Abstract
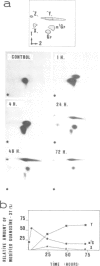
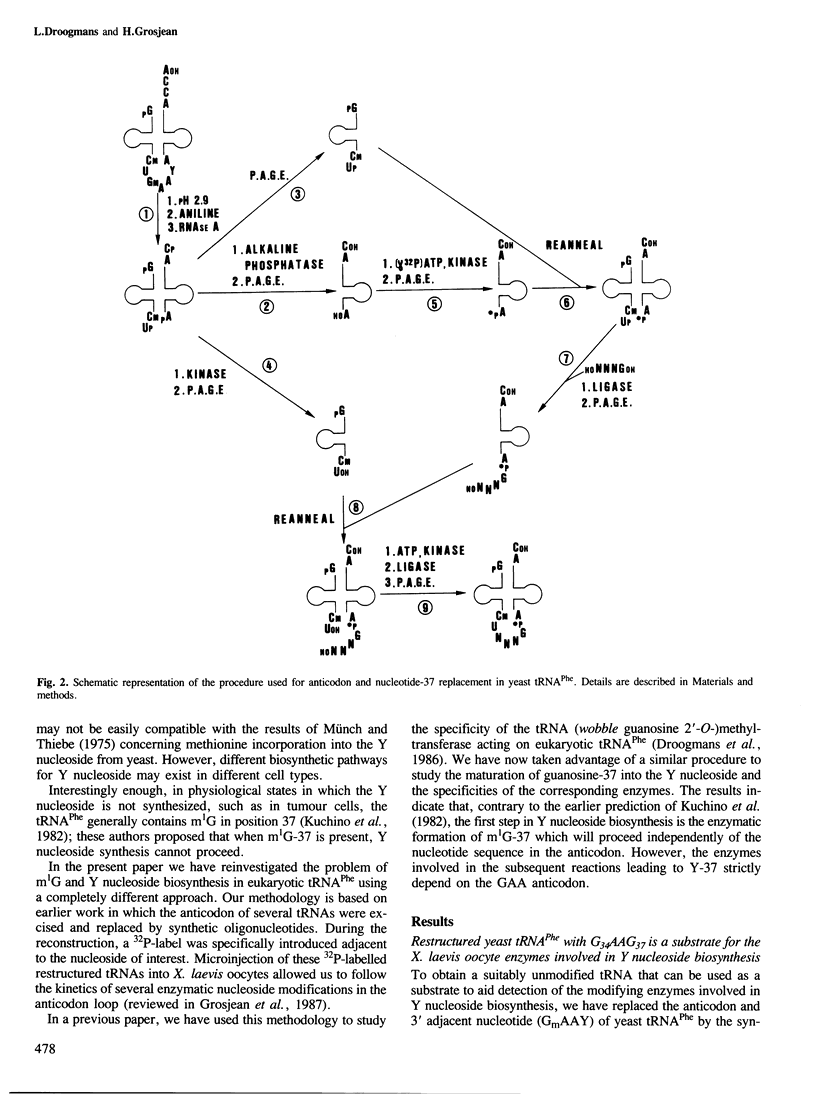
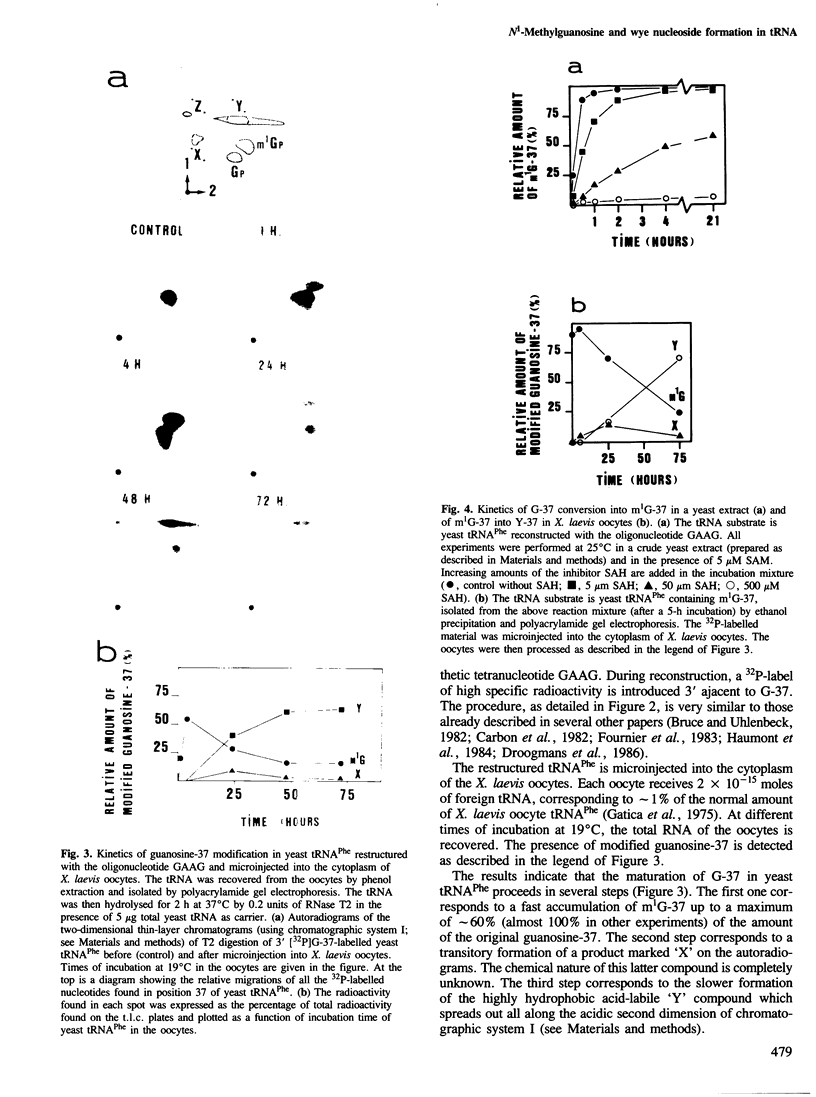
N1-Methylguanosine (m1G) or wye nucleoside (Y) are found 3' adjacent to the anticodon (position 37) of eukaryotic tRNAPhe. The biosynthesis of these two modified nucleosides has been investigated. The importance of the type of nucleosides in the anticodon of yeast tRNAPhe on the potentiality of this tRNA to be a substrate for the corresponding maturation enzyme has also been studied. This involved microinjection into Xenopus laevis oocytes and incubation in a yeast extract of restructured yeast tRNAPhe in which the anticodon GmAA and the 3' adjacent Y nucleoside were substituted by various tetranucleotides ending with a guanosine. The results obtained by oocyte microinjection indicate: that all the restructured yeast tRNAsPhe are efficient substrates for the tRNA (guanosine-37 N1)methyltransferase. This means that the anticodon sequence is not critical for the tRNA recognition by this enzyme; in contrast, for Y nucleoside biosynthesis, the anticodon sequence GAA is an absolute requirement; the conversion of G-37 into Y-37 nucleoside is a multienzymatic process in which m1G-37 is the first obligatory intermediate; all the corresponding enzymes are cytoplasmic. In a crude yeast extract, restructured yeast tRNAPhe with G-37 is efficiently modified only into m1G-37; the corresponding enzyme is a S-adenosyl-L-methionine-dependent tRNA methyltransferase. The pure Escherichia coli tRNA (guanosine-37 N1) methyltransferase is unable to modify the guanosine-37 of yeast tRNAPhe.
Full text
PDF

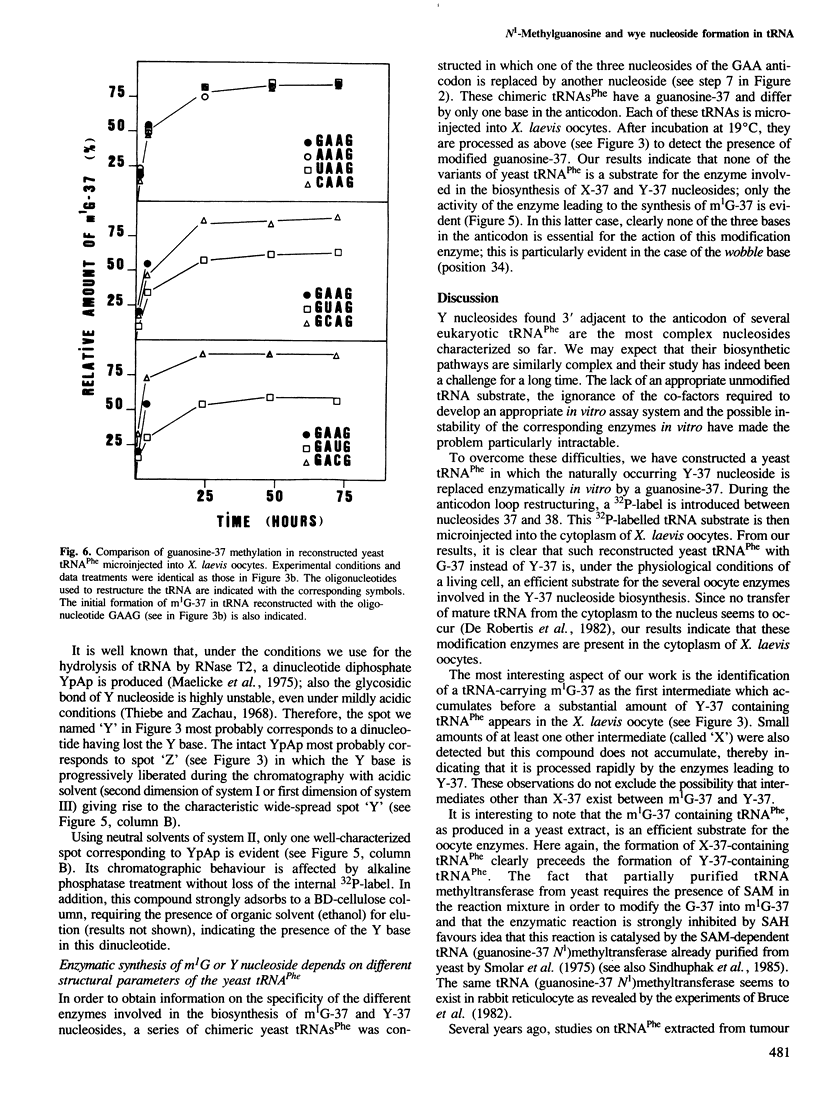


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blobstein S. H., Gebert R., Grunberger D., Nakanishi K., Weinstein I. B. Structure of the fluorescent nucleoside of yeast phenylalanine transfer ribonucleic acid. Arch Biochem Biophys. 1975 Apr;167(2):668–673. doi: 10.1016/0003-9861(75)90510-x. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Atkins J. F., Wills N., Uhlenbeck O., Gesteland R. F. Replacement of anticodon loop nucleotides to produce functional tRNAs: amber suppressors derived from yeast tRNAPhe. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7127–7131. doi: 10.1073/pnas.79.23.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982 Mar 2;21(5):855–861. doi: 10.1021/bi00534a007. [DOI] [PubMed] [Google Scholar]
- Carbon P., Haumont E., De Henau S., Keith G., Grosjean H. Enzymatic replacement in vitro of the first anticodon base of yeast tRNAAsp: application to the study of tRNA maturation in vivo, after microinjection into frog oocytes. Nucleic Acids Res. 1982 Jun 25;10(12):3715–3732. doi: 10.1093/nar/10.12.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Haumont E., Fournier M., de Henau S., Grosjean H. Site-directed in vitro replacement of nucleosides in the anticodon loop of tRNA: application to the study of structural requirements for queuine insertase activity. EMBO J. 1983;2(7):1093–1097. doi: 10.1002/j.1460-2075.1983.tb01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis E. M., Lienhard S., Parisot R. F. Intracellular transport of microinjected 5S and small nuclear RNAs. Nature. 1982 Feb 18;295(5850):572–577. doi: 10.1038/295572a0. [DOI] [PubMed] [Google Scholar]
- Droogmans L., Haumont E., de Henau S., Grosjean H. Enzymatic 2'-O-methylation of the wobble nucleoside of eukaryotic tRNAPhe: specificity depends on structural elements outside the anticodon loop. EMBO J. 1986 May;5(5):1105–1109. doi: 10.1002/j.1460-2075.1986.tb04329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. M., Nakanishi K., Barciszewski J., Rafalski A. J., Augustyniak H., Wiewiórowski M. Isolation and characterization of peroxy-Y base from phenylalanine transfer ribonucleic acid of the plant, Lupinus luteus. J Am Chem Soc. 1974 Dec 11;96(25):7797–7780. doi: 10.1021/ja00832a029. [DOI] [PubMed] [Google Scholar]
- Feldmann H., Falter H. Transfer ribonucleic acid from Mycoplasma laidlawii A. Eur J Biochem. 1971 Feb;18(4):573–581. doi: 10.1111/j.1432-1033.1971.tb01278.x. [DOI] [PubMed] [Google Scholar]
- Fournier M., Haumont E., de Henau S., Gangloff J., Grosjean H. Post-transcriptional modification of the wobble nucleotide in anticodon-substituted yeast tRNAArgII after microinjection into Xenopus laevis oocytes. Nucleic Acids Res. 1983 Feb 11;11(3):707–718. doi: 10.1093/nar/11.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica M., Tarragó A., Allende C. C., Allende J. E. Aminoacylation of transfer RNA microinjected into Xenopus laevis oocytes. Nature. 1975 Aug 21;256(5519):675–678. doi: 10.1038/256675a0. [DOI] [PubMed] [Google Scholar]
- Gillam I., Millward S., Blew D., von Tigerstrom M., Wimmer E., Tener G. M. The separation of soluble ribonucleic acids on benzoylated diethylaminoethylcellulose. Biochemistry. 1967 Oct;6(10):3043–3056. doi: 10.1021/bi00862a011. [DOI] [PubMed] [Google Scholar]
- Haumont E., Fournier M., de Henau S., Grosjean H. Enzymatic conversion of adenosine to inosine in the wobble position of yeast tRNAAsp: the dependence on the anticodon sequence. Nucleic Acids Res. 1984 Mar 26;12(6):2705–2715. doi: 10.1093/nar/12.6.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjalmarsson K. J., Byström A. S., Björk G. R. Purification and characterization of transfer RNA (guanine-1)methyltransferase from Escherichia coli. J Biol Chem. 1983 Jan 25;258(2):1343–1351. [PubMed] [Google Scholar]
- Kasai H., Goto M., Ikeda K., Zama M., Mizuno Y., Takemura S., Matsuura S., Sugimoto T., Goto T. Structure of wye (Yt base) and wyosine (Yt) from Torulopsis utilis phenylalanine transfer ribonucleic acid. Biochemistry. 1976 Feb 24;15(4):898–904. doi: 10.1021/bi00649a027. [DOI] [PubMed] [Google Scholar]
- Kasai H., Yamaizumi Z., Kuchino Y., Nishimura S. Isolation of hydroxy-Y base from rat liver tRNAPhe. Nucleic Acids Res. 1979 Mar;6(3):993–999. doi: 10.1093/nar/6.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Borek E., Grunberger D., Mushinski J. F., Nishimura S. Changes of post-transcriptional modification of wye base in tumor-specific tRNAPhe. Nucleic Acids Res. 1982 Oct 25;10(20):6421–6432. doi: 10.1093/nar/10.20.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. J., Nakanishi K., Grunberger D., Weinstein I. B. Biosynthetic studies of the Y base in yeast phenylalanine tRNA. Incorporation of guanine. Biochem Biophys Res Commun. 1973 Dec 10;55(3):818–823. doi: 10.1016/0006-291x(73)91217-5. [DOI] [PubMed] [Google Scholar]
- Maelicke A., von der Haar F., Sprinzl M., Cramer F. The structure of the anticodon loop of tRNAPhe from yeast as deduced from spectroscopic studies on oligonucleotides. Biopolymers. 1975 Jan;14(1):155–171. doi: 10.1002/bip.1975.360140112. [DOI] [PubMed] [Google Scholar]
- Mazabraud A. The nucleotide sequence of phenylalanine tRNA of Xenopus laevis. Biochimie. 1982 Oct;64(10):955–960. doi: 10.1016/s0300-9084(82)80359-3. [DOI] [PubMed] [Google Scholar]
- Münch H. J., Thiebe R. Biosynthesis of the nucleoside Y in yeast tRNAPhe: incorporation of the 3-amino-3-carboxypropyl-group from methionine. FEBS Lett. 1975 Mar 1;51(1):257–258. doi: 10.1016/0014-5793(75)80900-8. [DOI] [PubMed] [Google Scholar]
- Nakanishi K., Blobstein S., Funamizu M., Furutachi N., Van Lear G., Grunberger D., Lanks K. W., Weinstein I. B. Structure of the "peroxy-Y base" from liver tRNA Phe . Nat New Biol. 1971 Nov 24;234(47):107–109. doi: 10.1038/newbio234107b0. [DOI] [PubMed] [Google Scholar]
- Pergolizzi R. G., Engelhardt D. L., Grunberger D. Incorporation of lysine into Y base of phenylalanine tRNA in Vero cells. Nucleic Acids Res. 1979;6(6):2209–2216. doi: 10.1093/nar/6.6.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Sindhuphak T., Hellman U., Svensson I. Site specificities of three transfer RNA methyltransferases from yeast. Biochim Biophys Acta. 1985 Jan 29;824(1):66–73. doi: 10.1016/0167-4781(85)90030-2. [DOI] [PubMed] [Google Scholar]
- Smith C., Schmidt P. G., Petsch J., Agris P. F. Nuclear magnetic resonance signal assignments of purified [13C]methyl-enriched yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1985 Mar 12;24(6):1434–1440. doi: 10.1021/bi00327a023. [DOI] [PubMed] [Google Scholar]
- Smolar N., Hellman U., Svensson I. Two transfer RNA (1-methylguanine) methylases from yeast. Nucleic Acids Res. 1975 Jun;2(6):993–1004. doi: 10.1093/nar/2.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Moll J., Meissner F., Hartmann T. Compilation of tRNA sequences. Nucleic Acids Res. 1985;13 (Suppl):r1–49. doi: 10.1093/nar/13.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebe R., Zachau H. G. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur J Biochem. 1968 Sep 24;5(4):546–555. doi: 10.1111/j.1432-1033.1968.tb00404.x. [DOI] [PubMed] [Google Scholar]