Abstract
In the past, vast differences in ocular structure, development, and physiology throughout the animal kingdom led to the widely accepted notion that eyes are polyphyletic, that is, they have independently arisen multiple times during evolution. Despite the dissimilarity between vertebrate and invertebrate eyes, it is becoming increasingly evident that the development of the eye in both groups shares more similarity at the genetic level than was previously assumed, forcing a reexamination of eye evolution. Understanding the molecular underpinnings of cell type specification during Drosophila eye development has been a focus of research for many labs over the past 25 years, and many of these findings are nicely reviewed in Chapters 1 and 4. A somewhat less explored area of research, however, considers how these cells, once specified, develop into functional ocular structures. This review aims to summarize the current knowledge related to the terminal differentiation events of the retina, corneal lens, and pigmented epithelia in the fly eye. In addition, we discuss emerging evidence that the different functional components of the fly eye share developmental pathways and functions with the vertebrate eye.
1. Overview
The adult compound eye of the fruit fly, Drosophila melanogaster, is composed of a repeated array of ~800 individual unit eye, called ommatidia. Each adult ommatidium consists of approximately 20 cells (Fig. 5.1). Eight of these cells are photoreceptor (PR) neurons, photosensitive cells that project directly to the brain to transmit visual input. Immediately atop the PRs are six nonneuronal cells—four cone cells (CCs) and two primary pigment cells (PPCs)—that together secrete the corneal lens and an underlying crystalline structure known as the pseudocone. Approximately six secondary pigment cells (SPCs) and tertiary pigment cells (TPCs), also called interommatidial cells (IOCs), are then shared to form a boundary between ommatidia to limit light scattering. Finally, a mechanosensory bristle (interommatidial bristle, IOB) is present at every other apex of each ommatidium (Cagan and Ready, 1989a).
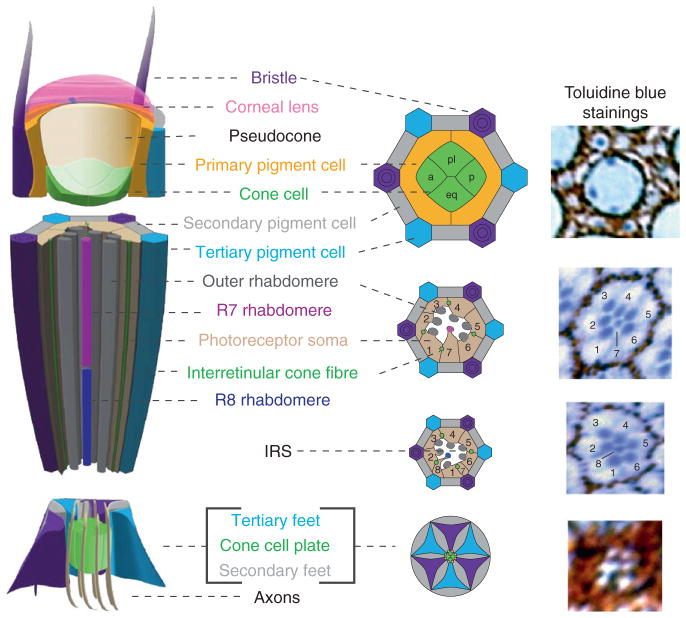
Figure 5.1.
Structure of an adult Drosophila ommatidium. Schematic of different regions of an adult ommatidium: the corneal lens region (top), the neural retina (middle), and the retinal floor (bottom). Corresponding regions from toluidine blue-stained semi-thin sections of an ommatidium are provided at the right. Color scheme is as follows: photoreceptor (PR) cell bodies, beige; PR rhabdomeres, dark gray cylinders (outer PRs), dark magenta cylinder (R7), or dark blue cylinder (R8); cone cells, green; primary pigment cells, yellow; secondary pigment cells, gray; tertiary pigment cells, turquoise; mechanosensory interommatidial bristle, purple hexagon; eye unit, longitudinal. The cone cells and primary pigment cells secrete the corneal lens (translucent pink) and a gelatinous pseudocone (translucent white). Each cone cell also extends an “interretinular fiber” between the photoreceptors, eventually expanding just proximal to the rhabdomeres to create a CC feet “plate” at the base of the retina. Based on the position within the ommatidia, the four cone cells are referred to as the apical (a), posterior (p), polar (pl), and equatorial (eq) cone cells. Secondary and tertiary pigment cells and the bristle form a characteristic hexagon around each ommatidia, with the pigment granules easily observed in the toludine blue stainings as reddish-brown (pteridine-containing) and black (xanthommatin-containing) vesicular-like structures. The apical surfaces of the secondary and tertiary pigment cells are tightly restricted, but the basal surfaces of these cells expand at the base of the retina to form a fenestrated membrane through which the axons project into the brain. The six outer photoreceptor rhabdomeres (gray from cells R1 through R6) form a trapezoid at the top of the eye and extend the length of the retina, enwrapping the IPR rhabdomeres—the R7 rhabdomere (Magenta) extends through the top two-thirds of the retina and the R8 rhabdomere (Blue) occupies the bottom third. In addition, the cell body of the R7 is positioned between the R1 and R6 cell, whereas the R8 cell body is located between the R1 and R2 cell, seen by cross section (middle diagrams and thin sections). The interhabdomeric space (white) that is important for preventing rhabdomere fusion is also seen. The entire central portion of the ommatidia is encapsulated by the cone cells—distally, with the rhabdomeres attached by “hemidesmosome-like” contacts, and proximally, with the rhabdomeres attached to the cone cell feet just below the end of the rhabdomere.
Drosophila undergoes a series of metamorphic processes before eclosing as an adult fly, 10 days after hatching. During each developmental stage, the eye undergoes discreet molecular and cellular changes. As an embryo, the organism sets aside small sets of cells that eventually produce adult external structures, such as the eye, wings, and legs. The cells specified to become ocular tissue are reserved in the larvae as part of the eye-antennal imaginal disc (see Fig. 5.2A and Chapter 1), a flat epithelial sheet that proliferates while the organism feeds and grows via three larval stages. At the end of the third and last larval stage, an epithelia-to-neuronal transition occurs at the anterior portion of the disc, marked by a physical change in the structure of the eye disc known as the morphogenetic furrow (MF). The MF migrates posterior to anterior through the eye disc, leaving behind cell clusters that ultimately mature into the highly regular lattice of ommatidia that forms the adult compound eye (Fig. 5.2A and B; Cagan and Ready, 1989a; Tomlinson and Ready, 1987b; Wolff and Ready, 1993).
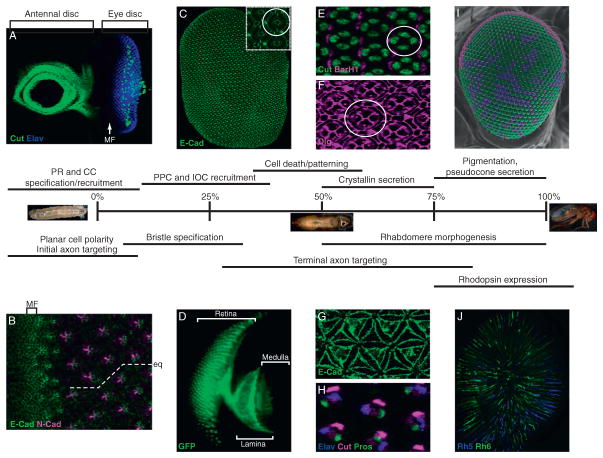
Figure 5.2.
Time course of Drosophila eye development. A summary of various developmental processes that occur during Drosophila pupal eye development (0–100%). Prior to pupation, in late third instar larva, the antennal/eye disc (A) is easily recognized by strong Cut expression (green) in the antennal portion (anterior, left), and clusters of Elav-positive photoreceptor clusters (blue) in the eye portion (posterior, right) corresponding to individual ommatidial units. Cut-positive cells are also present in the eye-imaginal disc, which represent subretinal glia and CCs precursors. Nonstained cells anterior to the morphogenetic furrow (MF) are retinal progenitors that are still proliferating (see Chapters 1 and 4 for further description). (B) The constricted apical surface of cells within the MF is obvious with E-Cadherin staining (green). In addition, the boundary between the R3 and R4 cell, marked by intense N-cadherin staining (purple), reveals the rotation of the ommatidia relative to the equator that is important for establishing the chiral trapezoid of photoreceptors observed in the adult retina. (C) E-cadherin staining (green) of a whole retina isolated from pupa at ~50% pupation shows the highly regular organization of ommatidia. Inset: A single ommatidium is circled. (D) Photoreceptor-driven Moesin::GFP at 50% pupation shows outer PR axons projecting to the lamina and IPR axons projecting to the medulla. (E) Cut (green) and BarH1(Magenta) specifically recognize the four CC and two primary pigment cell (PPC) nuclei at 50% pupation. (F) Discs Large (Dlg, Purple) highlights the apical contacts of the CCs, PPCs and interommatidial cells in 50% pupal retinas. (G) E-cadherin (green) of the basal surface of the retina shows the petal-shaped distribution of the IOC feet. (H) The bristle cell lineage is composed of four cells which express the transcription factors Cut and Pros, and the neural factor Elav. These nuclei are present at the base of the retina during their development, and eventually move more apically. (I) A scanning electron micrograph of an adult eye pseudocolored to represent the distribution of the pale (blue), yellow(green) and Dorsal Rim Area (DRA; magenta) ommatidia in the eye. (J) Whole mounted adult retina immunostained with Rhodopsin 5 (blue) and Rhodopsin 6 (green) in R8 rhabdomeres. Note the enrichment of Rh6 in the dorsal portion of the retina, corresponding to the dy ommatidia (see text for more detail).
Neuronal specification is the initial step of ommatidia formation and involves a stereotypical recruitment of the eight PR cells, R1–R8, through the reiterative use of EGF and Notch signaling (Brennan and Moses, 2000; Doroquez and Rebay, 2006; also see Chapter 4). The R8 cell arises first, followed by pairwise recruitment of R2/5, R3/4, and R1/6, and ending with R7 recruitment. Next, four nonneuronal CCs (also known as Semper cells) are recruited, and these cells are the last to be added during larval development. During early pupation, two PPCs then join each ommatidial cluster and fully enwrap the CCs (Fig. 5.2C–F). The light-isolating pigmented IOCs and the IOB are also recruited at this time and adopt a highly regular organization at the apical surface of the pupal retina (Fig. 5.2C and E–H; Cagan and Ready, 1989a; Ready et al., 1976; Waddington and Perry, 1960; Wolff and Ready, 1993).
All of the initial specification and patterning of the PRs, CCs, PPCs, IOCs, and IOBs occurs within a flat epithelial sheet and is complete within the first half of pupation. It is only during the last half of pupation that this flat retinal surface reshapes into the complex three-dimensional adult eye (Fig 5.2). During this latter half of development, the PRs extend their light-gathering apical surface, establish appropriate connections in the brain, and express the necessary proteins for phototransduction. In addition, the pseudocone and corneal lens are secreted, and pigmentation is established. Below, we highlight some of the molecular events that are known to drive these terminal differentiation steps. As will become obvious from this discussion, PR differentiation has been well studied, whereas studies on corneal lens and pigmented epithelia development remain considerably less explored.
2. The Retina: Drosophila PR Differentiation
2.1. General overview of fly PR subtypes
Historically, two classes of PRs have been defined in the fly eye, called outer photoreceptors (OPRs) and inner photoreceptors (IPRs). Functionally, these classes largely correspond to vertebrate rod and cone PRs, respectively. As such, OPRs and IPRs differ in several respects, including their position within the ommatidium, cell shape, rhodopsin gene expression, axonal projections, and physiological function.
Similar to the ciliary-based outer segments of vertebrate PRs, an expanded apical membrane compartment, known as a rhabdomere, houses the light-sensitive Rhodopsin proteins and the phototransduction machinery in fly PRs. In the fly, however, this compartment is not ciliary based, but instead, is comprised of organized microvilli that form a long cylindrical structure.
Six of the eight PRs found within an ommatidium, the R1 through R6 cells, represent the rod-like OPRs, and each of these cells develops a large rhabdomere that spans the depth of the retina. Together, their rhabdomeres form an asymmetric trapezoid whose chirality is determined by the planar cell polarity pathway active within the R3 and R4 cells (Adler, 2002; Mlodzik, 1999; Strutt, 2008). At the equator of the eye, the chirality of the trapezoid changes, allowing mirror symmetry of the eye (Fig. 5.2B). OPRs are highly sensitive to a broad spectrum of wavelengths of light and are important for motion detection and vision under dim light conditions (Hardie, 1985; Meinertzhagen and Hanson, 1993). The remaining two PRs, R7 and R8, represent the cone-like IPRs. The rhabdomeres of these cells are shorter and more slender than OPRs, and function under bright light conditions for color discrimination, R7 cells detecting UV wavelengths (345–375 nm), and the majority of R8 cells being sensitive to blue (437 nm) or green (508 nm) wavelengths (Fig. 5.2J; Feiler et al., 1992; Hardie, 1985; Salcedo et al., 1999; Yamaguchi et al., 2010). An exceptional subset of one to two rows of ommatidia is present in the dorsal half of the eye in which the IPRs are not involved in color discrimination, but instead are involved in detecting the vector of polarized light important for navigation (Fig. 5.2I; Hardie, 1985; Labhart and Meyer, 1999; Wernet et al., 2003). These ommatidia are referred to as Dorsal Rim Area (DRA) ommatidia (see Fig 5.2).
2.2. Terminal differentiation of fly PRs
Fly PR development occurs in two major steps: PR cell specification and terminal differentiation (Mollereau et al., 2001). PR cell specification occurs during the latest stages of larval development, and has been a topic of extensive study (for a review, see Chapter 4). PR terminal differentiation occurs during pupal development when PRs form their rhabdomeres, establish proper axonal projections into the brain, and begin expressing the rhodopsin genes that will in part determine their adult function (see Fig. 5.2 timeline). Below, we briefly review some of these events and several of the molecular players that promote these processes.
2.2.1. Rhabdomere development
The rhabdomere is an elongated apical structure of tightly packed and highly organized microvilli which is supported by a stalk membrane and zonula adherens (Fig. 5.3; Izaddoost et al., 2002; Longley and Ready, 1995; Pellikka et al., 2002). Although the rhabdomere is the apical surface of the PR, it extends perpendicular to the cell body. This orientation results from the CCs rising above the PRs during early pupation, causing the PR apical membranes to turn 90° and appose one another (Cagan and Ready, 1989a; Longley and Ready, 1995). At ~55% pupation, microvillus projections begin to emerge and delineate the apical membrane surface into two functional units—the rhabdomere and the stalk (Cagan and Ready, 1989a; Longley and Ready, 1995). Simultaneously, an extracellular matrix forms in the interrhabdomeric space that surrounds each developing rhabdomere and contributes to the exact spacing and positioning of each rhabdomere within an ommatidium (Husain et al., 2006; Zelhof et al., 2006). This process continues rather rapidly, and by 78% pupation, the interrhabdomeric space is well established, the microvilli have elongated, and the rhabdomeres have an elliptical cross section that becomes progressively more round throughout development (Cagan and Ready, 1989a; Longley and Ready, 1995) see also see Supplemental Movie in Sang and Ready (2002). By adulthood, the OPR rhabdomeres have expanded from an initial length of ~1 μm to occupy the full depth of the retina of ~100 μm. The R7 rhabdomeres only occupy the distal two-thirds of the retina while the R8 rhabdomeres fill the proximal one-third (Hardie, 1985).
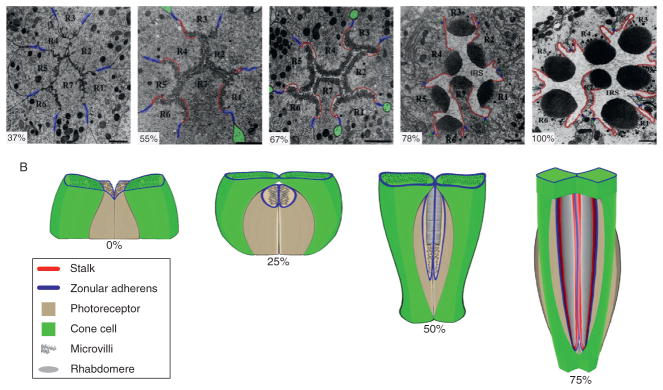
Figure 5.3.
Rhabdomere morphogenesis. (A) Coronal TEMs showing the apical membrane elaborations of photoreceptors R1 through R7 at different stages of development. The zonula adherens are marked with blue, the stalk region is highlighted in red, and the interrhabdomeric space (IRS) is the clear space between rhabdomeres that are obvious by 78% pupation (modified from Longley and Ready, 1995, with permission from Elsevier). Some of the interretinular fibers from cone cells, found directly adjacent to the zonula adherens are highlighted in green. (B) Diagram of the 90° turn of the photoreceptor apical surfaces during early pupation and elongation of the rhabdomeres (gray), the stalk region (red), and zonula adherens (blue) at later stages of development. Only two cone cells (green) and two photoreceptors are shown for clarity.
Rhodopsin contributes over 50% of a membrane proteins of the rhabdomere, and is required for the building and maintenance of the rhabdomere structure (Kumar and Ready, 1995). In fact, almost all proteins involved in the phototransduction pathway are similarly required to maintain photoreceptor integrity (Wang and Montell, 2007), indicating that like vertebrate PRs, form and function are tightly linked. Perhaps not surprisingly, transcription factors that regulate the expression of different components of the phototransduction machinery are also important for regulating rhabdomere morphogenesis. Two such factors are the homeodomain transcription factors Orthodenticle (Otd) and Pph13/Hazy (Mishra et al., 2010; Ranade et al., 2008; Tahayato et al., 2003; Vandendries et al., 1996; Zelhof et al., 2003). Interestingly, Otd and Pph13 individually regulate subsets of rhodopsins and phototransduction-encoding genes, and mutations in either factor cause rhabdomere defects; however, PRs lacking both factors fail to form any rhabdomeric structure, providing evidence that these factors control two independent PR morphogenetic pathways (Mishra et al., 2010). How these pathways are integrated, however, remains to be determined.
Besides the phototransduction machinery, a number of actin-binding proteins are critical for building the microvilli-rich rhabdomeric membrane, including Amphiphysin, WASp, Rac1, Moesin, Myo-II, MyoIII, and MyoV (Baumann, 2004; Chang and Ready, 2000; Deretic et al., 2004; Hicks et al., 1996; Li et al., 2007; Zelhof and Hardy, 2004; Zelhof et al., 2001). Moreover, molecules present in the stalk region, the zonula adherens, and the interrhabdomeric space play critical roles in rhabdomere elongation and maintenance. The stalk region expresses apical complex proteins such as Crumbs, Dpatj, and Par-6, and mutations in these factors lead to shortened and/or bifurcated rhabdomeres (Bachmann et al., 2008; Izaddoost et al., 2002; Nam and Choi, 2003, 2006; Nam et al., 2007; Pellikka et al., 2002; Richard et al., 2006a). The zonula adherens also recruit members of the Par protein complex (e.g., Par3), and mutations in these factors disturb distal, but not proximal, rhabdomere formation (Pinal et al., 2006). In contrast, components of the interrhabdomere space, including spacemaker (also known as eyes shut), prominin, and chaoptin, are important for maintaining distinct rhabdomeres between PRs, with mutations leading to the coalescence of rhabdomeres, a phenotype reminiscent of the fused rhabdoms commonly found in other invertebrate compound eyes (Husain et al., 2006; Van Vactor et al., 1988; Zelhof et al., 2006). Finally, factors associated with microtubule-based vesicle transport are critical for rhabdomere formation, including the small GTPases Rab1, Rab6, and Rab11 (Satoh et al., 1997, 2005; Shetty et al., 1998) as well as the Dynein/Dynactin complex (Fan, 2004; Fan and Ready, 1997; Tai et al., 1999), likely by transporting membrane-associated proteins such as Rhodopsin and TRP channels to the rhabdomeres, as well as regulating the endocytic recycling of these factors. As will be discussed later, many of these same proteins are also important for the formation and maintenance of vertebrate PRs, suggesting that studies of fly PR morphogenesis will be an important resource for understanding events related to retinal degeneration in vertebrates.
Because of the large rhabdomeres of OPRs and their preponderance in an ommatidium, much of what is understood about rhabdomere morphogenesis derives from studies of R1–R6 cells. While many of the same factors are also important in IPR morphogenesis, IPRs do exhibit differences that raise questions as to whether these cells require distinct regulatory pathways for their differentiation. For example, how is the smaller diameter of IPR rhabdomeres achieved, how is the length of their shorter elongation controlled, and how is the R7 rhabdomere positioned distally to the R8 rhabdomere? Similarly, how do the IPRs in DRA ommatidia (see below) acquire the same diameter as OPRs, and how do these cells form the distinct untwisted organization of their microvilli required for light polarization sensitivity, in contrast to all other rhabdomeres? Several factors involved in OPR morphogenesis are different in IPRs. For instance, Myo-II is critical for OPR rhabdomere formation, yet its expression in IPRs is weaker (Baumann, 2004)—could this account for the smaller size of their rhabdomere? In addition, several transcription factors originally identified for their ability to regulate IPR rhodopsin gene expression also control distinct aspects of IPR-specific morphogenetic processes. The zinc finger transcription factor, Senseless (Sens), and the homeodomain protein Otd, for example, are important to preserve the proximal position of the R8 cell (Tahayato et al., 2003; Xie et al., 2007), while the TALE homeodomain transcription factor Homothorax is critical for mediating all aspects of DRA ommatidia development, including their unique rhabdomere structure (Wernet et al., 2003). The target genes that these factors regulate to control these events are entirely unexplored, but should be useful for uncovering additional pathways that are important during PR morphogenesis.
2.2.2. Nuclear position
During specification, PR nuclei show a stereotypical basal-to-apical position as they are recruited: the nuclei from previously recruited cells are forced basally, so that eventually, the latest “born” cell nuclei are most apically position, and the oldest “born” cells have more basally located nuclei (Fig 5.7). One exception to this rule occurs with the R3/R4 nuclei, which maintain apical contacts even after the R1/R6 cells have been recruited (Tomlinson and Ready, 1987b); however, the functional consequence of this difference is currently unknown.
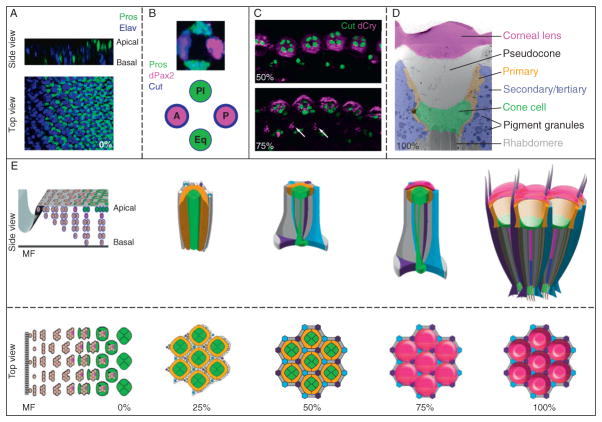
Figure 5.7.
Events leading to Drosophila corneal lens formation. (A) A third instar imaginal disc, stained with Elav (blue) to mark specified photoreceptors and the transcription factor Prospero (green), to mark the R7 photoreceptor and the cone cell precursors. The side view shows that the nuclei of cell move from a basal to apical position as they are recruited. (B) A high magnification of the cone cell layer from a single ommatidium shows that distinct subpopulations of cells that express different levels of Prospero (green), dPax2 (magenta), and Cut (blue) exist. This also is represented diagrammatically, with high Pros expression in equatorial (eq) and polar(pl) CCs, and high dPax2/Cut expression in anterior (a) and posterior (p) CCs. (C) Drosocrystallin (magenta) begins to be made in CCs, marked with Cut (green) at 50% pupation and is secreted from the cells by 75%. Drosocrystallin is also expressed at lower levels in the interommatidial bristle lineage (arrows). (D) A transmission electron micrograph of an adult ommatidium, pseudocolored to highlight the striated corneal lens (magenta), the clear pseudocone (gray), the primary pigment cells (PPCs, yellow), the cone cells (CCs, green), and the secondary/tertiary pigment cells (IOCs, purple). Note the abundant, large pigment granules in the IOCs, that the PPCs outline the CCs and pseudocone, and that the CCs lie between the pseudocone and the tips of the photoreceptor rhabdomeres. (E) Top and side view schemata of lens development, beginning from the imaginal disc through different stages of pupation using the same color scheme as in Fig. 5.1. The apical surface contacts change between the a/pCCs and eq/pl CC during pupation, patterning, and pruning of the IOCs occur prior to 30% pupation, and the corneal lens is secreted by ~75%. Afterward, the pseudocone is secreted and pushes the cone cells away from corneal lens.
In the adult eye, the PR nuclei also occupy characteristic positions: the OPR nuclei are positioned most distally, the R7 nucleus lies slightly below these, and the R8 nucleus occupies the most proximal portion of the retina. This means that the rhabdomeres of R1–R7 project proximally from their nuclei, whereas the rhabdomere of the R8 projects distally from its nucleus, suggesting this cell adopts a distinct cell polarity. Consistent with cell polarity being involved in proper nucleus localization, microtubule- and actin-associated proteins such as Glued/Dynactin and Klarischt/Marbles/Laminin A affect nuclear position of most PRs, even in the imaginal disc (Fan and Ready, 1997; Fischer-Vize and Mosley, 1994; Whited et al., 2004). PR-specific factors also can control nuclear position. Prospero (Pros), for instance, is expressed in the R7 cell, and Pros mutants develop IPRs whose rhabdomeres still retain their distal R7 position, but whose nuclei are proximally located, a characteristic unique to R8s (Cook et al., 2003). Likewise, overexpressing the R8-specific transcription factor Sens in R7 cells fails to change the position of the R7 rhabdomere, but does lead to a proximally positioned nucleus (Xie et al., 2007). Since Pros and Sens also influence other aspects of R7 versus R8 cell fates (see below), these data suggest that the nuclear position differences between the R7 and R8 are intimately linked to their cell fate choice. Why this is the case is currently not understood, but studies aimed at this question are likely to uncover additional developmental differences between these two related cell types.
2.2.3. PR projections
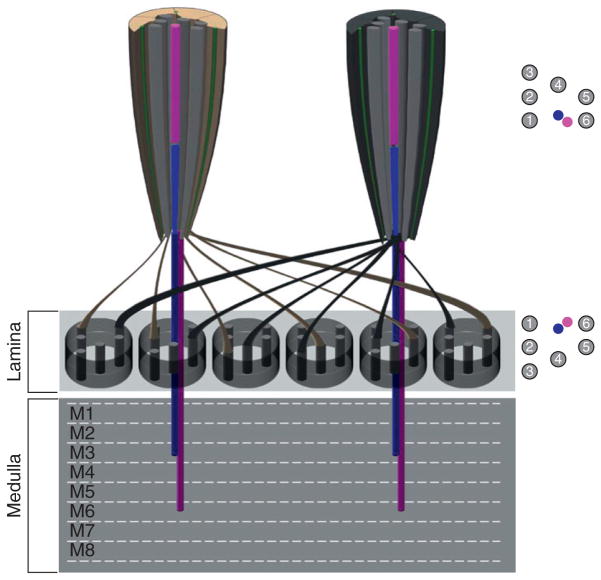
Axons from OPRs and IPRs project to two distinct optic ganglia beneath the retina: OPRs project to the first optic ganglion, the lamina, whereas R7 and R8 IPRs segregate to distinct layers in the second optic ganglion, the medulla (Fig. 5.4). Similar to other aspects of retinogenesis, PR axonal projection patterning occurs through two distinct processes and is distinct for OPRs versus IPRs. This topic has been extensively and elegantly reviewed recently, and thus will only be briefly summarized here (for reviews see Chiba, 2001; Mast et al., 2006; Matthews et al., 2008; Sanes and Zipursky, 2010; Tayler and Garrity, 2003).
Figure 5.4.
Axonal targeting differences between outer and inner photoreceptors. Diagram representing two ommatidia sharing lamina cartridges. The axons from the six outer PRs from each ommatidium turn 180° and project to six different cartridges present in the lamina neuropil present directly underneath the retina. R1–R6 positions within the lamina represent a mirror image of the outer photoreceptor arrangement found in the retina. The R7 (magenta) and R8 (blue) axons bypass the lamina and project to layers M3 and M6 respectively in the adult medulla.
PRs begin projecting their axons immediately after their specification in the eye imaginal disc. As the R8 is the first cell to be recruited during eye development, it initiates PR axonal projections into the developing optic lobes, to the top layer of the medulla, the M1 layer. As the R8 passes through the lamina, it activates the proliferation and differentiation of lamina neurons, which then recruit additional glia (Dearborn and Kunes, 2004; Perez and Steller, 1996; Winberg et al., 1992). The OPRs follow the R8 axon fascicles into the lamina, where they encounter two rows of glia, called the lamina plexus, that prevent OPRs from projecting beyond this point (Poeck et al., 2001). The R7 cell then projects its axon through the lamina and terminates in a layer slightly below the R8 projection. Interestingly, projections to the medulla appear to be a default choice, because many factors important for R1–R6 projections cause misprojections into this optic neuropil, and play permissive rather than instructive roles (Cafferty et al., 2004; Garrity et al., 1996; Hing et al., 1999; Kaminker et al., 2002; Newsome et al., 2000; Ruan et al., 2002; Suh et al., 2002).
These initial axonal projections are maintained until approximately 30% pupation, and afterward, undergo further refinement. At this time, the OPR axons from individual ommatidia begin to establish lateral contacts with other ommatidia in a process known as neural superposition (for reviews see Hardie, 1985; Meinertzhagen, 1975). This is an important process in Drosophila, because the rhabdomeres of different OPRs within a single ommatidium point to different directions whereas OPRs from adjacent ommatidia do converge on the same point, due to the curvature of the eye. Thus, to integrate the visual input from photoreceptors in separate ommatidia that converge on the same point, OPR axons twist 180° and project outward into six different “lamina cartridges,” maintaining a spatial pattern that replicates their position within the ommatidia (Fig. 5.4). For instance, the R1 PR axon projects to an R1 position within one cartridge, while the R2 projects to the R2 position in different cartridge. This convergence of visual information across six ommatidia leads to increased sensitivity and providing input important for motion detection. Interestingly, just like during the establishment of rhabdomere polarity in the retina, the R3/R4 PRs also determine the orientation of projections during neural superposition (Clandinin and Zipursky, 2000). Moreover, the atypical cadherin molecule Flamingo that controls the Frizzled-dependent asymmetric localization of the R3/R4 rhabdomeres is also critical for directing OPR axon growth cones to the correct cartridges (Lee et al., 2003; Usui et al., 1999). These data highlight the coordinated use of the same factors to establish proper positioning and function for PRs during fly retinogenesis.
Unlike the OPRs, the R7 and R8 within an ommatidium share the same light path, and since their axons pass directly through the lamina layer into the medulla, they provide a perfect retinotopic map of the eye. Immediately after reaching the medulla, R7 projects slightly deeper than R8. At ~17% pupation, the medulla begins to laminate, which allows further separation of the R7 and R8 axonal terminals. By 35% pupation, R7 has reached the M3 layer in the medulla while R8 remains in the M1 layer. At approximately 50% pupation, both the R7 and R8 axons project deeper into the medulla, with R7 reaching its final destination in the M6 layer and R8 terminating at the M3 layer (Fig. 5.4). similar to Flamingo functioning redundantly to control OPR patterning and axonal projections, factors important for controlling several aspects of R7 versus R8 PR patterning in the retina are also important for their proper targeting in the medulla. For instance, the transcription factor Sens not only regulates R8 specification, R8 cell position, and rhodopsin expression, but it is critical for M6-specific targeting of the R8 axon (Frankfort et al., 2001; Morey et al., 2008; Xie et al., 2007). Likewise, the homeodomain protein Pros, involved in R7-specific rhodopsin expression and nuclear position, is partly responsible for the targeting of R7 cells to the M3 layer (Cook et al., 2003; Kauffmann et al., 1996; Morey et al., 2008). Interestingly, mutation of either Sens or Pros lead to a reciprocal switch in R7 vs R8 projections—that is, sens mutants project to the R7 layer, while pros mutants project to the R8 layer (Morey et al., 2008), arguing that IPRs share factors that mediate medulla projections, with Sens and Pros further refining this projection pattern to distinct layers. To date, such a molecule has not yet been identified, but interesting candidates for this are Runt and Spalt, two transcription factors whose expression is restricted to the R7 and R8 shortly after their neural specification in the eye imaginal disc. Indeed, misexpressing Runt in OPRs does lead to mistargeting to the medulla. However, both Runt and Spalt loss of function IPRs maintain their appropriate targeting in the medulla, suggesting that other factors must also be involved or that these factors function redundantly (Kaminker et al., 2002; Mollereau et al., 2001).
2.2.4. Rhodopsin gene expression
OPRs are important for motion detection, whereas IPRs are important for color discrimination under bright light conditions (Bicker and Reichert, 1978; Hardie and Kirschfeld (1983); Hardie (1979); Hu and Stark, 1980; Menne and Spatz, 1977; Yamaguchi et al., 2010). In order to capture as much light information as possible, all OPRs express the same broad wavelength-sensitive Rhodopsin, Rhodopsin 1 (Rh1; O’Tousa et al., 1985; Stark et al., 1976; Zuker et al., 1985). In contrast, IPRs express a complex pattern of Rhodopsin-encoding genes in order to maximize the range of wavelengths they detect—R7 cells express UV-sensitive opsins, Rh3 and/or Rh4, while R8 cells can express Rh3, the blue-sensitive Rh5, or the green-sensitive Rh6 (Chou et al., 1996, 1999; Fortini and Rubin, 1990; Fryxell and Meyerowitz, 1987; Huber et al., 1997; Mazzoni et al., 2008; Mismer and Rubin, 1989; Montell et al., 1987; Zuker et al., 1987; Fig. 5.6). Rh gene expression begins during late (79–84%) pupation, with OPR-specific Rh1 being expressed first, and IPR-specific Rhs 3–6 being expressed shortly thereafter (Earl and Britt, 2006). Here, we will discuss the genetic pathways relevant for establishing the cell-specific expression of the Rhodopsin-encoding genes, as these have helped elucidate a better understanding of the genetic relationships among different color-sensitive photoreceptors.
Figure 5.6.
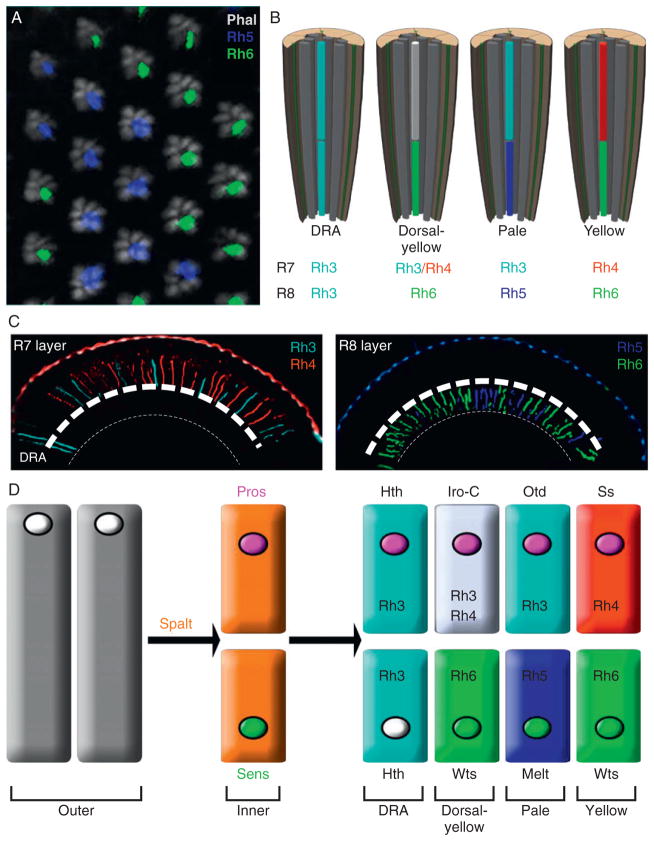
Ommatidial subtypes express different inner photoreceptor Rhodopsins. (A) A whole-mount staining of an adult retina stained with phalloidin (gray) shows the trapezoidal arrangement of the actin-rich rhabdomeres of the six outer photoreceptors and the random distribution of the pale and yellow ommatidia are revealed by immunostaining for Rh5 (blue) and Rh6 (green) that are expressed in the central R8 cells. (B) Diagram of the Dorsal Rim Area (DRA), dorsal yellow, pale, and yellow subsets found in the Drosophila eye, defined by the Rhodopsins expressed in the R7 and R8 inner photoreceptors. All outer photoreceptors express the same Rhodopin, Rhodopsin 1. (C) Transverse sections of adult eyes, dorsal left, stained with R7 Rhodopsins (left), Rh3 (cyan) and Rh4 (red), or R8 Rhodopsins (right), Rh5 (blue) and Rh6 (green). Note that two rows of ommatidia at the dorsal side of the eye express Rh3 in the R7 and R8 layers, representing the DRA ommatidia. Rh3 and Rh4 expression in the dy ommatidia are weaker than in the remainder of the eye. (D) Schematic representing the factors that direct inner photoreceptor identity, differentiation, and rhodopsin expression. The relative position of the nuclei that would be in the cell body for the different cell types are also indicated. See text for detail.
2.2.4.1. Rhodopsin promoters are multipartite
The expression of each Rhodopsin protein can be properly recapitulated by <250 bp of regulatory sequence upstream of the TATA box (Fortini and Rubin, 1990; Papatsenko et al., 2001; Tahayato et al., 2003). Sequence analysis of these promoters revealed a Rhodopsin Conserved Sequence I (RCSI) that is shared by all six Drosophila Rhodopsin promoters (Fig. 5.5). This sequence is an inverted repeat of a homeodomain-binding site separated by 3 nucleotides, and matches the P3 site previously identified as a perfect recognition sequence for a subset of paired-related homeodomain-containing proteins that include Pax6, the “master control gene” for eye development (Czerny and Busslinger, 1995; Sheng et al., 1997; Wilson et al., 1993; also see Chapter 1). Consistent with the possibility that Pax6 may regulate rhodopsin gene expression through this site, multimerization of the RCSI (3XP3) is sufficient to drive PR-specific gene expression in a wide range of animals, suggesting that the RCSI is recognized by an evolutionarily conserved paired-like transcription factor present in PRs like Pax6 (Berghammer et al., 1999; Gonzalez-Estevez et al., 2003; Sheng et al., 1997). However, whether Pax6 is the only factor responsible for this function currently remains unclear since recent studies indicate that another homeodomain factor, Hazy/Pph13, may also be critical for regulating RCSI-dependent Rh gene expression (Mishra et al., 2010; Punzo et al., 2001).
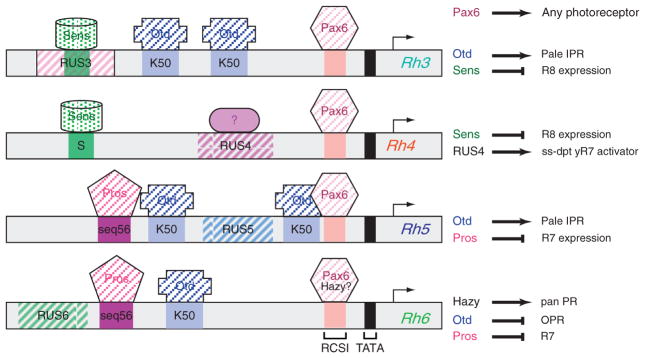
Figure 5.5.
Regulatory sequences of the inner photoreceptor Rhodopsin-encoding genes. Schematic of the minimal promoters for Rh3 through Rh6 that recapitulate expression of the endogenous genes. Senseless binding sites (S) are green, Otd binding sites (K50) are light blue, Pax6/RSCI sites (Rhodopsin Conserved Sequence I) are pale pink and Pros sites (seq56) are dark magenta. Rhodopsin Unique Sequences (RUS) 3, 4, 5, and 6 are represented by striped boxes. The summary of the role of each transcription factor is highlighted to the right. Otd activates Rh3 and Rh5, the two Rhodopsins expressed the pale ommatidia, and represses Rh6 in outer photoreceptors (Tahayato et al., 2003). Pros represses the R8 Rhodopsins, Rh5 and Rh6, in R7 photoreceptors (Cook et al., 2003), while Sens represses the R7 Rhodopsins, Rh3 and Rh4, in R8 photoreceptors (Xie et al., 2007). A transcription factor that is predicted to be activated by Spineless in yellow R7 cells to activate Rh4 is indicated by a ? on the Rh4 promoter. In addition, Hazy has recently been shown to be necessary and sufficient for Rh6 expression and bind to the RCSI, making it possible that Hazy, and not Pax6, is responsible for activating the Rh6 promoter in the fly eye (Mishra et al., 2010).
Outside the RCSI, each Rh promoter contains unique upstream sequences (Rhodopsin Unique Sequences, RUS) that show strong homology across multiple Drosophila species (Fig. 5.5), suggesting that these elements are responsible for directing gene-specific regulation (Fortini and Rubin, 1990; Papatsenko et al., 2001; Tahayato et al., 2003). This led to the model that Rhodopsin promoters are bipartite, with the RCSI providing generic PR specificity, and RUS elements providing subtype specificity (Fortini and Rubin, 1990). However, additional sequences have since been identified that are shared between different “classes” of rhodopsin promoters: for instance, both R7-specific rhodopsin promoters contain a conserved R8 repression element (S box, Fig. 5.5), and both R8 rhodopsin promoters share a conserved R7 repression element (seq56, Fig. 5.5; Cook et al., 2003; Tahayato et al., 2003; Xie et al., 2007). This suggests that a more complex combinatorial regulation leads to the diverse patterns of IPR rhodopsin gene expression. While the factors that recognize these shared sequences have largely been identified, and are discussed in more detail below, the factors that recognize the RUS sequences remain surprisingly elusive.
2.2.4.2. OPR versus IPR decisions
The Spalt (Sal) genes define IPR cell fate. Based on the fact that PR specification begins with the R8 and culminates with the R7, mature R7 and R8 cells were believed to arise from genetically distinct cell types. However, using a genetic screen for PR-restricted enhancer traps in the adult eye, Mollereau et al. (2000, 2001) identified a zinc finger transcription factor complex (spalt genes SalM and SalR), that is specifically enriched in R7 and R8 PRs, suggesting for the first time that these cells may share genetic components. Consistent with the possibility that Spalt regulates the fate of both cells, when the sal complex is genetically removed, ommatidia develop eight to nine OPRs and no IPRs, while overexpressing SalM in OPRs is sufficient to convert all PRs into IPRs (Domingos et al., 2004; Mollereau et al., 2001) These findings led to the discovery that PRs in the eye imaginal disc are bipotential, requiring Sal expression in the R7 and R8 to define IPR versus OPR cell fates. These studies also revealed that PR development occurs in at least two developmental steps: recruitment in the imaginal disc, and IPR versus OPR cell fate choices later during development. Currently, the mechanisms by which the sal genes achieve their function remain unclear.
2.2.4.3. R7 versus R8 cell fate decisions: Pros and Sens coordinate Rhodopsin expression, axonal targeting, and cell morphology
Mature R7 and R8 IPRs differ in numerous respects, including rhabdomere position, nucleus location, axonal targeting, and opsin gene expression. Thus, the finding that Sal controlled IPR versus OPR decisions led to questions of how the R7 and R8 cell then later distinguish themselves from a common IPR precursor. Many answers to this question came from studies focused on understanding how the IPR Rhodopsin genes themselves are regulated. Pros, for instance, was identified in a yeast one-hybrid screen for its ability to bind to a conserved sequence in the Rh5 and Rh6 promoters, and was subsequently shown to be expressed in R7 cells to specifically repress the expression of these R8-specific opsins, as well as prevent R8-specific nuclear position and axonal projections (Cook et al., 2003; Kauffmann et al., 1996; Morey et al., 2008). In contrast, the transcription factor Sens is expressed in R8 cells, binds to and represses R7-specific opsin promoters through a common S-box sequence, and prevents R7-specific rhabdomere position and axon targeting (Cook et al., 2003; Morey et al., 2008; Xie et al., 2007). Sens also contributes to positively activating both R8-specific opsins, likely as a non-DNA-binding coactivator (Xie et al., 2007). Together, these data suggest that Sal specifies a “generic” or default IPR that can express all IPR opsins, has an R8-like nuclear position, and has an R7-like rhabdomere position. Subsequently, Pros in the R7 and Sens in the R8 then repress the characteristics that are incompatible with their proper function in the adult eye. While their ability to regulate Rhodopsin gene targets is clear, how Pros and Sens control other aspects of R7 versus R8 cell fates, and whether other factors also participate in refining IPR differences remains an open question.
2.2.4.4. Ommatidial subtype specification: Hth, IroC, Otd, Ss, Melt, and Wts
Four distinct subtypes of ommatidia, called pale (p), yellow (y), dorsal yellow (dy), and dorsal rim area (DRA) ommatidia, are present in the adult eye and are defined based on which rhodopsins are expressed the R7 and R8 cells (Fig. 5.6B). DRA ommatidia are the least abundant of the subtypes, and these are restricted to one to two rows of ommatidia along the dorsal half of the eye. Unlike the other three subtypes, DRA ommatidia are not involved in color discrimination, but instead are involved in discerning the vector of polarized light to aid during navigation (Hardie, 1985; Labhart and Meyer, 1999; Wernet et al., 2003; Wunderer and Smola, 1982). This is facilitated by the fact that both IPRs express the same UV Rhodopsin, Rhodopsin 3, and because the membranes of the two IPR rhabdomeres form two crossed-over polarizing filters (Labhart and Meyer, 1999; Wernet et al., 2003; Wunderer and Smola, 1982). The TALE homeoprotein, Homothorax (Hth), is necessary and sufficient to induce all known DRA characteristics (Wernet et al., 2003); however, the responsible mechanisms and the target genes utilized by Hth to accomplish this function remains unexplored.
Distinction of p and y ommatidia was originally observed by the presence of the random distribution of a screening pigment in ~70% of ommatidia that appeared yellow under white light illumination versus the pale appearance in the remaining 30% of ommatidia (Kirschfeld et al., 1978). Later molecular analysis of Rh gene expression in Drosophila noted that the 30:70 ratio corresponded to the ratio of R7 cells expressing Rh3 and Rh4, (Fortini and Rubin, 1990) and R8 cells expressing Rh5 and Rh6, respectively (Chou et al., 1996; Papatsenko et al., 1997). Indeed, ~30% of ommatidia (“pale” ommatidia) express coupled Rh3:Rh5 expression in the R7 and R8, respectively, while the remaining ~70% of ommatidia (“yellow” ommatidia) express coupled Rh4:Rh6 in the R7 and R8 together with an additional screening pigment that gives the yellow color under white illumination (Chou et al., 1996, 1999; Mazzoni et al., 2008; Papatsenko et al., 1997; Stark and Thomas, 2004; Fig. 5.6A–C). Interestingly, Mazzoni et al. (2008) recently noted that a subset of “yellow” ommatidia that are restricted to the dorsal third of the eye coexpress Rh3 and Rh4 in the R7, but still express Rh6 in the underlying R8. Thus, these dorsal-restricted ommatidia are referred to as dorsal yellow (dy) ommatidia. These are a particularly curious subset of ommatidia, as they do not adhere to the normal “one sensory receptor per sensory cell” paradigm commonly adopted in sensory systems to avoid overlapping signals (Mazzoni et al., 2004), and are not distributed throughout the eye, but instead are regionally localized. Molecularly, the Iroquois complex of transcription factors (Iro-C) specify the dy ommatidia, consistent with the fact that Iro-C factors are repeatedly used during other dorsal–ventral patterning events in the fly eye (Cavodeassi et al., 2000; Mazzoni et al., 2008; Singh and Choi, 2003). Functionally, these ommatidia are likely to recognize a broader spectrum of wavelengths in the UV (Feiler et al., 1992), and are positioned to a region of the eye that is most commonly found facing the sky. Behaviorally, how the fly takes advantage of this subtype, however awaits exploration although it has been proposed that it serves to detect the solar orientation. Yamaguchi et al. (2010) recently established a useful method for testing the contribution of different IPRs to wavelength discrimination in Drosophila, which could be applied to address this exciting question in the near future.
Over the past few years, several factors have been identified that are necessary for creating the Drosophila retinal mosaic (summarized in Fig. 5.6D). These studies indicate that the p versus y decision is first made in R7 cells, and requires the stochastic activation of the transcription factor Spineless in yR7s (Ss; Wernet et al., 2006). Spineless is necessary and sufficient to activate Rh4 if expressed in IPRs or OPRs, and Ss-negative R7s (pR7s) express Rh3 by default (Wernet et al., 2006). However, mutation of a potential binding site for Ss in the Rh4 promoter does not affect reporter expression in vivo, and Ss is not able to regulate Rh4 promoter activity in vitro (T. Cook, unpublished results), indicating that Ss is likely to activate another factor to directly control Rh4 expression. Once the p versus y decision in R7s is made, pR7s sends an inductive signal to the underlying R8 (pR8s) to activate Rh5. In the absence of this signal, such as in eyes lacking all R7 cells, R8 cells express Rh6 by default (Chou et al., 1999). Therefore, although the pale fate in R7s is the default decision, the default decision in R8 cells is the yellow fate. Currently, the “pale” signaling molecule in pR7s remains unknown, but what is clear is that the activation of both pale opsins, Rh3 and Rh5, is directly controlled through K50 homeodomain binding sites by the transcription factor Otd (Fig. 5.5; Tahayato et al., 2003). Since Otd is expressed in all PRs, this suggests that a pale-specific coactivator is critical for this function.
Although the R7-dependent pale signaling pathway is not known, some of the signaling molecules that are required for mediating Rh5 versus Rh6 expression in the receiving R8 cell have been identified. These include the membrane-associated pleckstrin homology-containing protein Melted and the serine/threonine cytoplasmic kinase Warts (Wts, a.k.a. Lats; Mikeladze-Dvali et al., 2005). Melt expression is necessary and sufficient to induce Rh5 expression and to repress Wts expression in pR8s, whereas Wts is necessary and sufficient to induce Rh6 expression and repress Melt expression in yR8s. The bistable repression loop between Melt and Wts thereby ensures the mutually exclusive expression of Rh5 and Rh6 in different R8 subtypes. Consistent with Rh6 being the default R8 opsin, however, Wts appears to mediate the final output of the loop, while Melt is primarily involved in preventing Wts expression in pR8s. Since neither Melt nor Wts are DNA-binding factors, current work is focused on identifying the transcriptional mediators of the Melt/Wts pathway. This is a particularly interesting question, because Melt and Wts are most recognized for their roles in two independent growth regulatory pathways—the TOR and Hippo pathways, respectively (Harvey and Tapon, 2007; Hergovich and Hemmings, 2009; Reis and Hariharan, 2005; Teleman et al., 2005; Yin and Pan, 2007). Thus, further clarification of the role of these proteins in fly PR specification may have far-reaching implications in other fields of biology. Other questions that remain unanswered relate to how the initial stochastic decision for p versus y cell fate is made in the R7 layer, and what signaling pathway transmits this decision to the underlying R8 cell.
3. The Corneal Lens
In comparison with PR differentiation, much less is known regarding corneal lens formation in the Drosophila eye. This is somewhat surprising, because the lens is the most obvious structure in the fly eye for anatomical observation (Fig. 5.8A). The fly dioptic system comprises two distinct components: the corneal lens, a convex lamellated structure containing electron-dense microfibrils, and the underlying pseudocone, a fluid filled cavity seen by TEM cross section (Fig. 5.7D; Tomlinson, 1988; Youssef and Gardner, 1975). Early studies demonstrated that the corneal lens has a refractive index of 1.49, which determines a focal length that closely matches the distance from the corneal surface to the tip of the rhabdomeres (Youssef and Gardner, 1975). The pseudocone, however, has a lower refractive index of 1.3, suggesting that it may only have limited focusing power. Indeed, whether the pseudocone serves to focus light, similar to a lens, or merely creates the necessary distance between the cornea and the PRs remains unclear. Regardless, these data indicate that, like many land-dwelling animals, the Drosophila corneal surface is likely to be largely responsible for focusing light on the retina.
Figure 5.8.
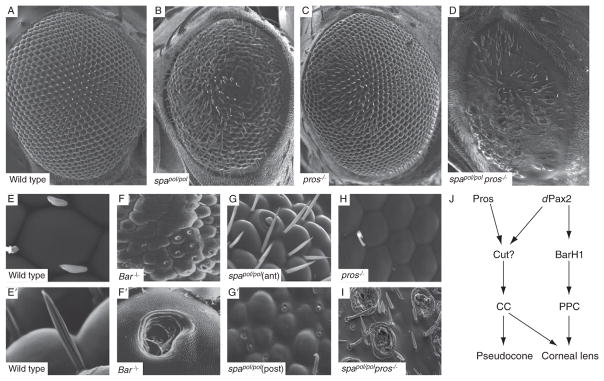
Scanning electron micrographs of adult eyes from wild type (A,E, E′), dPax2 spapol mutants (B,G, G′), whole eye prospero (pros) mutants (C,H), pros/spapol double mutants, and BarH1 mutants (F,F′; modified from (Higashijima et al., 1992), with permission from Genes and Development). The spapol mutant is a dPax2 hypomorph that shows differences in lens phenotypes between the anterior (G) and posterior (G′) portions of the eye. All images are oriented with anterior to the left. A summary of the role for these various CC and PPC expressed transcription factors is shown in J. See text for more detail.
3.1. Cone and PPC recruitment and patterning
The CCs are the first nonneuronal cells to be recruited in the eye imaginal disc, and this occurs immediately after PR specification is complete (Fig. 5.7A). In fact, CCs are derived from a common precursor pool of 5 cells, known as the R7 equivalence group, which gives rise to both the R7 PR and the four CCs (Dickson et al., 1992; Tomlinson et al., 1987). Cells within the R7 equivalence group express the Sevenless tyrosine kinase receptor, the EGF receptor, and the Notch receptor (Cagan and Ready, 1989b; Fortini et al., 1993; Jennings et al., 1994; Rebay et al., 1993; Tomlinson and Struhl, 2001; Tomlinson et al., 1987). Each of these cells require EGF and Notch signaling to form. However, only one of these cells differentiates into the R7 neuron due to the fact that only a single cell comes in direct contact with the Sevenless ligand, membrane-bound Boss, which is expressed on the previously specified R8 precursor. Since the Sevenless receptor signals through the same Ras/MAPK pathway as the EGF receptor, this Boss-receiving cell receives higher Ras signaling, and becomes specified as a neuron, while the remaining 4 cells adopt the default fate, that of cone (or Semper) cell (Cagan et al., 1992; Hart et al., 1990; Kramer et al., 1991; Reinke and Zipursky, 1988; Van Vactor et al., 1991). Over-activating Sevenless receptor signaling or overexpressing activated Ras in CC precursors can transform them into ectopic R7 PRs, and removing Sev signaling from the eye causes a failure in R7 differentiation but maintains the normal complement of four CCs (Basler et al., 1991; Dickson et al., 1992; Tomlinson and Ready, 1986). Together, these data led to the model that cells within the R7 equivalence group are all similarly capable of becoming R7 or CCs, and that this fate choice merely requires Sev-activated signaling. While these findings have been critical for defining the components of the Ras signaling pathway, the molecular mechanisms that mediate the dose-dependent neural (R7) versus nonneural (CC) fate decision remain unclear. Interestingly, however, not all cells within the R7 equivalence group respond the same to different mutants affecting R7/CC fate decisions instead, only one to two cells are generally affected (Basler et al., 1991; Bhattacharya and Baker, 2009; Dickson et al., 1992; Flores et al., 2000; Hayashi et al., 1998; Lai and Rubin, 1992; Matsuo et al., 1997; Tsuda et al., 2002). These data suggest that cells within the R7 equivalence group are actually not equivalent and that some bias toward R7 or CC fate exists in among these cells. Consistent with this idea, we have recently found that differential expression of two transcription factors, Pros and dPax2, in different CC precursors are important for establishing this bias (Fig. 5.7B), and that concurrent regulation of these factors is necessary to completely convert cells within the R7 equivalence group into R7 or CC fates (Charlton-Perkins and Cook, submitted).
Even though CCs are specified from the same precursor pool, they are recruited pairwise: first, the anterior and posterior CCs (aCC and pCC), followed, one or two ommatidial rows later, by the equatorial and polar CCs (eqCC and plCC; Tomlinson, 1988; Tomlinson and Ready, 1987a; Wolff and Ready, 1993). These two sets of CCs have also been referred to as primary and accessory CCs, respectively (Tomlinson and Ready, 1987a). Once recruited, the apical surfaces of the a/p CC contact each other, pushing aside the eq/pl CC surfaces. Soon after pupation, however, the apical contacts switch to the eq/pl CCs, as these cells rise apically above the a/p CCs. At ~18% pupation, the CCs then recruit two PPCs via Notch signaling, which ascend along the a/p CCs surfaces, wrap around the CC cluster, and meet in the middle of the pl/eq CCs. The PPCs remain anchored to the retinal floor until the retina begins to elongate, at which time they detach and fully wrap the CC bodies. Thus, the PPCs are the only cells in the retina that are not attached to the retinal floor (Cagan and Ready, 1989a). After the CCs and PPCs are recruited, both cell types provide EGF and Notch signals that are required for proper patterning of the remaining IOCs (Cagan and Ready, 1989b; Flores et al., 2000; Freeman, 1996; Miller and Cagan, 1998; Nagaraj and Banerjee, 2007; Voas and Rebay, 2004; Wech and Nagel, 2005; Yu et al., 2002; also see Chapter 4).
3.2. Lens terminal differentiation
The events that ultimately form a functional corneal lens can be separated into two developmental stages: one at ~50% pupation during which a “wispy lens material” is secreted that will later comprise the outermost corneal lens structure, and a later stage at ~75% pupation during which a gelatinous substance is secreted into the pseudocone (Cagan and Ready, 1989a). Together, the PPCs and CCs secrete the majority of the corneal lens, but SPCs also appear to contribute a darker material that is present at the tapered ends of the cornea between ommatidia. CCs, however, seem to solely contribute to the pseudocone (Cagan and Ready, 1989a; Waddington and Perry, 1960). Thus, it is likely that CCs switch developmental processes to contribute to the corneal lens versus the pseudocone. Because the corneal lens is a hard structure that is continuous with the cuticle of the fly head, as the CCs secrete the pseudocone, the CC cell bodies are compressed into a thin layer against the apical surfaces of the PRs. The PPCs, in contrast, surround the walls of the pseudocone (Fig. 5.7D).
The contents of the corneal lens and pseudocone remain largely unknown. Indeed, the only protein identified to date, named Drosocrystallin, was purified from a large-scale extraction of isolated corneal lens almost two decades ago (Komori et al., 1992). Biochemical analysis of Drosocrystallin revealed that it is calcium-binding glycoprotein, and sequence analysis suggests that it may be a member of a large class of cuticular proteins in insects (Janssens and Gehring, 1999; Komori et al., 1992). More recent immunohistochemistry analysis has demonstrated that Drosocrystallin is also present in mechanosensory organs, including the IOB (Dziedzic et al., 2009). Thus, like other developmental systems, Drosophila may have coopted a gene product involved in other cellular processes to be expressed in the lens simply because of its ability to form a clear crystalline material when expressed at high concentrations (Piatigorsky, 2003). Figure 5.7C shows that Drosocrystallin is almost exclusively synthesized in CCs at 50% pupation, and is secreted into the corneal lens structure at ~75% pupation. Drosocrystallin expression is also observed in the IOB lineage by this time (Fig. 5.7C, arrows). In the mature cornea, this protein distributes into fine lines that correspond to the striations that are observed in the structure by TEM (Fig. 5.7D; Komori et al., 1992). Two other abundant calcium-binding proteins were isolated at the same time as Drosocrystallin, but their identity remains unspecified.
With regard to proteins present in the pseudocone, even less is known. Two antibodies that specifically recognize this structure have been described (Edwards and Meyer, 1990; Fujita et al., 1982), but neither reagent remains available and the protein products recognized by these antibodies were not determined. However, one of these antibodies, 3G6, was originally identified as a glial cell marker in grasshoppers, and was only later shown to recognize crystalline cones from a variety of insects (Edwards and Meyer, 1990). Curiously, this observation supports a hypothesis that has been suggested several times by other investigators: that CCs may exhibit some glial-like features. This conjecture is partially based on the fact that CCs express Cut, dPax2, and Pros, transcription factors that are regularly associated with glia in other parts of the fly nervous system. In addition, PR morphology is severely disrupted in mutants that affect CC development (Banerjee et al., 2008; Daga et al., 1996; Fu and Noll, 1997; Siddall et al., 2003; Yan et al., 2003). Consistent with CCs serving as potential glia for PRs, CCs do fully enwrap the PRs in the retina: distally, the CCs form a “rhabdomere cap” that holds the most apical portion of the cell, interretinular fibers intercalate along the length of the PRs, and a bulbous cluster of the CC end feet wrap the basal portion of the rhabdomeres and cell body (Banerjee et al., 2008; Cagan and Ready, 1989a) (Figs. 5.1, 5.3). Thus, although it remains to be shown definitively, it is possible that CCs not only contribute to lens formation but may also function to maintain retinal integrity.
Many questions remain regarding the developmental regulation of CC and PPC differentiation and what the functional consequences of their development may be. Some insight into this has come from the analysis of homeodomain-containing transcription factors that are expressed in these cell types: BarH1/2, dPax2, Pros, and Cut (Blochlinger et al., 1993; Fu and Noll, 1997; Higashijima et al., 1992; Kauffmann et al., 1996). BarH1/2 is restricted to PPCs and the bristle lineage in the pupal eye, and loss of Bar function leads to fusion of some ommatidia and the appearance of a hole in the center of the corneal lens, previously described as a “blueberry” phenotype (Higashijima et al., 1992; Fig. 5.8F). A somewhat similar phenotype is observed in the most affected regions of spapol mutants, an eye-specific allele of dPax2, named for its sparkling (spa), polished (pol) appearance by light microscopy (Rickenbacher, 1954; Fig. 5.8B and G). dPax2 is expressed in both CCs and PPCs, but its primary phenotype appears to be loss of BarH1-positive PPCs (Fu and Noll, 1997). Recent studies, however, also suggest that dPax2 regulates Drosocrystallin expression (Dziedzic et al., 2009), which is largely synthesized in CCs (see above). Since lenses still form in spapol mutants, these data indicate that Drosocrystallin is not required for the formation of the crystallin lens structure per se.
Both Pros and dPax2 are transcriptional targets of the same pathways required for CC development (Flores et al., 2000; Hayashi et al., 2008; Xu et al., 2000). However, Pros is only expressed in CC nuclei during early CC recruitment, is turned off during mid-pupation, and its expression is then reinitiated in the CC cytoplasm at ~70% pupation (Charlton-Perkins and Cook, submitted; Cook et al., 2003; Kauffmann et al., 1996). Interestingly, because Pros expression is reinitiated at the same time that pseudocone formation begins in CCs, further understanding pros gene regulation in the eye may provide insight into the genetic pathways involved in this latter stage of lens development. Similarly to dPax2 mutants, removal of Pros during CC development causes relatively subtle changes in lens formation (Charlton-Perkins and Cook, submitted; also see Fig. 5.8B, C, G, and H). In contrast, removing both Pros and dPax2 causes a complete loss in lens formation and no CCs form (Fig. 5.8D and I) and (Charlton-Perkins and Cook, submitted). Thus, Pros and dPax2 combinatorially participate in CC formation. Surprisingly, despite the fact that Cut is expressed in all CCs from early specification through adulthood, little is known regarding its role in CC specification (Daga et al., 1996), and no role for Cut in lens genesis has been reported. However, its expression correlates strongly with properly specified CCs, suggesting that it will play an important role in CC function. Collectively, these data suggest that Pros and dPax2 regulate CC-specific targets and contribute to pseudocone and corneal lens formation, while dPax2 controls BarH1 expression in PPCs and that these cells contribute to maintaining separate ommatidia and aids in completing the formation of the corneal lens (Fig. 5.8J).
4. SPCs and TPCs: The Fly Retinal Pigment Epithelia?
Besides the PPCs, two additional nonneuronal pigmented cell types are present in the adult fly eye, known as SPCs and TPCs. These are also often referred to as IOCs, as they are shared between ommatidia, and arise from the same pool of interommatidial precursor cells (IPCs). Morphologically, SPCs and TPCs differ by the number of cell contacts they establish in the mature retina: SPCs contact two cells, whereas TPCs contact three. In addition, SPCs are involved in the secretion of at least a portion of the corneal lens, while TPCs (but not SPCs) must establish alternating positions at the vertices of each ommatidia with the bristle cell lineage (Cagan and Ready, 1989a), suggesting that the development of these two cell types may be somewhat different. It is also intriguing that SPCs and TPCs may be the default state of cells within the eye imaginal disc, since in eyeless mutants, the few cells that do survive differentiate into IOCs, and in ommatidia localized to the eye margin, all PRs, CCs, and PPCs are induced to die, yet SPC/TPCs are retained (Lim and Tomlinson, 2006).
Both the initial recruitment and patterning of SPCs and TPCs is well documented and nicely reviewed in Chapter 4. Briefly, after PPC recruitment, any remaining unspecified cells in the eye begin vying for contacts between PPCs. Approximately 70% of these cells will form SPC/TPCs, while the remaining 30% (~2,000 cells/eye) will be eliminated by programmed cell death. The oblique SPCs are established first, based on contacts with two PPCs, followed by formation of the TPCs and horizontal SPCs, which contact three and four PPCs, respectively (Cagan and Ready, 1989a). By ~37% pupation, IOC recruitment and patterning is largely complete, although at least some cell death continues until ~62% pupation (Cagan and Ready, 1989a).
After recruitment, the apical surfaces of the SPCs/TPCs gradually tighten via a process involving the transcription factor Escargot, a member of the Snail-related family of zinc finger transcription factors (Lim and Tomlinson, 2006). In contrast, the basal surfaces of the SPC/TPCs expand to form a nice petal shaped lattice, or spokes of a wheel (Fig. 5.2H), that ultimately forms the fenestrated membrane of the retina. This membrane may functionally represent the blood–retina barrier, and is rich in stress fibers and septate junctions (Banerjee et al., 2008; Longley and Ready, 1995). The molecular regulators of these complex morphogenetic changes, however fascinating, remain unexplored.
By ~62.5% pupation, the IOCs begin to generate two major types of pigmented granules. Type I granules are large and filled with the brown-colored pigment xanthommatin, also referred to as ommachrome. These granules are present in SPCs/TPCs, PPCs, PRs, and CC feet (Cagan and Ready, 1989a; Shoup, 1966; Wolff and Ready, 1993). Type II granules are small and contain xanthommatin and drosopterin, a red pigment also known as pteridine, and these are predominantly found in SPCs/TPCs. Over 85 different eye color mutants have been identified in the past 100 years, and these have not only leant insight into how these pigment granules are formed, but have also led to a better understanding of a wide range of protein sorting processes (Lloyd et al., 1998). These eye color mutants have been categorized into three functional subclasses: (1) the granule group, (2) the pigmentation synthesis group, and (3) the ABC transporter group. The “granule group” primarily encodes factors involved in protein sorting/biogenesis, and includes members of the AP3 adaptor complex, the VPS sorting complex, and several members of the Rab family of small GTPases (Kretzschmar et al., 2000; Ma et al., 2004; Mullins et al., 1999; Ooi et al., 1997; Simpson et al., 1997; Warner et al., 1998). The “pigment synthesis group” encodes enzymes that are involved in the processing of intracellular tryptophan required for the formation of xanthommatin and drosopterin (Summers et al., 1982). Finally, the “ABC transporter group” includes complexes associated with transmembrane transport (Jones and George, 2004). Members of this group are White, Brown, and Scarlet, all part of the ABC-G subfamily of ABC transporters. They represent “halves” of a complete ABC transporter and become active by heterodimerization. White/Brown dimers transport drosopterin pigments, while White/Scarlet dimers transport xanthommatin pigments (Dreesen et al., 1988; Ewart et al., 1994; Pepling and Mount, 1990). Originally, these transporters were thought to be localized to the IOC cell membranes, but later studies suggest that White and Scarlet transport xanthommatin precursors directly into the pigment granules, thus using a mechanism analogous to that used for melanin transport into melanosomes (Mackenzie et al., 2000; Tearle et al., 1989).
Functionally, eye pigmentation is important for limiting light scattering between ommatidia. However, it is also important for maintaining PR integrity, protecting them from light-induced damage. For instance, mutations in the White gene, and other mutations that lead to white-eyed flies, causes severe rhabdomere degeneration when flies are exposed to constant light for 10 days, whereas wild-type, red-eyed flies are unaffected under identical conditions (Lee and Montell, 2004). This evidence is compelling in light of the neuroprotective function assigned to eye pigmentation in humans and vertebrate models, which show retinal degeneration acceleration in situations of retinal hypopigmentation.
Another critical role of SPC/TPCs is in the formation of the Rhodopsin chromophore 11-cis-retinal (Wang and Montell, 2005, 2007; Wang et al., 2007). This important function of the SPC/TPCs was only recently discovered, with the majority of the vitamin A processing pathway in the fly being expressed outside the retina. NINA-B (neither inactivation nor after-potential B), the functional ortholog of RPE-65 in Drosophila (Oberhauser et al., 2008), for instance, is expressed in neurons within the brain, while NINA-D, a scavenger receptor required for dietary B-carotene absorption, is expressed in the midgut (Gu et al., 2004; Wang et al., 2007). Montell and coworkers reasoned, however, that since Drosophila Rhodopsin maturation requires chromophore binding, screening for mutants that disrupted specific aspects of Rhodopsin function may lead to the identification of additional proteins involved in the Vitamin A processing pathways. Indeed, from such a screen, the retinoid binding protein PINTA (prolonged depolarization after-potential is not apparent) was identified. PINTA is specifically expressed in IOCs, placing these cells for the first time into the phototransduction pathway, and establishing that IOCs are more similar to the vertebrate retinal pigment epithelia (RPE) than previously thought. While PINTA remains the only protein involved in chromophore production that has currently been localized to IOCs, the oxidoreductase NINA-G functions downstream of PINTA (Ahmad et al., 2006; Sarfare et al., 2005), making it likely this factor, as well as other factors involved in chromophore production, are also expressed in these cells.
5. Comparison of the Drosophila Eye with the Vertebrate Eye
One of the first breakthroughs into the idea of a common origin of eye development came from studies on the shared function of Pax6 in regulating eye formation (Glaser et al., 1992; Halder et al., 1995; Quiring et al., 1994). Since then, the majority of genes in the retinal determination cascade originally identified in Drosophila have been shown to have homologs in vertebrates that are comparably critical during early eye specification. These include the transcription factors Pax6, Dac, Eya, and So/Six3/Six6 (Gehring and Ikeo, 1999; Treisman, 1999; Wawersik and Maas, 2000; Wawersik et al., 2000; also see Chapter 1). Patterning factors are also conserved among different eye types: Hedgehog in flies and Sonic hedgehog in vertebrates, for instance, each provides a moving wave of morphogenesis during early retinogenesis (Jarman, 2000; Wallace, 2008). In addition, proteins such as Ato/Ath5 and Pros/Prox1 have been shown to play evolutionarily conserved roles in generating different neuronal cell types during fly/vertebrate retinogenesis (Brown et al., 2001; Cook, 2003; Cook et al., 2003; Dyer, 2003; Dyer et al., 2003; Wang et al., 2001; White and Jarman, 2000). Determining what other similarities exist among visual systems across separate phyla will be an ongoing enterprise and is nicely covered in several recent reviews (Arendt, 2003; Cook and Zelhof, 2008; Gehring, 2005; Jonasova and Kozmik, 2008; Sanes and Zipursky, 2010; Vopalensky and Kozmik, 2009). In the following section, we highlight some of the accumulating evidence suggesting that PR differentiation between vertebrates and flies share many developmental features.
5.1. The neural retina
Initially, the idea that vertebrate and invertebrate PRs are developmentally related was highly debated because of the many obvious differences between these cell types. For instance, light stimulation produces opposite electric potentials in these two PR cell types—in vertebrates they hyperpolarize via a phosphodiesterase cascade, while invertebrate PRs depolarize, using a phospholipase C cascade. The expanded PR apical surfaces used to concentrate light absorption also use different strategies—vertebrates have microtubule-based ciliary outer segments while Drosophila has actin-rich rhadomeric membranes. Vertebrate PR cells indirectly transfer visual information to the brain via retinal interneurons and ganglion cells, whereas Drosophila PRs are the only retina-specific cell types and project to functionally equivalent interneurons located in the underlying optic lobes (Sanes and Zipursky, 2010). Despite these marked anatomical and physiological differences, fly and vertebrate PR share striking similarities in the factors required for their morphogenesis. These include Otd/Crx (Furukawa et al., 1997; Ranade et al., 2008; Rivolta et al., 2001; Swaroop et al., 1999; Tahayato et al., 2003; Vandendries et al., 1996), Crumbs/CRB1 (Izaddoost et al., 2002; Kowalczyk and Moses, 2002; Mehalow et al., 2003; Pellikka et al., 2002; Richard et al., 2006b), Arrestin (Chen et al., 1999; Lee and Montell, 2004; Nakazawa et al., 1998), Prominin (Maw et al., 2000; Yang et al., 2008; Zelhof et al., 2006), Spacemaker (Eyes Shut)/RP25 (Abd El-Aziz et al., 2008; Collin et al., 2008; Husain et al., 2006; Osorio, 2007; Zelhof et al., 2006), MyoIII (Hicks et al., 1996; Porter and Montell, 1993; Redowicz, 2002; Walsh et al., 2002), and Rab proteins (Deretic, 1998; Deretic et al., 1995; Kwok et al., 2008; Li et al., 2007; Marzesco et al., 2001; Moritz et al., 2001; Shetty et al., 1998). Moreover, the majority of these factors are associated with retinal degenerative diseases, further emphasizing the evolutionary importance of these factors for maintaining an intact adult visual system.
Not only does PR morphogenesis in fly and vertebrate share similar factors, but other differentiation events also make use of common regulators. For example, many of the same factors that are critical for PR axon guidance in flies are also used for patterning neuronal connections in the vertebrate retina, raising the exciting possibility that the fly eye will provide an effective paradigm for deciphering the relatively intricate axonal patterning present in the vertebrate visual system (for a more complete review of this topic, see Sanes and Zipursky, 2010). In addition, PR-specific gene regulation also involves conserved factors between vertebrates and invertebrates. Drosophila Otd and its vertebrate orthologs Otx2 and Crx, for instance, are essential for regulating many common PR-specific target genes (Chen et al., 1997; Furukawa et al., 1997; Hsiau et al., 2007; Koike et al., 2007; Livesey et al., 2000; Nishida et al., 2003; Peng and Chen, 2005; Ranade et al., 2008; Tahayato et al., 2003). Combined, compelling evidence is beginning to emerge that suggest that similar genetic pathways are involved in building and/or maintaining multiple PR cell types. Thus, as has been postulated (Arendt, 2003; Cook and Zelhof, 2008; Erclik et al., 2009; Gehring, 2005; Vopalensky and Kozmik, 2009), eyes in Urbilateria (Bilateria’s last common ancestor) may have had PR cells that already expressed a number of interacting factors that have been maintained in the PR cell types found today, while structural and functional aspects have specialized to meet particular life conditions.
5.2. The cornea and lens
To date, few studies have addressed whether the genetic pathways involved in lens morphogenesis are conserved between vertebrates and invertebrates. However, like vertebrates, flies have a corneal structure that is largely responsible for focusing, and a crystalline region interposed between the cornea and the retina (the pseudocone in flies and the lens in mammals) that is likely to also contribute to the focusing power. Because a crystalline structure is necessary for a functional dioptic system, identifying proteins that regulate crystallin expression is a useful avenue for exploring conservation in lens development. Indeed, although many crystallins are recruited from ancestral proteins with distinct functions from their refractive function in the eye, their transcriptional regulation is relatively conserved (Cvekl and Duncan, 2007; Kozmik et al., 2003; Piatigorsky, 2003, 2006; Tomarev and Piatigorsky, 1996)
An impressive demonstration of this conservation came from studies that revealed that the chicken δ1-crystallin enhancer can direct expression specifically in the lens-secreting cells in Drosophila (Blanco et al., 2005). In vertebrates, this enhancer relies on binding sites for Sox2 and Pax6; similarly, Blanco and collaborators demonstrated that fly SoxN and dPax2 perform these same functions in Drosophila. The observation that dPax2 and Pax6 are functional homologs in this context is exciting, because both factors arise from a common ancestral factor known as PaxB (Kozmik et al., 2003). Thus, these data suggests that upon the divergence of PaxB into two separate factors, Pax2 “claimed” lens function in invertebrates whereas Pax6 claimed this function in vertebrates. Besides Pax and Sox factors sharing functions during crystallin regulation, our recent findings that Pros is important during the differentiation of lens-secreting cells in Drosophila (Charlton-Perkins and Cook, submitted) parallels findings that vertebrate Pros, Prox1, is important for fiber cell elongation (Wigle et al., 1999) and lends further support for genetic pathways being shared to form highly diverse lens structures.
In addition to transcription factors that may be functionally conserved, a number of signaling pathways may also serve overlapping functions during vertebrate and invertebrate lens genesis. For instance, high levels of FGF signaling are critical for many aspects of vertebrate lens development, and strong redundancy in this system appears to have been maintained to ensure correct signaling (Robinson, 2006; Zhao et al., 2008). Similarly, as discussed earlier, proper levels of EGF signaling are essential for multiple aspects of Drosophila CC and PPC differentiation, and only slight variations in these levels have dramatic effects on lens development (Flores et al., 2000; Fortini et al., 1992; Freeman, 1996; Hayashi et al., 2008; Miller and Cagan, 1998; Nagaraj and Banerjee, 2007; Tsuda et al., 2002; Voas and Rebay, 2004; Wech and Nagel, 2005). Since both pathways mediate their functions through the Ras/MAPK pathway, it is possible that vertebrate lenses adopted the FGF receptor whereas the fly adopted the EGF receptor to mediate the same events, much like the diverged functions of Pax6 and dPax2 described above. Interestingly, while Notch signaling has long been known to be critical for lens morphogenesis in flies, only recently has Notch signaling only recently been recognized for its contribution to vertebrate lens development (Cagan and Ready, 1989b; Jia et al., 2007; Le et al., 2009; Miller and Cagan, 1998; Rowan et al., 2008; Saravanamuthu et al., 2009). Fortunately, in flies, only one EGF receptor and one Notch receptor are present, whereas in the mouse, knocking out three of the four FGF receptors was necessary to reveal the extent to which this signaling pathway contributes to lens formation (Zhao et al., 2008). Similarly, it is likely that multiple Notch receptors and Notch ligands are going to participate in vertebrate lens formation (Bao and Cepko, 1997; Le et al., 2009; Saravanamuthu et al., 2009; Zecchin et al., 2005). Once more, the possibility of using the fly as a genetic model for understanding lens formation and maintenance should be an advantageous approach for addressing future questions related to normal and diseased states affecting the eye anterior segment development.
5.3. The pigmented epithelia
Evidence for whether the Drosophila IOCs are the functional equivalent to the vertebrate RPE remains particularly sparse. The vertebrate RPE accomplishes complex and diverse functions that make it essential for visual function, including light absorption, water and ionic balancing to guarantee PR excitability, maintenance of immune privilege, nutrient uptake and delivery to PRs, cycling of retinal, and recycling of outer segments (Rosenthal et al., 2005; Strauss, 2005). Moreover, malfunction of any one of these functions leads to vision failure and/or retinopathies. Interestingly, at least a subset of these functions has now been shown to be present in Drosophila, including light absorption and the Rhodopsin chromophore production. Likewise, both of these functions are necessary for retinal normal function. Another similarity between vertebrate RPE cells and Drosophila IOCs is the use ABC transporters to generate their pigmented granules. Although the vertebrate ABCRs are members of the A subfamily of transporters, while the fly’s belong to the G subfamily, the ABC-A and G groups share the strongest conservation among the other subgroups (Jones and George, 2004; Jones et al., 2009). Interestingly, ABCA4 mutations are associated with human retinal degenerative diseases, and albinos are recognized for their sensitivity to light-induced retinal damage. This parallels nicely with the fact that flies lacking eye pigmentation, either through specific mutations in ABC transporters or through other depigmentation mutations, show drastic light-induced PR degeneration (Lee and Montell, 2004; Xu et al., 2004; TC, unpublished observations). Finally, the recent postulation that Drosophila IOCs may be essential for creating the fly blood–retina barrier (Banerjee et al., 2008) harkens strongly to the role of the RPE providing the first line of protection from the surrounding choroidal blood supply. Despite these parallels, it is fairly certain that not all functions of the vertebrate and invertebrate RPE are conserved. For instance, in vertebrates, the RPE is critical for phagocytosing the constantly growing PR outer segments, whereas in Drosophila, no convincing evidence suggests that rhabdomeres shed their membranes into the IOC compartment. Nevertheless future studies aimed at detecting other possible similarities between the fly and vertebrate pigment epithelial cells are likely to gain a deeper understanding of at least some aspects of RPE function.
6. Summary
This age of high throughput gene expression profiling is an exciting time in biology and has led to a better appreciation of the striking degree to which developmental processes have been conserved to create different body plans. The eye is no exception, and as we have attempted to summarize here, a remarkable symmetry is found between vertebrate and fly eyes. Based on the rapid progress we have recently made in this area, there is no doubt that continuing such efforts, taking full advantage of the genetic tools now available in both mouse and fly models, will identify additional shared developmental processes that generate a diversity of cell types and visual structures.
Acknowledgments
The authors wish to thank members of the Cook, Gebelein, Brown, and Lang labs, Ross Cagan, Claude Desplan, Justin Kumar, Don Ready, and Andy Zelhof for discussions and input. We also apologize to those whose work has contributed to our current understanding of eye development that could not be included. This work was supported by NEI/NIH grant R01-EY017907 to TAC.
Abbreviations
- CC
cone cell
- IOB
interommatidial bristle
- IOC
interommatidial cell
- IPR
inner photoreceptor
- MF
morphogenetic furrow
- OPR
outer photoreceptor
- PPC
primary pigment cell
- PR
photoreceptor
- Pros
Prospero
- Rh
Rhodopsin
- RPE
retinal pigment epithelia
- Sens
Senseless
- SPC
secondary pigment cell
- TPC
tertiary pigment cell
References
- Abd El-Aziz MM, et al. EYS, encoding an ortholog of Drosophila spacemaker, is mutated in autosomal recessive retinitis pigmentosa. Nat Genet. 2008;40:1285–1287. doi: 10.1038/ng.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler PN. Planar signaling and morphogenesis in Drosophila. Dev Cell. 2002;2:525–535. doi: 10.1016/s1534-5807(02)00176-4. [DOI] [PubMed] [Google Scholar]
- Ahmad ST, et al. The role of Drosophila ninaG oxidoreductase in visual pigment chromophore biogenesis. J Biol Chem. 2006;281:9205–9209. doi: 10.1074/jbc.M510293200. [DOI] [PubMed] [Google Scholar]
- Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47:563–571. [PubMed] [Google Scholar]
- Bachmann A, et al. Drosophila Lin-7 is a component of the Crumbs complex in epithelia and photoreceptor cells and prevents light-induced retinal degeneration. Eur J Cell Biol. 2008;87:123–136. doi: 10.1016/j.ejcb.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Banerjee S, et al. Septate junctions are required for ommatidial integrity and blood-eye barrier function in Drosophila. Dev Biol. 2008;317:585–599. doi: 10.1016/j.ydbio.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler K, et al. Ligand-independent activation of the sevenless receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell. 1991;64:1069–1081. doi: 10.1016/0092-8674(91)90262-w. [DOI] [PubMed] [Google Scholar]
- Baumann O. Spatial pattern of nonmuscle myosin-II distribution during the development of the Drosophila compound eye and implications for retinal morphogenesis. Dev Biol. 2004;269:519–533. doi: 10.1016/j.ydbio.2004.01.047. [DOI] [PubMed] [Google Scholar]
- Berghammer AJ, et al. A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE. The HLH protein Extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Dev Biol. 2009;327:288–300. doi: 10.1016/j.ydbio.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker G, Reichert H. Visual learning in a photoreceptor degeneration mutant of Drosophila melanogaster. J Comp Physiol. 1978;A127:29–38. [Google Scholar]
- Blanco J, et al. Functional analysis of the chicken delta1-crystallin enhancer activity in Drosophila reveals remarkable evolutionary conservation between chicken and fly. Development. 2005;132:1895–1905. doi: 10.1242/dev.01738. [DOI] [PubMed] [Google Scholar]
- Blochlinger K, et al. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development. 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- Brennan CA, Moses K. Determination of Drosophila photoreceptors: Timing is everything. Cell Mol Life Sci. 2000;57:195–214. doi: 10.1007/PL00000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, et al. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferty P, et al. The receptor tyrosine kinase Off-track is required for layer-specific neuronal connectivity in Drosophila. Development. 2004;131:5287–5295. doi: 10.1242/dev.01406. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989a;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989b;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- Cagan RL, et al. The bride of sevenless and sevenless interaction: Internalization of a transmembrane ligand. Cell. 1992;69:393–399. doi: 10.1016/0092-8674(92)90442-f. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, et al. The Iroquois homeobox genes function as dorsal selectors in the Drosophila head. Development. 2000;127:1921–1929. doi: 10.1242/dev.127.9.1921. [DOI] [PubMed] [Google Scholar]
- Chang HY, Ready DF. Rescue of photoreceptor degeneration in rhodopsin-null Drosophila mutants by activated Rac1. Science. 2000;290:1978–1980. doi: 10.1126/science.290.5498.1978. [DOI] [PubMed] [Google Scholar]
- Chen S, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. Increased susceptibility to light damage in an arrestin knockout mouse model of Oguchi disease (stationary night blindness) Invest Ophthalmol Vis Sci. 1999;40:2978–2982. [PubMed] [Google Scholar]
- Chiba A. Precision networking: A look through the eyes of a fly. Neuron. 2001;32:381–384. doi: 10.1016/s0896-6273(01)00498-6. [DOI] [PubMed] [Google Scholar]
- Chou WH, et al. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–1115. doi: 10.1016/s0896-6273(00)80243-3. [DOI] [PubMed] [Google Scholar]
- Chou WH, et al. Patterning of the R7 and R8 photoreceptor cells of Drosophila: Evidence for induced and default cell-fate specification. Development. 1999;126:607–616. doi: 10.1242/dev.126.4.607. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Afferent growth cone interactions control synaptic specificity in the Drosophila visual system. Neuron. 2000;28:427–436. doi: 10.1016/s0896-6273(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Collin RW, et al. Identification of a 2 Mb human ortholog of Drosophila eyes shut/spacemaker that is mutated in patients with retinitis pigmentosa. Am J Hum Genet. 2008;83:594–603. doi: 10.1016/j.ajhg.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook T. Cell diversity in the retina: More than meets the eye. Bioessays. 2003;25:921–925. doi: 10.1002/bies.10356. [DOI] [PubMed] [Google Scholar]
- Cook B, Zelhof AC. Photoreceptors in evolution and disease. Nat Genet. 2008;40:1275–1276. doi: 10.1038/ng1108-1275. [DOI] [PubMed] [Google Scholar]
- Cook T, et al. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Duncan MK. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: Three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga A, et al. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996;10:1194–1205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- Dearborn R, Jr, Kunes S. An axon scaffold induced by retinal axons directs glia to destinations in the Drosophila optic lobe. Development. 2004;131:2291–2303. doi: 10.1242/dev.01111. [DOI] [PubMed] [Google Scholar]
- Deretic D. Post-Golgi trafficking of rhodopsin in retinal photoreceptors. Eye (Lond) 1998;12(Pt 3b):526–530. doi: 10.1038/eye.1998.141. [DOI] [PubMed] [Google Scholar]
- Deretic D, et al. rab8 in retinal photoreceptors may participate in rhodopsin transport and in rod outer segment disk morphogenesis. J Cell Sci. 1995;108(Pt 1):215–224. doi: 10.1242/jcs.108.1.215. [DOI] [PubMed] [Google Scholar]
- Deretic D, et al. Phosphoinositides, ezrin/moesin, and rac1 regulate fusion of rhodopsin transport carriers in retinal photoreceptors. Mol Biol Cell. 2004;15:359–370. doi: 10.1091/mbc.E03-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson B, et al. Prepattern in the developing Drosophila eye revealed by an activated torso—Sevenless chimeric receptor. Genes Dev. 1992;6:2327–2339. doi: 10.1101/gad.6.12a.2327. [DOI] [PubMed] [Google Scholar]
- Domingos PM, et al. Spalt transcription factors are required for R3/R4 specification and establishment of planar cell polarity in the Drosophila eye. Development. 2004;131:5695–5702. doi: 10.1242/dev.01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroquez DB, Rebay I. Signal integration during development: Mechanisms of EGFR and Notch pathway function and cross-talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- Dreesen TD, et al. The brown protein of Drosophila melanogaster is similar to the white protein and to components of active transport complexes. Mol Cell Biol. 1988;8:5206–5215. doi: 10.1128/mcb.8.12.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA. Regulation of proliferation, cell fate specification and differentiation by the homeodomain proteins Prox1, Six3, and Chx10 in the developing retina. Cell Cycle. 2003;2:350–357. [PubMed] [Google Scholar]
- Dyer MA, et al. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Dziedzic K, et al. The transcription factor D-Pax2 regulates crystallin production during eye development in Drosophila melanogaster. Dev Dyn. 2009;238:2530–2539. doi: 10.1002/dvdy.22082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl JB, Britt SG. Expression of Drosophila rhodopsins during photoreceptor cell differentiation: Insights into R7 and R8 cell subtype commitment. Gene Expr Patterns. 2006;6:687–694. doi: 10.1016/j.modgep.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Edwards JS, Meyer MR. Conservation of antigen 3G6: A crystalline cone constituent in the compound eye of arthropods. J Neurobiol. 1990;21:441–452. doi: 10.1002/neu.480210306. [DOI] [PubMed] [Google Scholar]
- Erclik T, et al. Eye evolution at high resolution: The neuron as a unit of homology. Dev Biol. 2009;332:70–79. doi: 10.1016/j.ydbio.2009.05.565. [DOI] [PubMed] [Google Scholar]
- Ewart GD, et al. Mutational analysis of the traffic ATPase (ABC) transporters involved in uptake of eye pigment precursors in Drosophila melanogaster Implications for structure-function relationships. J Biol Chem. 1994;269:10370–10377. [PubMed] [Google Scholar]
- Fan SS. Dynactin affects extension and assembly of adherens junctions in Drosophila photoreceptor development. J Biomed Sci. 2004;11:362–369. doi: 10.1007/BF02254441. [DOI] [PubMed] [Google Scholar]
- Fan SS, Ready DF. Glued participates in distinct microtubule-based activities in Drosophila eye development. Development. 1997;124:1497–1507. doi: 10.1242/dev.124.8.1497. [DOI] [PubMed] [Google Scholar]
- Feiler R, et al. Ectopic expression of ultraviolet-rhodopsins in the blue photoreceptor cells of Drosophila: Visual physiology and photochemistry of transgenic animals. J Neurosci. 1992;12:3862–3868. doi: 10.1523/JNEUROSCI.12-10-03862.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Vize JA, Mosley KL. Marbles mutants: Uncoupling cell determination and nuclear migration in the developing Drosophila eye. Development. 1994;120:2609–2618. doi: 10.1242/dev.120.9.2609. [DOI] [PubMed] [Google Scholar]
- Flores GV, et al. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Rubin GM. Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev. 1990;4:444–463. doi: 10.1101/gad.4.3.444. [DOI] [PubMed] [Google Scholar]
- Fortini ME, et al. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- Fortini ME, et al. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, et al. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron. 2001;32:403–414. doi: 10.1016/s0896-6273(01)00480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Fryxell KJ, Meyerowitz EM. An opsin gene that is expressed only in the R7 photoreceptor cell of Drosophila. EMBO J. 1987;6:443–451. doi: 10.1002/j.1460-2075.1987.tb04774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita SC, et al. Monoclonal antibodies against the Drosophila nervous system. Proc Natl Acad Sci USA. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, et al. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Garrity PA, et al. Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered. 2005;96:171–184. doi: 10.1093/jhered/esi027. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Glaser T, et al. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Estevez C, et al. Transgenic planarian lines obtained by electroporation using transposon-derived vectors and an eye-specific GFP marker. Proc Natl Acad Sci USA. 2003;100:14046–14051. doi: 10.1073/pnas.2335980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, et al. Drosophila ninaB and ninaD act outside of retina to produce rhodopsin chromophore. J Biol Chem. 2004;279:18608–18613. doi: 10.1074/jbc.M400323200. [DOI] [PubMed] [Google Scholar]
- Hardie RC. Electrophysiological analysis of the fly retina. I Comparative properties of R1–6 and R7 and R8. J Comp Physiol. 1979;129:19–33. [Google Scholar]
- Hardie RC, Kirschfeld K. Ultraviolet sensitivity of fly photoreceptors R7 and R8: Evidence for a sensitizing function. Biophys Struct Mech. 1983;9:171–180. [Google Scholar]
- Halder G, et al. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hardie R. In: Functional organization of the fly retina. Ottoson D, editor. Vol. 5. Springer; Berlin/Heidelberg/New York/Toronto: 1985. pp. 1–79. [Google Scholar]
- Hart AC, et al. Induction of cell fate in the Drosophila retina: The bride of sevenless protein is predicted to contain a large extracellular domain and seven transmembrane segments. Genes Dev. 1990;4:1835–1847. doi: 10.1101/gad.4.11.1835. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway—An emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Hayashi T, et al. Specification of primary pigment cell and outer photoreceptor fates by BarH1 homeobox gene in the developing Drosophila eye. Dev Biol. 1998;200:131–145. doi: 10.1006/dbio.1998.8959. [DOI] [PubMed] [Google Scholar]
- Hayashi T, et al. Cell-type-specific transcription of prospero is controlled by combinatorial signaling in the Drosophila eye. Development. 2008;135:2787–2796. doi: 10.1242/dev.006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Hemmings BA. Mammalian NDR/LATS protein kinases in hippo tumor suppressor signaling. Biofactors. 2009;35:338–345. doi: 10.1002/biof.47. [DOI] [PubMed] [Google Scholar]
- Hicks JL, et al. Role of the ninaC proteins in photoreceptor cell structure: Ultrastructure of ninaC deletion mutants and binding to actin filaments. Cell Motil Cytoskeleton. 1996;35:367–379. doi: 10.1002/(SICI)1097-0169(1996)35:4<367::AID-CM8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Higashijima S, et al. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 1992;6:50–60. doi: 10.1101/gad.6.1.50. [DOI] [PubMed] [Google Scholar]
- Hing H, et al. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Hsiau TH, et al. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE. 2007;2:e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KG, Stark WS. The roles of Drosophila ocelli and compound eye in phototaxis. J Comp Physiol. 1980;135:85–95. [Google Scholar]
- Huber A, et al. Molecular cloning of Drosophila Rh6 rhodopsin: The visual pigment of a subset of R8 photoreceptor cells. FEBS Lett. 1997;406:6–10. doi: 10.1016/s0014-5793(97)00210-x. [DOI] [PubMed] [Google Scholar]
- Husain N, et al. The agrin/perlecan-related protein eyes shut is essential for epithelial lumen formation in the Drosophila retina. Dev Cell. 2006;11:483–493. doi: 10.1016/j.devcel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Izaddoost S, et al. Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature. 2002;416:178–183. doi: 10.1038/nature720. [DOI] [PubMed] [Google Scholar]
- Janssens H, Gehring WJ. Isolation and characterization of drosocrystallin, a lens crystallin gene of Drosophila melanogaster. Dev Biol. 1999;207:204–214. doi: 10.1006/dbio.1998.9170. [DOI] [PubMed] [Google Scholar]
- Jarman AP. Developmental genetics: Vertebrates and insects see eye to eye. Curr Biol. 2000;10:R857–R859. doi: 10.1016/s0960-9822(00)00821-6. [DOI] [PubMed] [Google Scholar]
- Jennings B, et al. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- Jia J, et al. The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol Cell Biol. 2007;27:7236–7247. doi: 10.1128/MCB.00780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonasova K, Kozmik Z. Eye evolution: Lens and cornea as an upgrade of animal visual system. Semin Cell Dev Biol. 2008;19:71–81. doi: 10.1016/j.semcdb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Jones PM, George AM. The ABC transporter structure and mechanism: Perspectives on recent research. Cell Mol Life Sci. 2004;61:682–699. doi: 10.1007/s00018-003-3336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PM, et al. ABC transporters: A riddle wrapped in a mystery inside an enigma. Trends Biochem Sci. 2009;34:520–531. doi: 10.1016/j.tibs.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Kaminker JS, et al. Control of photoreceptor axon target choice by transcriptional repression of Runt. Nat Neurosci. 2002;5:746–750. doi: 10.1038/nn889. [DOI] [PubMed] [Google Scholar]
- Kauffmann RC, et al. Ras1 signaling and transcriptional competence in the R7 cell of Drosophila. Genes Dev. 1996;10:2167–2178. doi: 10.1101/gad.10.17.2167. [DOI] [PubMed] [Google Scholar]
- Kirschfeld K, et al. A photostable pigment within the rhabdomeres of fly photoreceptors no. 7. J Comp Physiol. 1978;125:275–284. [Google Scholar]
- Koike C, et al. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007;27:8318–8329. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori N, et al. Drosocrystallin, a major 52 kDa glycoprotein of the Drosophila melanogaster corneal lens. Purification, biochemical characterization, and subcellular localization. J Cell Sci. 1992;102(Pt 2):191–201. doi: 10.1242/jcs.102.2.191. [DOI] [PubMed] [Google Scholar]
- Kowalczyk AP, Moses K. Photoreceptor cells in flies and mammals: Crumby homology? Dev Cell. 2002;2:253–254. doi: 10.1016/s1534-5807(02)00138-7. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, et al. Role of Pax genes in eye evolution: A cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev Cell. 2003;5:773–785. doi: 10.1016/s1534-5807(03)00325-3. [DOI] [PubMed] [Google Scholar]
- Kramer H, et al. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature. 1991;352:207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- Kretzschmar D, et al. Defective pigment granule biogenesis and aberrant behavior caused by mutations in the Drosophila AP-3beta adaptin gene ruby. Genetics. 2000;155:213–223. doi: 10.1093/genetics/155.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Ready DF. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development. 1995;121:4359–4370. doi: 10.1242/dev.121.12.4359. [DOI] [PubMed] [Google Scholar]
- Kwok MC, et al. Proteomics of photoreceptor outer segments identifies a subset of SNARE and Rab proteins implicated in membrane vesicle trafficking and fusion. Mol Cell Proteomics. 2008;7:1053–1066. doi: 10.1074/mcp.M700571-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart T, Meyer EP. Detectors for polarized skylight in insects: A survey of ommatidial specializations in the dorsal rim area of the compound eye. Microsc Res Tech. 1999;47:368–379. doi: 10.1002/(SICI)1097-0029(19991215)47:6<368::AID-JEMT2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Rubin GM. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell. 1992;70:609–620. doi: 10.1016/0092-8674(92)90430-k. [DOI] [PubMed] [Google Scholar]
- Le TT, et al. Jagged 1 is necessary for normal mouse lens formation. Dev Biol. 2009;328:118–126. doi: 10.1016/j.ydbio.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Montell C. Suppression of constant-light-induced blindness but not retinal degeneration by inhibition of the rhodopsin degradation pathway. Curr Biol. 2004;14:2076–2085. doi: 10.1016/j.cub.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Lee RC, et al. The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat Neurosci. 2003;6:557–563. doi: 10.1038/nn1063. [DOI] [PubMed] [Google Scholar]
- Li BX, et al. Myosin V, Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J Cell Biol. 2007;177:659–669. doi: 10.1083/jcb.200610157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HY, Tomlinson A. Organization of the peripheral fly eye: The roles of Snail family transcription factors in peripheral retinal apoptosis. Development. 2006;133:3529–3537. doi: 10.1242/dev.02524. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, et al. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- Lloyd V, et al. Not just pretty eyes: Drosophila eye-colour mutations and lysosomal delivery. Trends Cell Biol. 1998;8:257–259. doi: 10.1016/s0962-8924(98)01270-7. [DOI] [PubMed] [Google Scholar]
- Longley RL, Jr, Ready DF. Integrins and the development of three-dimensional structure in the Drosophila compound eye. Dev Biol. 1995;171:415–433. doi: 10.1006/dbio.1995.1292. [DOI] [PubMed] [Google Scholar]
- Ma J, et al. Lightoid and Claret: A rab GTPase and its putative guanine nucleotide exchange factor in biogenesis of Drosophila eye pigment granules. Proc Natl Acad Sci USA. 2004;101:11652–11657. doi: 10.1073/pnas.0401926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie SM, et al. Sub-cellular localisation of the white/scarlet ABC transporter to pigment granule membranes within the compound eye of Drosophila melanogaster. Genetica. 2000;108:239–252. doi: 10.1023/a:1004115718597. [DOI] [PubMed] [Google Scholar]
- Marzesco AM, et al. Properties of Rab13 interaction with rod cGMP phosphodiesterase delta subunit. Meth Enzymol. 2001;329:197–209. doi: 10.1016/s0076-6879(01)29080-6. [DOI] [PubMed] [Google Scholar]
- Mast JD, et al. The mechanisms and molecules that connect photoreceptor axons to their targets in Drosophila. Semin Cell Dev Biol. 2006;17:42–49. doi: 10.1016/j.semcdb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Matsuo T, et al. Regulation of cone cell formation by Canoe and Ras in the developing Drosophila eye. Development. 1997;124:2671–2680. doi: 10.1242/dev.124.14.2671. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, et al. Of cartridges and columns: New roles for cadherins in visual system development. Neuron. 2008;58:1–3. doi: 10.1016/j.neuron.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Maw MA, et al. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet. 2000;9:27–34. doi: 10.1093/hmg/9.1.27. [DOI] [PubMed] [Google Scholar]
- Mazzoni EO, et al. ‘One receptor’ rules in sensory neurons. Dev Neurosci. 2004;26:388–395. doi: 10.1159/000082281. [DOI] [PubMed] [Google Scholar]
- Mazzoni EO, et al. Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol. 2008;6:e97. doi: 10.1371/journal.pbio.0060097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehalow AK, et al. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet. 2003;12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA. The development of neuronal connection patterns in the visual systems of insects. Ciba Found Symp. 1975:265–288. doi: 10.1002/9780470720110.ch13. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The Development of the Optic Lobe. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. [Google Scholar]
- Menne D, Spatz HC. Colour vision in Drosophila melanogaster. J Comp Physiol. 1977;A114:301–312. [Google Scholar]
- Mikeladze-Dvali T, et al. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Miller DT, Cagan RL. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 1998;125:2327–2335. doi: 10.1242/dev.125.12.2327. [DOI] [PubMed] [Google Scholar]
- Mishra M, Oke A, Lebel C, McDonald EC, Plummer Z, Cook TA, Zelhof AC. Pph13 and Orthodenticle define a dual regulatory pathway for photoreceptor cell morphogenesis and function. Development. 2010;137:2895–2904. doi: 10.1242/dev.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mismer D, Rubin GM. Definition of cis-acting elements regulating expression of the Drosophila melanogaster ninaE opsin gene by oligonucleotide-directed mutagenesis. Genetics. 1989;121:77–87. doi: 10.1093/genetics/121.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M. Planar polarity in the Drosophila eye: A multifaceted view of signaling specificity and cross-talk. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollereau B, et al. A green fluorescent protein enhancer trap screen in Drosophila photoreceptor cells. Mech Dev. 2000;93:151–160. doi: 10.1016/s0925-4773(00)00287-2. [DOI] [PubMed] [Google Scholar]
- Mollereau B, et al. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412:911–913. doi: 10.1038/35091076. [DOI] [PubMed] [Google Scholar]
- Montell C, et al. A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster. J Neurosci. 1987;7:1558–1566. doi: 10.1523/JNEUROSCI.07-05-01558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey M, et al. Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature. 2008;456:795–799. doi: 10.1038/nature07419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz OL, et al. Mutant rab8 Impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol Biol Cell. 2001;12:2341–2351. doi: 10.1091/mbc.12.8.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins C, et al. Defective expression of the mu3 subunit of the AP-3 adaptor complex in the Drosophila pigmentation mutant carmine. Mol Gen Genet. 1999;262:401–412. doi: 10.1007/s004380051099. [DOI] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 2007;134:825–831. doi: 10.1242/dev.02788. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, et al. Arrestin gene mutations in autosomal recessive retinitis pigmentosa. Arch Ophthalmol. 1998;116:498–501. doi: 10.1001/archopht.116.4.498. [DOI] [PubMed] [Google Scholar]
- Nam SC, Choi KW. Interaction of Par-6 and Crumbs complexes is essential for photoreceptor morphogenesis in Drosophila. Development. 2003;130:4363–4372. doi: 10.1242/dev.00648. [DOI] [PubMed] [Google Scholar]
- Nam SC, Choi KW. Domain-specific early and late function of Dpatj in Drosophila photoreceptor cells. Dev Dyn. 2006;235:1501–1507. doi: 10.1002/dvdy.20726. [DOI] [PubMed] [Google Scholar]
- Nam SC, et al. Antagonistic functions of Par-1 kinase and protein phosphatase 2A are required for localization of Bazooka and photoreceptor morphogenesis in Drosophila. Dev Biol. 2007;306:624–635. doi: 10.1016/j.ydbio.2007.03.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, et al. Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell. 2000;101:283–294. doi: 10.1016/s0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- Nishida A, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Oberhauser V, et al. NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proc Natl Acad Sci USA. 2008;105:19000–19005. doi: 10.1073/pnas.0807805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi CE, et al. Altered expression of a novel adaptin leads to defective pigment granule biogenesis in the Drosophila eye color mutant garnet. EMBO J. 1997;16:4508–4518. doi: 10.1093/emboj/16.15.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio D. Spam and the evolution of the fly’s eye. Bioessays. 2007;29:111–115. doi: 10.1002/bies.20533. [DOI] [PubMed] [Google Scholar]
- O’Tousa JE, et al. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, et al. A new rhodopsin in R8 photoreceptors of Drosophila: Evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, et al. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech Dev. 2001;101:143–153. doi: 10.1016/s0925-4773(00)00581-5. [DOI] [PubMed] [Google Scholar]
- Pellikka M, et al. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature. 2002;416:143–149. doi: 10.1038/nature721. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: New findings and challenges. Vis Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- Pepling M, Mount SM. Sequence of a cDNA from the Drosophila melanogaster white gene. Nucleic Acids Res. 1990;18:1633. doi: 10.1093/nar/18.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez SE, Steller H. Migration of glial cells into retinal axon target field in Drosophila melanogaster. J Neurobiol. 1996;30:359–373. doi: 10.1002/(SICI)1097-4695(199607)30:3<359::AID-NEU5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Crystallin genes: Specialization by changes in gene regulation may precede gene duplication. J Struct Funct Genomics. 2003;3:131–137. [PubMed] [Google Scholar]
- Piatigorsky J. Evolutionary genetics: Seeing the light: The role of inherited developmental cascades in the origins of vertebrate lenses and their crystallins. Heredity. 2006;96:275–277. doi: 10.1038/sj.hdy.6800793. [DOI] [PubMed] [Google Scholar]
- Pinal N, et al. Regulated and polarized PtdIns(3, 4, 5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr Biol. 2006;16:140–149. doi: 10.1016/j.cub.2005.11.068. [DOI] [PubMed] [Google Scholar]
- Poeck B, et al. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron. 2001;29:99–113. doi: 10.1016/s0896-6273(01)00183-0. [DOI] [PubMed] [Google Scholar]
- Porter JA, Montell C. Distinct roles of the Drosophila ninaC kinase and myosin domains revealed by systematic mutagenesis. J Cell Biol. 1993;122:601–612. doi: 10.1083/jcb.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo C, et al. The eyeless homeodomain is dispensable for eye development in Drosophila. Genes Dev. 2001;15:1716–1723. doi: 10.1101/gad.196401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring R, et al. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Ranade SS, et al. Analysis of the Otd-dependent transcriptome supports the evolutionary conservation of CRX/OTX/OTD functions in flies and vertebrates. Dev Biol. 2008;315:521–534. doi: 10.1016/j.ydbio.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready DF, et al. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rebay I, et al. Analysis of phenotypic abnormalities and cell fate changes caused by dominant activated and dominant negative forms of the Notch receptor in Drosophila development. C R Acad Sci III. 1993;316:1097–1123. [PubMed] [Google Scholar]
- Redowicz MJ. Myosins and pathology: Genetics and biology. Acta Biochim Pol. 2002;49:789–804. [PubMed] [Google Scholar]
- Reinke R, Zipursky SL. Cell–cell interaction in the Drosophila retina: The bride of sevenless gene is required in photoreceptor cell R8 for R7 cell development. Cell. 1988;55:321–330. doi: 10.1016/0092-8674(88)90055-4. [DOI] [PubMed] [Google Scholar]
- Reis T, Hariharan IK. Melting fat away in flies. Cell Metab. 2005;2:82–84. doi: 10.1016/j.cmet.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Richard M, et al. DPATJ plays a role in retinal morphogenesis and protects against light-dependent degeneration of photoreceptor cells in the Drosophila eye. Dev Dyn. 2006a;235:895–907. doi: 10.1002/dvdy.20595. [DOI] [PubMed] [Google Scholar]
- Richard M, et al. Towards understanding CRUMBS function in retinal dystrophies. Hum Mol Genet. 2006b;15(Spec2):R235–R243. doi: 10.1093/hmg/ddl195. [DOI] [PubMed] [Google Scholar]
- Rickenbacher J. Polished (pol), a new eye mutant in the 4th chromosome in Drosophila melanogaster. Z Indukt Abstamm Vererbungsl. 1954;86:61–68. [PubMed] [Google Scholar]
- Rivolta C, et al. Dominant Leber congenital amaurosis, cone-rod degeneration, and retinitis pigmentosa caused by mutant versions of the transcription factor CRX. Hum Mutat. 2001;18:488–498. doi: 10.1002/humu.1226. [DOI] [PubMed] [Google Scholar]
- Robinson ML. An essential role for FGF receptor signaling in lens development. Semin Cell Dev Biol. 2006;17:726–740. doi: 10.1016/j.semcdb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal R, et al. The fibroblast growth factor receptors, FGFR-1 and FGFR-2, mediate two independent signalling pathways in human retinal pigment epithelial cells. Biochem Biophys Res Commun. 2005;337:241–247. doi: 10.1016/j.bbrc.2005.09.028. [DOI] [PubMed] [Google Scholar]
- Rowan S, et al. Notch signaling regulates growth and differentiation in the mammalian lens. Dev Biol. 2008;321:111–122. doi: 10.1016/j.ydbio.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan W, et al. Bifocal is a downstream target of the Ste20-like serine/threonine kinase misshapen in regulating photoreceptor growth cone targeting in Drosophila. Neuron. 2002;36:831–842. doi: 10.1016/s0896-6273(02)01027-9. [DOI] [PubMed] [Google Scholar]
- Salcedo E, et al. Blue- and green-absorbing visual pigments of Drosophila: Ectopic expression and physiological characterization of the R8 photoreceptor cell-specific Rh5 and Rh6 rhodopsins. J Neurosci. 1999;19:10716–10726. doi: 10.1523/JNEUROSCI.19-24-10716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang TK, Ready DF. Eyes closed, a Drosophila p47 homolog, is essential for photoreceptor morphogenesis. Development. 2002;129:143–154. doi: 10.1242/dev.129.1.143. [DOI] [PubMed] [Google Scholar]
- Saravanamuthu SS, et al. Notch signaling is required for lateral induction of Jagged1 during FGF-induced lens fiber differentiation. Dev Biol. 2009;332(1):166–176. doi: 10.1016/j.ydbio.2009.05.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfare S, et al. The Drosophila ninaG oxidoreductase acts in visual pigment chromophore production. J Biol Chem. 2005;280:11895–11901. doi: 10.1074/jbc.M412236200. [DOI] [PubMed] [Google Scholar]
- Satoh A, et al. In situ inhibition of vesicle transport and protein processing in the dominant negative Rab1 mutant of Drosophila. J Cell Sci. 1997;110(Pt 23):2943–2953. doi: 10.1242/jcs.110.23.2943. [DOI] [PubMed] [Google Scholar]
- Satoh AK, et al. Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development. 2005;132:1487–1497. doi: 10.1242/dev.01704. [DOI] [PubMed] [Google Scholar]
- Sheng G, et al. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: Evidence for a conserved function in photoreceptors. Genes Dev. 1997;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- Shetty KM, et al. Rab6 regulation of rhodopsin transport in Drosophila. J Biol Chem. 1998;273:20425–20430. doi: 10.1074/jbc.273.32.20425. [DOI] [PubMed] [Google Scholar]
- Shoup JR. The development of pigment granules in the eyes of wild type and mutant Drosophila melanogaster. J Cell Biol. 1966;29:223–249. doi: 10.1083/jcb.29.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddall NA, et al. Mutations in lozenge and D-Pax2 invoke ectopic patterned cell death in the developing Drosophila eye using distinct mechanisms. Dev Genes Evol. 2003;213:107–119. doi: 10.1007/s00427-003-0295-y. [DOI] [PubMed] [Google Scholar]
- Simpson F, et al. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Choi KW. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development. 2003;130:6351–6360. doi: 10.1242/dev.00864. [DOI] [PubMed] [Google Scholar]
- Stark WS, Thomas CF. Microscopy of multiple visual receptor types in Drosophila. Mol Vis. 2004;10:943–955. [PubMed] [Google Scholar]
- Stark SW, et al. Spectral sensitivities and photopigments in adaptation of fly visual receptors. Naturwissenschaften. 1976;63:513–518. doi: 10.1007/BF00596847. [DOI] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Strutt D. The planar polarity pathway. Curr Biol. 2008;18:R898–R902. doi: 10.1016/j.cub.2008.07.055. [DOI] [PubMed] [Google Scholar]
- Suh GS, et al. Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron. 2002;33:35–46. doi: 10.1016/s0896-6273(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Summers KM, et al. Biology of eye pigmentation in insects. Adv In Insect Physiol. 1982;16:119–166. [Google Scholar]
- Swaroop A, et al. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: Direct evidence for the involvement of CRX in the development of photoreceptor function. Hum Mol Genet. 1999;8:299–305. doi: 10.1093/hmg/8.2.299. [DOI] [PubMed] [Google Scholar]
- Tahayato A, et al. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- Tai AW, et al. Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Tayler TD, Garrity PA. Axon targeting in the Drosophila visual system. Curr Opin Neurobiol. 2003;13:90–95. doi: 10.1016/s0959-4388(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Tearle RG, et al. Cloning and characterization of the scarlet gene of Drosophila melanogaster. Genetics. 1989;122:595–606. doi: 10.1093/genetics/122.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, et al. Drosophila Melted modulates FOXO and TOR activity. Dev Cell. 2005;9:271–281. doi: 10.1016/j.devcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Piatigorsky J. Lens crystallins of invertebrates—Diversity and recruitment from detoxification enzymes and novel proteins. Eur J Biochem. 1996;235:449–465. doi: 10.1111/j.1432-1033.1996.00449.x. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. Cellular interactions in the developing Drosophila eye. Development. 1988;104:183–193. doi: 10.1242/dev.104.2.183. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Sevenless: A cell-specific homeotic mutation of the Drosophila eye. Science. 1986;231:400–402. doi: 10.1126/science.231.4736.400. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Cell fate in the Drosophila ommatidium. Dev Biol. 1987a;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready D. Neuronal differentiation in the Drosophila ommatidium. Dev Biol. 1987b;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Struhl G. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol Cell. 2001;7:487–495. doi: 10.1016/s1097-2765(01)00196-4. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, et al. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell. 1987;51:143–150. doi: 10.1016/0092-8674(87)90019-5. [DOI] [PubMed] [Google Scholar]
- Treisman JE. A conserved blueprint for the eye? Bioessays. 1999;21:843–850. doi: 10.1002/(SICI)1521-1878(199910)21:10<843::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Tsuda L, et al. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Usui T, et al. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Van Vactor D, Jr, et al. Analysis of mutants in chaoptin, a photoreceptor cell-specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell. 1988;52:281–290. doi: 10.1016/0092-8674(88)90517-x. [DOI] [PubMed] [Google Scholar]
- Van Vactor DL, Jr, et al. Induction in the developing compound eye of Drosophila: Multiple mechanisms restrict R7 induction to a single retinal precursor cell. Cell. 1991;67:1145–1155. doi: 10.1016/0092-8674(91)90291-6. [DOI] [PubMed] [Google Scholar]
- Vandendries ER, et al. orthodenticle is required for photoreceptor cell development in the Drosophila eye. Dev Biol. 1996;173:243–255. doi: 10.1006/dbio.1996.0020. [DOI] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: Insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Vopalensky P, Kozmik Z. Eye evolution: Common use and independent recruitment of genetic components. Philos Trans R Soc Lond B Biol Sci. 2009;364:2819–2832. doi: 10.1098/rstb.2009.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH, Perry MM. The ultra-structure of the developing eye of Drosophila. Proc R Soc Lond, B, Biol Sci. 1960;153:155–178. [Google Scholar]
- Wallace VA. Proliferative and cell fate effects of Hedgehog signaling in the vertebrate retina. Brain Res. 2008;1192:61–75. doi: 10.1016/j.brainres.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Walsh T, et al. From flies’ eyes to our ears: Mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Montell C. Rhodopsin formation in Drosophila is dependent on the PINTA retinoid-binding protein. J Neurosci. 2005;25:5187–5194. doi: 10.1523/JNEUROSCI.0995-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- Wang SW, et al. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, et al. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J Cell Biol. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner TS, et al. The light gene of Drosophila melanogaster encodes a homologue of VPS41, a yeast gene involved in cellular-protein trafficking. Genome. 1998;41:236–243. [PubMed] [Google Scholar]
- Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- Wawersik S, et al. Pax6 and the genetic control of early eye development. Results Probl Cell Differ. 2000;31:15–36. doi: 10.1007/978-3-540-46826-4_2. [DOI] [PubMed] [Google Scholar]
- Wech I, Nagel AC. Mutations in rugose promote cell type-specific apoptosis in the Drosophila eye. Cell Death Differ. 2005;12:145–152. doi: 10.1038/sj.cdd.4401538. [DOI] [PubMed] [Google Scholar]
- Wernet MF, et al. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- Wernet MF, et al. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NM, Jarman AP. Drosophila atonal controls photoreceptor R8-specific properties and modulates both receptor tyrosine kinase and Hedgehog signalling. Development. 2000;127:1681–1689. doi: 10.1242/dev.127.8.1681. [DOI] [PubMed] [Google Scholar]
- Whited JL, et al. Dynactin is required to maintain nuclear position within postmitotic Drosophila photoreceptor neurons. Development. 2004;131:4677–4686. doi: 10.1242/dev.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Wilson D, et al. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- Winberg ML, et al. Generation and early differentiation of glial cells in the first optic ganglion of Drosophila melanogaster. Development. 1992;115:903–911. doi: 10.1242/dev.115.4.903. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern Formation in the Drosophila Retina. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. [Google Scholar]
- Wunderer H, Smola U. Morphological differentiation of the central visual cells R7/8 in various regions of the blowfly eye. Tissue Cell. 1982;14:341–358. doi: 10.1016/0040-8166(82)90032-5. [DOI] [PubMed] [Google Scholar]
- Xie B, et al. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development. 2007;134:4243–4253. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
- Xu C, et al. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell. 2000;103:87–97. doi: 10.1016/s0092-8674(00)00107-0. [DOI] [PubMed] [Google Scholar]
- Xu H, et al. A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen. EMBO J. 2004;23:811–822. doi: 10.1038/sj.emboj.7600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, et al. Contribution of photoreceptor subtypes to spectral wavelength preference in Drosophila. Proc Natl Acad Sci USA. 2010;107:5634–5639. doi: 10.1073/pnas.0809398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, et al. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Yang Z, et al. Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J Clin Invest. 2008;118:2908–2916. doi: 10.1172/JCI35891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Pan D. Fat flies expanded the hippo pathway: A matter of size control. Sci STKE. 2007;2007:pe12. doi: 10.1126/stke.3802007pe12. [DOI] [PubMed] [Google Scholar]
- Youssef NN, Gardner EJ. The compound eye in the opaque-eye phenotype of Drosophila melanogaster. Tissue Cell. 1975;7:655–668. doi: 10.1016/0040-8166(75)90034-8. [DOI] [PubMed] [Google Scholar]
- Yu SY, et al. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- Zecchin E, et al. Expression analysis of jagged genes in zebrafish embryos. Dev Dyn. 2005;233:638–645. doi: 10.1002/dvdy.20366. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Hardy RW. WASp is required for the correct temporal morphogenesis of rhabdomere microvilli. J Cell Biol. 2004;164:417–426. doi: 10.1083/jcb.200307048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelhof AC, et al. Drosophila Amphiphysin is implicated in protein localization and membrane morphogenesis but not in synaptic vesicle endocytosis. Development. 2001;128:5005–5015. doi: 10.1242/dev.128.24.5005. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, et al. Mutation of the photoreceptor specific homeodomain gene Pph13 results in defects in phototransduction and rhabdomere morphogenesis. Development. 2003;130:4383–4392. doi: 10.1242/dev.00651. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, et al. Transforming the architecture of compound eyes. Nature. 2006;443:696–699. doi: 10.1038/nature05128. [DOI] [PubMed] [Google Scholar]
- Zhao H, et al. Fibroblast growth factor receptor signaling is essential for lens fiber cell differentiation. Dev Biol. 2008;318:276–288. doi: 10.1016/j.ydbio.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker CS, et al. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- Zuker CS, et al. A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: Homologies with other signal-transducing molecules. J Neurosci. 1987;7:1550–1557. doi: 10.1523/JNEUROSCI.07-05-01550.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]