Abstract
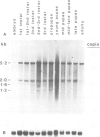
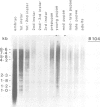
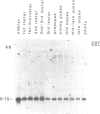
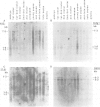
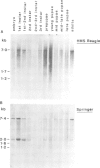
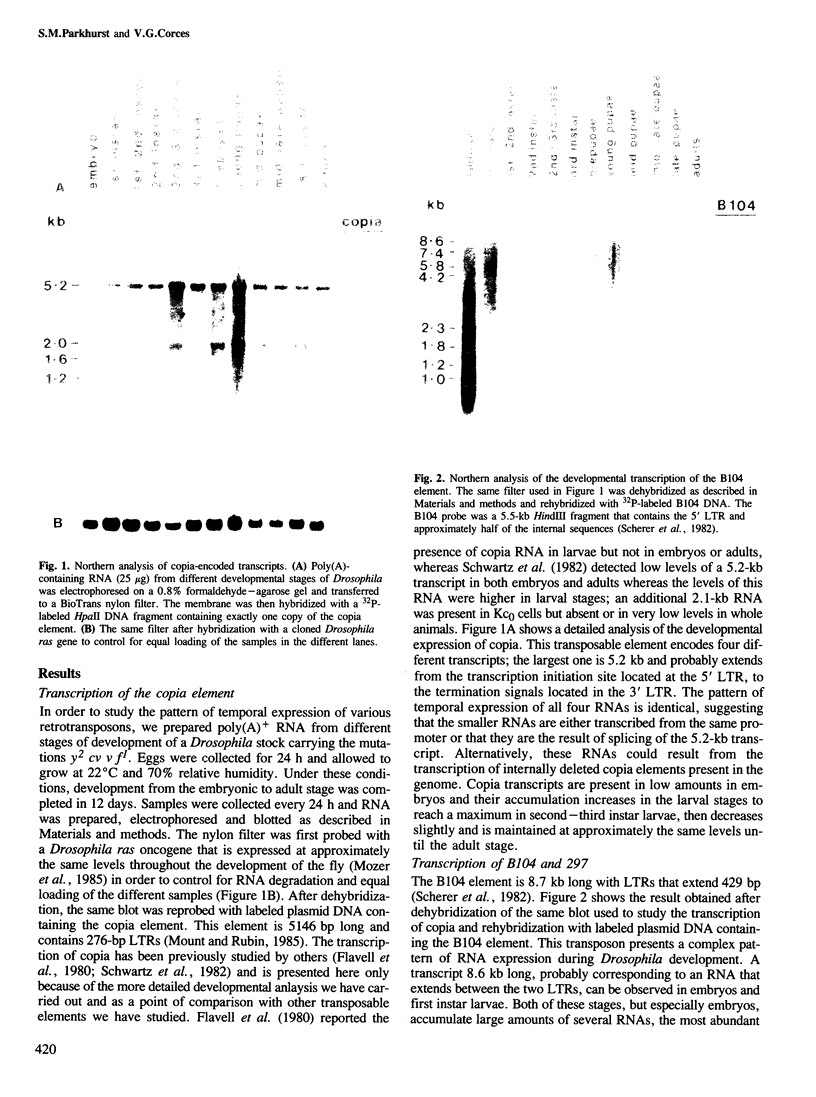
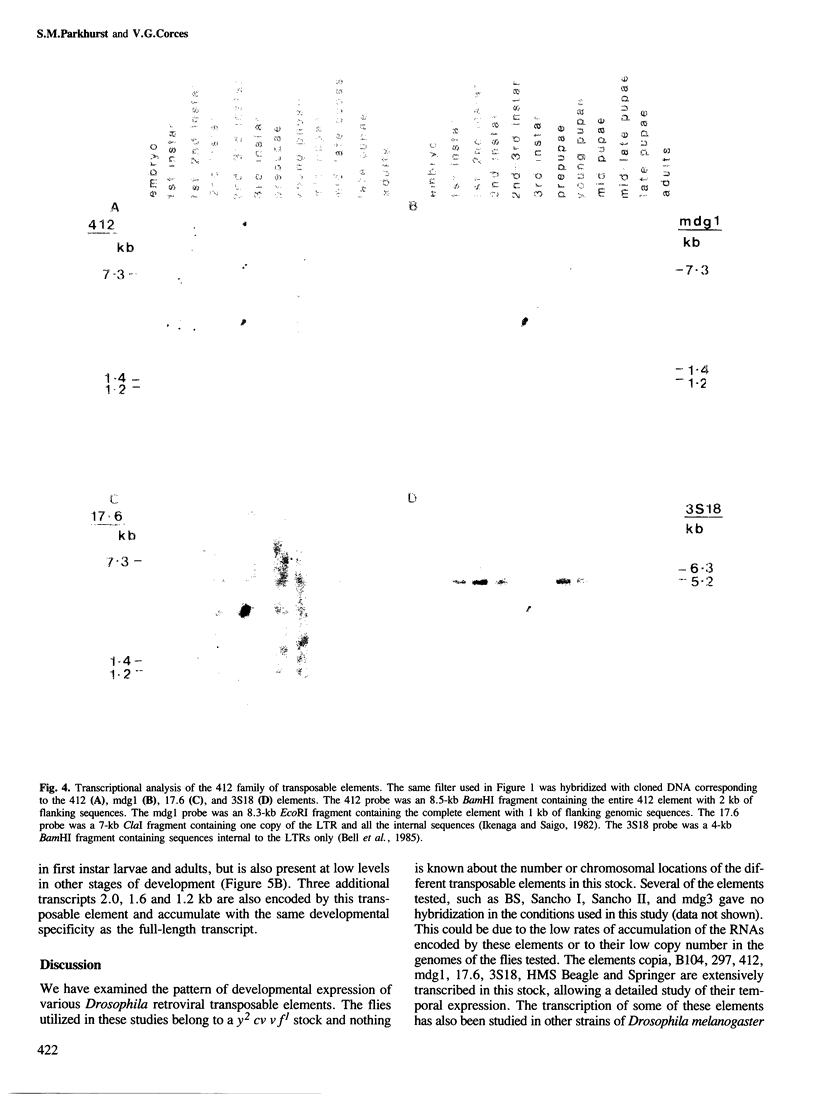
We have determined the pattern of temporal expression of several Drosophila retrovirus-like transposable elements. Some of these elements can be grouped into classes whose members show a similar profile of developmental transcription. The members of the 412 class, which includes 412, mdg1, 17.6 and 3S18, are transcribed mainly in the early larval and pupal stages of development, with small differences among the various members. HMS Beagle and Springer constitute another class where RNA accumulation in the larval stages is higher than in pupae and the adult flies accumulate more RNA than any other stage of development. Finally, the transcription of other elements such as copia, 297 and B104 follows a specific and individual pattern distinct from those described above. These results suggest the existence of evolutionary relationships among different transposable elements in Drosophila and the involvement of different cellular genes in the control of their expression.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arkhipova I. R., Mazo A. M., Cherkasova V. A., Gorelova T. V., Schuppe N. G., Llyin Y. V. The steps of reverse transcription of Drosophila mobile dispersed genetic elements and U3-R-U5 structure of their LTRs. Cell. 1986 Feb 28;44(4):555–563. doi: 10.1016/0092-8674(86)90265-5. [DOI] [PubMed] [Google Scholar]
- Bell J. R., Bogardus A. M., Schmidt T., Pellegrini M. A new copia-like transposable element found in a Drosophila rDNA gene unit. Nucleic Acids Res. 1985 Jun 11;13(11):3861–3871. doi: 10.1093/nar/13.11.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Carbonare B. D., Gehring W. J. Excision of copia element in a revertant of the white-apricot mutation of Drosophila melanogaster leaves behind one long-terminal repeat. Mol Gen Genet. 1985;199(1):1–6. doi: 10.1007/BF00327501. [DOI] [PubMed] [Google Scholar]
- Cullen B. R., Lomedico P. T., Ju G. Transcriptional interference in avian retroviruses--implications for the promoter insertion model of leukaemogenesis. Nature. 1984 Jan 19;307(5948):241–245. doi: 10.1038/307241a0. [DOI] [PubMed] [Google Scholar]
- Emerman M., Temin H. M. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell. 1984 Dec;39(3 Pt 2):449–467. [PubMed] [Google Scholar]
- Finnegan D. J., Fawcett D. H. Transposable elements in Drosophila melanogaster. Oxf Surv Eukaryot Genes. 1986;3:1–62. [PubMed] [Google Scholar]
- Flavell A. J. Role of reverse transcription in the generation of extrachromosomal copia mobile genetic elements. Nature. 1984 Aug 9;310(5977):514–516. doi: 10.1038/310514a0. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Ruby S. W., Toole J. J., Roberts B. E., Rubin G. M. Translation and developmental regulation of RNA encoded by the eukaryotic transposable element copia. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7107–7111. doi: 10.1073/pnas.77.12.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenaga H., Saigo K. Insertion of a movable genetic element, 297, into the T-A-T-A box for the H3 histone gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4143–4147. doi: 10.1073/pnas.79.13.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlik C. C., Fyrberg E. A. An insertion within a variably spliced Drosophila tropomyosin gene blocks accumulation of only one encoded isoform. Cell. 1985 May;41(1):57–66. doi: 10.1016/0092-8674(85)90061-3. [DOI] [PubMed] [Google Scholar]
- Kugimiya W., Ikenaga H., Saigo K. Close relationship between the long terminal repeats of avian leukosis-sarcoma virus and copia-like movable genetic elements of Drosophila. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3193–3197. doi: 10.1073/pnas.80.11.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor R. L., Parkhurst S. M., Corces V. G. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol. 1986 Apr;6(4):1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrokhi L. J., Obolenkova L. A., Priimägi A. F., Ilyin Y. V., Gerasimova T. I., Georgiev G. P. The nature of unstable insertion mutations and reversions in the locus cut of Drosophila melanogaster: molecular mechanism of transposition memory. EMBO J. 1985 Dec 30;4(13B):3781–3787. doi: 10.1002/j.1460-2075.1985.tb04148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossie K. G., Young M. W., Varmus H. E. Extrachromosomal DNA forms of copia-like transposable elements, F elements and middle repetitive DNA sequences in Drosophila melanogaster. Variation in cultured cells and embryos. J Mol Biol. 1985 Mar 5;182(1):31–43. doi: 10.1016/0022-2836(85)90025-7. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Rubin G. M. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol. 1985 Jul;5(7):1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozer B., Marlor R., Parkhurst S., Corces V. Characterization and developmental expression of a Drosophila ras oncogene. Mol Cell Biol. 1985 Apr;5(4):885–889. doi: 10.1128/mcb.5.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Corces V. G. Forked, gypsys, and suppressors in Drosophila. Cell. 1985 Jun;41(2):429–437. doi: 10.1016/s0092-8674(85)80016-7. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Corces V. G. Interactions among the gypsy transposable element and the yellow and the suppressor of hairy-wing loci in Drosophila melanogaster. Mol Cell Biol. 1986 Jan;6(1):47–53. doi: 10.1128/mcb.6.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Corces V. G. Retroviral elements and suppressor genes in Drosophila. Bioessays. 1986 Aug;5(2):52–57. doi: 10.1002/bies.950050203. [DOI] [PubMed] [Google Scholar]
- Scherer G., Tschudi C., Perera J., Delius H., Pirrotta V. B104, a new dispersed repeated gene family in Drosophila melanogaster and its analogies with retroviruses. J Mol Biol. 1982 May 25;157(3):435–451. doi: 10.1016/0022-2836(82)90470-3. [DOI] [PubMed] [Google Scholar]
- Schwartz H. E., Lockett T. J., Young M. W. Analysis of transcripts from two families of nomadic DNA. J Mol Biol. 1982 May 5;157(1):49–68. doi: 10.1016/0022-2836(82)90512-5. [DOI] [PubMed] [Google Scholar]
- Snyder M. P., Kimbrell D., Hunkapiller M., Hill R., Fristrom J., Davidson N. A transposable element that splits the promoter region inactivates a Drosophila cuticle protein gene. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7430–7434. doi: 10.1073/pnas.79.23.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B. M., Bayev A. A., Finnegan D. J. Nucleotide sequence of terminal repeats of 412 transposable elements of Drosophila melanogaster. A similarity to proviral long terminal repeats and its implications for the mechanism of transposition. J Mol Biol. 1981 Dec 25;153(4):897–915. doi: 10.1016/0022-2836(81)90458-7. [DOI] [PubMed] [Google Scholar]
- Winston F., Durbin K. J., Fink G. R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984 Dec;39(3 Pt 2):675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]