ABSTRACT
Mutational escape of HIV-1 from HIV-1-specific CD8+ T lymphocytes (CTLs) is a major barrier for effective immune control. Each epitope typically is targeted by multiple clones with distinct T cell receptors (TCRs). While the clonal repertoire may be important for containing epitope variation, determinants of its composition are poorly understood. We investigate the clonal repertoire of 29 CTL responses against 23 HIV-1 epitopes longitudinally in nine chronically infected untreated subjects with plasma viremia of <3,000 RNA copies/ml over 17 to 179 weeks. The composition of TCRs targeting each epitope varied considerably in stability over time, although clonal stability (Sorensen index) was not significantly time dependent within this interval. However, TCR stability inversely correlated with epitope variability in the Los Alamos HIV-1 Sequence Database, consistent with TCR evolution being driven by epitope variation. Finally, a robust inverse correlation of TCR breadth against each epitope versus epitope variability further suggested that this variability drives TCR repertoire diversification. In the context of studies demonstrating rapidly shifting HIV-1 sequences in vivo, our findings support a variably dynamic process of shifting CTL clonality lagging in tandem with viral evolution and suggest that preventing escape of HIV-1 may require coordinated direction of the CTL clonal repertoire to simultaneously block escape pathways.
IMPORTANCE Mutational escape of HIV-1 from HIV-1-specific CD8+ T lymphocytes (CTLs) is a major barrier to effective immune control. The number of distinct CTL clones targeting each epitope is proposed to be an important factor, but the determinants are poorly understood. Here, we demonstrate that the clonal stability and number of clones for the CTL response against an epitope are inversely associated with the general variability of the epitope. These results show that CTLs constantly lag epitope mutation, suggesting that preventing HIV-1 escape may require coordinated direction of the CTL clonal repertoire to simultaneously block escape pathways.
KEYWORDS: cytotoxic T lymphocytes, human immunodeficiency virus
INTRODUCTION
The major histocompatibility class I (MHC-I)-restricted CD8+ T lymphocyte (CTL) response is a critical arm of immunity for clearance or chronic control of many viral infections. The central protective role of human immunodeficiency virus type 1 (HIV-1)-specific CTLs in the pathogenesis of HIV-1 infection is clear from studies such as in vivo CD8 depletion in the simian immunodeficiency virus (SIV)-macaque model (1–3) and the consistent observation that the MHC-I locus is the strongest correlate to immune control of HIV-1 infection in multiple genetic screening studies (4–6). However, the CTL response fails to contain infection in the vast majority of infected persons, who eventually progress to severe immunodeficiency and death without treatment. The high mutation rate and genetic plasticity of HIV-1 likely are major contributors to this failure; viral adaptation to CTL responses is the major driver of viral sequence evolution in infected persons (7–9).
The CTL response against any given epitope is typically polyclonal, comprised of CTLs with distinct T cell receptors (TCRs). While these CTL clones have expanded in response to the same epitope, different TCRs may differ significantly in their recognition of different epitope sequence variants (10). Moreover, an epitope variant that escapes recognition by a CTL response can be recognized by a de novo variant-specific response that does not recognize the original epitope sequence (11). Clonal breadth has therefore been raised as a potentially important parameter for CTL containment of HIV-1 infection through broader coverage of epitope variation (12, 13). Although prior studies have suggested clonal stability (14) or shifting clonotypes (15) of HIV-1-specific CTL responses, the determinants of the clonal repertoire of CTL responses are poorly understood. Here, we examine the TCR repertoire over time for several HIV-1 epitopes in persons with chronic untreated infection who all maintained plasma viremia of <3,000 RNA copies/ml.
RESULTS
Quantitative spectratyping defines CTL clonal composition and breadth against HIV-1 epitopes longitudinally in persons with chronic HIV-1 infection.
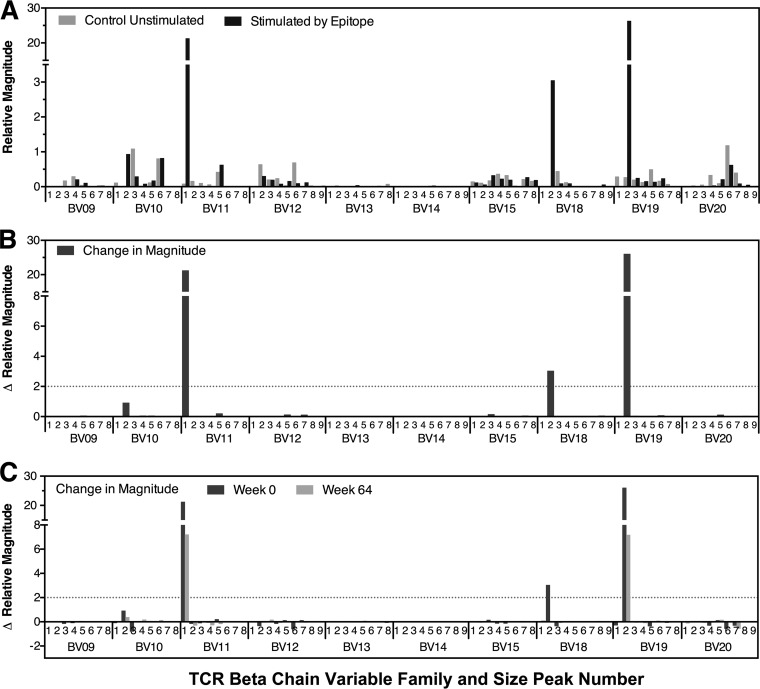
Quantitative spectratyping was utilized to assess the longitudinal clonal composition and breadth of 29 CTL responses against 23 epitopes in 9 chronically HIV-1-infected subjects, all of whom spontaneously maintained plasma viremia of <3,000 RNA copies/ml without receiving antiretroviral therapy during observation (Table 1). The time intervals of follow-up ranged from 17 to 179 weeks. Peripheral blood mononuclear cells (PBMCs) were cultured in the presence and absence of the epitopes of interest, followed by quantitative spectratyping as previously described (16) to identify epitope-specific clonal peak expansions within beta variable (BV) gene families (Fig. 1).
TABLE 1.
HIV-1-infected participants and CTL responses evaluated longitudinallya
| Participant | MHC class I |
Duration of infection | Epitope |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | Sequence (abbreviated) | MHC- I type | Location | Diversity | Sampling interval (wk) | Similarity | Mean breadth | ||
| S00009 | 03 | 15 | 03 | >14 yr | GLNKIVRMY (GY9) | B*15 | Gag 269–277 | 0.06 | 114 | 0.73 | 7.0 |
| 26 | 38 | 12 | ALVEICTEMEK (AK11) | A*03 | Pol 188–198 | 0.48 | 114 | 0.32 | 1.5 | ||
| AIFQSSMTK (AK9) | A*03 | Pol 313–321 | 0.57 | 114 | 0.00 | 2.5 | |||||
| IKLEPVHGVY (IY10) | B*15 | Pol 464–473 | 0.44 | 114 | 0.72 | 4.5 | |||||
| S00016 | 03 | 18 | 02 | >4 mo | KIRLRPGGK (KK9) | A*03 | Gag 18–26 | 0.40 | 179 | 0.14 | 4.5 |
| 32 | 40 | 07 | RLRPGGKKKY (RY10-G) | A*03 | Gag 20–29 | 0.60 | 179 | 0.35 | 2.5 | ||
| KELYPLASL (KL9-G) | B*40 | Gag 481–489 | 0.83 | 179 | 0.39 | 4.0 | |||||
| LEKHGAITS (LS9) | B*40 | Nef 37–45 | 0.86 | 179 | 0.00 | 2.5 | |||||
| KEKGGLEGL (KL9-N) | B*40 | Nef 92–100 | 0.51 | 179 | 0.05 | 2.5 | |||||
| RRQDILDLWIY (RY11) | C*07 | Nef 105–115 | 0.83 | 179 | 0.47 | 4.5 | |||||
| S00024 | 02 | 13 | 06 | >13 yr | RLRPGGKKKY (RY10-G) | B*15 | Gag 20–29 | 0.60 | 76 | 0.25 | 1.5 |
| 15 | 07 | SLYNTVATL (SL9) | A*02 | Gag 77–85 | 0.86 | 76 | 0.26 | 3.0 | |||
| GLNKIVRMY (GY9) | B*15 | Gag 269–277 | 0.06 | 76 | 0.57 | 7.0 | |||||
| RQANFLGKI (RI9) | B*13 | Gag 429–437 | 0.24 | 76 | 0.22 | 4.5 | |||||
| GQGQWTYQI (GI9) | B*13 | Pol 488–496 | 0.74 | 76 | 0.17 | 1.5 | |||||
| S00031 | 02 | 13 | 03 | >8 yr | RLRDLLLIV (RV9) | A*02 | Env 770–778 | 0.69 | 132 | 0.00 | 3.0 |
| 15 | 06 | ||||||||||
| S00036 | 02 | 39 | 06 | >12 yr | SLYNTVATL (SL9) | A*02 | Gag 77–85 | 0.86 | 111 | 0.00 | 1.0 |
| 11 | 57 | 07 | ISPRTLNAW (IW9-G) | B*57 | Gag 147–155 | 0.47 | 176 | 0.52 | 3.7 | ||
| KAFSPEVIPMF (KF11) | B*57 | Gag 162–172 | 0.14 | 64 | 0.43 | 3.5 | |||||
| IVLPEKDSW (IW9-P) | B*57 | Pol 399–407 | 0.73 | 176 | 0.00 | 0.7 | |||||
| VLEWRFDSR (VR9) | A*02 | Nef 180–188 | 0.91 | 111 | 0.00 | 2.0 | |||||
| S00039 | 31 | 13 | 04 | >3 mo | RQANFLGKI (RI9) | B*13 | Gag 429–437 | 0.24 | 43 | 0.55 | 6.0 |
| 32 | 35 | 06 | GQGQWTYQI (GI9) | B*13 | Pol 488–496 | 0.74 | 43 | 0.00 | 1.0 | ||
| S00052 | 01 | 18 | 06 | >14 yr | KAFSPEVIPMF (KF11) | B*57 | Gag 162–172 | 0.14 | 107 | 0.63 | 4.0 |
| 25 | 57 | 12 | AVRHFPRIW (AW9) | B*57 | Vpr 30–38 | 0.86 | 161 | 0.68 | 2.0 | ||
| RTVRLIKLLY (RY10-R) | B*57 | Rev 14–23 | 0.98 | 30 | 0.52 | 1.0 | |||||
| S00085 | 02 | 57 | 08 | >15 yr | ILKEPVHGV (IV9) | A*02 | Pol 464–472 | 0.43 | 17 | 0.91 | 2.5 |
| 33 | 65 | 18 | |||||||||
| S00096 | 01 | 08 | 07 | >9 yr | YRLDQQLLGIWGC (YC13) | C*07 | Env 586–598 | 0.70 | 20 | 0.56 | 1.0 |
| 32 | 51 | 14 | RRGWEALKY (RY9) | A*01 | Env 787–796 | 0.81 | 20 | 0.61 | 1.0 | ||
All the participants were men who did not receive antiretroviral therapy during the study duration. The MHC-I types, duration of infection at study onset, epitope sequence and restriction, epitope diversity across all HIV-1 subtype B sequences in the Los Alamos HIV Sequence Database, time intervals analyzed, Sorenson similarity index between time points, and mean clonal breadth as determined by spectratyping across time points are indicated.
FIG 1.
Delineation of the clonal profile of CTL responses targeting HIV-1 epitopes. An example of quantitative spectratyping is shown. PBMCs were cultured in the presence or absence of the epitope of interest, followed by spectratyping of 24 BV gene families within isolated CD8+ T lymphocytes, using quantitative PCR to determine the copy numbers of each family. The relative concentration of each BV family was calculated as the ratio of its copy number to the median copy number across all families. The relative magnitude of each spectratype peak was calculated as the fraction of the peak area within the summed area of all peaks in its family multiplied by the relative concentration of the family. The last 10 of the 24 analyzed families are shown as representative examples, because they contained a mixture of families with and without epitope-specific responses. (A) Results are shown for unstimulated and peptide-stimulated peak profiles, demonstrating some families with epitope-specific expansions (BV11, BV18, and BV19). (B) The magnitude of change of each peak in response to epitope stimulation was calculated by subtracting the relative magnitude of unstimulated spectratypes from that of epitope-stimulated spectratypes. Results for the same 10 BV families are shown, quantifying the epitope-specific expansions (defined as increases of ≥2 units) in families BV11, BV18, and BV19 from panel A. (C) Results for the magnitude changes of each peak in response to epitope stimulation for the same 10 BV families for two different time points.
Clonal stability of CTL responses against HIV-1 epitopes varied independently of time over the period of observation.
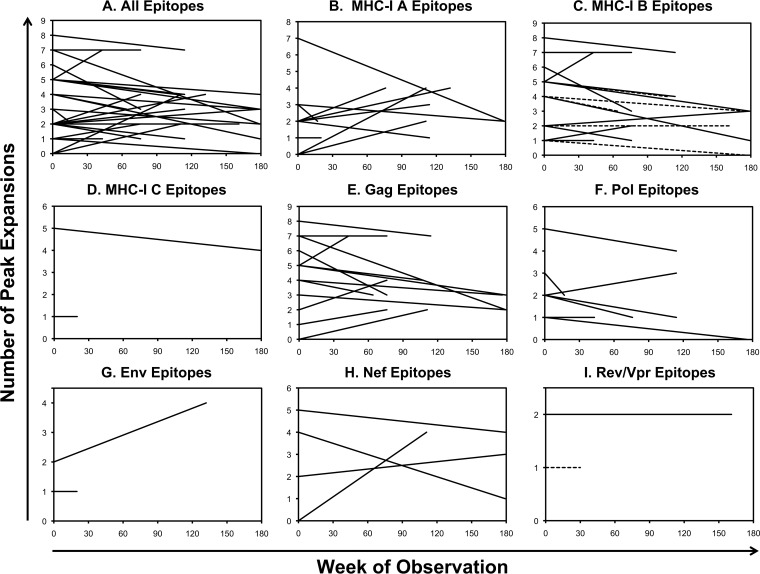
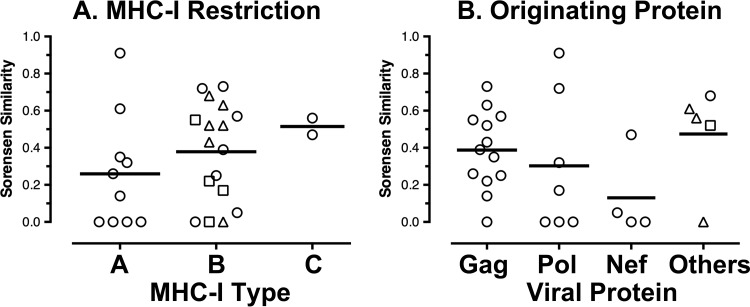
Examination of epitope-specific TCRs showed generally stable breadth over time (Fig. 2A). There was little variation in the number of clones targeting each epitope over time, and breadth and stability were similar between epitopes restricted by MHC-I A and B versus C types (Fig. 2B to D) or from different viral proteins (Fig. 2E to I). However, there were varying patterns of clonal dominance ranging from highly stable to shifting profiles. In some instances, the TCR clones targeting an epitope showed shifts in composition (Fig. 3A), whereas other responses showed stability (Fig. 3B) within the same person. To assess whether the varying degree of clonal stability between epitopes was due to variation in the duration of longitudinal observation, clonal similarity was compared to the duration between time points (Fig. 4A). This analysis showed that there was no significant relationship between the time elapsed and TCR clonal similarity between time points for the period of observation ranging from 17 to 176 weeks, suggesting that time was not a significant factor in the degree of clonal variation over the duration of observation.
FIG 2.
Generally stable clonal breadth of HIV-1 epitope-specific CTL responses over time. The clonal breadths of responses to the HIV-1 epitopes in Table 1 (as delineated in Fig. 1) are plotted over time for all epitopes (A), epitopes divided according to MHC-I restriction (B to D), and epitopes divided according to viral protein source (E to I). In panel C, the dashed lines indicate B*57-restricted responses. In panel I, the dashed line indicates a Rev epitope, while the solid line indicates a Vpr epitope.
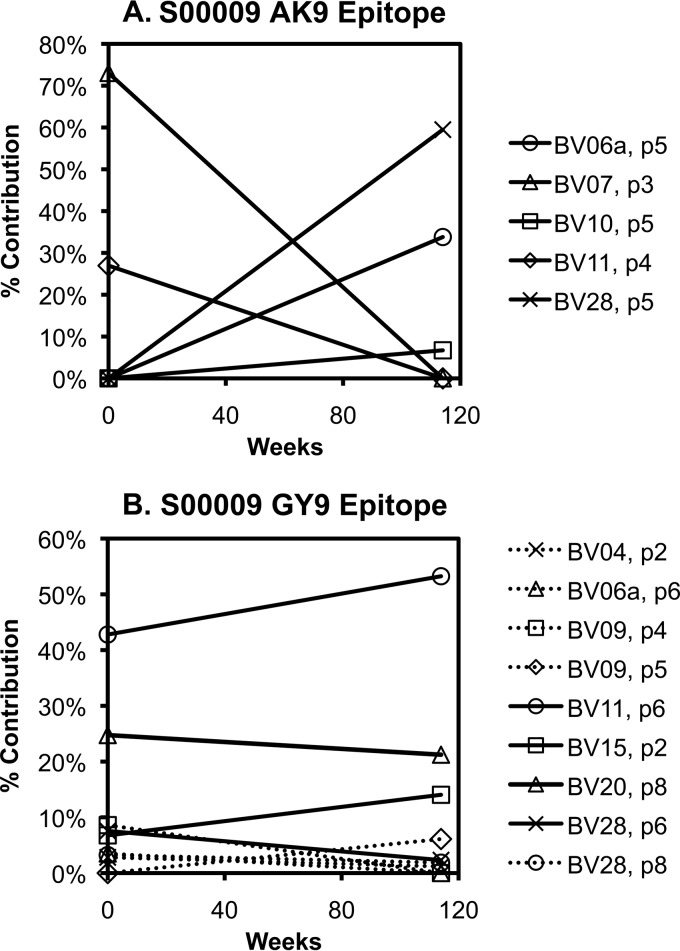
FIG 3.
Varying clonal stability of CTL responses over time. An example of a CTL response exhibiting changing clonal composition over time is shown (A). The response to AK9 in subject S00009 was comprised of clonal expansions in BV7 (within peak 3) and BV11 (peak 4) at the baseline evaluation, but then clonal expansions in BV6a (peak 6), BV10 (peak 5), and BV28 (peak 5) 114 weeks later. (B) In contrast, another response (against the GY9 epitope) in the same person showed a relatively stable clonal composition over the same span of time.
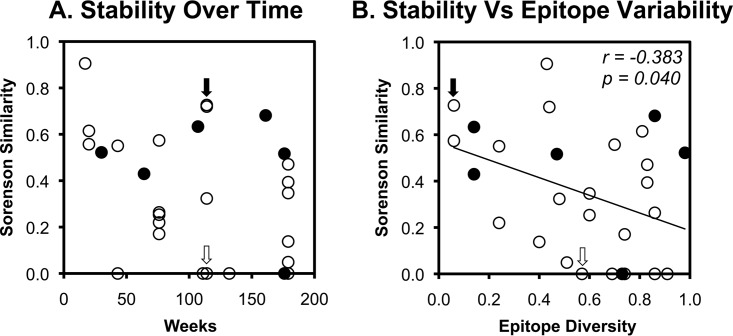
FIG 4.
Correlates of clonal stability of CTL responses against HIV-1 epitopes. The clonal similarity of the CTL responses in Table 1 over time is plotted against time (A) and epitope variability (B) as approximated by sequence diversity among all subtype B sequences in the Los Alamos HIV Sequence Database. B*57-restricted epitopes are highlighted as filled circles. For reference, the examples in Fig. 3 are indicated by open (AK9 epitope) and solid (GY9 epitope) arrows. Correlations were evaluated using Spearman's rank test.
TCR clonal stability inversely correlates with epitope variability.
Because CTL persistence is driven by antigen recognition, epitope variability was compared to TCR clonal stability. Global epitope sequence diversity across all subtype B sequences in the Los Alamos HIV Sequence Database was calculated as a surrogate marker for epitope variability, given that most of the subjects had viremia below the limits of detection. This revealed a modest but statistically significant inverse correlation of TCR clonal stability and epitope variability (Fig. 4B). Of note, epitopes restricted by B*57, which is associated with superior immune containment of HIV-1 infection in some persons, spanned the range of epitope diversity and stability. Also, clonal stabilities were similar between epitopes restricted by MHC-I A and B versus C types and epitopes from different viral proteins (Fig. 5). Overall, these data suggested that the least variable epitopes were associated with greater TCR clonal stability and that this process is MHC-I independent.
FIG 5.
Clonal stability of CTL responses subdivided by MHC-I restriction and targeted HIV-1 proteins. The longitudinal similarity measurements plotted in Fig. 4 were divided according to MHC-I restriction (A) or targeted viral protein (B). (A) Triangles represent B*57, and squares represent B*13. (B) Triangles represent Env, circles represent Rev, and the square represents Vpr.
TCR clonal breadth inversely correlates with epitope variability.
Epitope variability, again reflected by the surrogate marker of epitope sequence diversity, was compared to TCR clonal breadth to investigate whether greater variability in epitope sequences might drive more clonal responses. Comparison of epitope diversity to TCR clonal breadth against each epitope revealed a robust inverse correlation (Fig. 6). Again, epitopes restricted by MHC-I B*57 spanned the spectrum of epitope diversity and clonal breadth and did not appear distinct from other epitope responses. These data in the context of the above-mentioned findings are consistent with a bidirectional interrelationship between epitope variability and the clonal composition of CTL responses against HIV-1 infection.
FIG 6.
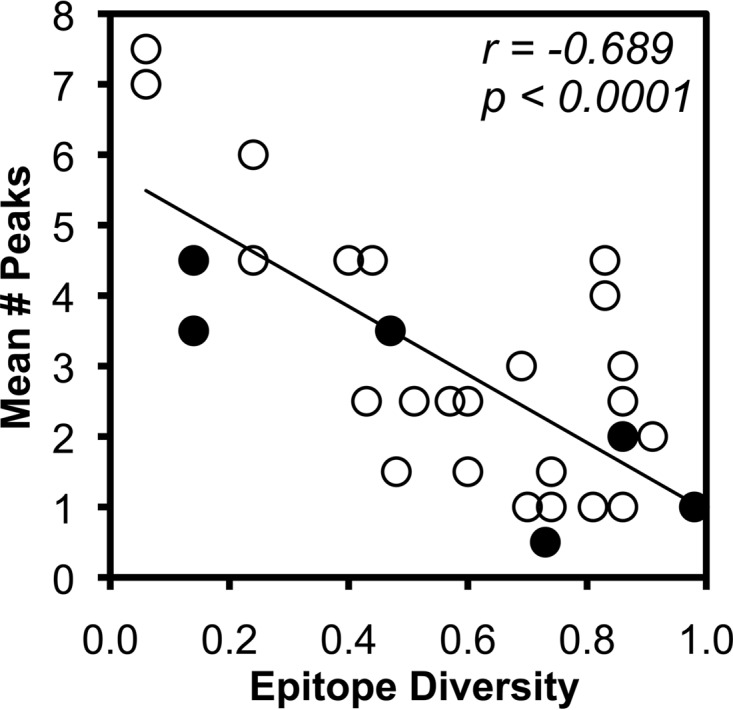
Inverse correlation of epitope variability and CTL clonal breadth. The average clonal breadth of each HIV-1 epitope-specific CTL response (shown in Fig. 2) is plotted against epitope variability as approximated by sequence diversity among all subtype B sequences in the Los Alamos HIV Sequence Database. B*57-restricted epitopes are highlighted as filled circles. Correlations were evaluated using Spearman's rank test.
DISCUSSION
As adaptive immunity, CTL responses arise and are maintained by the presence of the epitopes that they recognize. As occurs physiologically when a viral infection is cleared and effector CTLs are no longer needed, HIV-1-specific CTL frequencies decay to low resting memory levels after virus replication is suppressed by antiretroviral drug treatment (17, 18). At the level of CTLs recognizing a single epitope, the same process occurs if a targeted epitope mutates to become a completely unrecognized variant (19, 20), indicating that the composition of the CTL response is driven by viral epitope sequence evolution. Conversely, however, the CTL response applies selective pressure that drives viral epitope evolution (7–9), individual clones within the response can select for different epitope escape variants (10), and HIV-1 tends to revert epitope mutations when a prior CTL response is absent (21).
Our data support the concept of a dynamic bidirectional interaction between the CTL response and HIV-1 sequence variation in vivo, and our prior observation of “partial escape” was demonstrated as persistence of viral epitopes with reduced fitness under CTL pressure in vivo (22). It has been demonstrated that the HIV-1 quasispecies in vivo has ongoing shifts in the frequency of individual CTL epitope mutants (23, 24). In the context of CTL expansion and contraction being driven by epitope recognition and nonrecognition, respectively, and differential recognition of epitope variants by different CTL clones recognizing the same epitope, the observed variation in CTL clonal frequencies over time is consistent with different clones expanding and contracting according to varying epitope variants within the quasispecies. In turn, the differential recognition of epitope variants that drives variation between frequencies of individual CTL clones also exerts differential antiviral pressure between different epitope variants, driving viral evolution. This process is analogous to the genetic coevolution of broadly neutralizing antibodies and HIV-1 Env in vivo (25), in which the neutralizing antibody response continuously lags behind viral evolution (26). This lag prevents the efficacy of antibodies in the infected persons in whom they arise, yet administration of a broadly neutralizing antibody to another person in whom HIV-1 has not coevolved can yield a potent antiviral effect (27–30), presumably by blocking escape pathways in advance of their evolution.
Our results provide a likely explanation for varying prior observations of TCR clonotype stability (14) versus variability (15) in HIV-1-infected persons. HIV-1 CTL epitopes vary markedly in their sequence constraint, and thus in the breadth of their mutation landscape. Therefore, HIV-1 has more options for epitope variation in some epitopes than others, which consequently drives more options to escape CTL responses. The inverse correlation between CTL clonal stability and epitope diversity supports a scenario where greater epitope variability allows more dynamic shifting in the population of epitope variants, followed by greater shifting in the CTL clones responding to the variants.
Finally, the robust inverse correlation between CTL clonal breadth and epitope diversity is novel evidence for an important determinant of the CTL response against HIV-1; we previously demonstrated that clonal breadth is not correlated with MHC-I restriction or the frequency of the response (16). Again, this is consistent with a bidirectional interaction of viral sequence evolution and the CTL response at the clonal level, in which greater epitope variability allows exposure of the CTL response to more epitope variants, driving greater CTL clonal breadth. This finding also suggests an explanation for the observation that “public TCR” clonotypes shared between multiple persons with a common MHC-I allele have mostly been observed for highly conserved epitopes, such as the B*57-restricted KF11 epitope in Gag (13, 31, 32). Additionally, it supports the rationale for using mixtures of sequences in vaccines to encompass diversity that drives greater recognition of epitope variation by CTL responses, such as the “mosaic” strategy (33, 34).
Although not a focus of this report, it is interesting that the “protective” MHC-I type B*57 presents epitopes that span the observed range of variability in sequence diversity, stability of CTL responses, and clonal breadth of CTL responses. Although indirectly relevant, this finding is less consistent with the proposed mechanism of B*57 protection via thymic selection of more promiscuous TCRs (35) versus the concept that the protective contribution of CTL responses is epitope specific (36).
There are inherent limitations to our findings. Epitope sequence diversity in the Los Alamos HIV Sequence Database is an indirect approximation of the mutational space available for epitope variation in a given person, which may be context dependent in terms of the strain of HIV-1 and host factors. Moreover, there are two major reasons for using this parameter as opposed to diversity within the participants' endogenous HIV-1 epitope sequences. First, the low levels of viremia in these persons is a significant technical barrier to ensuring acquisition of enough clonal sequences for accurate assessment of epitope diversity in their viral quasispecies. Second, and more important, the interaction of epitope sequences and CTLs is bidirectional. While the key parameter of interest is epitope plasticity, i.e., options for mutation, the observed diversity in each individual is the net outcome of the interaction, i.e., the options remaining after CTL targeting. Moreover, CTL responses against different epitopes likely vary significantly in their antiviral activity (37, 38), and thus, the selective pressure driving epitope diversity and resulting TCR diversity may not be uniform. Thus, while imperfect, sequence diversity in the Los Alamos HIV Sequence Database is a less biased indicator of epitope plasticity, reflecting the general variability of the epitope across all persons, most of whom do not have the CTL response studied here.
Finally, our methodology for defining epitope-specific TCRs depends on the capacity of CTLs to proliferate in response to an epitope, which could be biased by varying proliferative capacity or the use of a fixed epitope sequence, both factors that could underestimate clonal breadth. Because all our subjects were persons with stable controlled viremia off antiretroviral therapy, CTL functionality should have been relatively intact and similar between individuals.
In conclusion, we observed inverse correlations between epitope variability and CTL clonal stability over time and between epitope variability and CTL clonal breadth in persons with chronic stable untreated HIV-1 infection and plasma viremia of <3,000 RNA copies/ml. These findings provide evidence for a bidirectional interaction of the CTL response and HIV-1 sequence evolution where immunity lags virus sequence evolution. This suggests the potential of coordinating the CTL response in vaccine or immunotherapeutic approaches to block escape pathways in advance of viral evolution.
MATERIALS AND METHODS
Participants and peripheral blood mononuclear cells.
The participants (Table 1) were persons with chronic HIV-1 infection who were not receiving antiretroviral treatment during the study period. All the participants were spontaneous “controllers” of infection with persistent maintenance of viremia at <3,000 HIV-1 RNA copies/ml plasma during observation. MHC-I typing was performed by the clinical immunogenetics laboratory at UCLA Medical Center. All participants provided informed consent under a UCLA Institutional Review Board-approved protocol. PBMCs were isolated by Ficoll-Hypaque gradient and viably cryopreserved until use.
Detection of HIV-1-specific CTL responses by IFN-γ ELISpot assay.
CTL responses were identified by gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assay using polyclonally expanded CD8+ T lymphocytes, as previously described (39–41). Screening was performed using previously described minimal epitopes for the MHC-I types of the individuals (42).
Delineation of the HIV-1 epitope-specific CTL clonal repertoires.
For confirmed CTL responses against minimal epitopes, the clonal breadth of CTL clones recognizing selected HIV-1 epitopes was assessed by a quantitative spectratyping assay as previously described (16). In brief, PBMCs were cultured with the epitope to enrich the CTLs recognizing that epitope, with a parallel control cultured without the epitope. CD8+ T lymphocytes were then purified for TCR analysis of cDNA using real-time PCR for each of 24 BV gene families. The relative concentration of each family was calculated as the ratio of its mean number of copies to the median number of copies across all families (relative units). For each family, capillary electrophoresis was performed on the real-time PCR product to resolve the size distributions of TCRs, and the concentration of each size population within a family was calculated using its percent contribution to the whole BV family. Epitope-specific TCRs were identified by comparing epitope-stimulated to control unstimulated spectratype profiles for peaks expanded by more than 2 relative units by epitope stimulation (an arbitrary cutoff based on control data showing that most nonspecific peak variations with this method are <1 unit).
TCR repertoire analyses.
Comparison of epitope-specific TCR similarities between time points was performed using the abundance-based Sorensen Index (43): similarity = 2UV/U + V, where UV represents the summed magnitudes of shared spectratype peak expansions between time points and U and V represent the summed magnitudes of all expansions at each time point (thus, a value of 1 indicates all identical expansions of the same magnitudes, whereas a value of 0 indicates no shared expansions).
Epitope sequence variability analyses.
The variability of epitopes was assessed by calculating the Simpson diversity (Ds) index (44) using the set of all clade B sequences (excluding unresolved amino acids or deletions) returned by the QuickAlign tool at the Los Alamos National Laboratory HIV Sequence Database (http://www.hiv.lanl.gov) for each epitope, and diversity was calculated as follows: , where, N is the total number of sequences included for the analysis, n is the frequency of the ith epitope variant sequence, and c is the total number of epitope variants (thus, a value of 1 indicates an infinite number of different sequences, whereas a value of 0 indicates that all sequences are identical).
Statistical tests.
For evaluation of correlations between two variables (Fig. 4 and 6), Spearman's rank test was utilized. For evaluation of differences between groups, the Mann-Whitney test was utilized (Fig. 5).
ACKNOWLEDGMENTS
This work was supported by PHS grants AI043203, DE025166, and AI028697 (UCLA AIDS Institute and Center for AIDS Research) and a grant from the AIDS Healthcare Foundation.
We thank our study participants for their generous sample donations.
REFERENCES
- 1.Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med 189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matano T, Shibata R, Siemon C, Connors M, Lane HC, Martin MA. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol 72:164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 4.International HIV Controllers Study, Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld M, Henn MR, Le Gall S, Streeck H, Haas DW, Kuritzkes DR, Robbins GK, Shafer RW, Gulick RM, Shikuma CM, Haubrich R, Riddler S, Sax PE, Daar ES, Ribaudo HJ, Agan B, Agarwal S, Ahern RL, Allen BL, Altidor S, Altschuler EL, Ambardar S, Anastos K, Anderson B, Anderson V, Andrady U, Antoniskis D, Bangsberg D, Barbaro D, Barrie W, Bartczak J, Barton S, Basden P, Basgoz N, Bazner S, Bellos NC, Benson AM, Berger J, Bernard NF, Bernard AM, Birch C, Bodner SJ, Bolan RK, Boudreaux ET, Bradley M, Braun JF, Brndjar JE, Brown SJ, Brown K, Brown ST, Burack J, Bush LM, Cafaro V, Campbell O, Campbell J, Carlson RH, Carmichael JK, Casey KK, Cavacuiti C, Celestin G, Chambers ST, Chez N, Chirch LM, Cimoch PJ, Cohen D, Cohn LE, Conway B, Cooper DA, Cornelson B, Cox DT, Cristofano MV, Cuchural G Jr, Czartoski JL, Dahman JM, Daly JS, Davis BT, Davis K, Davod SM, DeJesus E, Dietz CA, Dunham E, Dunn ME, Ellerin TB, Eron JJ, Fangman JJ, Farel CE, Ferlazzo H, Fidler S, Fleenor-Ford A, Frankel R, Freedberg KA, French NK, Fuchs JD, Fuller JD, Gaberman J, Gallant JE, Gandhi RT, Garcia E, Garmon D, Gathe JC Jr, Gaultier CR, Gebre W, Gilman FD, Gilson I, Goepfert PA, Gottlieb MS, Goulston C, Groger RK, Gurley TD, Haber S, Hardwicke R, Hardy WD, Harrigan PR, Hawkins TN, Heath S, Hecht FM, Henry WK, Hladek M, Hoffman RP, Horton JM, Hsu RK, Huhn GD, Hunt P, Hupert MJ, Illeman ML, Jaeger H, Jellinger RM, John M, Johnson JA, Johnson KL, Johnson H, Johnson K, Joly J, Jordan WC, Kauffman CA, Khanlou H, Killian RK, Kim AY, Kim DD, Kinder CA, Kirchner JT, Kogelman L, Kojic EM, Korthuis PT, Kurisu W, Kwon DS, LaMar M, Lampiris H, Lanzafame M, Lederman MM, Lee DM, Lee JM, Lee MJ, Lee ET, Lemoine J, Levy JA, Llibre JM, Liguori MA, Little SJ, Liu AY, Lopez AJ, Loutfy MR, Loy D, Mohammed DY, Man A, Mansour MK, Marconi VC, Markowitz M, Marques R, Martin JN, Martin HL Jr, Mayer KH, McElrath MJ, McGhee TA, McGovern BH, McGowan K, McIntyre D, Mcleod GX, Menezes P, Mesa G, Metroka CE, Meyer-Olson D, Miller AO, Montgomery K, Mounzer KC, Nagami EH, Nagin I, Nahass RG, Nelson MO, Nielsen C, Norene DL, O'Connor DH, Ojikutu BO, Okulicz J, Oladehin OO, Oldfield EC III, Olender SA, Ostrowski M, Owen WF Jr, Pae E, Parsonnet J, Pavlatos AM, Perlmutter AM, Pierce MN, Pincus JM, Pisani L, Price LJ, Proia L, Prokesch RC, Pujet HC, Ramgopal M, Rathod A, Rausch M, Ravishankar J, Rhame FS, Richards CS, Richman DD, Rodes B, Rodriguez M, Rose RC III, Rosenberg ES, Rosenthal D, Ross PE, Rubin DS, Rumbaugh E, Saenz L, Salvaggio MR, Sanchez WC, Sanjana VM, Santiago S, Schmidt W, Schuitemaker H, Sestak PM, Shalit P, Shay W, Shirvani VN, Silebi VI, Sizemore JM Jr, Skolnik PR, Sokol-Anderson M, Sosman JM, Stabile P, Stapleton JT, Starrett S, Stein F, Stellbrink HJ, Sterman FL, Stone VE, Stone DR, Tambussi G, Taplitz RA, Tedaldi EM, Telenti A, Theisen W, Torres R, Tosiello L, Tremblay C, Tribble MA, Trinh PD, Tsao A, Ueda P, Vaccaro A, Valadas E, Vanig TJ, Vecino I, Vega VM, Veikley W, Wade BH, Walworth C, Wanidworanun C, Ward DJ, Warner DA, Weber RD, Webster D, Weis S, Wheeler DA, White DJ, Wilkins E, Winston A, Wlodaver CG, van't Wout A, Wright DP, Yang OO, Yurdin DL, Zabukovic BW, Zachary KC, Zeeman B, Zhao M. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limou S, Le Clerc S, Coulonges C, Carpentier W, Dina C, Delaneau O, Labib T, Taing L, Sladek R, Deveau C, Ratsimandresy R, Montes M, Spadoni JL, Lelievre JD, Levy Y, Therwath A, Schachter F, Matsuda F, Gut I, Froguel P, Delfraissy JF, Hercberg S, Zagury JF. 2009. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J Infect Dis 199:419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 6.Pelak K, Goldstein DB, Walley NM, Fellay J, Ge D, Shianna KV, Gumbs C, Gao X, Maia JM, Cronin KD, Hussain SK, Carrington M, Michael NL, Weintrob AC. 2010. Host determinants of HIV-1 control in African Americans. J Infect Dis 201:1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 8.Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O'Sullivan KM, Desouza I, Feeney ME, Eldridge RL, Maier EL, Kaufmann DE, Lahaie MP, Reyor L, Tanzi G, Johnston MN, Brander C, Draenert R, Rockstroh JK, Jessen H, Rosenberg ES, Mallal SA, Walker BD. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol 79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, McNevin J, Cao J, Zhao H, Genowati I, Wong K, McLaughlin S, McSweyn MD, Diem K, Stevens CE, Maenza J, He H, Nickle DC, Shriner D, Holte SE, Collier AC, Corey L, McElrath MJ, Mullins JI. 2006. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J Virol 80:9519–9529. doi: 10.1128/JVI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang OO, Sarkis PT, Ali A, Harlow JD, Brander C, Kalams SA, Walker BD. 2003. Determinants of HIV-1 mutational escape from cytotoxic T lymphocytes. J Exp Med 197:1365–1375. doi: 10.1084/jem.20022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen TM, Yu XG, Kalife ET, Reyor LL, Lichterfeld M, John M, Cheng M, Allgaier RL, Mui S, Frahm N, Alter G, Brown NV, Johnston MN, Rosenberg ES, Mallal SA, Brander C, Walker BD, Altfeld M. 2005. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J Virol 79:12952–12960. doi: 10.1128/JVI.79.20.12952-12960.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol 168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- 13.Simons BC, Vancompernolle SE, Smith RM, Wei J, Barnett L, Lorey SL, Meyer-Olson D, Kalams SA. 2008. Despite biased TRBV gene usage against a dominant HLA B57-restricted epitope, TCR diversity can provide recognition of circulating epitope variants. J Immunol 181:5137–5146. doi: 10.4049/jimmunol.181.7.5137. [DOI] [PubMed] [Google Scholar]
- 14.Kalams SA, Johnson RP, Trocha AK, Dynan MJ, Ngo HS, D'Aquila RT, Kurnick JT, Walker BD. 1994. Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire. J Exp Med 179:1261–1271. doi: 10.1084/jem.179.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer-Olson D, Brady KW, Bartman MT, O'Sullivan KM, Simons BC, Conrad JA, Duncan CB, Lorey S, Siddique A, Draenert R, Addo M, Altfeld M, Rosenberg E, Allen TM, Walker BD, Kalams SA. 2006. Fluctuations of functionally distinct CD8+ T-cell clonotypes demonstrate flexibility of the HIV-specific TCR repertoire. Blood 107:2373–2383. doi: 10.1182/blood-2005-04-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balamurugan A, Ng HL, Yang OO. 2010. Rapid T cell receptor delineation reveals clonal expansion limitation of the magnitude of the HIV-1-specific CD8+ T cell response. J Immunol 185:5935–5942. doi: 10.4049/jimmunol.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogg GS, Jin X, Bonhoeffer S, Moss P, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Hurley A, Markowitz M, Ho DD, McMichael AJ, Nixon DF. 1999. Decay kinetics of human immunodeficiency virus-specific effector cytotoxic T lymphocytes after combination antiretroviral therapy. J Virol 73:797–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalams SA, Goulder PJ, Shea AK, Jones NG, Trocha AK, Ogg GS, Walker BD. 1999. Levels of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte effector and memory responses decline after suppression of viremia with highly active antiretroviral therapy. J Virol 73:6721–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamieson BD, Yang OO, Hultin L, Hausner MA, Hultin P, Matud J, Kunstman K, Killian S, Altman J, Kommander K, Korber B, Giorgi J, Wolinsky S. 2003. Epitope escape mutation and decay of human immunodeficiency virus type 1-specific CTL responses. J Immunol 171:5372–5379. doi: 10.4049/jimmunol.171.10.5372. [DOI] [PubMed] [Google Scholar]
- 20.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med 3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 21.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med 10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 22.Lewis MJ, Dagarag M, Khan B, Ali A, Yang OO. 2012. Partial escape of HIV-1 from cytotoxic T lymphocytes during chronic infection. J Virol 86:7459–7463. doi: 10.1128/JVI.06724-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, Zody MC, Erlich RL, Green LM, Berical A, Wang Y, Casali M, Streeck H, Bloom AK, Dudek T, Tully D, Newman R, Axten KL, Gladden AD, Battis L, Kemper M, Zeng Q, Shea TP, Gujja S, Zedlack C, Gasser O, Brander C, Hess C, Gunthard HF, Brumme ZL, Brumme CJ, Bazner S, Rychert J, Tinsley JP, Mayer KH, Rosenberg E, Pereyra F, Levin JZ, Young SK, Jessen H, Altfeld M, Birren BW, Walker BD, Allen TM. 2012. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog 8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer W, Ganusov VV, Giorgi EE, Hraber PT, Keele BF, Leitner T, Han CS, Gleasner CD, Green L, Lo CC, Nag A, Wallstrom TC, Wang S, McMichael AJ, Haynes BF, Hahn BH, Perelson AS, Borrow P, Shaw GM, Bhattacharya T, Korber BT. 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One 5:e12303. doi: 10.1371/journal.pone.0012303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Program NCS, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A 100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW, Learn GH, Proschan MA, Kreider EF, Blazkova J, Bardsley M, Refsland EW, Messer M, Clarridge KE, Tustin NB, Madden PJ, Oden K, O'Dell SJ, Jarocki B, Shiakolas AR, Tressler RL, Doria-Rose NA, Bailer RT, Ledgerwood JE, Capparelli EV, Lynch RM, Graham BS, Moir S, Koup RA, Mascola JR, Hoxie JA, Fauci AS, Tebas P, Chun TW. 2016. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med 375:2037–2050. doi: 10.1056/NEJMoa1608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP Jr, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O'Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun TW, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE, Team VRC 601 Study Team . 2015. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PubMed] [Google Scholar]
- 30.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu CL, Lorenzi JC, Feldmann A, Braunschweig M, Nogueira L, Oliveira T, Shimeliovich I, Patel R, Burke L, Cohen YZ, Hadrigan S, Settler A, Witmer-Pack M, West AP Jr, Juelg B, Keler T, Hawthorne T, Zingman B, Gulick RM, Pfeifer N, Learn GH, Seaman MS, Bjorkman PJ, Klein F, Schlesinger SJ, Walker BD, Hahn BH, Nussenzweig MC. 2016. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535:556–560. doi: 10.1038/nature18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillespie GM, Stewart-Jones G, Rengasamy J, Beattie T, Bwayo JJ, Plummer FA, Kaul R, McMichael AJ, Easterbrook P, Dong T, Jones EY, Rowland-Jones SL. 2006. Strong TCR conservation and altered T cell cross-reactivity characterize a B*57-restricted immune response in HIV-1 infection. J Immunol 177:3893–3902. doi: 10.4049/jimmunol.177.6.3893. [DOI] [PubMed] [Google Scholar]
- 32.Yu XG, Lichterfeld M, Chetty S, Williams KL, Mui SK, Miura T, Frahm N, Feeney ME, Tang Y, Pereyra F, Labute MX, Pfafferott K, Leslie A, Crawford H, Allgaier R, Hildebrand W, Kaslow R, Brander C, Allen TM, Rosenberg ES, Kiepiela P, Vajpayee M, Goepfert PA, Altfeld M, Goulder PJ, Walker BD. 2007. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J Virol 81:1619–1631. doi: 10.1128/JVI.01580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barouch DH, O'Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, Sun YH, La Porte A, Riggs AM, Lynch DM, Clark SL, Backus K, Perry JR, Seaman MS, Carville A, Mansfield KG, Szinger JJ, Fischer W, Muldoon M, Korber B. 2010. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med 16:319–323. doi: 10.1038/nm.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santra S, Liao HX, Zhang R, Muldoon M, Watson S, Fischer W, Theiler J, Szinger J, Balachandran H, Buzby A, Quinn D, Parks RJ, Tsao CY, Carville A, Mansfield KG, Pavlakis GN, Felber BK, Haynes BF, Korber BT, Letvin NL. 2010. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med 16:324–328. doi: 10.1038/nm.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. 2012. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol 13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mothe B, Llano A, Ibarrondo J, Daniels M, Miranda C, Zamarreno J, Bach V, Zuniga R, Perez-Alvarez S, Berger CT, Puertas MC, Martinez-Picado J, Rolland M, Farfan M, Szinger JJ, Hildebrand WH, Yang OO, Sanchez-Merino V, Brumme CJ, Brumme ZL, Heckerman D, Allen TM, Mullins JI, Gomez G, Goulder PJ, Walker BD, Gatell JM, Clotet B, Korber BT, Sanchez J, Brander C. 2011. Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med 9:208. doi: 10.1186/1479-5876-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang OO, Kalams SA, Rosenzweig M, Trocha A, Jones N, Koziel M, Walker BD, Johnson RP. 1996. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J Virol 70:5799–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang OO, Kalams SA, Trocha A, Cao H, Luster A, Johnson RP, Walker BD. 1997. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J Virol 71:3120–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ibarrondo FJ, Anton PA, Fuerst M, Ng HL, Wong JT, Matud J, Elliott J, Shih R, Hausner MA, Price C, Hultin LE, Hultin PM, Jamieson BD, Yang OO. 2005. Parallel human immunodeficiency virus type 1-specific CD8+ T-lymphocyte responses in blood and mucosa during chronic infection. J Virol 79:4289–4297. doi: 10.1128/JVI.79.7.4289-4297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabado RL, Kilpatrick S, Ali A, Dagarag M, Ng HL, Cao H, Yang OO. 2005. Detection of HIV-1-specific CTL responses in clade B infection with clade C peptides and not clade B consensus peptides. J Immunol Methods 296:1–10. doi: 10.1016/j.jim.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Yang OO, Daar ES, Ng HL, Shih R, Jamieson BD. 2011. Increasing CTL targeting of conserved sequences during early HIV-1 infection is correlated to decreasing viremia. AIDS Res Hum Retroviruses 27:391–398. doi: 10.1089/aid.2010.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frahm N, Baker B, Brander C. 2008. Identification and optimal definition of HIV-derived cytotoxic T lymphocyte (CTL) epitopes for the study of CTL escape, functional avidity, and viral evolution, p 3–24. In Korber BTM, Brander C, Haynes B, Koup R, Moore JP, Walker BD, Watkins DI (ed), HIV Molecular immunology 2008. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM. [Google Scholar]
- 43.Chao A, Chazdon RL, Colwell RK, Shen TJ. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecology Lett 8:148–159. doi: 10.1111/j.1461-0248.2004.00707.x. [DOI] [Google Scholar]
- 44.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. 2007. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods 321:182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]