Abstract
Humans and other mammals are limited in their natural abilities to regenerate lost body parts. In contrast, many salamanders are highly regenerative and can spontaneously replace lost limbs even as adults. As salamander limbs are anatomically similar to human limbs, knowing how they regenerate should provide important clues for regenerative medicine. Though interest in understanding the mechanics of this process has never waivered, until recently, researchers have been vexed by seemingly impenetrable logistics of working with these creatures at a molecular level. Chief among the problems has been the very large salamander genomes, and not a single salamander genome has been fully sequenced to date. Recently, the enormous gap in sequence information has been bridged by approaches that leverage mRNA as the starting point. Together with functional experimentation, this data is rapidly enabling researchers to finally uncover the molecular mechanisms underpinning the incredible biological process of limb regeneration.
Keywords: axolotl, Ambystoma mexicanum, limb, regeneration, transcriptomics
Why use the axolotl?
The phenomenon of limb regeneration in salamanders has been a part of the formal scientific canon for nearly 250 years [1], but salamanders are a large and diverse group. Most studies have focused on either newts or axolotls or both. “Axolotl” is the common name for Ambystoma mexicanum, a species of salamander native to a few lakes near Mexico City (Figure 1A). They can be bred year-round in the lab with relative ease (Figure 1B). Axolotls are neotenic, meaning they grow and become sexually mature without undergoing the final stage of the canonical amphibian life cycle; they are permanently aquatic and outfitted with external gills and other accouterments to support this lifestyle (Figure 1C; full axolotl staging series in [2, 3]). The axolotl generation time, just under one year, is still much shorter than many other salamanders. In the past eleven years, both transgenesis [4] and genome editing [5–8] have been successfully performed in axolotls in several laboratories. Retroviral infection, which results in genomic integration, has also been demonstrated in embryos and in larval and adult limbs [9, 10]. Localized genome editing has now been developed, allowing for study of mutant cells in vivo without necessarily waiting for homozygous loss-of-function mutants [10]. Genome engineering is poised to be the definitive tool for doing bona fide loss-of-function genetics in axolotls and to finally allow for direct attribution of wild-type gene function in the process of limb regeneration. Yet, this “reverse genetic” strategy is only productive if sequence data exists for the genes of interest, which is required for targeting. Additionally, without broad and unbiased screening of tissue-specific and time-specific expression data, a targeted gene candidate approach would still be required. Surely the limb development literature, as well as hints from diverse fields related to regeneration (cellular reprogramming, stem cell biology, among others) would provide starting points that could prove fruitful. However, transcriptomics and other mRNA-based techniques offer to illuminate the identities of transcripts that might not otherwise be considered. These newly-implicated transcripts might have orthologs in fully-sequenced species, but some may not, and some may be unique to salamanders. Using the mRNA from regenerating axolotl tissues as the experimental guide is a solid, unbiased approach to discovering the mechanisms of limb regeneration. Below, we review the impact of transcriptome studies towards revealing the genes that underlie the abilities of axolotls to regenerate limbs.
Figure 1. Axolotl basics and genome-modifying tools.
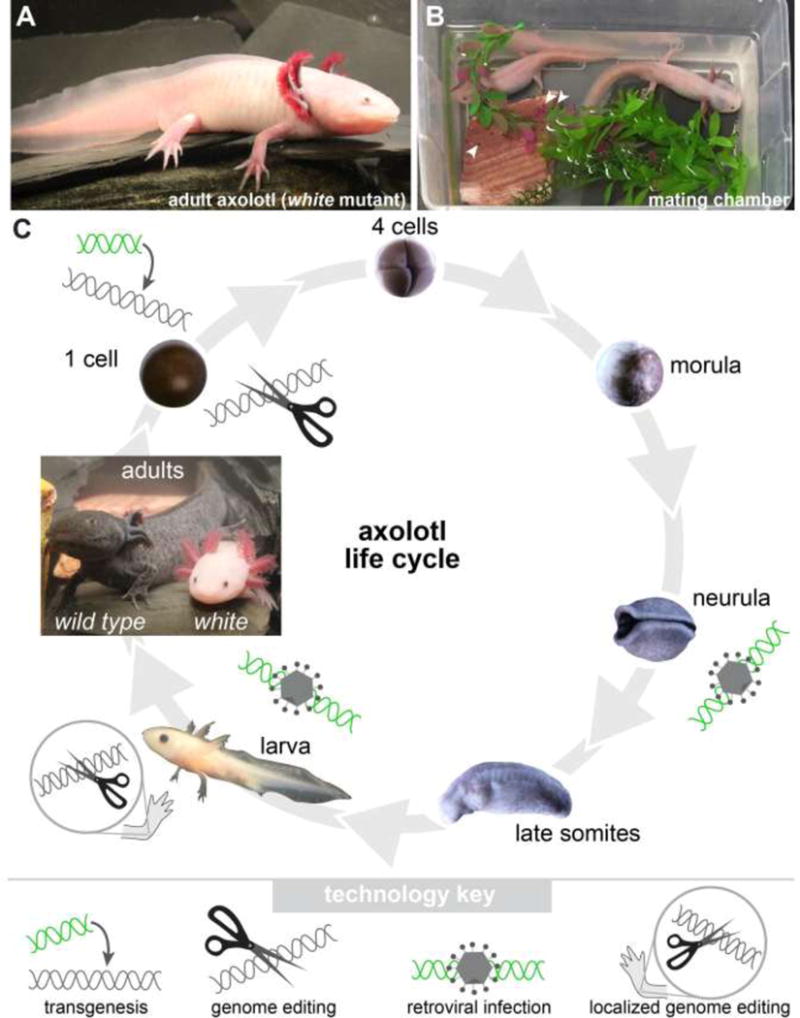
(A) Shown is an adult axolotl of the white genotype. Note the large limbs. Forelimbs have four digits, while hindlimbs have five digits. (B) Male and female axolotl in mating chamber with eggs. (C) Axoltol life cycle with validated points for genomic modification manipulations noted. Axolotl embryogenesis spans approximately 10–12 days; only some stages are shown. Embryos develop within a transparent jelly coat, which must be removed to permit injections. Two adults are shown. At left is the wild-type genotype whose skin is a darkly-pigmented mottled brownish-black. At right is the white mutant. Note that specimens are not drawn to scale. Techniques that modify the genome are noted at stages where the techniques have been employed to date.
Basics of Limb Regeneration
To appreciate the strides that have been made in elucidating the genetics of axolotl limb regeneration, a brief orientation to the process at the gross anatomical level is helpful (Figure 2, Key Figure, reviewed in [11]). Following amputation, axolotls shed very little blood at the site of injury. Within hours, the cut stump is ensheathed by a thin layer of epidermal cells, which migrate from stump epidermis. They collect at the cut end and proliferate, forming a wound epidermis. Wound epidermis is structurally and molecularly distinct from fully-differentiated, intact epidermis [12]. In the days following re-epithelialization, progenitor cells are activated within the tissues of the stump. The term activation encompasses both the re-entry of progenitor cells into the cell cycle as well as the accumulation of these cells at the tip of the stump, beneath the wound epidermis. Activated progenitor cells may originate from stem cells or by dedifferentiation; the relative contribution of these two mechanisms remains unclear and may differ across tissue types. Together, activated cells accumulated at the tip of the stump make up the blastema. Blastema cells are highly proliferative and while they are thought to be under the influence of factors generated by the overlying wound epidermis, unidirectional signaling is not the whole story. Blastemas are said to behave as autonomous units because they can be transplanted to receptive areas elsewhere on the body, where they give rise to limbs [13]. Furthermore, positional information is encoded within blastemas. For example, blastemas fated to produce only a foot do so even when transplanted elsewhere, while full leg blastemas produce full legs when transplanted elsewhere [13, 14]. While blastema cells appear rather similar to one another, and resemble fibroblasts in their morphology, data from transplantation studies implies they are actually heterogeneous in origin and potential [15, 16]. Once the blastema pool has reached a critical size, the bulbous structure flattens out at the “palette” stage, future cartilage cells coalesce and condense, and differentiation of the various tissue types takes place. Ultimately, the newly-regenerated limb achieves a perfect form outwardly indistinguishable from the lost limb. Importantly, limbs regenerate to the correct size regardless of the size or age of the animal. The entire regenerated limb is functionally integrated at the site of the original stump and with the entire body.
Figure 2. Outline of cellular events during limb regeneration.
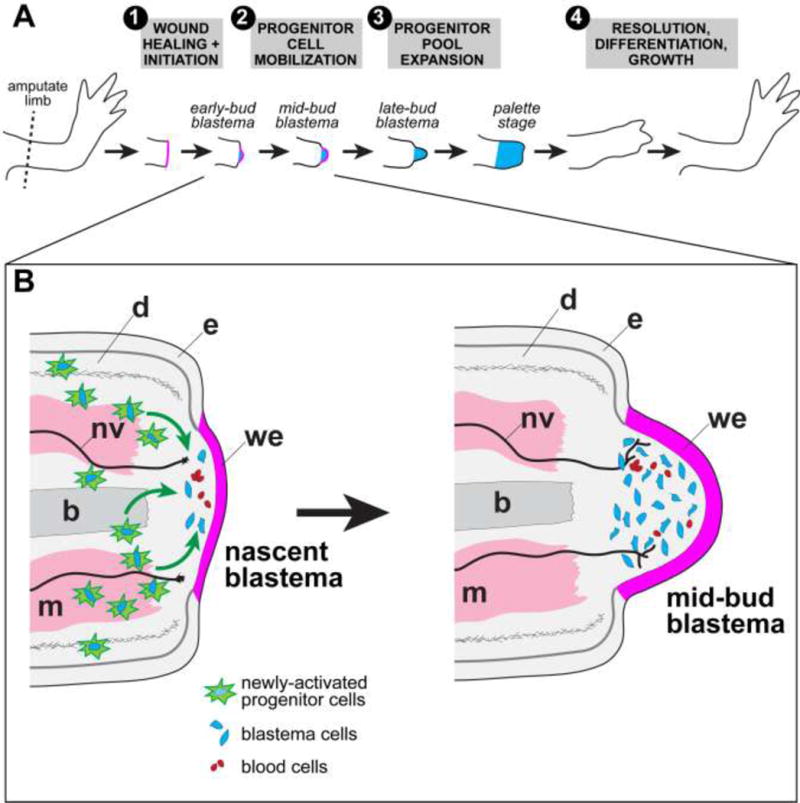
(A) General progression from unamputated to fully regenerated. (1) Immediately following amputation (within ~24 hours), a thin wound epidermis (magenta) forms across the cut stump via migration of stump epidermal cells. Wound epidermis thickens as cells within it proliferate. (2) A visible bump, called a blastema (blue), forms beneath the wound epidermis. Blastema cells are derived from activated progenitor cells within various stump tissues that migrate to the tip. (3) Blastema cells proliferate to expand the progenitor pool. (4) The initial regeneration response resolves, cells begin to undergo differentiation, and the limb continues to grow to the appropriate size. (B) Early steps are shown in more detail in inset. Architectures of tissues such as bone and muscle are locally deconstructed near the amputation plane and are therefore shown as jagged. Newly-activated progenitor cells, which give rise to future blastema cells, are depicted with bright green starburst outlines. These cells are cued to re-enter the cell cycle and some fraction of them presumably migrate to the space immediately below the wound epidermis. Blood cells, both red and white, are intermingled with blastema cells. A “nascent blastema” is equivalent to very early-bud stage blastema in other literature. Noted are: epidermis (e), wound epidermis (we), dermis (d), bone (b, medium gray), muscle (m, pink), nerves (nv, black). Migration of newly-activated progenitor cells to the tip of the stump during blastema formation is implied by the green arrows.
Genomics
Though the axolotl has an enormous genome—estimated to be ~32 GB (original estimate from A. tigrinum [17]; recent estimate in axolotl [18]) —it is a simple diploid with fourteen pairs of chromosomes [19]. There is evidence that the extremely large genome is highly repetitive and contains unusually long introns [20]. A linkage map has been produced [21] and since updated [22]. While axolotls exhibit extensive conservation in synteny between chickens and humans, homologous genome segments in axolotl are on average 51 and 14 times longer than their respective chicken and human counterparts [22]. This is an interesting finding that, combined with the evidence of unusually long introns, suggests gene regulatory elements may be spaced further apart in salamanders than in other vertebrates. The enormous axolotl genome is thought to derive from an ancient episode of genome expansion resulting from the activity of mobile elements, particularly gypsy and LINE 1/2 elements [18]. By leveraging the most modern technologies and techniques in sequencing and assembly [23–27], obtaining a reference genome for axolotl may soon be tractable. However, several critical issues remain to be resolved before the first full genome assembly is likely to be created, such as sufficient read length and improved methods for handling genomic complexity during assembly. Until then, transcriptomics will continue to provide many of our best insights into the genetics of limb regeneration.
Importantly, there is no reason to believe that traditional genetic approaches will not succeed in these animals. Two classical loss-of-function axolotl pigmentation mutants have recently been mapped, white (d/d,) and albino (a/a,) [8]. For both, linkage mapping was used to identify a small interval containing a handful of genes, and existing data from other systems for the genes within these intervals was used to narrow to a single candidate. Sequencing revealed predicted loss-of-function mutations in the mutant stocks, and both loci were rescued using transgenesis. This study revealed that white stocks harbor a homozygous mutation in endothelin 3, while albino stocks are mutant for tyrosinase [8]. Though the cloning of these mutants were major efforts, as the genetic linkage map is improved for axolotl, and genome editing and rescue strategies become more routine, the function of other genes will likely be illuminated with these gold-standard techniques. Among the genes awaiting this type of thorough analysis in the future are those directly controlling key components of the limb regeneration process.
EST Projects and Microarrays for Uncovering Regeneration Transcripts
Before RNA-seq became a widely-available methodology (circa 2009), several expressed sequence tag (EST) projects for axolotls yielded important sequence and expression information. This type of strategy converts mRNA from expressed genes into cDNA, clones the cDNAs en masse to create a library, and then sequences into the cDNA inserts, reading several hundreds of bases into the ends of the original transcripts. In 2004, 17,352 ESTs were sequenced from two sources—mid-development, neural-tube-stage, embryos, and tail blastemas from 6 days post-amputation [28]. This study assembled the ESTs into 6,377 contigs, with average length at 569 bp, which was estimated to represent perhaps 25% of axolotl gene expression [28]. The authors estimated 19% of transcript contigs to be unique to axolotl insofar as they did not match existing nucleotide or protein sequences then available in public databases. Notably, with this technology, few of the recovered contigs were likely to constitute full-length coding sequences or open reading frames (ORFs); here, they estimated only 5.4% of the contigs to include the entire ORF. While regenerating limbs were not included in the study, extensive overlap may exist between tail blastemas and limb blastemas, hence this study was an important contribution even to limb regeneration efforts. In that same year, a second axolotl EST project was reported comprising a more comprehensive set of sample tissues (blastemas from limbs and tails, one stage of embryos, brain, kidney, spleen, liver, heart, gills, and gonads) [29]. This study also sampled from the closely-related tiger salamander (Ambystoma tigrinum, ~11 million years diverged from axolotl lineage) [29].
Since these ground-breaking EST studies, several additional studies have employed differential hybridization techniques to evaluate gene expression that may underlie regenerative abilities. For example, the single four-days-post-amputation time point (very early blastema) was chosen to compare to unamputated limbs in a “suppression subtractive hybridization” screen performed in [30]. This screen yielded 279 sequence-able clones that were likely specific to or highly enriched in early blastema. Of the sequenced clones, only one was previously implicated in axolotl limb regeneration (mmp-3/10a), proving this was a viable regeneration gene discovery tool for the time. Subtractive hybridization also led to the identification of a cell-surface molecule, Prod1, critical for instructing the proximo-distal polarity of the regenerating limb in newts, and it has subsequently been implicated in the same process in axolotls [31, 32].
EST sequencing projects identified thousands of salamander transcripts including several with implicated roles in regeneration. Harnessing these sequences provided further opportunities for large-scale expression studies using microarray technologies. Microarrays have the advantage of providing standardization as they can be commercially fabricated, shared across labs, and processed and analyzed using a common protocol. They can be used to probe any condition of interest. The main drawback is that expressed genes not represented on the microarray will, of course, be lost to downstream analyses. Microarrays have shed valuable light on the genes driving limb regeneration. They were successfully employed to discover transcripts that differ in regenerating limbs versus those that fail to regenerate due to denervation [33]. This approach is imperative for understanding how limbs are dependent upon innervation for their successful regeneration, a fact that has been known for 200+ years but still remains poorly understood [34, 35]. Microarrays were also useful for delineating the gene expression relevant to the wound epidermis covering limb slated to regenerate versus epidermis engaged in simple wound repair [36]. This analysis is important because though limb regeneration does involve early wound healing, the wound healing it employs is necessarily different from that used to return the skin alone to homeostasis. They may share some attributes, but to understand how regenerative wound epidermis influences stump tissue dynamics, subtracting the transcripts engaged in the simpler healing closed task is likely a useful strategy. Because mammals do not typically respond to amputations by growing blastemas, and since blastemas are required for regeneration, understanding the process of how salamanders convert an initial wound-healing response into blastema production and regeneration will be essential.
Another microarray study was designed to uncover transcripts that are enriched in limbs undergoing full regeneration as compared to limbs more simply healing lateral wounds that are not undergoing full limb regeneration [37]. Furthermore, this study compared the gene expression in these two cases to gene expression in developing limb buds – the initial tissue outgrowths filled with limb progenitor cells that fuel the first development of the limb in immature animals. They can therefore be considered analogous in their potential to blastemas on amputated limbs. However, since limb buds arise programmatically during the normal course of animal development and from a standard-sized starting point, they are categorically different from blastemas. Blastemas, instead, arise from tissues that only moments earlier were fully differentiated and engaged in functions such as movement, structural support, and sensation. An animal can be challenged to regenerate a limb at any point in its life and from any point along the proximo-distal axis of the original limb. This study nicely resolved the waves of transcriptional activity corresponding to three major phases of limb regeneration involving initial wound healing, subsequent blastema formation, and finally limb redevelopment, identifying approximately a hundred significant genes with functions associated with morphogenesis, organogenesis, and related roles. Future functional experimentation will be required to explore the specific contributions of these genes to regenerative processes. Some of the most tantalizing future work may reveal functional differences between how limbs develop and how they regenerate. Unlocking these differences could be crucial for stimulating regenerative responses in otherwise non-regenerative contexts, such as in humans.
Recently, an extensive microarray study aimed at describing global transcriptional changes over time in the local tissues following amputation was published [38]. This study examined unamputated limbs and regenerating limbs over the course of the first 28 days of regeneration post-amputation, with a 1-mm piece of tissue harvested from the tip of the regenerating limbs. Its power lies in the total number of time points sampled (20) and the number of biological replicates per time point (10), providing more granular resolution into these early events than earlier studies. The time points sampled were overlaid with existing morphological landmarks that develop in a stereotypical order during normal regeneration and have been characterized at a histological level [39]. Targeted transcript expression values were quantified at each time point and also compared to day 0 (unamputated), providing both a view of how expression changes over the course of regeneration as well as how each time point differs from the homeostatic state. This approach enabled identification of transcriptional changes that may underpin the transitions that occur during the early stages of the regenerative process. A further feature of the work is the deposition of all data at Sal-Site (www.ambystoma.org). This platform allows for simple searching of genes of interest based on gene name, gene symbol, or probe ID (custom Affymetrix GeneChip Amby002; ~20,000 unique probes).
Transcriptomics
The era of transcriptomics has revolutionized gene discovery in axolotls (Table 1 summarizes key genes, many of which were identified by transcriptomics). RNA-seq makes no presumptions about the identity of the transcripts at play except that they can be captured in the purification protocol and subject to reverse transcription, adapter ligation, and amplification. This more versatile method enables detection of a greater dynamic range for expression values for all individual transcripts, capturing both sequence and expression data simultaneously, all at single nucleotide resolution. The decision of how deeply to sequence samples is left to the researcher. For example, if sequenced sufficiently deep, even very lowly-expressed transcripts may be recoverable, and these may be predicted to control fundamental aspects of limb development (for example, transcription factors). With RNA-seq, the saturation effect that occurs with microarrays for very highly-expressed transcripts does not exist; therefore, an accurate representation of the relative expression values for very highly-expressed genes can be determined. Furthermore, recent advances in RNA-seq technology permit application of the technique to very low inputs of RNA (including individual cells), which now opens the door to further dissection of the specific tissues and cells integral to limb regeneration where only minute quantities of RNA may be available for study.
Table 1. Curated list of genes implicated in axolotl limb regeneration.
Shown are genes whose putative involvement was first realized through expression methods discussed here. However, notable genes identified via other means, such as candidate-based approaches, are also included. This list is not exhaustive, and to be included a gene either must have been identified in more than one study, must have been validated (for example, by qRT-PCR or mRNA in situ hybridization), or must have functional experimental data in vivo.
| gene symbol |
gene name | expression pattern | proposed function |
reference(s) | type of study |
|---|---|---|---|---|---|
| BMP-2 | bone morphogenic protein 2 | early, mid, and late regeneration; interdigital | promotes apoptosis | [38, 45] | candidate; microarray |
| cd38 | cluster of differentiation 38 | enriched in blastemas from proximal amputations over distal amputations | ? | [10] | RNA-seq |
| cirbp | cold-inducible RNA-binding protein | blastema-enriched | cytoprotective | [46](newt); [10, 38] | subtractive hybridizatio; RNA-seq; microarray |
| fen1 | flap structure- specific endonuclease 1 | enriched in regenerating over unamputated | genome stability? | [30, 33, 38] | subtractive hybridization ; microarray |
| fgf8 | fibroblast growth factor 8 | early wound epidermis/basal; early mesenchyme immediately beneath wound epidermis | promotes cellular proliferation? | [37, 47, 48] | microarray; candidate |
| fgf10 | fibroblast growth factor 10 | early regeneration; enriched in amputation wounds vs. lateral wounds | promotes cellular proliferation? | [37] | microarray |
| fus | FUS RNA binding protein | blastema-enriched | ? | [10, 38] | RNA-seq; microarray |
| hmox1 | heme oxygenase 1 | enriched in regenerating over unamputated | stress response/early wound healing? | [30, 33, 38] | subtractive hybridization ; microarray |
| hnRNP a1 | heterogeneous nuclear ribonucleoprotein A1 | blastema-enriched | ? | [10, 38] | RNA-seq; microarray |
| hoxa-9 | homeobox a9 | blastema (throughout), early through palette stage | patterning? | [49] | candidate |
| hoxa-13 | homeobox a13 | distal-most aspect of blastema, early through palette stage | patterning? | [49] | candidate |
| hoxb-13 | homeobox b13 | very early blastema through palette stage | patterning? | [50] | candidate |
| hoxc-8 | homeobox c8 | 0–1 days post-amputation | ? | [38] | microarray |
| hoxc-10 | homeobox c10 | very early blastema through palette stage | patterning? | [50] | candidate |
| hoxd-8 | homeobox d8 | very early stump/proximal; mid-bud blastema | patterning? | [51] | candidate |
| hoxd-10 | homeobox d10 | very early stump; mid-bud blastema | patterning? | [51] | candidate |
| hoxd-11 | homeobox d11 | early-bud blastema/posterior margin; proximal expression in regenerating portion through palette stage | patterning? | [51] | candidate |
| hsp70 | heat shock protein 70 | regeneration-enriched; all stages (early blastema through palette and digits) | ? | [52] | candidate |
| kazald1 | kazal type serine peptidase inhibitor domain 1 | blastema-enriched | promotes cellular proliferation | [10, 38, 53] | RNA-seq; microarray |
| lmx-1b | LIM homeobox transcription factor 1 beta | dorsal blastema; also enriched at 0–1 dpa over 2–3 dpa | dorsalizing factor? | [38, 54] | microarray; candidate |
| MARCKS -like | MARCKS-like protein | wound epidermis | promotes cellular proliferation | [55] | functional assay |
| meis1 | meis homeobox 1 | enriched in blastemas from proximal amputations versus distal amputations | proximal specification; downstream of retinoic acid signaling | [56] | candidate |
| meis2 | meis homeobox 1 | enriched in blastemas from proximal amputations versus distal amputations | proximal specification; downstream of retinoic acid signaling | [56] | candidate |
| mmp11 | matrix metalloproteinas e 11 | blastema-enriched | wound healing/matrix remodeling? | [10, 57] | RNA-seq; candidate |
| mmp1 | matrix metalloproteinas e 1 | enriched in 0–1 dpa over 2–3 dpa | wound healing/matrix remodeling? | [38] | microarray |
| mmp3 | matrix metalloproteinas e 3 | mid-blastema; enriched in 0–1 dpa over 2–3 dpa | wound healing/matrix remodeling? | [30, 33, 38, 57] | subtractive hybridization; microarray; candidate |
| mmp9 | matrix metalloproteinas e 9 | blastema (early-mid), wound epidermis/heterogeneous, possibly downstream of innervation | wound healing/matrix remodeling? | [57] | candidate |
| mmp10 | matrix metalloproteinas e 10 | enriched in 0–1 dpa over 2–3 dpa | wound healing/matrix remodeling? | [38] | microarray |
| msx1 | msh homeobox 1 | enriched in amputation wounds vs. lateral wounds; late bud blastema | inhibits differentiation? controls apoptosis? morphogenesis? | [37, 58]; newt [59, 60] | microarray; candidate |
| msx2 | msh homeobox 2 | enriched in amputation wounds versus lateral wounds; early bud blastema | cell activation? inhibits differentiation? | [37, 58, 61] | microarray; candidate |
| nrg1 | neuregulin-1 | innervated blastemas; also enriched at 0–1 dpa over 2–3 dpa | nerve- dependent blastema growth (newts and axolotls) | [38, 62–64] | candidate; microarray |
| p53 | tumor protein p53 | activity downregulated during blastema formation and upregulated during redifferentiation; however also upregulated at 0–1 dpa over 2–3 dpa | negative regulator of cell cycle re- entry, promotes redifferentiatio n later | [38, 44, 65] | microarray; RNA-seq; candidate |
| PL1 | piwi-like 1 | mid-blastema/heterogeneous; wound epidermis/heterogeneous | promotes cellular proliferation, cytoprotective | [66] | candidate |
| PL2 | piwi-like 2 | mid- blastema/heterogeneous; wound epidermis/heterogeneous | promotes cellular proliferation, cytoprotective | [66] | candidate |
| prod1 | prod1 | blastema/higher in proximal blastemas | regulation of proximo-distal axis | [31](newt) [32, 67] | subtractive hybridization |
| ptma | prothymosin alpha | blastema-enriched | ? | [10] | RNA-seq |
| rbp2a | retinol-binding protein 2a | blastema-enriched | proximal-distal axis specification? | [10] | RNA-seq |
| sfrs1 | serine and arginine rich splicing factor | blastema-enriched | ? | [10] | RNA-seq |
| shh | sonic hedgehog | posterior margin of blastema | proliferation and patterning | [47, 68, 69] | candidate |
| shox | short stature homeobox | enriched in blastemas from proximal amputations versus distal amputations | proximal-distal axis specification? | [10] | RNA-seq |
| smad2 | SMAD family member 2 | expression upregulated during late regeneration | TGF-β1 signal transduction | [70] | candidate |
| sox-9 | SRY-box 9 | esp. late/cartilage condensations | cartilage differentiation | [45] | candidate |
| tbx4 | T-box transcription factor 4 | blastema-enriched, specifically in hindlimbs | forelimb specification? | [71] | candidate |
| tbx5 | T-box transcription factor 5 | blastema-enriched, specifically in forelimbs | forelimb specification | [10, 71, 72] | RNA-seq; candidate |
| TGF-β1 | transforming growth factor beta 1 | early regeneration | promotes cellular proliferation and downstream gene expression | [38, 70, 73] | microarray; candidate |
| tsp-1 | thrombospondin- 1 | early wound epidermis/basal; blastema/heterogenous | wound healing + angiogenesis? | [38, 74] | microarray; candidate |
| tsp-4 | thrombospondin- 4 | blastema, connective tissues | extracellular matrix? | [38, 74, 75] | candidate |
| twist1 | twist family BHLH transcription factor 1 | blastema/heterogeneous | ? | [10, 76] | candidate |
| twist3 | twist family BHLH transcription factor 3 | blastema/heterogeneous | ? | [37, 76, 77] | microarray; candidate |
| wnt-5a | wingless-type MMTV integration site family, member 5a | early regeneration; enriched in amputation wounds vs. lateral wounds | promotes regeneration? —inhibiting canonical Wnt signaling blocks regeneration | [37, 78, 79] | microarray; candidate |
| wnt-5b | wingless-type MMTV integration site family, member 5b | early regeneration; enriched in amputation wounds vs. lateral wounds | promotes regeneration? —inhibiting canonical Wnt signaling blocks regeneration | [37, 78, 79] | microarray; candidate |
| wnt-7a | wingless-type MMTV integration site family, member 7a | blastema (diffuse) | ? | [54] | candidate |
Similar to the microarray experiments described above [38], but with an RNA-seq approach, another recent study profiled changes in gene expression over the course of limb regeneration [40]. The main finding highlighted by the authors was an “oncogene burst,” the enrichment of expression of genes with oncogenic activity in other organisms. Speculation about the role of oncogenes and tumor suppressors in limb regeneration has been longstanding (reviewed in [41]), so the hypothesis promoted by the authors piqued interest in this debate again. This approach should benefit from future functional experimentation to determine the extent to which activation of putative oncogenes drives aspects of limb regeneration. 2016 saw the publication of the most comprehensive transcriptome assembly for axolotl to date [42]. This work reported the gene expression over the full course of axolotl embryogenesis. Many of the transcripts reconstructed may also be active in limb regeneration, as embryos must expand progenitor pools, pattern fields of cells, and grow, and these tasks need also occur in limb regeneration. Furthermore, core signaling pathways are often used for multiple purposes by animals, so simply having sequence data for as many genes as possible is an empowering resource for the field.
Recently, a novel axolotl de novo transcriptome has been produced that is both very complete and quite comprehensive [10]. In this work, several tissues from within limbs, including muscle, cartilage, and bone, were dissected and separately sequenced. Additionally, deep sequencing of the mRNAs expressed by intact, unamputated limbs en masse was performed on samples from four distinct locations along the proximo-distal axis of the limbs so that transcription potentially relevant to positional memory could be explored. For blastemas, the stage of regeneration representing the peak of blastema growth prior to differentiation was chosen, which in an adult is about 23 days post-amputation. Blastemas arising from both proximal and distal amputations were sampled, which allowed us to examine gene expression differences between blastemas fated to regenerate an entire limb versus those fated to simply regenerate a hand/foot. Additional tissues sampled in this analysis included blood vessels, heart, testes, and ovaries. A cohort of 151 genes whose expression is highly enriched in both proximal and distal blastemas compared to every other tissue sampled was identified. Among these included a handful of genes previously implicated in limb regeneration, but also many genes that were not previously implicated, and many without any known homologs among publicly available sequence data. A handful of genes predicted to show highly enriched expression in particular tissues were further experimentally validated. Perhaps the most exciting aspect of this work is that since axolotl experimental techniques have caught up with data acquisition, we were able to perform direct functional studies with newly identified genes of interest. For example, cirbp, the axolotl ortholog of mammalian cold-inducible RNA binding protein, was found to be a cytoprotective factor for blastema cells; diminishing the expression of cirbp in blastemas caused an increase in cell death. We also discovered a growth-promoting role for kazald1, which is predicted to encode a secreted protein with a mammalian Kazal-type serine peptidase inhibitor domain, an insulin growth factor binding domain, and an immunoglobulin domain.
An interesting outcome of all of these studies has been the implication of transcripts potentially unique to salamanders as being important for limb regeneration. For example, ~40% of these blastema-enriched transcripts have not been found to have a significant match to known sequences [10]. This means that some subset of these unknown transcripts may be innovations in the salamander lineage, and a subset may represent ancestral genes that have been lost in other tetrapods or diverged to the point of being unrecognized. Although we can predict ORFs and in some cases protein domains, gaining more informative insights is more difficult without homologous gene information, and most all of these genes remain unexplored at the functional level. Future experimentation should produce exciting findings about whether these animals utilize some novel elements to enable their regenerative prowess. If so, activities for these putative salamander-specific gene products in a mammalian context should be considered.
Additionally, the field should further investigate the possibility that mammals have innate roadblocks to regeneration that salamanders either have overcome or simply do not possess. One identified example is the p16/ARF locus, which encodes two proteins that in mammals act in concert with Rb as tumor suppressors, but are very likely absent in salamanders [43]. In the absence of both Rb and p16/ARF activity, differentiated mammalian myocytes can be cued to re-enter the cell cycle and proliferate [43]. This work provides evidence that the approach of removing possible regenerative roadblocks, based on what is known about salamander genetics, is a viable way to improve regenerative responses in mammals. From the existing gene expression data, new hypotheses about what genes might need to be attenuated during regeneration can also be explored. For example, genes whose expression is specifically downregulated during the initiation stages might normally be repressing cell cycle re-entry (as in [44]) or dedifferentiation during homeostasis. Putative salamander “regeneration antagonizers” with mammalian orthologs can be investigated in the loss-of-function context in mice and other models to test the prediction that removing them might improve regeneration.
Concluding Remarks
Axolotls are a powerful model for peering into how a tetrapod limb can achieve full regeneration. The tools required to identify the strongest candidate drivers of this process and, importantly, to manipulate them and assess outcomes, have finally arrived. Through a combination of microarray and RNA-seq approaches, the field now has a solid foundation of transcriptional information on which to build. Several other approaches and extensions should provide an even more comprehensive understanding of the molecular factors enabling limb regeneration (See Outstanding Questions). Epigenetics is likely to play a role in cell state changes and cellular memories during regeneration, and technologies to probe this possibility now exist. Proteomic approaches will help answer questions concerning post-transcriptional output. Future RNA-seq studies are also likely to uncover roles for non-coding RNAs given the wide availability of strand-specific RNA-sequencing. RNA-seq at the single-cell level is destined to yield unprecedented resolution into the transcriptional changes that underly each phase of the regenerating limb, illuminating blastemal cell types, state transitions, and yield insights into critical interactions between cell types within the blastemal microenvironment. The next several years promise to be an extraordinarily exciting time to continue investigating the fascinating question of how salamanders regenerate limbs.
Outstanding Questions.
Are all signals acting locally, or are some systemic controls important for limb regeneration? Why do salamanders respond to amputation with blastema creation, while mammals usually do not? How much of the required response is linked to genes possibly unique to salamanders?
What is the relative contribution of stem cell activation versus dedifferentiation to blastema formation?
What is the cellular make-up of the blastema, and how might cells keep track of their lineages and potentials?
How do processes outside of transcription influence limb regeneration?
Do the principles of limb regeneration hold for other organs, and how do they manifest in other organisms, both regenerative and non-regenerative?
What are the limits to axolotl limb regeneration, and how might these help refine hypotheses about innate regenerative hurdles in mammals?
Can lessons learned from axolotl limb regeneration be translated into therapeutic approaches for regenerative medicine?
Trends.
The experimental toolset has now largely caught up to the interest in understanding limb regeneration, finally allowing for precise experimentation at a cellular and molecular level.
A huge amount of transcript data has emerged from which to gather clues about how limb regeneration occurs.
Differential gene expression analysis has enabled the identification of transcripts that are highly enriched, as well as highly repressed, in key tissues required for limb regeneration. These are prime starting points for hypothesis generation and functional experimentation.
Several genes whose involvement would not have been predicted by candidate gene approaches have now been implicated in limb regeneration, underscoring the need to take unbiased approaches to gene discovery.
De novo transcriptomes and reference tissue sequence data are important new resources for the field.
Acknowledgments
We thank NICHD (1R03HD083434 and 1DP2HD087953 to J.L.W) and NIAMS (1R03AR068126 to J.L.W.) for support and two anonymous reviewers for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spallanzani L. Reproductions of the legs in the aquatic salamander., in An Essay on Animal Reproductions [Prodromo di un opera da imprimersi sopra la riproduzioni anamali] Becket and de Hondt; London: 1769. pp. 68–72. [Italian: 1768] [Google Scholar]
- 2.Schreckenberg GM, Jacobson AG. Normal stages of development of the axolotl. Ambystoma mexicanum. Dev Biol. 1975;42(2):391–400. doi: 10.1016/0012-1606(75)90343-7. [DOI] [PubMed] [Google Scholar]
- 3.Bordzilovskaya NP, Dettlaff TA, Duhon ST, Malacinski GM. Developmental-stage series of axolotl embryos. In: Armstrong JB, Malacinski GM, editors. Developmental Biology of the Axolotl. Oxford University Press; New York: 1989. pp. 201–219. [Google Scholar]
- 4.Sobkow L, et al. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev Biol. 2006;290(2):386–97. doi: 10.1016/j.ydbio.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Flowers GP, et al. Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development. 2014;141(10):2165–71. doi: 10.1242/dev.105072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fei JF, et al. CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem Cell Reports. 2014;3(3):444–59. doi: 10.1016/j.stemcr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo TH, Kowalko JE, DiTommaso T, Nyambi M, Montoro DT, Essner JJ, Whited JL. TALEN-mediated gene editing of the thrombospondin-1 locus in axolotl. Regeneration. 2015;2(1):37–43. doi: 10.1002/reg2.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodcock MR, et al. Identification of Mutant Genes and Introgressed Tiger Salamander DNA in the Laboratory Axolotl, Ambystoma mexicanum. Sci Rep. 2017;7(1):6. doi: 10.1038/s41598-017-00059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whited JL, et al. Pseudotyped retroviruses for infecting axolotl in vivo and in vitro. Development. 2013;140(5):1137–46. doi: 10.1242/dev.087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant DM, et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017;18(3):762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka EM. The Molecular and Cellular Choreography of Appendage Regeneration. Cell. 2016;165(7):1598–608. doi: 10.1016/j.cell.2016.05.038. [DOI] [PubMed] [Google Scholar]
- 12.Campbell LJ, Crews CM. Wound epidermis formation and function in urodele amphibian limb regeneration. Cell Mol Life Sci. 2008;65(1):73–9. doi: 10.1007/s00018-007-7433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Both NJ. The developmental potencies of the regeneration blastema of the axolotl limb. Wilhelm Roux Arch Entwickl Mech Org. 1970;165(3):242–276. doi: 10.1007/BF01380787. [DOI] [PubMed] [Google Scholar]
- 14.Crawford K, Stocum DL. Retinoic acid proximalizes level-specific properties responsible for intercalary regeneration in axolotl limbs. Development. 1988;104(4):703–12. doi: 10.1242/dev.104.4.703. [DOI] [PubMed] [Google Scholar]
- 15.Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460(7251):60–5. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 16.Muneoka K, Fox WF, Bryant SV. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev Biol. 1986;116(1):256–60. doi: 10.1016/0012-1606(86)90062-x. [DOI] [PubMed] [Google Scholar]
- 17.Straus NA. Comparative DNA renaturation kinetics in amphibians. Proc Natl Acad Sci U S A. 1971;68(4):799–802. doi: 10.1073/pnas.68.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keinath MC, et al. Initial characterization of the large genome of the salamander Ambystoma mexicanum using shotgun and laser capture chromosome sequencing. Sci Rep. 2015;5:16413. doi: 10.1038/srep16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fankhauser G, Humphrey RR. Induction of triploidy and haploidy in axolotl eggs by cold treatment. Biological Bulletin. 1942;83(3):367–374. [Google Scholar]
- 20.Smith JJ, et al. Genic regions of a large salamander genome contain long introns and novel genes. BMC Genomics. 2009;10:19. doi: 10.1186/1471-2164-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voss SR, et al. Conserved vertebrate chromosome segments in the large salamander genome. Genetics. 2001;158(2):735–46. doi: 10.1093/genetics/158.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voss SR, et al. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res. 2011;21(8):1306–12. doi: 10.1101/gr.116491.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 24.Lam ET, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012;30(8):771–6. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng GX, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34(3):303–11. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuleshov V, Snyder MP, Batzoglou S. Genome assembly from synthetic long read clouds. Bioinformatics. 2016;32(12):i216–i224. doi: 10.1093/bioinformatics/btw267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillippy AM. New advances in sequence assembly. Genome Res. 2017;27(5):xi–xiii. doi: 10.1101/gr.223057.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habermann B, et al. An Ambystoma mexicanum EST sequencing project: analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries. Genome Biol. 2004;5(9):R67. doi: 10.1186/gb-2004-5-9-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putta S, et al. From biomedicine to natural history research: EST resources for ambystomatid salamanders. BMC Genomics. 2004;5(1):54. doi: 10.1186/1471-2164-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorsic M, Majdic G, Komel R. Identification of differentially expressed genes in 4-day axolotl limb blastema by suppression subtractive hybridization. J Physiol Biochem. 2008;64(1):37–50. doi: 10.1007/BF03168233. [DOI] [PubMed] [Google Scholar]
- 31.da Silva SM, Gates PB, Brockes JP. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev Cell. 2002;3(4):547–55. doi: 10.1016/s1534-5807(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 32.Shaikh N, Gates PB, Brockes JP. The Meis homeoprotein regulates the axolotl Prod 1 promoter during limb regeneration. Gene. 2011;484(1–2):69–74. doi: 10.1016/j.gene.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monaghan JR, et al. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol. 2009;7:1. doi: 10.1186/1741-7007-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Todd JT. On the process of reproduction of the members of the aquatic salamander. The Quarterly Journal of Science, Literature, and the Arts. 1823;16:84–96. [Google Scholar]
- 35.Kumar A, Brockes JP. Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 2012;35(11):691–9. doi: 10.1016/j.tins.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Campbell LJ, et al. Gene expression profile of the regeneration epithelium during axolotl limb regeneration. Dev Dyn. 2011;240(7):1826–40. doi: 10.1002/dvdy.22669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knapp D, et al. Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program. PLoS One. 2013;8(5):e61352. doi: 10.1371/journal.pone.0061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voss SR, et al. Gene expression during the first 28 days of axolotl limb regeneration I: Experimental design and global analysis of gene expression. Regeneration (Oxf) 2015;2(3):120–136. doi: 10.1002/reg2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tank PW, Carlson BM, Connelly TG. A staging system for forelimb regeneration in the axolotl, Ambystoma mexicanum. J Morphol. 1976;150(1):117–28. doi: 10.1002/jmor.1051500106. [DOI] [PubMed] [Google Scholar]
- 40.Stewart R, et al. Comparative RNA-seq analysis in the unsequenced axolotl: the oncogene burst highlights early gene expression in the blastema. PLoS Comput Biol. 2013;9(3):e1002936. doi: 10.1371/journal.pcbi.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomerantz JH, Blau HM. Tumor suppressors: enhancers or suppressors of regeneration? Development. 2013;140(12):2502–12. doi: 10.1242/dev.084210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang P, et al. Analysis of embryonic development in the unsequenced axolotl: Waves of transcriptomic upheaval and stability. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pajcini KV, et al. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7(2):198–213. doi: 10.1016/j.stem.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yun MH, Gates PB, Brockes JP. Regulation of p53 is critical for vertebrate limb regeneration. Proc Natl Acad Sci U S A. 2013;110(43):17392–7. doi: 10.1073/pnas.1310519110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guimond JC, et al. BMP-2 functions independently of SHH signaling and triggers cell condensation and apoptosis in regenerating axolotl limbs. BMC Dev Biol. 2010;10:15. doi: 10.1186/1471-213X-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang D,ZX, Zhao J, Zhou Y, Zhong C, Zhang J, Huang X. Subtractive screen of potential limb regeneration related genes from Pachytriton brevipes. Mol Biol Rep. 2014;41:1015–1026. doi: 10.1007/s11033-013-2946-z. [DOI] [PubMed] [Google Scholar]
- 47.Nacu E, et al. FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature. 2016;533(7603):407–10. doi: 10.1038/nature17972. [DOI] [PubMed] [Google Scholar]
- 48.Han MJ, An JY, Kim WS. Expression patterns of Fgf-8 during development and limb regeneration of the axolotl. Dev Dyn. 2001;220(1):40–8. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1085>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 49.Gardiner DM, et al. Regulation of HoxA expression in developing and regenerating axolotl limbs. Development. 1995;121(6):1731–41. doi: 10.1242/dev.121.6.1731. [DOI] [PubMed] [Google Scholar]
- 50.Carlson MR, et al. Expression of Hoxb13 and Hoxc10 in developing and regenerating Axolotl limbs and tails. Dev Biol. 2001;229(2):396–406. doi: 10.1006/dbio.2000.0104. [DOI] [PubMed] [Google Scholar]
- 51.Torok MA, et al. Expression of HoxD genes in developing and regenerating axolotl limbs. Dev Biol. 1998;200(2):225–33. doi: 10.1006/dbio.1998.8956. [DOI] [PubMed] [Google Scholar]
- 52.Levesque M, et al. Expression of heat-shock protein 70 during limb development and regeneration in the axolotl. Dev Dyn. 2005;233(4):1525–34. doi: 10.1002/dvdy.20458. [DOI] [PubMed] [Google Scholar]
- 53.Athippozhy A, et al. Characterization of transcriptional responses of dorsal root ganglia cultured in the presence and absence of blastema cells from regenerating salamander limbs. Regeneration (Oxford) 2014;1(2):1–10. doi: 10.1002/reg2.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimokawa T, et al. Lmx-1b and Wnt-7a expression in axolotl limb during development and regeneration. Okajimas Folia Anat Jpn. 2013;89(4):119–24. doi: 10.2535/ofaj.89.119. [DOI] [PubMed] [Google Scholar]
- 55.Sugiura T, et al. MARCKS-like protein is an initiating molecule in axolotl appendage regeneration. Nature. 2016;531(7593):237–40. doi: 10.1038/nature16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mercader N, Tanaka EM, Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132(18):4131–42. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- 57.Yang EV, et al. Expression of Mmp-9 and related matrix metalloproteinase genes during axolotl limb regeneration. Dev Dyn. 1999;216(1):2–9. doi: 10.1002/(SICI)1097-0177(199909)216:1<2::AID-DVDY2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 58.Koshiba K, et al. Expression of Msx genes in regenerating and developing limbs of axolotl. J Exp Zool. 1998;282(6):703–14. doi: 10.1002/(sici)1097-010x(19981215)282:6<703::aid-jez6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 59.Simon HG, et al. Differential expression of myogenic regulatory genes and Msx-1 during dedifferentiation and redifferentiation of regenerating amphibian limbs. Dev Dyn. 1995;202(1):1–12. doi: 10.1002/aja.1002020102. [DOI] [PubMed] [Google Scholar]
- 60.Crews L, et al. Expression and activity of the newt Msx-1 gene in relation to limb regeneration. Proc Biol Sci. 1995;259(1355):161–71. doi: 10.1098/rspb.1995.0024. [DOI] [PubMed] [Google Scholar]
- 61.Carlson MR, Bryant SV, Gardiner DM. Expression of Msx-2 during development, regeneration, and wound healing in axolotl limbs. J Exp Zool. 1998;282(6):715–23. doi: 10.1002/(sici)1097-010x(19981215)282:6<715::aid-jez7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 62.Farkas JE, et al. Neuregulin-1 signaling is essential for nerve-dependent axolotl limb regeneration. Development. 2016;143(15):2724–31. doi: 10.1242/dev.133363. [DOI] [PubMed] [Google Scholar]
- 63.Brockes JP, Kintner CR. Glial growth factor and nerve-dependent proliferation in the regeneration blastema of Urodele amphibians. Cell. 1986;45(2):301–6. doi: 10.1016/0092-8674(86)90394-6. [DOI] [PubMed] [Google Scholar]
- 64.Wang L, Marchionni MA, Tassava RA. Cloning and neuronal expression of a type III newt neuregulin and rescue of denervated, nerve-dependent newt limb blastemas by rhGGF2. J Neurobiol. 2000;43(2):150–8. doi: 10.1002/(sici)1097-4695(200005)43:2<150::aid-neu5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 65.Villiard E, et al. Urodele p53 tolerates amino acid changes found in p53 variants linked to human cancer. BMC Evol Biol. 2007;7:180. doi: 10.1186/1471-2148-7-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu W, et al. Activation of germline-specific genes is required for limb regeneration in the Mexican axolotl. Dev Biol. 2012;370(1):42–51. doi: 10.1016/j.ydbio.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Echeverri K, Tanaka EM. Proximodistal patterning during limb regeneration. Dev Biol. 2005;279(2):391–401. doi: 10.1016/j.ydbio.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 68.Roy S, Gardiner DM, Bryant SV. Vaccinia as a tool for functional analysis in regenerating limbs: ectopic expression of Shh. Dev Biol. 2000;218(2):199–205. doi: 10.1006/dbio.1999.9556. [DOI] [PubMed] [Google Scholar]
- 69.Roy S, Gardiner DM. Cyclopamine induces digit loss in regenerating axolotl limbs. J Exp Zool. 2002;293(2):186–90. doi: 10.1002/jez.10110. [DOI] [PubMed] [Google Scholar]
- 70.Denis JF, et al. Activation of Smad2 but not Smad3 is required to mediate TGF-beta signaling during axolotl limb regeneration. Development. 2016;143(19):3481–3490. doi: 10.1242/dev.131466. [DOI] [PubMed] [Google Scholar]
- 71.Khan P, Linkhart B, Simon HG. Different regulation of T-box genes Tbx4 and Tbx5 during limb development and limb regeneration. Dev Biol. 2002;250(2):383–92. [PubMed] [Google Scholar]
- 72.Shimokawa T, et al. Misexpression experiment of Tbx5 in axolotl (Ambystoma mexicanum) hindlimb blastema. Okajimas Folia Anat Jpn. 2013;89(4):113–8. doi: 10.2535/ofaj.89.113. [DOI] [PubMed] [Google Scholar]
- 73.Lévesque M,GS, Finnson K, Desmeules S, Villiard E, Pilote M, Philip A, Roy S. Transforming growth factor: beta signaling is essential for limb regeneration in axolotls. PLoS One. 2007;2(11) doi: 10.1371/journal.pone.0001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whited JL, et al. Dynamic expression of two thrombospondins during axolotl limb regeneration. Dev Dyn. 2011;240(5):1249–58. doi: 10.1002/dvdy.22548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whited JL, Lehoczky JA, Tabin CJ. Inducible genetic system for the axolotl. Proc Natl Acad Sci U S A. 2012;109(34):13662–7. doi: 10.1073/pnas.1211816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kragl M, et al. Muscle and connective tissue progenitor populations show distinct Twist1 and Twist3 expression profiles during axolotl limb regeneration. Dev Biol. 2013;373(1):196–204. doi: 10.1016/j.ydbio.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 77.Satoh A, Bryant SV, Gardiner DM. Regulation of dermal fibroblast dedifferentiation and redifferentiation during wound healing and limb regeneration in the Axolotl. Dev Growth Differ. 2008;50(9):743–54. doi: 10.1111/j.1440-169X.2008.01072.x. [DOI] [PubMed] [Google Scholar]
- 78.Ghosh S, et al. Analysis of the expression and function of Wnt-5a and Wnt-5b in developing and regenerating axolotl (Ambystoma mexicanum) limbs. Dev Growth Differ. 2008;50(4):289–97. doi: 10.1111/j.1440-169X.2008.01000.x. [DOI] [PubMed] [Google Scholar]
- 79.Kawakami Y, et al. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20(23):3232–7. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]


