the unpaired t test. P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
We thank the members of the avian immunosuppressive disease group for providing valuable suggestions and supporting the Harbin Veterinary Research Institute with their help.
This work was supported by the Major Project of the National Natural Science Foundation of China (grant 31430087), the earmarked fund for the Modern Agro-Industry Technology Research System (grant CARS-42-G07), and the State Key Laboratory of Veterinary Biotechnology Foundation (grant SKLVBP2015009).
REFERENCES
- 1.Kibenge FS, Dhillon AS, Russell RG. 1988. Biochemistry and immunology of infectious bursal disease virus. J Gen Virol 69:1757–1775. doi: 10.1099/0022-1317-69-8-1757. [DOI] [PubMed] [Google Scholar]
- 2.Muller H, Scholtissek C, Becht H. 1979. The genome of infectious bursal disease virus consists of two segments of double-stranded RNA. J Virol 31:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson PJ, McKern NM, Power BE, Azad AA. 1986. Genomic structure of the large RNA segment of infectious bursal disease virus. Nucleic Acids Res 14:5001–5012. doi: 10.1093/nar/14.12.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azad AA, Jagadish MN, Brown MA, Hudson PJ. 1987. Deletion mapping and expression in Escherichia coli of the large genomic segment of a birnavirus. Virology 161:145–152. doi: 10.1016/0042-6822(87)90180-2. [DOI] [PubMed] [Google Scholar]
- 5.von Einem UI, Gorbalenya AE, Schirrmeier H, Behrens SE, Letzel T, Mundt E. 2004. VP1 of infectious bursal disease virus is an RNA-dependent RNA polymerase. J Gen Virol 85:2221–2229. doi: 10.1099/vir.0.19772-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Lu Z, Zhang L, Gao L, Wang N, Gao X, Wang Y, Li K, Gao Y, Cui H, Gao H, Liu C, Zhang Y, Qi X, Wang X. 2016. Ribosomal protein L4 interacts with viral protein VP3 and regulates the replication of infectious bursal disease virus. Virus Res 211:73–78. doi: 10.1016/j.virusres.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Maraver A, Ona A, Abaitua F, Gonzalez D, Clemente R, Ruiz-Diaz JA, Caston JR, Pazos F, Rodriguez JF. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J Virol 77:6438–6449. doi: 10.1128/JVI.77.11.6438-6449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saugar I, Irigoyen N, Luque D, Carrascosa JL, Rodriguez JF, Caston JR. 2010. Electrostatic interactions between capsid and scaffolding proteins mediate the structural polymorphism of a double-stranded RNA virus. J Biol Chem 285:3643–3650. doi: 10.1074/jbc.M109.075994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garriga D, Navarro A, Querol-Audi J, Abaitua F, Rodriguez JF, Verdaguer N. 2007. Activation mechanism of a noncanonical RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 104:20540–20545. doi: 10.1073/pnas.0704447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjalmarsson A, Carlemalm E, Everitt E. 1999. Infectious pancreatic necrosis virus: identification of a VP3-containing ribonucleoprotein core structure and evidence for O-linked glycosylation of the capsid protein VP2. J Virol 73:3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton RM, Rodriguez JF. 2014. Rescue of infectious birnavirus from recombinant ribonucleoprotein complexes. PLoS One 9:e87790. doi: 10.1371/journal.pone.0087790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luque D, Saugar I, Rejas MT, Carrascosa JL, Rodriguez JF, Caston JR. 2009. Infectious bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J Mol Biol 386:891–901. doi: 10.1016/j.jmb.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delgui LR, Rodriguez JF, Colombo MI. 2013. The endosomal pathway and the Golgi complex are involved in the infectious bursal disease virus life cycle. J Virol 87:8993–9007. doi: 10.1128/JVI.03152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Pinto V, Guarinoa F, Guarnera A, Messina A, Reina S, Tomasello FM, Palermo V, Mazzoni C. 2010. Characterization of human VDAC isoforms: a peculiar function for VDAC3? Biochim Biophys Acta 1797:1268–1275. doi: 10.1016/j.bbabio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 15.Benz R. 1994. Permeation of hydrophilic solutes through mitochondrial outer membranes: review on mitochondrial porins. Biochim Biophys Acta 1197:167–196. doi: 10.1016/0304-4157(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 16.Blachly-Dyson E, Forte M. 2001. VDAC channels. IUBMB Life 52:113–118. doi: 10.1080/15216540152845902. [DOI] [PubMed] [Google Scholar]
- 17.Fiek CBR, Roos N, Brdiczka D. 1982. Evidence for identity between the hexokinase-binding protein and the mitochondrial porin in the outer membrane of rat liver mitochondria. Biochim Biophys Acta 688:629–640. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu S, Narita M, Tsujimoto Y. 1999. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature 399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 19.Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N. 2010. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med 31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Feng X, Ching CB, Chen WN. 2012. EBV up-regulates cytochrome c through VDAC1 regulations and decreases the release of cytoplasmic Ca2+ in the NPC cell line. Cell Biol Int 36:733–738. doi: 10.1042/CBI20110368. [DOI] [PubMed] [Google Scholar]
- 21.Feng XS, Chowbay B, Chen WN, Ching CB. 2011. ITRAQ-coupled 2-D LC-MS/MS analysis of differentially expressed serum proteins in nasopharyngeal carcinoma clinical samples: potential in biomarker discovery. J Med Imaging Health Informatics 1:177–183. doi: 10.1166/jmihi.2011.1022. [DOI] [Google Scholar]
- 22.Feng XS, Zhang JH, Chen WN, Ching CB. 2011. Proteome profiling of Epstein-Barr virus infected nasopharyngeal carcinoma cell line: identification of potential biomarkers by comparative iTRAQ-coupled 2D LC/MS-MS analysis. J Proteomics 74:567–576. doi: 10.1016/j.jprot.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Wang Y, Xue Y, Li X, Cao H, Zheng SJ. 2012. Critical role for voltage-dependent anion channel 2 in infectious bursal disease virus-induced apoptosis in host cells via interaction with VP5. J Virol 86:1328–1338. doi: 10.1128/JVI.06104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maraver A, Clemente R, Rodriguez JF, Lombardo E. 2003. Identification and molecular characterization of the RNA polymerase-binding motif of infectious bursal disease virus inner capsid protein VP3. J Virol 77:2459–2468. doi: 10.1128/JVI.77.4.2459-2468.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tacken MGJ, Peeters BPH, Thomas AAM, Rottier PJM, Boot HJ. 2002. Infectious bursal disease virus capsid protein VP3 interacts both with VP1, the RNA-dependent RNA polymerase, and with viral double-stranded RNA. J Virol 76:11301–11311. doi: 10.1128/JVI.76.22.11301-11311.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu F, Ren X, Wang Y, Qi X, Song J, Gao Y, Qin L, Gao H, Wang X. 2013. A single amino acid V4I substitution in VP1 attenuates virulence of very virulent infectious bursal disease virus (vvIBDV) in SPF chickens and increases replication in CEF cells. Virology 440:204–209. doi: 10.1016/j.virol.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 27.Gao L, Li K, Zhong L, Zhang L, Qi X, Wang Y, Gao Y, Wang X. 2017. Eukaryotic translational initiation factor 4AII reduces the replication of infectious bursal disease virus by inhibiting VP1 polymerase activity. Antiviral Res 139:102–111. doi: 10.1016/j.antiviral.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Shoshan-Barmatz V, Ben-Hail D. 2012. VDAC, a multi-functional mitochondrial protein as a pharmacological target. Mitochondrion 12:24–34. doi: 10.1016/j.mito.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Grills C, Jithesh PV, Blayney J, Zhang SD, Fennell DA. 2011. Gene expression meta-analysis identifies VDAC1 as a predictor of poor outcome in early stage non-small cell lung cancer. PLoS One 6:e14635. doi: 10.1371/journal.pone.0014635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang N, Kham SK, Koh GS, Suang Lim JY, Ariffin H, Chew FT, Yeoh AE. 2011. Identification of prognostic protein biomarkers in childhood acute lymphoblastic leukemia (ALL). J Proteomics 74:843–857. doi: 10.1016/j.jprot.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Xiao ZQ, Chen ZC, Li JL, Li C, Zhang PF, Li MY. 2007. Proteomic analysis of the aging-related proteins in human normal colon epithelial tissue. J Biochem Mol Biol 40:72–81. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh T, Pandey N, Maitra A, Brahmachari SK, Pillai B. 2007. A role for voltage-dependent anion channel Vdac1 in polyglutamine-mediated neuronal cell death. PLoS One 2:e1170. doi: 10.1371/journal.pone.0001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Godbole A, Varghese J, Sarin A, Mathew MK. 2003. VDAC is a conserved element of death pathways in plant and animal systems. Biochim Biophys Acta 1642:87–96. doi: 10.1016/S0167-4889(03)00102-2. [DOI] [PubMed] [Google Scholar]
- 34.Lu AJ, Dong CW, Du CS, Zhang QY. 2007. Characterization and expression analysis of Paralichthys olivaceus voltage-dependent anion channel (VDAC) gene in response to virus infection. Fish Shellfish Immunol 23:601–613. doi: 10.1016/j.fsi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Zaid H, Abu-Hamad S, Israelson A, Nathan I, Shoshan-Barmatz V. 2005. The voltage-dependent anion channel-1 modulates apoptotic cell death. Cell Death Differ 12:751–760. doi: 10.1038/sj.cdd.4401599. [DOI] [PubMed] [Google Scholar]
- 36.Boya P, Pauleau AL, Poncet D, Gonzalez-Polo RA, Zamzami N, Kroemer G. 2004. Viral proteins targeting mitochondria: controlling cell death. Biochim Biophys Acta 1659:178–189. doi: 10.1016/j.bbabio.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Boya P, Roumier T, Andreau K, Gonzalez-Polo RA, Zamzami N, Castedo M, Kroemer G. 2003. Mitochondrion-targeted apoptosis regulators of viral origin. Biochem Biophys Res Commun 304:575–581. doi: 10.1016/S0006-291X(03)00630-2. [DOI] [PubMed] [Google Scholar]
- 38.Shirakata Y, Koike K. 2003. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J Biol Chem 278:22071–22078. doi: 10.1074/jbc.M301606200. [DOI] [PubMed] [Google Scholar]
- 39.Zamarin D, Garcia-Sastre A, Xiao X, Wang R, Palese P. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog 1:e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castedo M, Perfettini JL, Andreau K, Roumier T, Piacentini M, Kroemer G. 2003. Mitochondrial apoptosis induced by the HIV-1 envelope. Ann N Y Acad Sci 1010:19–28. doi: 10.1196/annals.1299.004. [DOI] [PubMed] [Google Scholar]
- 41.Lin W, Zhang Z, Xu Z, Wang B, Li X, Cao H, Wang Y, Zheng SJ. 2015. The association of receptor of activated protein kinase C 1 (RACK1) with infectious bursal disease virus viral protein VP5 and voltage-dependent anion channel 2 (VDAC2) inhibits apoptosis and enhances viral replication. J Biol Chem 290:8500–8510. doi: 10.1074/jbc.M114.585687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller H, Nitschke R. 1987. The two segments of the infectious bursal disease virus genome are circularized by a 90,000-Da protein. Virology 159:174–177. doi: 10.1016/0042-6822(87)90363-1. [DOI] [PubMed] [Google Scholar]
- 43.Pizzi M. 1950. Sampling variation of the fifty percent end-point, determined by the Reed-Muench (Behrens) method. Hum Biol 22:151–190. [PubMed] [Google Scholar]
- 44.Qi X, Gao Y, Gao H, Deng X, Bu Z, Wang X, Fu C, Wang X. 2007. An improved method for infectious bursal disease virus rescue using RNA polymerase II system. J Virol Methods 142:81–88. doi: 10.1016/j.jviromet.2007.01.021. [DOI] [PubMed] [Google Scholar]