Abstract
Background and Objectives:
Endophytic actinobacteria colonize inside the plant tissues without causing damages to the host plant. Since these microorganisms colonize in the different parts of plants and can stop plant disease, they have been applied as biological agents for controlling human diseases. The aim of this study was molecular identification and measuring the antimicrobial activity of endophytic Actinomycetes isolated from medicinal plants of Iran.
Materials and Methods:
The total of 23 medicinal plant samples were collected, sterilized, and crushed. Small pieces of the crushed samples were then cultured directly on three selective media. Grown colonies were identified by 16S rRNA gene sequencing method. Each isolate was cultured in TSB medium and then antimicrobial compound was extracted using ethyl acetate and tested against the target bacteria.
Results:
Sixteen out of 23 bacterial isolates (69%) exhibited antimicrobial activity against the selected pathogenic bacteria, such as Bacillus cereus, Staphylococcus aureus, Bacillus subtilis, Klebsiella pneumoniae, Citrobacter freundii, Proteus mirabilis, Shigella flexneri and Escherichia coli.
Conclusion:
Our Study showed a high phylogenetic diversity and the potent antibiotic activity of endophytic bacteria in medicinal plants of Iran.
Keywords: Endophytic, Actinomycets, Medicinal plants, Antimicrobial
INTRODUCTION
Natural sources of drugs have played important roles in medicine during the last decades. Since 1981 to 2006 nearly 70% of new drugs and chemical agents had natural sources (1). Over 22,000 natural products are isolated from microorganisms. Actinobacteria alone produce more than 45% of antibiotics in the world. Actinobacteria are Gram-positive bacteria with high guanine and cytosine content in their DNA. Some of them form filaments that resemble mycelia of the unrelated fungi (2, 3). Actinobacteria are frequently found in soil microflora and produce bioactive compounds including antibiotics (4, 5), antitumor compounds (6, 7), enzymes (8), and immunosuppressive agents (9). Due to the increasing emergence of bacterial resistance and replacing potent antimicrobial medicines, it is very important to focus on new antimicrobial sources. Actinomycete bioactive compounds are used safely in human and veterinary medicine products (3, 9). Many actinobacteria enter the inner tissues of plants and act either as pathogen or endophytic (6). The actinomycetes that reside in the inner tissues of living plants are known as endophytic actinobacteria (10, 11). They live in roots, stems, flowers, fruits, seeds or in many other tissues of plants (12) where they can stimulate growth of the host plants under adverse conditions and also fight plant diseases (13). Many of metabolites produced by these bacteria have antibacterial activities such as munumbicins A–D (13–15), celastramycins A–B (16), kakadumycins (17), and dimethyl novobiocins (18), which are isolated from Sterptomyces spp. (19). The aim of this study was to isolate and identify antimicrobial activity of endophytic actinomycets in medicinal plants.
MATERIALS AND METHODS
Sample collection.
Twenty three plant samples, including leaves, flowers, and fruits, were collected from four different provinces of Iran (Khuzestan, Tehran, Khoramabad, and Ilam) between 2013 and 2104. Plants were identified by Department of Botany at Chamran University, Iran. For bacterial isolation, each part of the plants was placed into a sterile plastic bag and transferred to the microbiology laboratory of Ahvaz Jundishapur University of Medical Sciences.
Surface sterilization.
The collected samples were washed by tap water, dried, and processed by a five step surface sterilization procedure, which included 4–10 min washing with 5% NaOCl, 10 min wash with 2.5% Na2S2O3, 5 min wash with 75% ethanol, one wash with sterile water, and final rinse with 10% NaS2O3 for 10 min. All samples were then dried at 100°C for 10 min (10). To check the sterilization, randomly surface-sterilized tissues were imprinted on blood agar (Merck Germany), incubated at 28°C for 2 days, and examined for microbial growth (10, 14).
Isolation of endophytes.
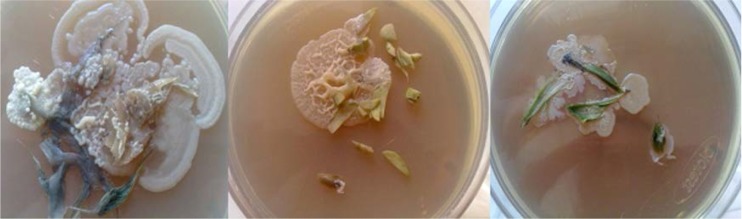
Three samples of each plant were aseptically crushed into small fragments and placed directly on the international streptomyces project 2 (ISP2) agar (HI Media, India), R3A agar (1g yeast extract, 1g protease-peptone, 1g casamino acid, 1g glucose, 1g starch, 0.5 g sodium pyruvate, 0.6 g K2HPO4, 1g MgSO4 7H2O, and 18g agar in 1 liter distilled water), and blood agar. Plates were incubated at 28°C for 2 to 4 days. Bacterial isolates were identified by their morphology and characteristics of their colonies such as size, shape, colour, and growth rate at different temperatures (Fig. 1). Colonies with similar morphological features were grouped into the same species (15).
Fig. 1.
Colonies of endophytic bacteria grown on R3A culture medium
Molecular identification.
16S rRNA gene was amplified using specific primer for selected isolates. The PCR products were sequenced using 27F (5′-AGAGTTTGATCCTGGCTCAG-3′), 1525-R (5′AAGGAGGTGWTCCARCC-3′) and 907-R (5′-CCGTCAATTCMTTTRAGTTT-3′) with an ABI 3100 genetic analyser. The PCR was carried out with an initial denaturation at 94°C for 2 min, 30 cycles of 94°C for 1 min, 60°C for 1 min, 72°C for 3 min, and final extension at 72°C for 10 min. Dye Terminator Cycle Sequencing Genome Lab™ was used to determine the sequence of PCR products according to the manufacturer’s instructions (Biometra Co, Germany).
Phylogenetic analysis.
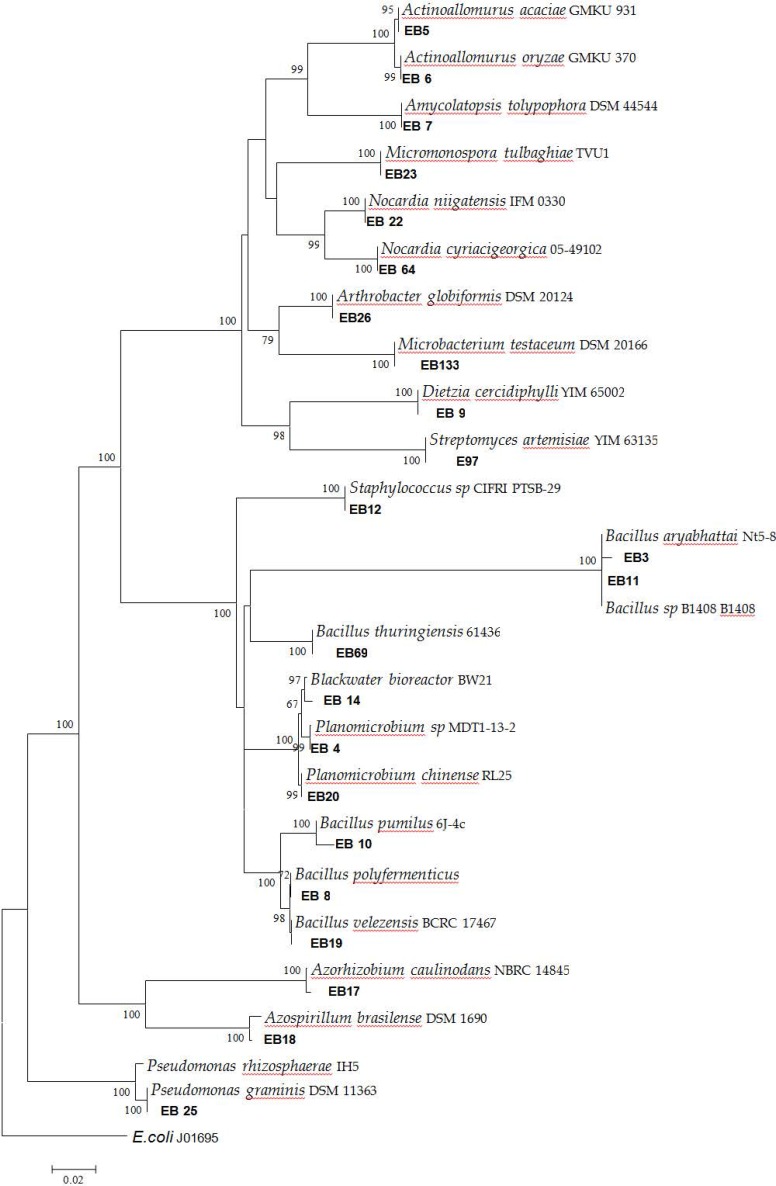
Multiple alignments of the 16S rRNA were generated using sequences of our bacterial isolates and the bacterial strains from Gen-Bank database with the jPhydit program (16). MEGA version 4 (17) was used to construct a neighbour-joining phylogenetic tree using the Kimura two-parameter. The reliability of the branching and clustering pattern was estimated from 1000 bootstrap replicates. The tree was rooted with E. coli 16S rRNA.
Extraction of antimicrobial compounds.
The isolates were cultured in TSB (Tryptic Soy Broth, Merck Germany) and incubated at 28°C for 14 to 16 days, after which we proceeded with extraction of antimicrobial compounds using 3 methods. In the first method, one part of the cultured media was mixed with one part ethyl acetate (1:1 ration) and stirred with magnetic stirrer for 6 h. The organic supernatant was separated and centrifuged at 5,000 rpm for 10 min. The ethyl acetate layer was transferred into a clean flask, and dried with rota-evaporator (Heidolph, Germany) at 50°C. The dry extract was dissolved in 2 ml of ethanol. To test antimicrobial susceptibility of the extracts, 25 μl of each extract in ethanol was used to soak blank discs (7 mm in diameters) (18). In the second method, another part of media was incubated in boiling water for 5 min and in cold water for 5 min then added one part of cultured media were mixed with one part ethyl acetate (1:1 ratio). As described for the first method, samples were then processed. In the third method, the last part of media, which contained bacteria, was sonicated for 3 min at 160W and extraction method was completed as described for the first method. Bacterial isolates obtained using the 3 methods were tested by disc diffusion against pathogenic bacteria, including Staphylococcus aurous, Bacillus subtilis, Pseudomonas aeruginosa, Citrobacter freundii, Shigella flexneri, Escherichia coli, Klebsiella pneumoniae, Bacillus cereus, and Proteus mirabilis. The inhibition zone was measured for each bacterial species separately.
RESULTS
Antimicrobial susceptibility.
Using the plant tissues crushed and cultured in Petri dishes, plant-associated bacteria were successfully isolated (Fig. 1). The antibacterial activity of the isolated bacteria was examined using the antibiotic disc diffusion method. All isolated bacteria displayed anti-bacterial activities against the selected bacteria for antibiotic susceptibility. Among the isolated bacteria, 16 isolates (69.56%) obtained by the second method (hot method) showed strong antiviral activities by producing large inhibition zones. Using the third method (ultrasonic method) 13 isolates (56.52%) were obtained that all displayed antiviral activities. Among the isolates of the third method, EB7 (isolated from the root of Stachys lavandulifolia) and EB69 (isolated from the root of Physalis alkekengi) displayed a broad antibacterial activity against all the target bacteria (Table 1, Fig. 2).
Table 1.
Antimicrobial activity* of endophytic bacteria extracted by 2 methods against pathogenic bacteria
| Antimicrobial compounds extracted with hot method | Antimicrobial compounds extracted with ultrasonic method | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate number | B. cereus | S. aureus | B. subtilis | K. pneumoniae | C. freundii | P. mirabilis | Sh. Flexneri | E. coli | B. cereus | S. aureus | B. subtilis | K.pneumoniae | C. freundii | P. mirabilis | Sh. flexneri | E. coli | |
| EB4 | +++ | − | − | − | − | − | − | − | − | − | − | − | − | − | ++ | − | |
| EB5 | +++ | +++ | +++ | − | ++ | − | + | − | − | − | − | − | +++ | − | − | − | |
| EB14 | − | − | − | ++ | ++ | − | ++ | − | − | − | − | ++ | ++ | − | ++ | − | |
| EB18 | − | − | +++ | + | − | + | +++ | − | − | − | − | +++ | − | − | − | − | |
| EB22 | − | − | − | − | − | − | + | + | − | − | − | ++ | +++ | − | +++ | − | |
| EB23 | +++ | − | − | + | − | − | + | − | − | − | − | − | +++ | − | − | − | |
| EB25 | + | + | ++ | + | − | − | + | − | − | − | − | +++ | − | − | ++ | − | |
| EB26 | +++ | − | +++ | + | − | − | + | − | − | − | − | ++ | − | − | ++ | − | |
| EB97 | − | − | − | − | − | − | − | − | − | − | +++ | − | +++ | − | − | − | |
| EB 133 | + | − | + | − | − | + | − | + | − | − | − | − | ++ | − | − | − | |
| EB3 | − | ++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| EB 6 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| EB 7 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | + | +++ | +++ | − | +++ | ||
| EB 8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| EB9 | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | |
| EB 10 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| EB11 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| EB12 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| EB17 | − | − | ++ | − | − | − | ++ | − | − | − | +++ | ++ | − | − | +++ | − | |
| EB 19 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| EB20 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| EB64 | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| EB69 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − | +++ | +++ | − | ++ | − | |
Inhibition zone, EB: Endophytic Beiranvand (Beiranvand is surname of the first author), − : No activity, +: weak activity, ++: moderate activity, +++: high activity
Fig. 2.
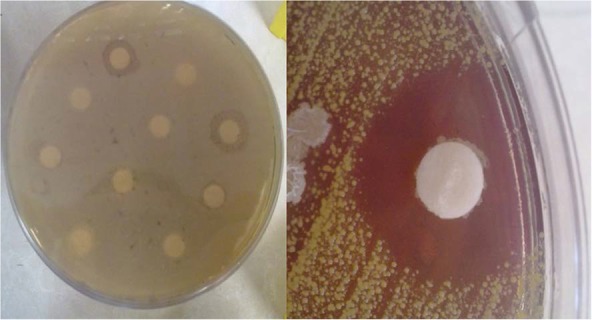
Antimicrobial susceptibility test of endophytic bacteria
Identification of bacteria.
We sequenced and performed phylogenetic analysis of 23 bacterial isolates obtained from 23 plants. The results of phylogenetic analysis of 16S rRNA (Table 2 and Fig. 3) showed that our isolates belonged to different bacterial species, including Gram negative (such as Pseudomonas graminis) and Gram positive bacteria (such as Bacillus thuringiensis). Among our isolates, there were also three isolates whose species could not be identified. These isolates, whose genera were identified, included isolate EB4 (Planomicrobium Sp), EB11 (Bacillus sp), EB12 (Staphylococcus Sp), and EB23 (Bacillus Sp). Taken together, our results suggested that there is wide range of diversity among endophytic bacteria.
Table 2.
Taxonomic characterisation of actinobacteria isolated from different plant organs.
| Isolate | Host plant | Sampling Location | Organ | Species | Genbank accession | similarity |
|---|---|---|---|---|---|---|
| EB 3 | Allium schoenoprasum | Khorramabad | Root | Bacillus aryabhattai | KP209388 | 99/44 |
| EB 4 | Mentha pulegium | Ahvaz, Mullah Agha | Root | Planomicrobium Sp | KP324949 | 99/34 |
| EB 5 | Marrubium vulgare | Ahvaz, Salnd Mountain | Root | Actinoallomuru acacia | KP209377 | 100 |
| EB 6 | Falcaria vulgaris | Khorramabad | Root | Actinoallomurus oryzae | KP209378 | 100 |
| EB 7 | Stachys lavandulifolia | Tehran, Damavand | Root | Amycolatopsis tolypophora | KP209379 | 100 |
| EB 8 | Ocimum basilicum | Ahvaz | Root | Bacillus polyfermenticus | KP209385 | 100 |
| EB 9 | Alcea amcheri | Ahvaz, Salnd Mountain | Stem | Dietzia cercidiphylli | KP209389 | 99/29 |
| EB 10 | Chenopodium album | Khorramabad | Root | Bacillus pumilus | KP209384 | 99/29 |
| EB 11 | Gundelia tournefortii | Khorramabad | Rood | Bacillus. Sp | KP324948 | 100 |
| EB 12 | Achillea millefolium | Ahvaz, Imam Abdullah | Root | Staphylococcus Sp | KP324950 | 99/93 |
| EB14 | Borago officinalis | Ahvaz | Leaf | Blackwater bioreactor | KP209383 | 98/96 |
| EB 17 | Allium ursinum | Tehran, Damavand | Stem | Azorhizobium caulinodans | KP209380 | 99/79 |
| EB 18 | Zataria multiflora | Tehran, Damavand | Root | Azospirillum brasilense | KP209381 | 98/55 |
| EB19 | Chenopodium album | Khorramabad | Root | Bacillus velezensis | KP209386 | 100 |
| EB20 | Lavandula angustifolia | Tehran, Damavand | Root | Planomicrobium chinense | KP209393 | 100 |
| EB22 | Cymbopogon oliviery | Ahvaz, Haftcal road | Rood | Nocardia niigatensis | KP209393 | 100 |
| EB 23 | Phasaeolous vulgaris | Khormabad | Stem | Bacillus Sp | KP209390 | 100 |
| EB 25 | Teucrium polium | Ahvaz, Salnd Mountain | Root | Pseudomonas graminis | KP209394 | 100 |
| EB 26 | Aloe vera | Khorramabad | Leaf | Arthrobacter globiformis | KP209382 | 100 |
| EB64 | Cucumis sativus | Khorramabad | Fruit | Nocardia cyriacigeorgica | KP209396 | 100 |
| EB 69 | physalis alkekengi | Ahvaz | Root | Bacillus thuringiensis | KP209387 | 100 |
| EB97 | Rheum rhaponticum | Tehran, Damavand | Stem | Streptomyces artemisiae | KP209395 | 100 |
| EB 133 | Coriandrum sativum | Khorramabad | Root | Microbacterium testaceum | KP209391 | 100 |
Fig. 3.
Identification of endophytic bacteria using 16S rRNA
DISCUSSION
Since actinobacteria have been applied in producing different kinds of current medicines, more recent studies are investigating new bacterial sources. This study focused on actinobacteria, in particular actinomycets, which was found in a large number of the isolated bacteria obtained from selected plants. We isolated 23 endophytic bacterial isolates, among which species of 19 isolates were identified by 16S rRNA. The majority of the isolates could be considered for producing antibiotics against target bacteria. Two out of 23 endophytic bacterial isolates, EB4 and EB7, showed inhibitory activity against Bacillus cereus. In addition, EB9 showed inhibitory activity against Staphylococcus aureus, Citrobacter freundii and Shigella flexneri. Sixteen isolates (69%) obtained by hot method showed strong activity against selected pathogenic organisms and two of them (EB7 and EB69) had broad spectrum antibacterial activity (Table 1). Ultrasonic method showed that 13 out of 23 isolates (46%) inhibited microbial growth. Studies have shown that most of the isolated organisms were Bacillus (25, 26). Similarly to our study, Janso et al. reported that 105 out of 123 endophytic actinomycets isolated from tropical plants belonged to 17 different genera and also Sphaerisporangium and Planotetraspora as rare genera, which have not been reported previously. They had nearly 60% bioactive activities (25). Strobel et al. isolated Streptomyces, Microbispora, and Nocardiodes as endophytic bacteria and showed that their isolates produce antimicrobial compounds with inhibitory effects against Gram positive bacteria (11). Another study isolated 560 endophytic actinomycetes from 26 herbal species. Their isolates, which belonged to a wide range of bacterial species, showed strong antimicrobial properties (27).
Fig. 4.
Phylogenetic tree constructed using 16s rRNA gene sequences.
CONCLUSION
Overall, study revealed a high phylogenetic diversity among endophytic bacteria isolated from different areas of Iran. Our isolates showed considerable antimicrobial activities. This study demonstrated that hot method was more efficient than the other method for isolating endophytic bacteria. Endophytic actinobacteria can be used for producing new bioactive compounds. Such compounds can be used against pathogenic bacteria resistant to the current antibiotics.
ACKNOWLEDGEMENT
Authors thank the Health Research Institute, Infectious and Tropical Diseases Research Center, Jundishapur University of Medical Sciences, Ahvaz, Iran for financial support (Grant No. 91134). We also thank Abdolreza Namdarian for his helpful collaboration.
REFERENCES
- 1. Newman DJ. Natural products as leads to potential drugs: an old process or the new hope for drug discovery. J Med Chem 2008; 51: 2589– 2599. [DOI] [PubMed] [Google Scholar]
- 2. Costa FG, Zucchi TD, Melo ISd. Biological control of phytopathogenic fungi by endophytic actinomycetes isolated from maize (Zea mays L.). Braz Arch Biol Technol 2013; 56: 948– 955. [Google Scholar]
- 3. Wu Y, Lu C, Qian X, Huang Y, Shen Y. Diversities within genotypes, bioactivity and biosynthetic genes of endophytic actinomycetes isolated from three pharmaceutical plants. Curr Microbiol 2009; 59: 475– 482. [DOI] [PubMed] [Google Scholar]
- 4. Cragg GM, Newman DJ. Natural products: a continuing source of novel drug leads. Biochim Biophys Acta 2013; 1830: 3670– 3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchanan GO, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. Sporolides A and B: structurally unprecedented halogenated macrolides from the marine Actinomycete Salinispora tropica. Organic letters 2005; 7: 2731– 2734. [DOI] [PubMed] [Google Scholar]
- 6. Cragg GM, Newman DJ. Plants as a source of anti-cancer agents. J Ethnopharmacol 2005; 100: 72– 79. [DOI] [PubMed] [Google Scholar]
- 7. Kingston DG. Tubulin-interactive natural products as anticancer agents. J Nat Prod 2009; 72: 507– 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oldfield C, Wood NT, Gilbert SC, Murray FD, Faure FR. Desulphurisation of benzothiophene and dibenzothiophene by actinomycete organisms belonging to the genus Rhodococcus, and related taxa. Antonie Van Leeuwenhoek 1998; 74( 1–3): 119– 132. [DOI] [PubMed] [Google Scholar]
- 9. Mann J. Natural products as immunosuppressive agents. Nat Prod Rep 2001; 18: 417– 430. [DOI] [PubMed] [Google Scholar]
- 10. Qin S, Li J, Chen H-H, Zhao G-Z, Zhu W-Y, Jiang C-L, et al. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol 2009; 75: 6176– 6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rathod D, Dar M, Gade A, Shrivastava RB, Rai M, Varma A. ( 2013). Microbial endophytes: progress and challenges. In: Biotechnology for Medicinal Plants. Eds, Chandra S, Lata H, Varma A. Springer-Verlag GmbH, Wien; pp. 101– 121. [Google Scholar]
- 12. Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 2003; 67: 491– 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hasegawa S, Meguro A, Shimizu M, Nishimura T, Kunoh H. Endophytic actinomycetes and their interactions with host plants. Actinomycetologica 2006; 20: 72– 81. [Google Scholar]
- 14. Kang H-Y, Park D-J, Lee J-C, Kwon M-K, Kim S-B, Kim C-J. Isolation of Agrobacterium sp. BE516 from the root of Miscanthus sacchariflorus and its plant growth promoting activity. J Appl Biol Chem 2012; 55: 129– 133. [Google Scholar]
- 15. Castillo U, Harper JK, Strobel GA, Sears J, Alesi K, Ford E, et al. Kakadumycins novel antibiotics from Streptomyces sp. NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Microbiology Letters 2003; 224: 183– 190. [DOI] [PubMed] [Google Scholar]
- 16. Igarashi Y, Miura SS, Fujita T, Furumai T. Pterocidin, a cytotoxic compound from the endophytic Streptomyces hygroscopicus. J Antibiot (Tokyo) 2006; 59: 193– 5. [DOI] [PubMed] [Google Scholar]
- 17. Hasegava T, Takizava M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 1983; 29: 319– 322. [Google Scholar]
- 18. Jeon Y-S, Chung H, Park S, Hur I, Lee J-H, Chun J. jPHYDIT: a JAVA-based integrated environment for molecular phylogeny of ribosomal RNA sequences. Bioinformatics 2005; 21: 3171– 3173. [DOI] [PubMed] [Google Scholar]
- 19. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24: 1596– 1599. [DOI] [PubMed] [Google Scholar]
- 20. Amin M, Jahangirnezhad M, Rasaei N, Pipelzadeh MH, Rafiee M. Evaluation of the effect of Persian shallot (Allium hirtifolium, boiss) aqueous extract on mouth bacterial count compared with chlorhexidine mouth rinse. Afr J Microbiol Res 2012; 6: 5809– 5813. [Google Scholar]
- 21. Turenne CY, Tschetter L, Wolfe J, Kabani A. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol 2001; 39: 3637– 3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sessitsch A, Reiter B, Pfeifer U, Wilhelm E. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol Ecol 2002; 39: 23– 32. [DOI] [PubMed] [Google Scholar]
- 23. Garbeva P, Van Overbeek L, Van Vuurde J, Van Elsas J. Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA based PCR fragments. Microb Ecol 2001; 41: 369– 383. [DOI] [PubMed] [Google Scholar]
- 24. Tortoli E. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin Microbiol Rev 2003; 16( 2): 319– 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Janso JE, Carter GT. Biosynthetic potential of phylogenetically unique actinomycetes from tropical plants. Appl Environ Microbiol 2010; 76: 4377– 4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verma VC, Gond SK, Kumar A, Mishra A, Kharwar RN, Gange AC. Endophytic actinomycetes from Azadirachta indica A. Juss.: isolation, diversity, and anti-microbial activity. Microbial Ecology 2009; 57: 749– 756. [DOI] [PubMed] [Google Scholar]
- 27. K, Penttinen P, Guan T, Xiao J, Chen Q, Xu J, et al. The diversity and anti-microbial activity of endophytic actinomycetes isolated from medicinal plants in Panxi plateau, China. Curr Microbiol 2011; 62: 182– 190. [DOI] [PubMed] [Google Scholar]