Abstract
Basic helix–loop–helix (bHLH) transcription factors play critical roles in organism development and disease by regulating cell proliferation and differentiation. Transcriptional activity, whether by bHLH homo- or heterodimerization, is dependent on protein–protein and protein–DNA interactions mediated by α-helices. Thus, α-helical decoys have been proposed as potential targeted therapies for pathologic bHLH transcription. Here, we developed a library of stabilized α-helices of OLIG2 (SAH-OLIG2) to test the capacity of hydrocarbon-stapled peptides to disrupt OLIG2 homodimerization, which drives the development and chemoresistance of glioblastoma multiforme, one of the deadliest forms of human brain cancer. Although stapling successfully reinforced the α-helical structure of bHLH constructs of varying length, sequence-specific dissociation of OLIG2 dimers from DNA was not achieved. Re-evaluation of the binding determinants for OLIG2 self-association and stability revealed an unanticipated role of the C-terminal domain. These data highlight potential pitfalls in peptide-based targeting of bHLH transcription factors given the liabilities of their positively charged amino acid sequences and multifactorial binding determinants.
Graphical abstract
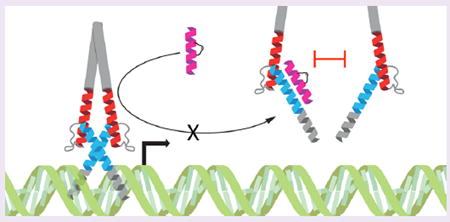
The human proteome is composed of 20 000 proteins, 3000 of which have been deemed druggable by conventional means using small molecule therapeutics.1 Transcription factors are largely classified as undruggable due to the large and complex surfaces that underlie their interactions with partner proteins and DNA. Thus, significant efforts have focused on developing nucleic acid mimetics2 and peptides3 as alternatives to small molecules for these formidable targets. The basic helix–loop–helix (bHLH) motif defines one such class of transcription factors that is essential to normal tissue development, cell lineage specification, and homeostatic control of cell proliferation.4 Perhaps the most notorious undrugged bHLH factors are the MYC proteins, which are implicated in a host of human cancers.5 The MYC-MAX heterodimer, for example, is structurally characterized by a C-terminal leucine zipper that engages as a coiled coil of α-helices, extending to a helix–loop–helix motif, which ultimately terminates in a basic region composed of lysines and arginines that bind to the major groove of DNA.6 Expression of a mutant, dominant negative form of the MYC bHLH-zip, termed omomyc, can disrupt the MYC-MAX-DNA interaction and block oncogenic cell proliferation, suggesting a strategy for directly disarming MYC through α-helical mimicry.7 Inhibitor of DNA binding (ID) proteins contain HLH domains that lack the DNA-binding basic region and represent natural dominant negative inhibitors of bHLH transcription, forming nonfunctional heterodimers with compatible bHLH targets.8 Efforts to capitalize on recapitulating various helical components of the bHLH domain to disrupt native dimers have thus far failed to yield cell active compounds for preclinical or clinical translation.
Classic liabilities of peptides as prototype therapeutics include the potential loss of bioactive structure, rapid proteolytic degradation in vivo, and limited intracellular access. We previously discovered that installing all-hydrocarbon cross-links or “staples” into peptide sequences that are structured in the native protein but unfold when synthesized in isolation can restore α-helical structure, confer striking protease resistance, and even promote cellular uptake by a pinocytic mechanism when appropriately designed.9,10 These novel attributes of peptide stapling have enabled the development of a series of anticancer peptides targeting a diversity of pathologic proteins, including members of the BCL-2 family,11,12 HDM2 and HDMX,13 β-catenin,14 EED,15 and KRAS.16 A notable commonality of these successful stapled peptide applications, including the first stapled peptide to advance to clinical testing in human cancer,17 is the helix-in-groove binding mechanism for protein interaction. Typically, the bioactive α-helices are amphipathic, with the hydrophobic face engaging a hydrophobic groove on the target protein, with complementary electrostatic and hydrophilic interactions at the amphipathic boundary providing further binding reinforcement.11,18 Whether or not peptide stapling is as effective in disrupting coiled coil or other types of α-helical interactions is either underexplored or underreported.
To examine the potential utility of developing dominant negative stapled peptides to block bHLH interactions, we focused our design efforts on targeting OLIG2, a bHLH transcription factor of the central nervous system (CNS), where it plays critical roles in neuronal differentiation during development and drives glioma cell proliferation.19 OLIG2 is uniformly expressed in some of the most malignant brain tumors, such as glioblastoma multiforme.20 Here, we developed a library of stabilized α-helices of OLIG2 (SAH-OLIG2) by inserting all-hydrocarbon staples into various portions of the bHLH motif, varying staple position, staple type, and peptide sequence length. Although initially encouraged by successful α-helical reinforcement and dose-responsive modulation of purified OLIG2 in electrophoretic mobility shift assays (EMSA), specificity testing revealed liabilities of the bHLH sequence composition and prompted a deeper exploration of the binding determinants for OLIG2 homodimerization. The new insights highlight important challenges to be overcome in deploying stapled peptides to target high priority bHLH transcription factors.
RESULTS AND DISCUSSION
Design and Biochemical Activity of Single-Helix Stapled OLIG2 bHLH Decoys
Since bHLH transcription factors such as OLIG2 require dimerization to stably bind their target DNA, we reasoned that structurally stabilized α-helical peptides mimicking this region could serve as competitive antagonists and potentially abrogate downstream transcription (Figure 1A). Thus, we synthesized a series of i, i+4 (cross-link spanning one helical turn) stabilized alpha-helices of OLIG2 (SAH-OLIG2) based on the sequences of bHLH helices 1 and 2 (SAH-OLIG2H1, SAH-OLIG2H2), varying peptide length, charge, and staple positioning (Figure 1B). Circular dichroism analysis revealed marked α-helical induction for a subset of SAH-OLIG2 peptides compared to the essentially unfolded template peptides, with the helix 2 series demonstrating especially striking structural stabilization (Figure 1C,D).
Figure 1.
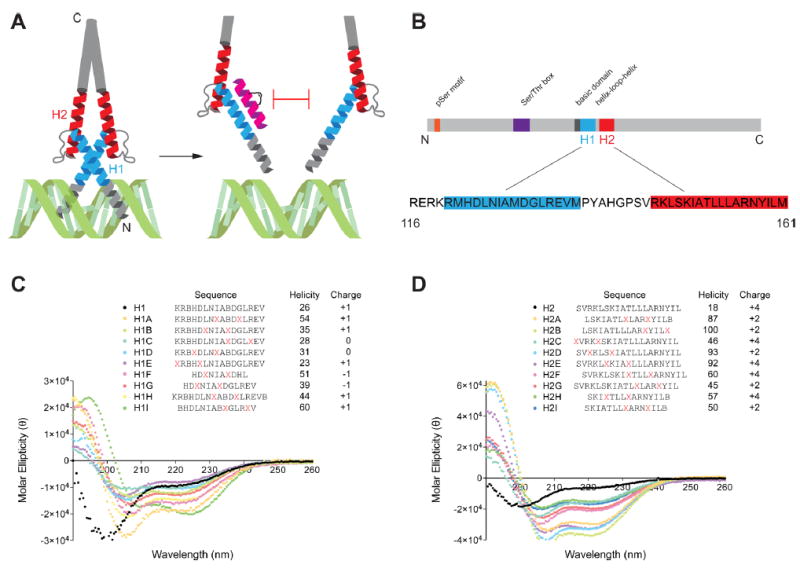
Design and α-helicity of SAH-OLIG2 peptides. (A) Design strategy for generating stapled peptides to mimic helices 1 or 2 of the OLIG2 bHLH domain to disrupt dimerization and E-box DNA engagement. (B) Domains of the OLIG2 protein, highlighting the bHLH region employed for stapled peptide design. (C,D) Sequence, α-helicity, and overall charge of SAH-OLIG2H1 A-I (C) and SAH-OLIG2H2 A-I (D) peptides. X, S-pentenyl alanine; B, norleucine.
To test the effect of SAH-OLIG2 peptides on formation of the OLIG2-DNA complex in vitro, which requires OLIG2 homodimerization21 (Supporting Information Figure S1), we performed EMSA screening assays. Human OLIG2 protein purified from COS7 cells was preincubated with SAH-OLIG2 peptides and then mixed with a 32P-labeled double-stranded DNA probe corresponding to the OLIG2 binding site of the HB9 promotor region, followed by polyacrylamide gel electrophoresis. Whereas SAH-OLIG2 peptides modeled after helix 1 had little to no effect on complex formation (Figure 2A, Supporting Information Figure S2A,B), several helix 2 decoys unexpectedly enhanced complex formation (Figure 2C), and the one construct that initially appeared to be inhibitory (SAH-OLIG2H2B) lacked dose-responsive activity upon further evaluation (Supporting Information Figure S2C). A notable difference between the helix 1 and 2 series of SAH-OLIG2 peptides was the extent of overall charge, with helix 1 constructs ranging from −1 to +1 and helix 2 derivatives having more positive charge, with a range of +2 to +4 (Figure 1C,D). To explore a sequence-independent basis for the observed complex-enhancing activity, we generated a scrambled version of SAH-OLIG2H2C for comparative EMSA testing and indeed found that both the wild-type and scrambled SAH-OLIG2 constructs similarly stabilized the OLIG2-DNA complex (Figure 2C). To further explore whether positive charge alone could account for this nonspecific activity, we tested two unrelated stapled peptides bearing an overall charge of +4 and +7. In each case, formation of the OLIG2-DNA complex was favored by addition of the positively charged stapled or unstapled peptides (Figure 2D).
Figure 2.
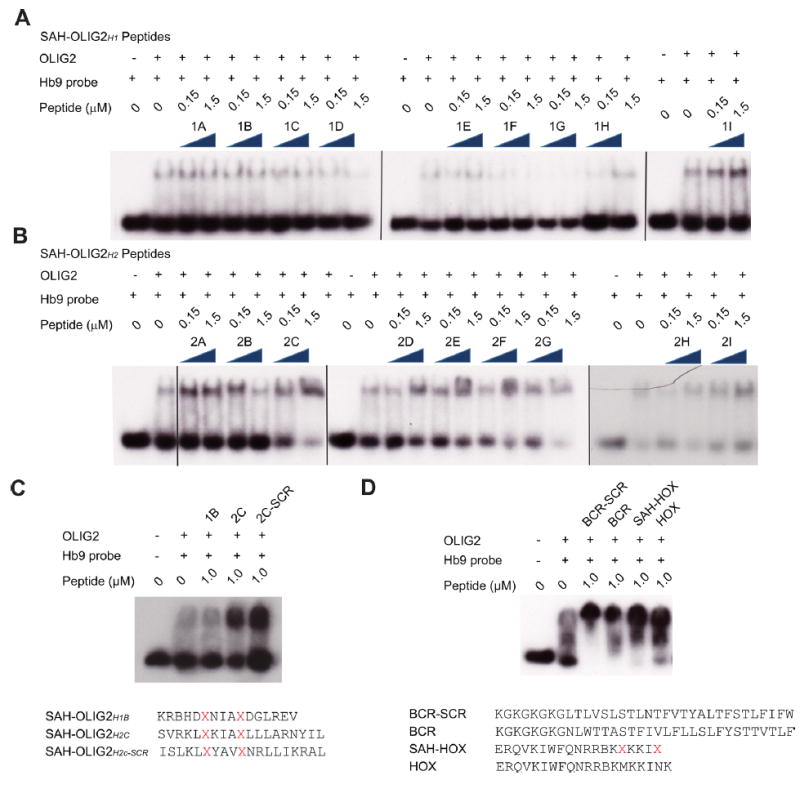
Biochemical activity of SAH-OLIG2 peptides in EMSA assays. (A,B) The effects of SAH-OLIG2H1 (A) and SAH-OLIG2H2 (B) peptides on OLIG2–DNA complex formation, as assessed by EMSA assay. (C,D) Sequence dependence of the SAH-OLIG2H2C effect was evaluated by EMSA using the corresponding scrambled stapled peptide (C) and unrelated constructs bearing +4 and +7 overall positive charge (D). SCR, scrambled.
Characterization of a Full-Length, Doubly Stapled OLIG2 bHLH Mimetic
In an effort to develop a bHLH decoy with sequence-dependent biochemical activity, we synthesized a stapled peptide corresponding to the entire bHLH domain, reasoning that dominant negative functionality may require a much larger construct for effective competition. To reinforce α-helical structure in each of the helical regions, we installed two i, i+7 staples (cross-links spanning two helical turns), yielding a 46-amino-acid-long stapled peptide—the longest construct we have generated to date (Figure 3A). Circular dichroism revealed striking transformation of the spectra from a random coil contour for the unmodified template peptide to a classical double minima for the doubly stapled peptide, reflective of α-helical structural stabilization (Figure 3B). However, in EMSA testing, SAH-OLIG2bHLH had no disruptive effect on formation of the OLIG2/DNA complex (Figure 3C). These data suggest that even tripling the size of the stapled peptide construct may be insufficient to inhibit bHLH–DNA interactions.
Figure 3.
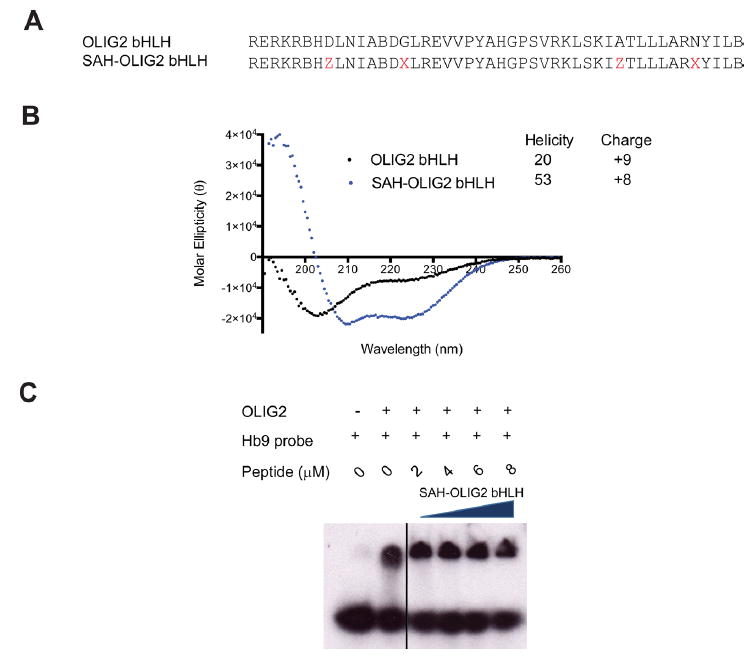
Synthesis and characterization of a doubly stapled OLIG2 bHLH domain. (A) Design of a full-length bHLH domain peptide bearing an (i, i+7) staple in each helical region. X, S-pentenyl alanine; Z, R-octenyl alanine; B, norleucine. (B) Alpha-helicity of a doubly stapled and full-length bHLH peptide. CD analysis demonstrated marked α-helical induction of SAH-OLIG2bHLH compared to the corresponding unmodified peptide. (C) Effect of SAH-OLIG2bHLH peptide on OLIG2–DNA complex formation, as assessed by EMSA assay.
Binding Determinants for OLIG2 Homodimerization and Stability
Given the inability of bHLH-based, SAH-OLIG2 peptides synthesized-to-date to disrupt OLIG2 homodimerization in vitro, we next examined by yeast two-hybrid screen whether additional domains of OLIG2 may be important binding determinants for self-association. Human wild-type OLIG2 and a series of deletion constructs and subdomains were cloned into yeast two-hybrid vectors containing either activating or DNA-binding domains, and each construct was evaluated for binding activity against the other using a high-stringency yeast two-hybrid method22 (Figure 4A). Whereas wild-type OLIG2 and its N-terminal deletion construct produced robust homodimerization activity with full-length OLIG2, deletion of the C-terminus—despite retention of the bHLH domain—showed no interaction signal (Figure 4A). Interestingly, the N-terminal deletion construct engaged with full-length OLIG2 but not itself, suggesting that truncation mutants may be less stable and, in this case, require full-length protein to maintain dimerization. Constructs bearing an extended bHLH domain (e bHLH), the discrete bHLH domain, or any of its subcomponents failed to induce a positive interaction signal for self-association or engagement with other longer forms of OLIG2 (Figure 4A). These data suggested that the C-terminal domain of OLIG2 may play a role in enforcing homodimerization and that the OLIG2 bHLH domain and its subcomponents may be insufficient for self-association or engagement of full-length OLIG2.
Figure 4.
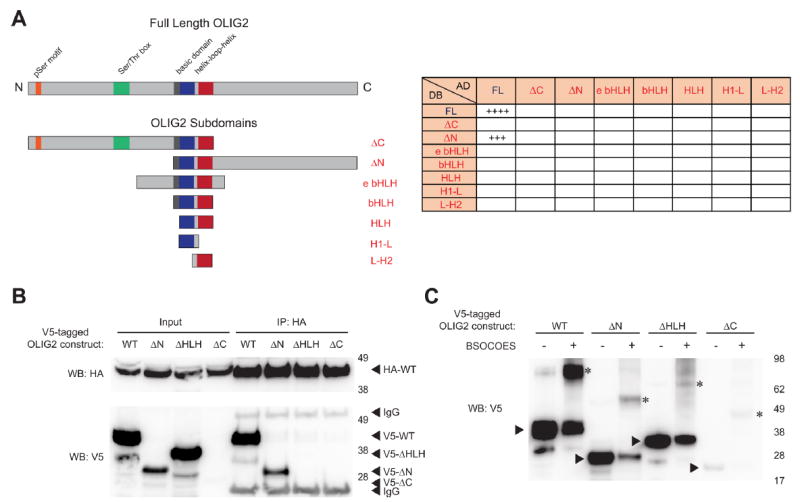
Binding determinants of OLIG2 homodimerization. (A) A series of OLIG2 constructs bearing discrete functional subdomains were generated for protein interaction analysis by yeast two-hybrid screening (left). In addition to self-association of wild-type OLIG2, only the OLIG2 ∆N construct, which bears both the bHLH and carboxy terminal domains, was capable of dimerizing with wild-type OLIG2 (right). (B) Protein expression levels of V5-tagged OLIG2 and the indicated deletion constructs were compared by V5 western analysis, and their capacity for association with HA-tagged, full-length OLIG2 evaluated by HA-immunoprecipitation from the corresponding mixtures of COS7 cell lysates followed by V5 Western blotting. (C) COS7 cell lysates containing the indicated V5-tagged OLIG2 constructs were subjected to BSOCOES cross-linking, and uncross-linked and cross-linked samples were subjected to SDS-PAGE and V5 western analysis. Except for OLIG2 ∆C samples, which contained insufficient V5-tagged protein, the quantity of protein loaded was adjusted to achieve near-equivalency of OLIG2 protein inputs. Arrowheads indicate the expressed OLIG2 constructs, and asterisks denote the corresponding dimeric species.
To evaluate the yeast two-hybrid findings in orthogonal protein interaction assays, we first compared the expression levels of V5-tagged wild-type (WT) and ∆N, ∆C, and ∆HLH deletion constructs of OLIG2 (lacking amino acids 1–89, 209–323, and 121–175, respectively) in transiently transfected COS7 cells. A clear hierarchy was observed, with WT demonstrating the highest level of expression followed by OLIG2 ∆HLH (Figure 4B). Whereas OLIG2 ∆N levels were considerably lower than those of WT and OLIG2 ∆HLH, little to no protein expression was observed for OLIG2 ∆C, a finding that could explain this construct’s negative yeast two-hybrid results and implicate the C-terminal domain in OLIG2 protein stability. We then mixed a COS7 cell lysate containing HA-tagged, full-length OLIG2 with the individual V5 lysates, followed by anti-HA immunoprecipitation and V5 western analysis. Consistent with the yeast two-hybrid results, HA-tagged full-length OLIG2 effectively dimerized with V5-tagged full-length OLIG2 and OLIG2 ∆N (Figure 4B). Little to no coimmunoprecipitation was observed for V5-tagged OLIG2 ∆HLH, and the interaction capacity of OLIG2 ∆C was not evaluable due to a lack of expression. Finally, as an alternate test for self-association, the lysates containing the individual V5-tagged constructs were subjected to BSOCOES cross-linking. Upon normalization for V5-tagged protein level in the lysates, WT–WT and ∆N–∆N dimeric species were clearly evident, with a low level of ∆HLH–∆HLH species even observed, suggesting that inclusion of the C-terminus supports protein expression and, at least in part, protein self-association (Figure 4C). For the minute amount of OLIG2 ∆C expression detected, a faint ∆C–∆C dimeric species was also observed, indicating that neither deletion of the HLH domain nor the C-terminal region alone fully abrogate OLIG2 self-association. Taken together, these data, along with the negative impact of HLH point mutations (Supporting Information Figure 1), generally reinforce the importance of the HLH domain in OLIG2 dimerization but also suggest a novel role for the OLIG2 C-terminus in protein stability and self-association.
Conclusions
During CNS development, the bHLH transcription factor OLIG2 is expressed in neural progenitor cells, regulating self-renewal and differentiation into neurons and oligodendrocytes by activating and repressing its transcriptional targets.20,23-26 OLIG2 expression in the postnatal brain is maintained in transit-amplifying cells of the subventricular zone, NG2-positive glia, and mature oligodendrocytes.27-29 In addition to its developmental functions, OLIG2 is expressed in 100% of diffuse human gliomas, where it identifies a subset of highly tumorigenic cells that are resistant to radiation and chemotherapy.20,30,31 The pro-mitotic role of OLIG2 is achieved in part by its N-terminal triple serine motif that, when phosphorylated, opposes p53 function in normal neural progenitor cells and their malignant equivalents.31,32 Indeed, OLIG2 is a high impact drug target for human brain cancer because of its exclusive CNS expression,27,29,33 largely dispensable role in mature CNS function,34 and proven link to the development, maintenance, and treatment resistance of malignant gliomas.23,31
Efforts to target bHLH transcription factors have yet to produce clinical candidates for inhibiting pathologic transcription.35-37 Here, we attempted to target OLIG2 by disrupting its dimeric interface using stapled peptides modeled after the individual bHLH helices and even the entire bHLH interaction domain. Whereas staple incorporation yielded constructs with marked α-helical induction, sequence-specific blockade of OLIG2-DNA interactions was not achieved. Instead, we observed nonspecific effects of peptide positive charge on in vitro stabilization of the OLIG2–DNA complex. Although we do not rule out the capacity of appropriately designed stapled peptide helices to engage bHLH targets and disrupt dimeric complexes, our studies revealed key pitfalls to consider in advancing such prototype inhibitors. Indeed, these challenges prompted us to re-evaluate the binding determinants for OLIG2 homodimerization. Whereas our yeast two-hybrid, coimmunoprecipitation, and cross-linking studies reinforced the importance of the bHLH domain as a key driver of OLIG2 self-association, the OLIG2 C-terminal region unexpectedly emerged as both an obligate component for stable protein expression and a contributor to self-association. Thus, in addition to highlighting the challenges associated with peptide sequence specificity in targeting bHLH domain–DNA interactions, our protein interaction data revealed that exclusive targeting of the bHLH domain may be insufficient for disrupting OLIG2 dimerization and potentially other bHLH transcription factors. Indeed, our comparative protein expression analysis of OLIG2 deletion constructs suggests that targeting the C-terminal domain could potentially destabilize OLIG2 and suppress its cellular levels.
To date, the majority of successful stapled peptide applications have relied on mimicry of structurally defined helix-in-groove interactions to induce a gain12,18 or loss11,13-15 of function. In the case of bHLH transcription factors, the strength and disposition of “helix-on-helix” dimeric interactions may be less amenable to peptide-based targeting. Our results further indicate that complementary regions outside of the bHLH interfaces may play a key role, limiting the potential efficacy of compounds designed to selectively target this region, including recently identified small molecule prototypes for OLIG2 bHLH inhibition.38 Given the importance of disarming OLIG2 in brain cancer, structural determination of OLIG2 homodimeric complexes that contain both the bHLH domain and C-terminal region will be critical to future, structure-based design of experimental inhibitors.
METHODS
Stapled Peptide Synthesis and Characterization
Synthetic peptides containing (i, i+4) and (i, i+7) staples were generated and purified to >95% homogeneity by LC/MS, quantified by amino acid analysis, and subjected to circular dichroism performed on an Aviv Biomedical spectrophotometer, as previously described.39,40
Purification of Cellular OLIG2
OLIG2-V5-HIS cDNA was transiently transfected into COS-7 cells using the pcDNA3.1 vector. Cells were lysed in hypotonic lysis buffer (50 mM Tris-HCl at pH 7.5, 0.2% [v/v] NP-40, 5 mM EDTA) and then high salt buffer (final concentration: 10 mM HEPES, 420 mM NaCl, 0.2 mM EDTA). OLIG2 protein was then purified from cellular lysates by immunoprecipiation with V5 antibody (Sigma), V5 peptide (Sigma) elution, affinity purification with Ni-NTA beads (Qiagen), and elution in 200 mM imidazole and Buffer A (50 mM Tris at pH 7.5, 150 mM KCl, 5 mM MgCl2, 10% [v/v] glycerol, 0.1% [v/v] NP40, and protease inhibitors).
Electrophoretic Mobility Shift Assays
Human OLIG2 protein, purified as described above, was quantified by silver stain using a serial dilution of quantified BSA protein control. The following DNA sequence based on the Hb9 E-box-containing promotor was employed: 5′ AGCTAATTTCCCAGATGGGCCAA 3′ and 3′ AGCTTTGGCCCATCTGGGAAATT 5′. Oligonucleotides were annealed in annealing buffer (10 mM Tris [pH 7.5], 50 mM NaCl, 1 mM EDTA) using a thermocycler for 5 min at 95 °C, and allowed to cool for approximately 1 h. The annealed probe was labeled with [α-32P] 6000 Ci mmol−1 dCTP (PerkinElmer) using Klenow (New England Biolabs). The radioactive DNA probe was incubated with 2 ng of OLIG2 protein in binding buffer (20 mM Tris [pH 7.5], 5 mM MgCl2, 0.1% [v/v] NP40, 0.5 mM DTT, 10% glycerol) and the indicated concentrations of SAH-OLIG2 peptides for 30 min at 4 °C. Protein/DNA complexes were analyzed by 4% nondenaturing polyacrylamide gel electrophoresis (PAGE). After drying the gel, radioactive signal was detected using a Typhoon 9400 (GE Healthcare Life Sciences).
Yeast Two-Hybrid Screening
Yeast two-hybrid screening was performed as previously described in detail.22 Briefly, Olig2 constructs were amplified by PCR and cloned into pDONR entry vectors, followed by recombination into pDEST destination vectors containing an activating or DNA-binding domain, respectively (Gateway cloning system, Invitrogen). Olig2 constructs and their nucleotide sequences included the following: Full-length Olig2 (1–972), Olig2∆C (1–510), Olig2∆N (277–972), Olig2 extended bHLH (277–510), Olig2 bHLH (325–489), Olig2 HLH (361–489), Olig2 H1-Loop (361–429), and Olig2 Loop-H2 (406–489). Plasmids were then transformed into yeast strains Y8800 MATa and Y8930 MATα. For primary screening, a single bait was mixed in liquid yeast extract peptone dextrose (YEPD) with a single prey. Diploids were then selected in liquid YEPD lacking leucine and tryptophan. Successful interaction of the bait and prey was identified by growth on YEPD plates containing 3-amino-1,2,4-triazole (3-AT). Autoactivators were discarded following growth on YEPD plates containing cycloheximide. True hit colonies were lysed, subjected to PCR, and submitted for Sanger sequencing to identify the relevant AD-X and DB-Y interacting constructs. All stages of screening were performed using each construct fused to both AD and DB domains as bait.
Cellular Expression and Co-immunoprecipitation of OLIG2 Constructs
COS-7 cells were transfected with WT (HA- or V5-tagged) or the indicated deletion constructs of V5-tagged OLIG2 (∆N, ∆HLH, and ∆C, lacking amino acids 1–89, 121–175, and 209–323, respectively) using Lipofectamine LTX. After 48 h, cells were lysed by incubation with a 1% (w/v) CHAPS lysis buffer (150 mM NaCl, 50 mM Tris at pH 7.4, 10 mM DTT) and protein concentration of the soluble fractions measured by BCA kit according to the manufacturer’s instructions (Thermo Scientific). Equivalent amounts (0.3 mg) of cell lysate samples containing HA-WT OLIG2 and the indicated V5 constructs were mixed and incubated overnight with anti-HA agarose beads (Pierce). The beads were washed three times with lysis buffer, eluted by boiling in LDS buffer, and subjected to western analysis with HA (Sigma-Aldrich, #12CA5) and V5 (Thermo, R960-25) antibodies.
OLIG2 Cross-Linking
COS-7 cells were transfected with WT or the above-described deletion constructs of V5-tagged OLIG2 using Lipofectamine LTX. After 48 h, cells were lysed by incubation with a 1% CHAPS lysis buffer (200 mM mannitol, 70 mM sucrose, 1 mM EGTA, 10 mM HEPES pH 7.5) and protein concentration of the soluble fractions measured by BCA kit. BSOCOES (bis(2-(sulfosuccinimido-oxycarbonyloxy)ethyl)sulfone, Pierce) was added to the lysates from a 10× stock solution to obtain a final concentration of 10 mM. After incubation at RT for 30 min, the cross-linker was quenched for 15 min with 1 M Tris-HCl (pH 7.5), which was added to a final concentration of 20 mM. The samples were then boiled in LDS buffer and subjected to western analysis using a V5 antibody (Thermo, R960–25).
Supplementary Material
Acknowledgments
We thank S. Lee and E. Smith for figure preparation, A. Dricot for technical assistance with the yeast two-hybrid screen, and Y. Nakatani for helpful discussions. This work was supported by National Institutes of Health grant 5P01CA142536 to C.D.S. and L.D.W., a Leukemia and Lymphoma Scholar Award to L.D.W., and an NSF predoctoral award to A.L.E.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.6b00465. Figures S1 and S2 and supplementary methods (PDF)
Notes
The authors declare the following competing financial interest(s): L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics.
References
- 1.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discovery. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Fechter EJ, Dervan PB. Allosteric inhibition of protein–DNA complexes by polyamide–intercalator conjugates. J Am Chem Soc. 2003;125:8476–8485. doi: 10.1021/ja030125e. [DOI] [PubMed] [Google Scholar]
- 3.Pescarolo MP, Bagnasco L, Malacarne D, Melchiori A, Valente P, Millo E, Bruno S, Basso S, Parodi S. A retro-inverso peptide homologous to helix 1 of c-Myc is a potent and specific inhibitor of proliferation in different cellular systems. FASEB J. 2001;15:31–33. doi: 10.1096/fj.00-0422fje. [DOI] [PubMed] [Google Scholar]
- 4.Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 6.Luscher B, Larsson LG. The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene. 1999;18:2955–2966. doi: 10.1038/sj.onc.1202750. [DOI] [PubMed] [Google Scholar]
- 7.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 9.Walensky LD, Bird GH. Hydrocarbon-stapled peptides: principles, practice, and progress. J Med Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaBelle JL, Katz SG, Bird GH, Gavathiotis E, Stewart ML, Lawrence C, Fisher JK, Godes M, Pitter K, Kung AL, Walensky LD. A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J Clin Invest. 2012;122:2018–2031. doi: 10.1172/JCI46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takada K, Zhu D, Bird GH, Sukhdeo K, Zhao JJ, Mani M, Lemieux M, Carrasco DE, Ryan J, Horst D, Fulciniti M, Munshi NC, Xu W, Kung AL, Shivdasani RA, Walensky LD, Carrasco DR. Targeted disruption of the BCL9/beta-catenin complex inhibits oncogenic Wnt signaling. Sci Transl Med. 2012;4:148ra117. doi: 10.1126/scitranslmed.3003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, Orkin SH. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leshchiner ES, Parkhitko A, Bird GH, Luccarelli J, Bellairs JA, Escudero S, Opoku-Nsiah K, Godes M, Perrimon N, Walensky LD. Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc Natl Acad Sci U S A. 2015;112:1761–1766. doi: 10.1073/pnas.1413185112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To KH, Olson KA, Kesavan K, Gangurde P, Mukherjee A, Baker T, Darlak K, Elkin C, Filipovic Z, Qureshi FZ, Cai H, Berry P, Feyfant E, Shi XE, Horstick J, Annis DA, Manning AM, Fotouhi N, Nash H, Vassilev LT, Sawyer TK. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci U S A. 2013;110:E3445–3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, Walensky LD. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meijer DH, Kane MF, Mehta S, Liu H, Harrington E, Taylor CM, Stiles CD, Rowitch DH. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci. 2012;13:819–831. doi: 10.1038/nrn3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, Louis DN, Stiles CD, Rowitch DH. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Paes de Faria J, Andrew P, Nitarska J, Richardson WD. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron. 2011;69:918–929. doi: 10.1016/j.neuron.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- 23.Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, Anderson DJ, Stiles CD, Rowitch DH. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 25.Satoh J, Asahina N, Kitano S, Kino Y. A Comprehensive Profile of ChIP-Seq-Based Olig2 Target Genes in Motor Neuron Progenitor Cells Suggests the Possible Involvement of Olig2 in the Pathogenesis of Amyotrophic Lateral Sclerosis. J Cent Nerv Syst Dis. 2015;7:1–14. doi: 10.4137/JCNSD.S23210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 27.Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 28.Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 30.Marie Y, Sanson M, Mokhtari K, Leuraud P, Kujas M, Delattre JY, Poirier J, Zalc B, Hoang-Xuan K. OLIG2 as a specific marker of oligodendroglial tumour cells. Lancet. 2001;358:298–300. doi: 10.1016/S0140-6736(01)05499-X. [DOI] [PubMed] [Google Scholar]
- 31.Mehta S, Huillard E, Kesari S, Maire CL, Golebiowski D, Harrington EP, Alberta JA, Kane MF, Theisen M, Ligon KL, Rowitch DH, Stiles CD. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y, Meijer DH, Alberta JA, Mehta S, Kane MF, Tien AC, Fu H, Petryniak MA, Potter GB, Liu Z, Powers JF, Runquist IS, Rowitch DH, Stiles CD. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906–917. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Miles DK, Hoang T, Shi J, Hurlock E, Kernie SG, Lu QR. The basic helix-loop-helix transcription factor olig2 is critical for reactive astrocyte proliferation after cortical injury. J Neurosci. 2008;28:10983–10989. doi: 10.1523/JNEUROSCI.3545-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fairman R, Beran-Steed RK, Anthony-Cahill SJ, Lear JD, Stafford WF, 3rd, DeGrado WF, Benfield PA, Brenner SL. Multiple oligomeric states regulate the DNA binding of helix-loop-helix peptides. Proc Natl Acad Sci U S A. 1993;90:10429–10433. doi: 10.1073/pnas.90.22.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh I, Chmielewski J. A beta-sheet peptide inhibitor of E47 dimerization and DNA binding. Chem Biol. 1998;5:439–445. doi: 10.1016/s1074-5521(98)90160-0. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh I, Issac R, Chmielewski J. Structure-function relationship in a beta-sheet peptide inhibitor of E47 dimerization and DNA binding. Bioorg Med Chem. 1999;7:61–66. doi: 10.1016/s0968-0896(98)00214-4. [DOI] [PubMed] [Google Scholar]
- 38.Tsigelny IF, Mukthavaram R, Kouznetsova VL, Chao Y, Babic I, Nurmemmedov E, Pastorino S, Jiang P, Calligaris D, Agar N, Scadeng M, Pingle SC, Wrasidlo W, Makale MT, Kesari S. Multiple spatially related pharmacophores define small molecule inhibitors of OLIG2 in glioblastoma. Oncotarget. 2015 doi: 10.18632/oncotarget.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bird GH, Bernal F, Pitter K, Walensky LD. Synthesis and biophysical characterization of stabilized a-helices of BCL-2 domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bird GH, Crannell WC, Walensky LD. Chemical synthesis of hydrocarbon-stapled peptides for protein interaction research and therapeutic targeting. Curr Protoc Chem Biol. 2011;3:99–117. doi: 10.1002/9780470559277.ch110042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


