Abstract
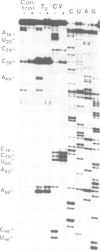
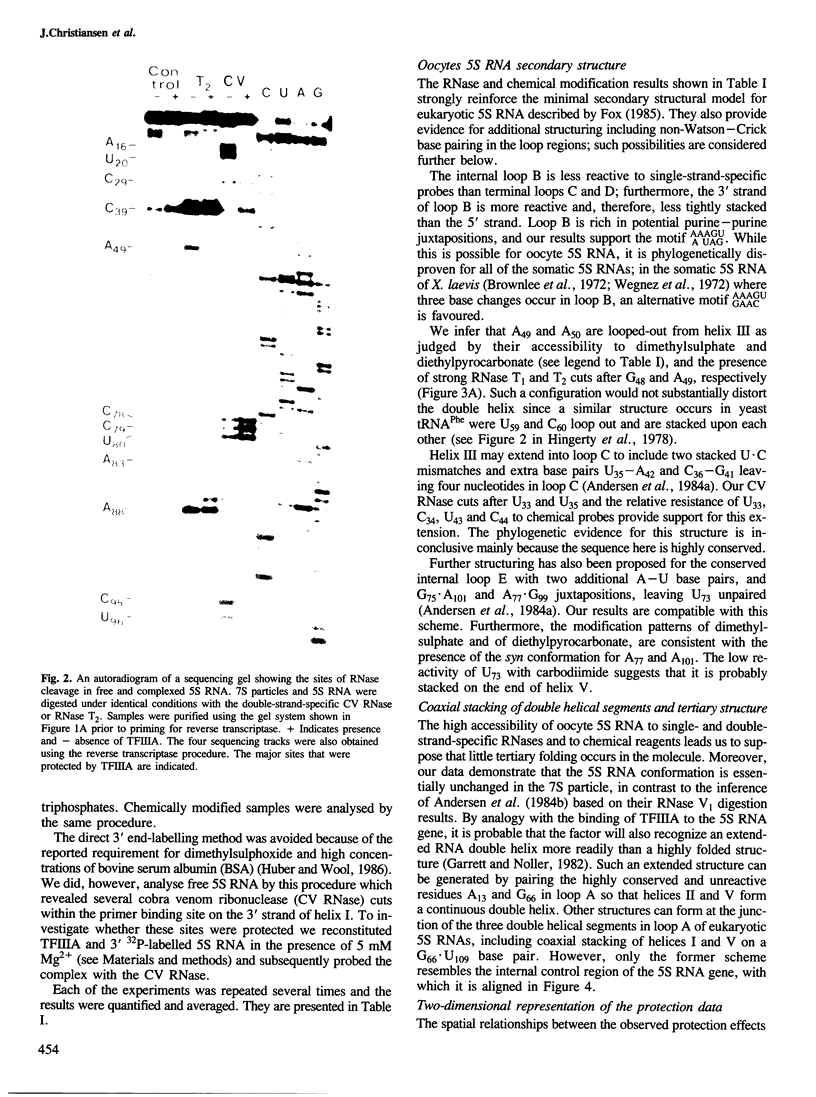
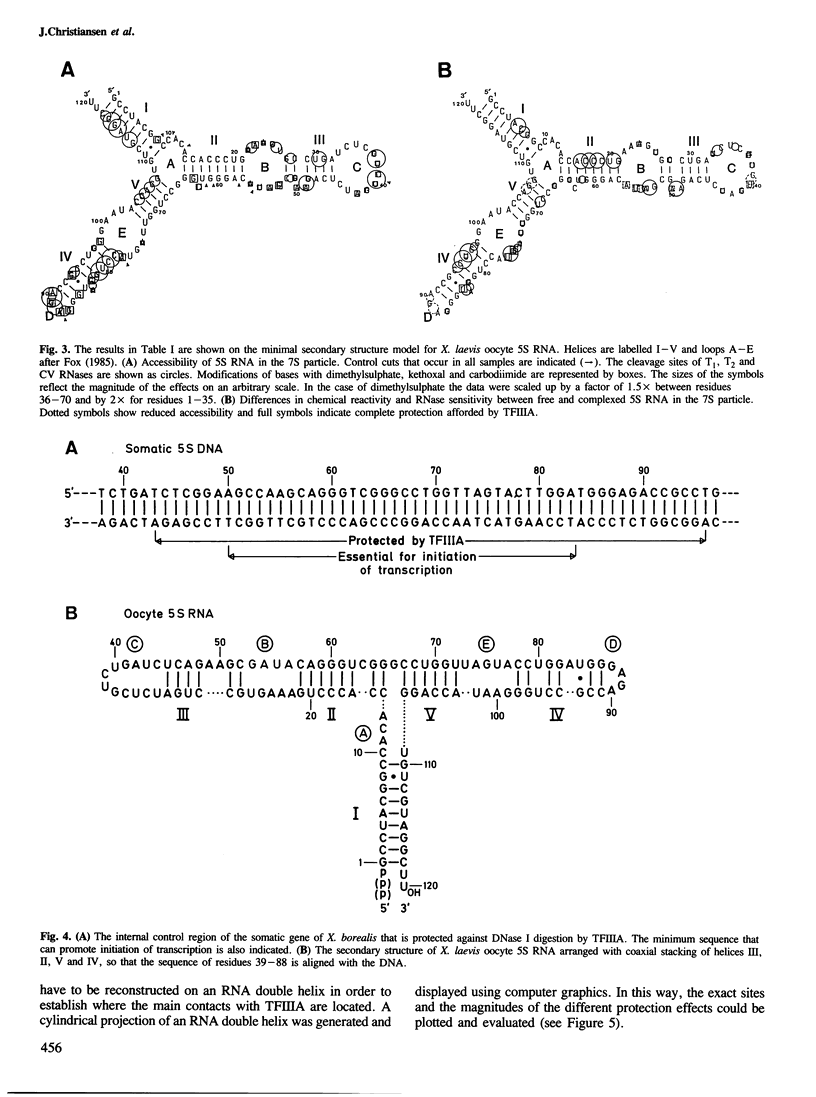
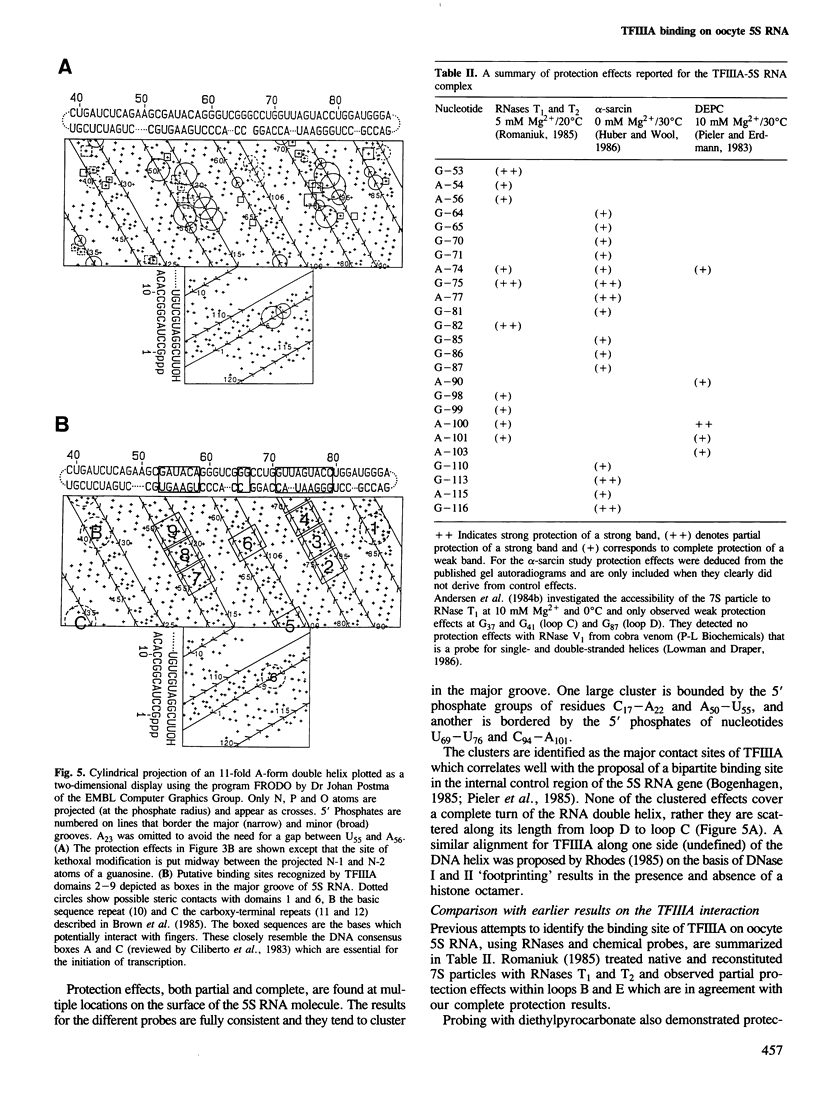
RNases and chemical probes were used to study the accessibility of each nucleotide of 5S RNA in the native and reconstituted 7S particle from Xenopus laevis oocytes. RNase or chemically treated 5S RNA from intact 7S particles was isolated and analysed using an oligodeoxynucleotide primer and reverse transcriptase. The results were superimposed on a cylindrical projection of an RNA double helix and the protection effects were shown to cluster at two regions on the molecular surface. A three-dimensional model is proposed for the 7S particle in which protein-RNA contacts occur mainly in the major groove of 5S RNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen J., Delihas N., Hanas J. S., Wu C. W. 5S RNA structure and interaction with transcription factor A. 1. Ribonuclease probe of the structure of 5S RNA from Xenopus laevis oocytes. Biochemistry. 1984 Nov 20;23(24):5752–5759. doi: 10.1021/bi00319a013. [DOI] [PubMed] [Google Scholar]
- Andersen J., Delihas N., Hanas J. S., Wu C. W. 5S RNA structure and interaction with transcription factor A. 2. Ribonuclease probe of the 7S particle from Xenopus laevis immature oocytes and RNA exchange properties of the 7S particle. Biochemistry. 1984 Nov 20;23(24):5759–5766. doi: 10.1021/bi00319a014. [DOI] [PubMed] [Google Scholar]
- Arnott S. Major groove or minor groove? 1986 Mar 27-Apr 2Nature. 320(6060):313–313. doi: 10.1038/320313a0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F. The intragenic control region of the Xenopus 5 S RNA gene contains two factor A binding domains that must be aligned properly for efficient transcription initiation. J Biol Chem. 1985 May 25;260(10):6466–6471. [PubMed] [Google Scholar]
- Brown R. S., Sander C., Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985 Jul 8;186(2):271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Cartwright E., McShane T., Williamson R. The nucleotide sequence of somatic 5 S RNA from Xenopus laevis. FEBS Lett. 1972 Sep 1;25(1):8–12. doi: 10.1016/0014-5793(72)80442-3. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. Gene regulation. Fingers and DNA half-turns. Nature. 1986 Aug 14;322(6080):597–598. doi: 10.1038/322597a0. [DOI] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Huber P. W., Wool I. G. Identification of the binding site on 5S rRNA for the transcription factor IIIA: proposed structure of a common binding site on 5S rRNA and on the gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1593–1597. doi: 10.1073/pnas.83.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Ladiges W. C., Raff R. F., Brown S., Deeg H. J., Storb R. The canine major histocompatibility complex. Supertypic specificities defined by the primed lymphocyte test (PLT). Immunogenetics. 1984;19(4):359–365. doi: 10.1007/BF00345410. [DOI] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- McCall M., Brown T., Hunter W. N., Kennard O. The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA. Nature. 1986 Aug 14;322(6080):661–664. doi: 10.1038/322661a0. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B., Wegnez M. Isolation of a 7S particle from Xenopus laevis oocytes: a 5S RNA-protein complex. Proc Natl Acad Sci U S A. 1979 Jan;76(1):241–245. doi: 10.1073/pnas.76.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Erdmann V. A. Isolation and characterization of a 7 S RNP particle from mature Xenopus laevis oocytes. FEBS Lett. 1983 Jul 4;157(2):283–287. doi: 10.1016/0014-5793(83)80562-6. [DOI] [PubMed] [Google Scholar]
- Pieler T., Oei S. L., Hamm J., Engelke U., Erdmann V. A. Functional domains of the Xenopus laevis 5S gene promoter. EMBO J. 1985 Dec 30;4(13B):3751–3756. doi: 10.1002/j.1460-2075.1985.tb04144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J. Characterization of the RNA binding properties of transcription factor IIIA of Xenopus laevis oocytes. Nucleic Acids Res. 1985 Jul 25;13(14):5369–5387. doi: 10.1093/nar/13.14.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D., Engelke D., Ng S. Y., Shastry B. S., Roeder R. G. The binding of a transcription factor to deletion mutants of a 5S ribosomal RNA gene. Cell. 1981 Mar;23(3):665–669. doi: 10.1016/0092-8674(81)90429-3. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Sproat B. S., Brown D. M. A new linkage for solid phase synthesis of oligodeoxyribonucleotides. Nucleic Acids Res. 1985 Apr 25;13(8):2979–2987. doi: 10.1093/nar/13.8.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegnez M., Monier R., Denis H. Sequence heterogeneity of 5 S RNA in Xenopus laevis. FEBS Lett. 1972 Sep 1;25(1):13–20. doi: 10.1016/0014-5793(72)80443-5. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Zueva V. S., Mankin A. S., Bogdanov A. A., Baratova L. A. Specific fragmentation of tRNA and rRNA at a 7-methylguanine residue in the presence of methylated carrier RNA. Eur J Biochem. 1985 Feb 1;146(3):679–687. doi: 10.1111/j.1432-1033.1985.tb08704.x. [DOI] [PubMed] [Google Scholar]