Abstract
Genome-wide association studies (GWAS) have identified hundreds of cardiometabolic disease (CMD) risk loci. However, they contribute little to genetic variance, and most downstream gene-regulatory mechanisms are unknown. We genotyped and RNA-sequenced vascular and metabolic tissues from 600 coronary artery disease patients in the STARNET study. Gene expression traits associated with CMD risk SNPs identified by GWAS were more extensively found in STARNET than in tissue- and disease-unspecific gene-tissue expression studies, indicating sharing of downstream cis-/trans-gene regulation across tissues and CMDs. In contrast, the regulatory effects of other GWAS risk SNPs were tissue-specific; abdominal fat emerged as an important gene-regulatory site for blood lipids, such as for the LDL-cholesterol and coronary artery disease risk-gene PCSK9. STARNET provides insights into gene-regulatory mechanisms for CMD risk loci, facilitating their translation into opportunities for diagnosis, therapy and prevention.
In 2012, cardiovascular disease accounted for 17.5 million deaths, nearly one-third of all deaths worldwide, and >80% (14.1 million) were from coronary artery disease (CAD) and stroke. CAD is preceded by cardiometabolic diseases (CMDs) such as hypertension, impaired lipid and glucose metabolism, and systemic inflammation (1, 2). Genome-wide association studies (GWAS) have identified hundreds of DNA variants associated with risk for CAD (3), hypertension (4), blood lipid levels (5), markers of plasma glucose metabolism (6–10), type 2 diabetes (6, 11), body mass index (12), rheumatoid arthritis (13), systemic lupus erythematosus (14), ulcerative colitis (15) and Crohn’s disease (16). However, identifying susceptibility genes responsible for these loci has proven difficult.
GWAS loci typically span large, noncoding, intergenic regions with numerous single-nucleotide polymorphisms (SNPs) in strong linkage disequilibrium. These regions are enriched in cis-regulatory elements (17) and expression quantitative trait loci (eQTLs) (18–20), suggesting that gene regulation is the principal mechanism by which risk loci affect complex disease etiology. However, it is largely unknown whether this gene-regulatory effect includes one or several genes acting in one or multiple tissues and whether risk loci for different diseases share cis- and trans-gene regulation. A better understanding of gene regulation may also shed light on why known GWAS risk loci explain only ~10% of expected heritable variance in CMD risk (21). Possibly, multiple risk loci, acting through common cis- and trans-genes, contribute synergistically to heritability (22, 23).
In the Stockholm-Tartu Atherosclerosis Reverse Networks Engineering Task study (STARNET) (fig. S1), we recruited 600 well-characterized (table S1, fig. S2) CAD patients, genotyped DNA (6,245,505 DNA variant calls with minor allele frequency >5%, fig. S3), and sequenced RNA isolated from blood, atherosclerotic-lesion-free internal mammary artery (MAM), atherosclerotic aortic root (AOR), subcutaneous fat (SF), visceral abdominal fat (VAF), skeletal muscle (SKLM), and liver (LIV) (15–30 million reads per sample, figs. S4–S11, table S2).
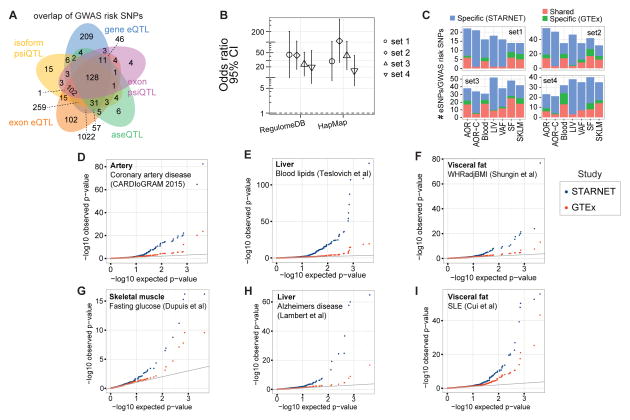
In total, ~8 million cis-eQTLs were identified, and nearly half were unique SNP-gene pairs (figs. S12–S26, tables S3–S7). The STARNET cis-eQTLs were enriched in genetic associations established by GWAS for CAD, CMDs and Alzheimer’s disease (AD) (3–16, 24) (figs. S27–S33) and were further enriched after epigenetic filtering (figs. S34–S39). Of 3,326 genome-wide significant risk SNPs identified by GWAS to date (25), 2,047 (61%) had a matching cis-QTL in STARNET (Fig. 1A). Of the 54 lead risk SNPs verified in meta-analyses of CAD GWAS (3), 38 cis-eQTLs with a regulatory trait concordance score (RTC) >0.9 and at least one candidate gene were identified in STARNET (table S8, fig. S27). Compared to large datasets of cis-eQTL isolated only from blood, cis-eQTLs across all tissues in STARNET matched >10-fold more CAD and CMD-related GWAS risk SNPs (Fig. 1B). STARNET cis-eQTLs isolated from CAD-affected tissues also matched several-fold more CAD and CMD-related GWAS risk SNPs than cis-eQTLs from corresponding tissues isolated from predominantly healthy individuals in GTEx (18) (Fig. 1C). Thus, not all gene-regulatory effects of disease risk SNPs are identifiable in blood or healthy tissues. This notion was further underscored by comparing the statistical significances of cis-eQTLs for GWAS risk SNPs in STARNET with corresponding associations in GTEx (Fig. 1D). In STARNET, gene fusions (table S9) and CAD-related loss of function mutations (table S10) were also detected.
Fig. 1. QTLs and disease-associated risk SNPs identified by GWAS.
(A) Venn diagram showing 2,047/3,326 disease-associated risk SNPs from the NHGRI GWAS catalog overlapping with at least one form of STARNET e/psi/aseQTLs. (B) Odds ratios that STARNET eQTLs coincide with CAD-associated risk SNPs (Set 1, CARDIoGRAM-C4D, n=53; Set 2, CARDIoGRAM extended, n=150) (3), blood lipids (Set 3, n=35) (5), and metabolic traits (Set 4, n=132) (6, 8, 10, 12) versus blood eQTLs from RegulomeDB and HapMap. The y-axis shows odds ratios. Error bars, 95% confidence intervals. (C) Stacked bar plots comparing tissue-specific eQTLs from STARNET and GTEx (18) coinciding with disease-associated risk SNPs in the same Sets 1–4 as in (B). (D–I). Q-Q plots showing associations of tissue-specific STARNET (blue) and GTEx (18) (red) cis-eQTLs of disease-associated risk SNPs identified by GWAS for CAD (3) (D), blood lipids (5) (E), waist-hip ratio (12) (F), fasting glucose (6) (G), AD (24) (H), and SLE (14) (I).
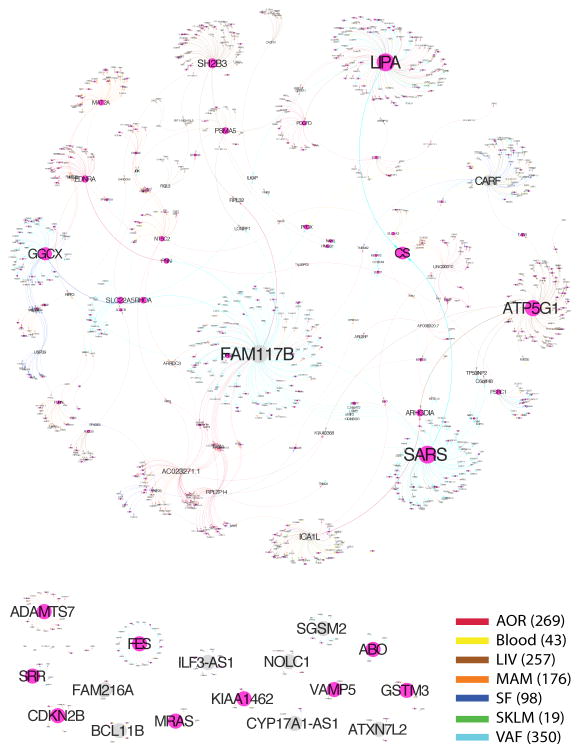
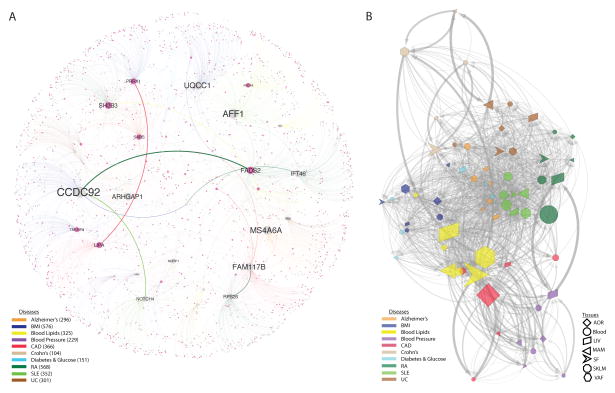
The cis effects of disease-associated risk loci identified by GWAS are central for understanding downstream molecular mechanisms of disease. However, these cis-genes likely also affect downstream trans-genes. To identify possible trans effects, we ran a targeted analysis to call both cis- and trans-genes for lead risk SNPs identified by GWAS. After assigning cis-eQTLs for 562 risk SNPs for CAD, CMDs and AD (3–16, 24), we used a causal inference test (26) to conservatively call causal correlations between the cis-genes and trans-genes by assessing the probability that an interaction was causal (SNP→cis-gene→trans-gene, false discovery rate [FDR]<1%) and not reactive (SNP→trans-gene→cis-gene, P>0.05) (26) (table S11). We found extensive sharing of cis- and trans-gene regulation by GWAS risk loci across tissues and CMDs. In CAD, 28 risk loci with at least one causal interaction (FDR <1%, P>0.05) had a total of 51 cis-genes and 1040 trans-genes. Of these, 26 risk loci, 37 cis-genes (including 27 key drivers (27)), and 994 trans-genes were connected in a main CAD regulatory gene network acting across all 7 tissues (Fig. 2). The trans-genes in this network were enriched with genes previously associated with CAD and atherosclerosis (Fisher’s test, 1.54-fold, P=8E-10, table S11). Sharing of cis/trans-genes downstream of complex disease risk loci also emerged for other CMDs and AD (3–16, 24) (fig. S40). In fact, we identified 33 cis-genes regulated by risk SNPs across all CMDs, including CAD and AD, acting as key drivers in a pan-disease cis/trans-gene regulatory network (Fig. 3A).
Fig. 2. A cis/trans gene-regulatory network of CAD risk SNPs.
A main gene-regulatory network of cis-and trans-genes associated with 21/46 index SNPs for risk loci identified for CAD by meta-analysis in the CARDIoGRAM GWAS of CAD (3) inferred using a causal inference test (26).
Fig. 3. Cis and trans gene regulation across CMDs and Alzheimer’s disease.
(A) A pan-disease risk SNP cis/trans-gene regulatory network. Thirty-six top key disease drivers, including 33 cis-genes for risk SNPs identified for CMDs including CAD and AD by GWAS (3–16, 24) were identified as having >100 downstream genes in any disease-specific network or belonging to the top 5 key drivers in the main regulatory gene network for each disease (table S11). Node (gene) and edge color indicate disease belonging. Edge thickness represents how frequent an edge is the shortest path between all pairs of network nodes. Node size reflects the number of downstream nodes in the network. RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; UC, ulcerative colitis. (B) cis and trans gene regulation across disease/tissue pairs. Nodes represent unique disease-tissue pairs. Edges occur when a cis-gene in one node have downstream trans-genes present also in another node. Edge thickness defined as in (A). Node size reflects its centrality in the network: The position of the nodes in the network (i.e., layout) was derived from an edge weighted spring layout algorithm. The “weight” is defined as the number of trans genes that have a connection from the upstream node’s cis genes, normalized by the total number of trans genes between two connecting nodes — resulting in that highly connected nodes are positioned in the center of the network.
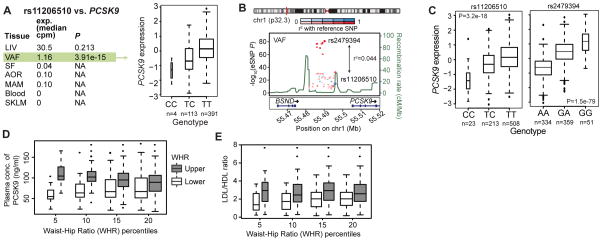
Among CMDs, cis/trans-genes of GWAS risk SNPs for blood lipid levels (5) emerged as central (Fig. 3B) where tissue-specific down-stream effects were beside LIV (46 cis- and 150 trans-genes) observed in the fat tissues (SF; 45 cis- and 372 trans-genes: VAF; 38 cis- and 465 trans-genes) (fig. S41, table S11). Visceral abdominal fat examples included ABCA8/ABCA5 (rs4148008) associated with 36 downstream trans-genes in VAF and HDL; EVI5 (rs7515577) associated with 32 VAF trans-genes and total cholesterol; and STARD3 (rs11869286) associated with 7 VAF trans-genes and HDL. In addition, the cis-gene TMEM258 (rs174546) with 22 trans-genes in abdominal fat surfaced as a parallel/alternative regulatory site of plasma LDL to the proposed FADS-1,2,3 in LIV (5) (fig. S41). Other risk SNPs with VAF-specific cis-genes had few or even no trans-genes (fig. S41). For example, two risk SNPs—rs11206510 for CAD and rs12046679 for LDL cholesterol level (3, 5)—regulate PCSK9 in VAF, not in LIV (Fig. 4A, B). The VAF-specificity of these eQTLs PCSK9 in were confirmed in an independent gene expression dataset from morbidly obese patients (28) (Fig. 4C, fig. S30) suggesting that PCSK9 is secreted from VAF into the portal vein to affect hepatic LDL receptor degradation, LDL plasma levels and risk for CAD (29). Interestingly and as previously suggested (30), we observed that STARNET patients in the upper, compared to the lower, 5th–20th percentiles of waist–hip ratio, (i.e., patients with and without “male fat”) had higher levels of circulating PCSK9 (Fig. 4D) and LDL/HDL ratio (Fig. 4E).
Fig. 4. PCSK9 regulation in VAF, not LIV, increases risk for elevated LDL/HDL ratio.
(A) PCSK9 was expressed in STARNET LIV and VAF but only associated with the CAD risk SNP rs11206510 in VAF (FDR<0.001). Box plot of allelic PCSK9 expression of the CAD risk SNP rs11206510 showing dosage effect of the T allele (P=3.91e-15; FDR=4e-04). (B) Regional plot of the PCSK9 locus. rs2479394, linked to plasma LDL levels by GWAS (5), acts independently of rs11206510 as the lead eQTL of PCSK9 expression in VAF. rs2479394 was not an eQTL of PCSK9 in STARNET LIV. (C) Box plots of allelic PCSK9 expression in VAF of rs11206510 and rs2479394 in a gene-tissue expression study of morbidly obese patients (fig. S29) (28). Box plots of PCSK9 levels (D) and ratios of LDL/HDL (E) in plasma isolated from the STARNET patients within the upper and lower 5th–20th percentile of waist-hip ratio (WHR) (PCSK9; 5th, P=8.0e-11; 10th, P=1.9e-11; 15th, P=5.9e-05; 20th, P=0.004: LDL/HDL ratio; 5th, P=0,007; 10th, P=0.001; 15th P=0.0005; 20th, P=0.0009.
STARNET provides new insights into tissue-specific gene-regulatory effects of disease-associated risk SNPs identified by GWAS, as exemplified by abdominal fat for blood lipids, and will be a complementary resource for exploring GWAS findings moving forward. Furthermore, STARNET also revealed unexpected sharing of cis- and trans-genes downstream of risk loci for CMDs across both tissues and diseases. We anticipate that the identified cis/trans-gene regulatory networks will help elucidate the complex downstream effects of risk loci for common complex diseases, including possible epistatic effects that could shed light on the missing heritability of CMD risk. Given the detailed phenotypic data on STARNET patients, we can begin to identify how genetic variability interacts with environmental perturbations across tissues to cause pathophysiological alterations and complex diseases.
Supplementary Material
Acknowledgments
The STARNET study was supported by the University of Tartu (SP1GVARENG (JLMB)), the Estonian Research Council (ETF grant #8853 (AR and JLMB)), the Astra-Zeneca Translational Science Centre-Karolinska Institutet (a joint research program in translational science, (JLMB)), Clinical Gene Networks AB (CGN) as an SME of the FP6/FP7 EU-funded integrated project CVgenes@target (HEALTH-F2-2013-601456), the Leducq transatlantic networks; CAD Genomics (CG, EES and JLMB) and Sphingonet (CB), the Torsten and Ragnar Söderberg Foundation (CB), the Knut and Alice Wallenberg Foundation (CB), the American Heart Association (A14SFRN20840000, JK, EES and JLMB), the National Institutes of Health (NIH NHLBI, R01HL125863, JLMB; NIH NHLBI R01HL71207, EES; R01AG050986, Roussos; NIH NHLBI K23HL111339, CG; NIH NHLBI K08HL111330, JK) and the Veterans Affairs (Merit grant BX002395, PR). The DNA genotyping and RNA sequencing were in part performed by the SNP&SEQ technology platform at Science for Life, the National Genomics Infrastructure (NGI) in Uppsala and Stockholm supported by Swedish Research Council (VR-RF1), Knut and Alice Wallenberg Foundation and UPPMAX. CGN has financially contributed to the STARNET study. JLMB is the founder and chairman of CGN. JLMB, EES and AR are on the board of directors for CGN. JLMB, TM and AR own equity in CGN and receive financial compensation from CGN. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. The STARNET data is accessible through dbGAP.
Footnotes
Supplementary online materials
Material and Methods Figures S1–S41 Tables S1–S11
References and Notes
- 1.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 2.Shulman GI. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N Engl J Med. 2014;371:2237–2238. doi: 10.1056/NEJMc1412427. §. [DOI] [PubMed] [Google Scholar]
- 3.Nikpay M, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soranzo N, et al. Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strawbridge RJ, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning AK, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An P, et al. Genome-wide association study identifies common loci influencing circulating glycated hemoglobin (HbA1c) levels in non-diabetic subjects: The Long Life Family Study (LLFS) Metabolism. 2014;63:461–468. doi: 10.1016/j.metabol.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Y, Sheng Y, Zhang X. Genetic susceptibility to SLE: Recent progress from GWAS. J Autoimmun. 2013;41:25–33. doi: 10.1016/j.jaut.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foroughi Asl H, et al. Expression quantitative trait Loci acting across multiple tissues are enriched in inherited risk for coronary artery disease. Circ Cardiovasc Genet. 2015;8:305–315. doi: 10.1161/CIRCGENETICS.114.000640. [DOI] [PubMed] [Google Scholar]
- 20.Talukdar HA, et al. Cross-Tissue Regulatory Gene Networks in Coronary Artery Disease. Cell Syst. 2016;2:196–208. doi: 10.1016/j.cels.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei WH, Hemani G, Haley CS. Detecting epistasis in human complex traits. Nat Rev Genet. 2014;15:722–733. doi: 10.1038/nrg3747. [DOI] [PubMed] [Google Scholar]
- 23.Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. doi: 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang IM, et al. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol Syst Biol. 2012;8:594. doi: 10.1038/msb.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenawalt DM, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leander K, et al. Circulating Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) Predicts Future Risk of Cardiovascular Events Independently of Established Risk Factors. Circulation. 2016;133:1230–1239. doi: 10.1161/CIRCULATIONAHA.115.018531. [DOI] [PubMed] [Google Scholar]
- 30.Yusuf S, et al. Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. Lancet. 2005;366:1640–1649. doi: 10.1016/S0140-6736(05)67663-5. [DOI] [PubMed] [Google Scholar]
- 31.Serruys PW, et al. Assessment of the SYNTAX score in the Syntax study. EuroIntervention. 2009;5:50–56. doi: 10.4244/eijv5i1a9. [DOI] [PubMed] [Google Scholar]
- 32.Sianos G, et al. The SYNTAX Score: an angiographic tool grading the complexity of coronary artery disease. EuroIntervention. 2005;1:219–227. [PubMed] [Google Scholar]
- 33.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otsuka F, Yahagi K, Sakakura K, Virmani R. Why is the mammary artery so special and what protects it from atherosclerosis? Ann Cardiothorac Surg. 2013;2:519–526. doi: 10.3978/j.issn.2225-319X.2013.07.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adler Y, et al. Spiral computed tomography evidence of close correlation between coronary and thoracic aorta calcifications. Atherosclerosis. 2004;176:133–138. doi: 10.1016/j.atherosclerosis.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 36.Björkegren JLM, et al. Plasma cholesterol-induced lesion networks activated before regression of early, mature, and advanced atherosclerosis. PLoS Genet. 2014;10:e1004201. doi: 10.1371/journal.pgen.1004201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hägg S, et al. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm Atherosclerosis Gene Expression (STAGE) study. PLoS Genet. 2009;5:e1000754. doi: 10.1371/journal.pgen.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunderson KL, et al. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howie B, et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 44.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-Seq data. BMC Bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.‘t Hoen PAC, et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31:1015–1022. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stranger BE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Statist. 2001;29:1165–1188. [Google Scholar]
- 62.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simas AB, Barreto-Souza W, Rocha AV. Improved estimators for a general class of beta regression models. Comput Stat Data Anal. 2010;54:348–366. [Google Scholar]
- 64.Smithson M, Verkuilen J. A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychol Methods. 2006;11:54–71. doi: 10.1037/1082-989X.11.1.54. [DOI] [PubMed] [Google Scholar]
- 65.Foissac S, Sammeth M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007;35:W297–W299. doi: 10.1093/nar/gkm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sims D, et al. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 67.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Flutre T, Wen X, Pritchard J, Stephens M. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 2013;9:e1003486. doi: 10.1371/journal.pgen.1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roussos P, et al. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katoh K, Misawa K, Kuma K-i, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.The CARDIoGRAMplusC4D Consortium. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 78.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strawbridge RJ, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 82.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 83.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.C. Roadmap Epigenomics et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schork AJ, et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roussos P, et al. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deloukas P, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Strawbridge RJ, et al. Genome-Wide Association Identifies Nine Common Variants Associated With Fasting Proinsulin Levels and Provides New Insights Into the Pathophysiology of Type 2 Diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 92.Smyth GK, Phipson B. Permutation P-values should never be zero: calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol. 2010;9 doi: 10.2202/1544-6115.1585. [DOI] [PubMed] [Google Scholar]
- 93.Binns D, et al. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Skogsberg J, et al. Transcriptional Profiling Uncovers a Network of Cholesterol-Responsive Atherosclerosis Target Genes. PLoS Genet. 2008;4:e1000036. doi: 10.1371/journal.pgen.1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Osborne JD, et al. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10:S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lovely RS, et al. Assessment of genetic determinants of the association of γ′ fibrinogen in relation to cardiovascular disease. Atertio Thromb Vasc Biol. 2011;31:2345–2352. doi: 10.1161/ATVBAHA.111.232710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.