Abstract
Energy dependent proteolysis is essential for all life, but uncontrolled degradation leads to devastating consequences. In bacteria, oligomeric AAA+ proteases are responsible for controlling protein destruction and are regulated in part by adaptor proteins. Adaptors are regulatory factors that shape protease substrate choice by either restricting or enhancing substrate recognition in several ways. In some cases, protease activity or assembly itself requires adaptor binding. Adaptors can also alter specificity by acting as scaffolds to tether particular substrates to already active proteases. Finally, hierarchical assembly of adaptors can use combinations of several activities to enhance the protease’s selectivity. Because the lifetime of the constituent proteins directly affects the duration of a particular signaling pathway, regulated proteolysis impacts almost all cellular responses. In this review, we describe recent progress in regulated protein degradation, focusing on fundamental principles of adaptors and how they perform critical biological functions, such as promoting cell cycle progression and quality control.
Introduction
The regulated degradation of proteins both governs and responds to signaling pathways. For example, many damaging conditions elicit the upregulation of response proteins that are harmful during normal conditions (e.g., cell division inhibitors upon DNA damage). By degrading those factors after the insult has passed, cells limit the potential toxic consequences of persistent upregulation. Damaged or poor quality proteins themselves must be eliminated from cellular pools as accumulating these products could lead to harmful impacts on the protein homeostasis network. Normal growth also requires regulated protein degradation. For example, cell cycle progression in Caulobacter crescentus requires the coordinated degradation of transcription factors and signaling proteins [1]. Similarly, developmental changes such as stationary phase entry in E. coli and sporulation in Bacillus subtilis also relies on regulated protein degradation [2,3]. Despite these different pathways, regulated protein degradation by energy dependent proteases shares many common features, notably the need for highly specific engagement of target substrates, often through the use of adaptor proteins.
The initial engagement of substrates by proteases determines selectivity
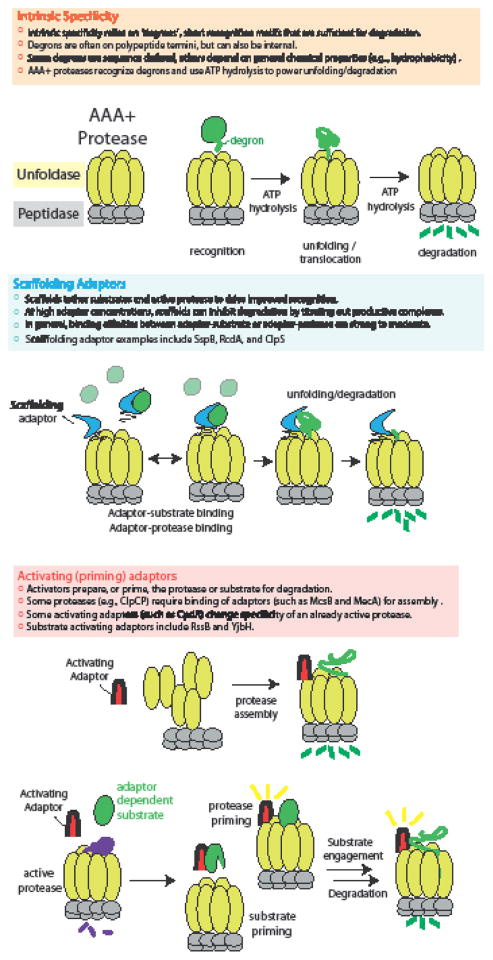
AAA+ proteases are composite enzymes with two conserved functional modules [4]. An ATPase recognizes a target substrate and fuels unfolding of the substrate through cycles of ATP hydrolysis. The unfolded protein is translocated to a relatively nonspecific peptidase that then destroys the protein (Figure 1). AAA+ proteases are extraordinarily processive. Once engaged, these machines are capable of translocating and degrading polypeptides hundreds to thousands of residues long with little regard for the specific sequences of the translocated substrates [5]. Therefore, the governing step for protease specificity appears to be the initial recognition and engagement of the substrate by the ATPase.
Figure 1. Specificity of AAA+ proteases.
Regulated protein degradation relies on both the intrinsic specificity of the proteases and the presence of adaptor proteins that alter specificity. In the most general sense, adaptors can act as simple scaffolds or as activators.
Proteases themselves have certain intrinsic specificity. For example, the ClpXP protease recognizes nonpolar C-terminal residues with high stringency where a single mutation in this recognition site can dramatically alter recognition [6]. Similarly, the Lon protease has a preference for stretches of hydrophobic residues, but, unlike ClpXP recognition, tolerates many single amino acid changes in these regions [7]. In addition, there are cases where proteases have overlapping specificity, such as recent work showing that ClpAP and Lon both recognize and degrade the replication initiator DnaA [8].
Despite this intrinsic specificity, additional regulation is needed to allow for the precise degradation of the wide range of substrates found in the bacterial proteome. Synthesis of the proteases can restrict activity to specific times and localization of the proteases can limit activity to a specific space. However, it appears that many instances of protease regulation use auxiliary factors known as adaptors that promote or inhibit protease activity and selectivity. Although there are many different adaptor proteins known, they can be grouped into two nonexclusive functional classes: those that activate the protease and those that deliver substrates to an already active protease (Figure 1).
Adaptor-dependent protease activation
The ClpCP protease in Bacillus subtilis regulates competency and sporulation, degrading targets such as the competence inhibitor ComS [9]. Like other AAA+ unfoldases, ClpC must assemble into an oligomer to be active. However, in contrast to most other AAA+ unfoldases, ClpC assembly requires an additional step, such as binding of an adaptor protein like MecA [10,11], which delivers substrates such as ComK and ComS. In this model, MecA binding to ClpC causes oligomerization and activation of ATPase function. This assembled ClpC can then template the assembly of ClpP, allowing it to oligomerize and producing the final active protease complex (Figure 1). Interestingly, MecA is itself degraded by ClpCP [9,12] and because MecA is required for protease assembly, this linking of protease activity to a requisite adaptor allows for self-limitation of the protease complex. Structural insights reveal how MecA binds the N-terminal domain of ClpC to form an interlaced ring that induces oligomerization of ClpC [13] and clarifies elements within MecA itself is recognized for degradation [14]. Another known adaptor of ClpCP is McsB, a nonconventional kinase, which was shown to activate ClpCP and promote degradation of CtsR [15].
Interestingly, recent reports now suggest that McsB is an arginine kinase that performs noncanonical phosphorylation of arginine residues in target proteins, including ClpC itself [16,17] [18]. A most intriguing recent observation is that proteins with phosphorylated arginines are readily degraded by ClpCP alone, suggesting that recognition of a phosphorylated arginine is sufficient to activate ClpCP even without an additional adaptor [19]. Because McsB activity is needed during heat stress [20], a tempting speculation is that arginine phosphorylation of various targets during this stress condition leads to activation of ClpCP. This activated protease then destroys potentially proteotoxic species or performs regulated proteolysis to support appropriate signaling responses. Therefore, McsB seems to play dual roles in acting as a MecA-type adaptor [15] and in generating adaptor-independent phosphorylated substrates [19].
The Lon protease also presents a situation where protease activation is driven by the presence of an accessory factor. The Lon protease has been long known to be an allosterically regulated protease, where addition of a protein substrate can increase ATPase and peptide hydrolysis activity. More recently, it was found that this same mechanism explains how the Lon protease destroys the replication initiator DnaA during proteotoxic stress [21]. Here, the model is that misfolded proteins generated during proteotoxic stress are recognized and cleared by the Lon protease. This recognition of misfolded substrates also activates Lon in trans to degrade DnaA, delaying the energetically costly process of DNA replication until the proteotoxic stress has cleared. Reducing DnaA activity is crucial to prevent overreplication and similar deactivation through degradation by Lon and ClpAP have also been observed during entry into stationary phase [8,22]. Interestingly, since Lon activity responds to other biological molecules such as nucleic acids[23,24], activation of DnaA degradation may be a feature of other responses in addition to proteotoxic conditions.
Adaptor-dependent substrate delivery
In many cases, adaptors are not required for AAA+ protease activity but instead tune the specificity of the attendant protease by acting as passive scaffolds or selective activators. By combining these different adaptor types, the cell can also ensure robust, hierarchical degradation as seen in the bacterial cell cycle.
Scaffold functions for adaptors
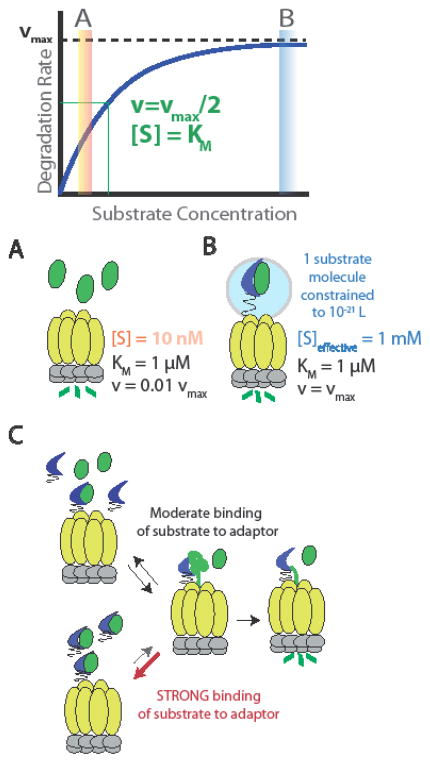
The most straightforward mechanism for adaptor function is to operate as a simple scaffold. Just as increasing reactant concentrations drives product formation in chemical reactions, increasing substrate concentration drives recognition and proteolysis by AAA+ proteases. In the simplest case, adaptors can bind to substrates and proteases simultaneously, tethering substrates to the protease to increase local concentration and promote proteolysis. As shown by example in Figure 2, if the KM of degradation of substrate A by the protease is 1 uM, but A is only present at 10 nM, then the degradation rate of A will be ~1% of maximum (0.01 vmax). Let us assume that the adaptor binds a single molecule of A and scaffolds it to the protease with an effective tether length of 100 A (~25 unstructured residues). This would result in one molecule in an occupied volume of ~10^-21 L, or ~1 mM, an effective concentration well above the KM. In this case, the substrate would be recognized and degraded by the protease at the maximum rate possible (vmax). It is important to note that in these cases, the proper binding affinity between substrate and adaptor is crucial (Figure 2): The lifetime of the substrate-adaptor complex must be long enough to ensure handoff of the substrate to the protease. However, if the affinity is too strong, then the adaptor will restrict protein degradation by failing to let go of the substrate so that it can be engaged by the protease. That said, in some cases, the directional nature of protein unfolding may allow for forceful extraction of the substrate from the adaptor despite strong binding.
Figure 2. Mechanisms and consequences of scaffolding adaptors.
Schematized Michaelis-Menten kinetics of substrate degradation illustrating two regimes of substrate concentration: [S] is well below the KM (A) and well above the KM (B). If [S] is 100-fold below the KM, then the degradation rate is ~1% of the maximum velocity (A). Scaffolding adaptors can tether substrates to the protease and increase local concentration by this leashing. As shown in B, a tether length of ~25 residues would result in constraining a single molecule to a volume of ~10−21 L, which yields an effective concentration of ~1 mM, sufficient to drive degradation at the maximum rate. (C) Substrate recognition by the adaptor must be of at least moderate affinity in order to ensure specificity. However, excessively tight binding of the adaptor restricts delivery of the substrate and inhibits overall degradation.
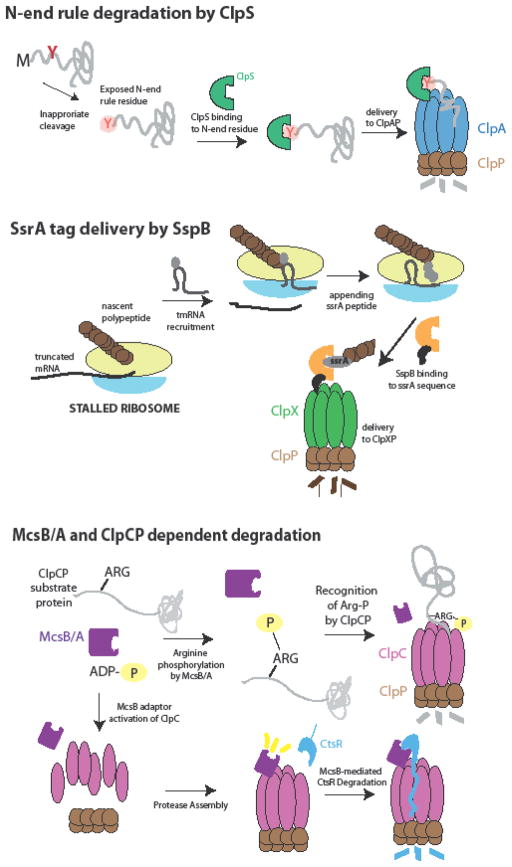
Adaptors that appear to function in this manner are present across protease systems (Figure 3). For example, the SspB adaptor binds the ssrA peptide, delivering proteins tagged with this peptide to ClpXP for degradation [25–27], a well-characterized event that is currently being used as a method to control protein degradation in vivo [28,29]. ClpS binds N-end rule residues (L,Y,W,F), tethering substrates containing these residues to the ClpAP protease which recognizes portions of the substrate adjacent to this tag[30–32]. Interestingly, recent work suggests that remodeling of the ClpS adaptor by the ClpA unfoldase promotes handoff of substrate from the adaptor to the pore of ClpA [33,34]. More recently, it was found that some ClpS orthologs have either restricted or expanded specificity, suggesting that ClpS delivery may be further tuned by selection of different N-end rule substrate pools [35] [36]. ClpS can also inhibit degradation of some substrates by ClpAP, including ssrA-tagged substrates and DnaA [37] [8]. This demonstrates how tight control of ClpS levels can dramatically shift the balance of substrates degraded in a ClpAP dependent manner.
Figure 3. Examples of scaffolding adaptors.
(a) Residues that are recognized by N-end rule degradation (L,F,W,Y) are shielded in normally synthesized proteins. Processing or cleavage can result in exposure of an N-end rule residue, which is bound by ClpS. As part of its mechanism, ClpS tethers these substrates to the ClpAP protease to deliver them for degradation. (b) SsrA-tagged proteins are products of stalled ribosomes resulting from translation of nonsense mRNAs. Stalled ribosomes are rescued by through the tmRNA pathway where the ssrA peptide is cotranslationally attached to the nascent polypeptide. The SspB adaptor recognizes the ssrA sequence and delivers proteins containing this tag to ClpXP for degradation. (c) Like MecA, the McsB kinase has been shown to act as an adaptor which drives ClpCP oligomerization to promote CtsR degradation. In addition, the kinase complex McsB/A was recently shown to target proteins susceptible to stress for degradation by phosphorylating arginine residues. In this case, the ClpCP protease is activated by the phosphorylated arginine and the modified protein is degraded.
The RssB adaptor binds strongly to the RpoS stationary sigma factor and delivers it to ClpXP, but seems to only weakly interact with ClpX alone - suggesting that simple tethering is not the only contributing element to this adaptor function [38, 44]. In the case of RssB, the Ira-family of anti-adaptors blocks delivery of RpoS in many ways, including competition for cargo and by inducing adaptor conformational changes. These effects contribute to the complex regulation of this critical sigma factor during growth and stress responses [39][40]. Finally, the RcdA adaptor in Caulobacter binds a number of targets and tethers them to the ClpXP protease. However, in this case, ClpXP must first be activated by an additional adaptor [41,42].
Priming functions for adaptors
Adaptors can also behave as more than simple scaffolds. In the case of the CpdR adaptor from Caulobacter crescentus, binding of this protein to the unique N-domain of the ClpX unfoldase prepares the protease for recognition of a class of substrates[42]. Here, the adaptor does not strongly interact with its cargo in the absence of the ClpX N-domain. This preparation, or priming, of the protease could arise from conformational changes in either CpdR or ClpX upon adaptor binding or could stem from a composite interaction surface that is only present in the complex. Regardless of mechanism, this type of priming function has features distinguishable from a simple scaffold (Figure 1). For example, high concentrations of scaffolds such as SspB actually inhibit substrate degradation due to the fact that partial occupancy of the scaffold restricts simultaneous binding of substrate and protease. By contrast, CpdR activity resists such inhibition as the substrate binding activity is only present when the adaptor binds the protease [42].
Similar types of priming functions have been observed with other adaptor systems. Binding of the YjbH adaptor is required for Spx degradation by ClpXP in gram-positive bacteria, but YjbH does not bind ClpX well on its own. Here, YjbH binding unveils a C-terminal degron in Spx that is in turn recognized by ClpXP [43]. The RssB adaptor binds RpoS directly, but its weak binding to ClpX suggests that priming of the substrate by the adaptor is needed for protease recognition [38,44]. Thus it seems that the ability of adaptors to activate either protease or substrate (Figure 1) for eventual degradation is a mechanism that has arisen in many systems.
Adaptor hierarchies
Combining adaptors results in adaptor hierarchies that can selectively degrade substrate classes dependent on the degree of assembly (Figure 4). In Caulobacter crescentus, the coordinated degradation of many proteins accompanies cell cycle progression. This includes chemoreceptors [45,46], transcription factors [45,47–49], metabolic enzymes [45,50,51], and replication factors[45,52]. The ClpXP protease was known to be responsible for degrading many of these proteins, but because levels of the protease do not fluctuate, there must be some regulatory control of ClpXP activity needed for the selective destruction of proteins during cell cycle progression. Genetic evidence pointed to a requirement for the proteins CpdR, RcdA, and PopA in degrading the essential transcription factor CtrA[53–56]. Interestingly, CpdR was also needed for degradation of the chemoreceptor McpA [55] and the phosphodiesterase PdeA [57]. However, loss of RcdA or PopA had no effect on the degradation of these substrates [54,57].
Figure 4. Selective substrate degradation from a hierarchical assembly of adaptors.
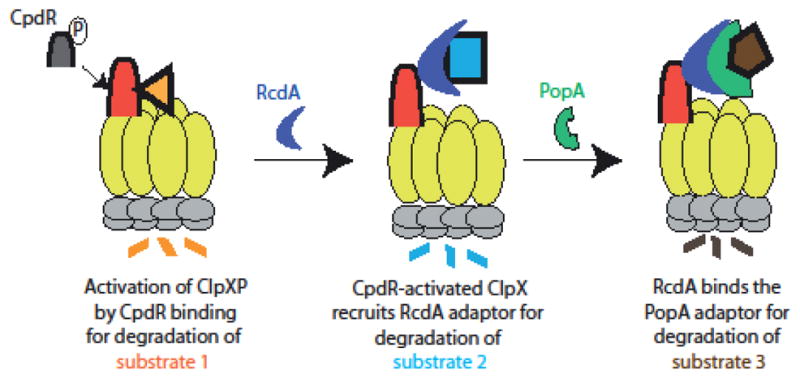
In Caulobacter crescentus, the degree of assembly of adaptors dictates which substrates are degraded. Substrate 1 is degraded by an active ClpXP following binding of a dephosphorylated CpdR adaptor, which primes ClpXP for selective substrate recognition. Substrate 2 is bound by the RcdA adaptor, which delivers cargos to a CpdR-activated protease and restricts degradation of substrate 1. Finally, the PopA adaptor can bind RcdA to form a complex that delivers Substrate 3. The hierarchical assembly of these adaptors during the Caulobacter cell cycle leads to ordered, progressive degradation of key regulators.
The requirement of CpdR as an adaptor for ClpXP-mediated degradation of PdeA and the reconstitution of this activity using purified components was a key breakthrough in the understanding of cell cycle regulated proteolysis [57]. CpdR is differentially phosphorylated during the cell cycle with dephosphorylation of CpdR during the G1-S transition concurrent with degradation of PdeA [53,55,57,58]. Consistent with this finding, phosphorylation of CpdR inactivated its adaptor activity [55,57] and subsequent work showed that this CpdR binding to ClpX was lost upon CpdR phosphorylation [21]. As described above, CpdR is a priming adaptor that activates the ClpXP protease for degradation of various substrates including PdeA and McpA [42]. Interestingly, primed ClpXP could also recruit the RcdA adaptor that was found to be an adaptor in its own right, tethering several substrates to the CpdR-activated ClpXP [41]. Finally, it was found that a complex of RcdA and the cyclic di-GMP bound form of PopA could bind CtrA, delivering it for degradation [41] [59]. Taken together, this model explains why some adaptors like CpdR are required for all protease substrates, but others, like RcdA, are only needed for a subset of these targets (Figure 4).
The Caulobacter cell cycle adaptors are the clearest experimental demonstration of an adaptor hierarchy. However, it seems likely that similar hierarchies will be found in other bacterial systems. These hierarchies are particularly well suited for controlling the rapid and coordinated degradation of many substrates in a sharply defined window of time, such as during developmental transitions or stress responses. The logic here is that cytoplasmic dilution through cell division during normal growth is sufficient to efficiently reduce protein levels in rapidly dividing bacteria. By contrast, a developmental transition between cell types is not accompanied by an increase in cell numbers. Therefore, any decrease in protein levels between the cell types must be governed by protein degradation. Similarly, if stress responses must occur at timescales faster than cell division, then regulated protein degradation must be used to dynamically control protein levels in this process. In line with this notion, recent work has shown a potential adaptor-mediated proteolysis pathway crucial for removal of unfit cells during sporulation in Bacillus subtilis [60].
Adaptor dependent proteolysis in regulation and quality control
Adaptors can target proteins for immediate degradation or can help stage proteolysis in a more regulated manner. Immediate delivery of substrates would be especially useful for eliminating aberrant proteins that fail protein quality control. Staged or timed degradation is critical for robust biological processes, such as during cell cycle progression or developmental changes. Given these different outcomes, it is interesting to consider how the biochemical features of specific adaptor-substrate pairs provide appropriate support for these outcomes.
As described earlier, the SspB protein binds the ssrA peptide to deliver ssrA-tagged proteins to the ClpXP protease[25] (Figure 3). Similarly, the ClpS adaptor binds to N-terminal residues (L,Y,W,F) that fall into the N-end rule class of rapidly degraded substrates. One common feature for both these adaptors is that they have relatively high affinity for their targets. For example, SspB binds the free ssrA peptide with a KD of 100–300 nM [61] and ClpS binds an N-end degron containing peptide with 100–400 nM affinity [31].
It is interesting to consider the SspB and ClpS adaptor dependent substrates in the context of the biological processes that lead to ssrA-tagging or destabilizing N-end rule residues. For example, the ssrA-tag is added to nascent polypeptides co-translationally during the process of trans-translation[27]. The dominant situation for trans-translation is when ribosomes encounter a truncated or damaged mRNA that results in stalling of the ribosome. Accumulation of these stalled ribosomes is deadly for cells due to the toxic decrease in translational capacity[62]. Trans-translation is the process by which stalled ribosomes recruit the small noncoding RNA tmRNA and the protein SmpB, which together bind the stalled ribosome causing it to shift onto a small open reading frame encoded on the tmRNA. Translation of this ORF results in appending of the ssrA peptide, which is delivered to ClpXP by SspB, followed by normal termination[27]. Therefore, most naturally occurring ssrA-tagged proteins result from the translation of a damaged or flawed template mRNA. In these cases, the most conservative approach for the cell is to eliminate these proteins immediately as it is unlikely that these partial proteins retain the normal biological function of originally intended full-length protein.
Similarly, all polypeptides that arise from normal translation contain methionine as their first residue. In bacteria, this methionine is removed by a specific aminopeptidase if the residue in the second position is G,A,P,S,T,V or C [63]. Importantly, natural full-length polypeptides would not contain residues at their N-termini that fall into the N-end rule primary degron class (Y,F,L,W). This suggests that most polypeptides containing N-end rule residues are generated from cleavage or processing of otherwise mature proteins. This processing can be regulated, as suggested for the targeted degradation of the DNA protection protein Dps and the Putrescine-Aminotransferase protein PATase in E. coli [64,65]. However, it is also possible that protein unfolding or damage could expose polypeptide regions that that would be subject to nonspecific cleavage in the cell. These new N-termini may themselves be destabilizing or may be further modified by amino acid transferases to generate N-end rule degrons [65,66]. Regardless of how these destabilizing N-termini are generated, the presence of these N-end rule degrons signal immediate degradation through ClpS/ClpAP.
A similar type of protease-mediated quality control mechanism was recently proposed in B. subtilis. As described earlier, phosphorylation of arginine residues results in substrates that activate the ClpCP protease for degradation of those modified proteins. The McsB kinase responsible for this atypical phosphorylation is upregulated during heat stress and a speculation is that arginine phosphorylation occurs on proteins that are most likely to misfold during proteotoxic stress[17]. This is functionally similar to the types of scenarios described above in that the modified substrates are immediately responded to by proteases, although there does not seem to be a role for an additional adaptor in this case.
The degrons (ssrA tags or N-end rule residues) delivered by SspB and ClpS are signals for immediate destruction of the target protein. Therefore, it stands to reason that the adaptor tightly binds the degron sequence on its own with high affinity without regard to the rest of the target as these tagged proteins must always be degraded. This is in contrast to situations where protein degradation must be controlled more precisely, such as during cell cycle progression where proteins must be selectively stabilized and degraded during different stages of the division cycle. In these cases, the simple presence or absence of adaptor binding motifs are insufficient to support these complex protein level dynamics and additional mechanisms such as adaptor hierarchies are needed.
Perspectives
All cells face the challenge of tightly regulating the contents of their proteome while maintaining dynamics. Essential cellular events are often triggered by the targeted destruction of important regulatory proteins. A high degree of substrate specificity must be achieved before proteolysis should take place, because proteolysis is an irreversible process. To ensure that only particular substrates are selected for degradation, eukaryotic and prokaryotic organisms have evolved methods of conferring substrate specificity to their proteolytic machinery. For example, eukaryotes confer protease specificity by directly altering the substrates themselves, making use of linear and branched ubiquitin chains to assign specificity [67,68] [69] In contrast, bacteria use regulatory adaptor proteins that act as scaffolds, tethers or priming factors for AAA+ proteases rather than altering the composition of the substrate itself.
Adaptor proteins serve both inhibitory and activating roles for the protease, preventing the premature degradation of cytosolic proteins and enhancing the degradation of a limited subset of critical targets. In some cases, these roles are further elaborated by the hierarchical assembly of several adaptors. We speculate that adaptor proteins should make ideal antibiotic targets because they only assemble on the protease during specific developmental stages or during times of environmental stress, rendering it difficult for the bacteria to develop antibiotic resistance. An outstanding objective is to now understand how adaptor proteins bind and deliver their substrate(s) given the sequence and structural differences between all these partners. By doing so, we will be able to predict new adaptor dependent protease pathways that regulate bacterial signaling.
Highlights.
Regulated protein degradation in bacteria relies on intrinsic protease specificity and adaptor-mediated elaboration of protease specificity.
Adaptors acting as simple scaffolds tether cargo with proteases and increase local effective concentration to drive substrate delivery.
Adaptors can be obligate activators of proteases or can prime already active proteases for selective substrate degradation.
Adaptor hierarchies can assemble from multiple adaptor types.
Poor quality proteins can be immediately targeted for degradation through constitutive recognition of degrons by adaptors.
Acknowledgments
The authors thank all members of the Chien lab and the larger working group of the Protein Homeostasis theme in the Institute for Applied Life Sciences for discussions. Work in the Chien lab is funded in part by the NIH (R01GM111706).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Joshi KK, Chien P. Regulated Proteolysis in Bacteria: Caulobacter. Annu Rev Genet. 2016;50:423–445. doi: 10.1146/annurev-genet-120215-035235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Molecular Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 3.Gerth U, Kock H, Kusters I, Michalik S, Switzer RL, Hecker M. Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol. 2008;190:321–331. doi: 10.1128/JB.01233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivares AO, Baker TA, Sauer RT. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol. 2016;14:33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide translocation by the AAA+ ClpXP protease machine. Chem Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci U S A. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA+ protease. Genes and Development. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Francis LI, Jonas K, Laub MT, Chien P. ClpAP is an auxiliary protease for DnaA degradation in Caulobacter crescentus. Mol Microbiol. 2016;102:1075–1085. doi: 10.1111/mmi.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. The EMBO journal. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirstein J, Schlothauer T, Dougan DA, Lilie H, Tischendorf G, Mogk A, Bukau B, Turgay K. Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. The EMBO journal. 2006;25:1481–1491. doi: 10.1038/sj.emboj.7601042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlothauer T, Mogk A, Dougan DA, Bukau B, Turgay K. MecA, an adaptor protein necessary for ClpC chaperone activity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2306–2311. doi: 10.1073/pnas.0535717100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persuh M, Mandic-Mulec I, Dubnau D. A MecA paralog, YpbH, binds ClpC, affecting both competence and sporulation. Journal of Bacteriology. 2002;184:2310–2313. doi: 10.1128/JB.184.8.2310-2313.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Mei Z, Qi Y, Yan C, Hu Q, Wang J, Shi Y. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature. 2011;471:331–335. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- 14.Mei Z, Wang F, Qi Y, Zhou Z, Hu Q, Li H, Wu J, Shi Y. Molecular determinants of MecA as a degradation tag for the ClpCP protease. Journal of Biological Chemistry. 2009;284:34366–34375. doi: 10.1074/jbc.M109.053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirstein J, Dougan DA, Gerth U, Hecker M, Turgay K. The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. The EMBO Journal. 2007;26:2061–2070. doi: 10.1038/sj.emboj.7601655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuhrmann J, Schmidt A, Spiess S, Lehner A, Turgay K, Mechtler K, Charpentier E, Clausen T. McsB Is a Protein Arginine Kinase That Phosphorylates and Inhibits the Heat-Shock Regulator CtsR. Science. 2009;324:1323 L. doi: 10.1126/science.1170088. P-1327. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt A, Trentini DB, Spiess S, Fuhrmann J, Ammerer G, Mechtler K, Clausen T. Quantitative Phosphoproteomics Reveals the Role of Protein Arginine Phosphorylation in the Bacterial Stress Response. Molecular & Cellular Proteomics : MCP. 2014;13:537–550. doi: 10.1074/mcp.M113.032292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsholz AK, Turgay K, Michalik S, Hessling B, Gronau K, Oertel D, Mader U, Bernhardt J, Becher D, Hecker M, et al. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc Natl Acad Sci U S A. 2012;109:7451–7456. doi: 10.1073/pnas.1117483109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trentini DB, Suskiewicz MJ, Heuck A, Kurzbauer R, Deszcz L, Mechtler K, Clausen T. Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature. 2016;539:48–53. doi: 10.1038/nature20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wozniak DJ, Tiwari KB, Soufan R, Jayaswal RK. The mcsB gene of the clpC operon is required for stress tolerance and virulence in Staphylococcus aureus. Microbiology. 2012;158:2568–2576. doi: 10.1099/mic.0.060749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonas K, Liu J, Chien P, Laub MT. Proteotoxic stress induces a cell cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell. 2013;154:623–636. doi: 10.1016/j.cell.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leslie DJ, Heinen C, Schramm FD, Thuring M, Aakre CD, Murray SM, Laub MT, Jonas K. Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA. PLoS Genet. 2015;11:e1005342. doi: 10.1371/journal.pgen.1005342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung CH, Goldberg AL. DNA stimulates ATP-dependent proteolysis and protein-dependent ATPase activity of protease La from Escherichia coli. Proc Natl Acad Sci U S A. 1982;79:795–799. doi: 10.1073/pnas.79.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu GK, Smith MJ, Markovitz DM. Bacterial protease Lon is a site-specific DNA-binding protein. J Biol Chem. 1997;272:534–538. [PubMed] [Google Scholar]
- 25.Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 26.Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes & Development. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science (New York, NY) 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 28.McGinness KE, Baker TA, Sauer RT. Engineering Controllable Protein Degradation. Molecular Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Griffith KL, Grossman AD. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol Microbiol. 2008;70:1012–1025. doi: 10.1111/j.1365-2958.2008.06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 31.Wang KH, Roman-Hernandez G, Grant RA, Sauer RT, Baker TA. The molecular basis of N-end rule recognition. Mol Cell. 2008;32:406–414. doi: 10.1016/j.molcel.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang KH, Sauer RT, Baker TA. ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev. 2007;21:403–408. doi: 10.1101/gad.1511907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivera-Rivera I, Román-Hernández G, Sauer RT, Baker TA. Remodeling of a delivery complex allows ClpS-mediated degradation of N-degron substrates. Proceedings of the National Academy of Sciences. 2014;111:E3853–E3859. doi: 10.1073/pnas.1414933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Román-Hernández G, Hou JY, Grant RA, Sauer RT, Baker TA. The ClpS adaptor mediates staged delivery of N-end-rule substrates to the AAA+ ClpAP protease. Molecular cell. 2011;43:217–228. doi: 10.1016/j.molcel.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein BJ, Grant RA, Sauer RT, Baker TA. Structural Basis of an N-Degron Adaptor with More Stringent Specificity. Structure. 2016;24:232–242. doi: 10.1016/j.str.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan JL, Ward L, Truscott KN, Dougan DA. The N-end rule adaptor protein ClpS from Plasmodium falciparum exhibits broad substrate specificity. FEBS Lett. 2016;590:3397–3406. doi: 10.1002/1873-3468.12382. [DOI] [PubMed] [Google Scholar]
- 37.Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a Substrate Modulator of the ClpAP Machine. Molecular Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets σ(S) for degradation by ClpXP. Genes & Development. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battesti A, Hoskins JR, Tong S, Milanesio P, Mann JM, Kravats A, Tsegaye YM, Bougdour A, Wickner S, Gottesman S. Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev. 2013;27:2722–2735. doi: 10.1101/gad.229617.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Battesti A, Majdalani N, Gottesman S. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl Acad Sci U S A. 2015;112:5159–5164. doi: 10.1073/pnas.1504639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi KK, Berg M, Radhakrishnan SK, Viollier PH, Chien P. An Adaptor Hierarchy Regulates Proteolysis during a Bacterial Cell Cycle. Cell. 2015;163:419–431. doi: 10.1016/j.cell.2015.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau J, Hernandez-Alicea L, Vass RH, Chien P. A Phosphosignaling Adaptor Primes the AAA+ Protease ClpXP to Drive Cell Cycle Regulated Proteolysis. Molecular cell. 2015;59:104–116. doi: 10.1016/j.molcel.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan CM, Hahn E, Zuber P. Adaptor bypass mutations of Bacillus subtilis spx suggest a mechanism for YjbH-enhanced proteolysis of the regulator Spx by ClpXP. Molecular microbiology. 2014;93:426–438. doi: 10.1111/mmi.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studemann A, Noirclerc-Savoye M, Klauck E, Becker G, Schneider D, Hengge R. Sequential recognition of two distinct sites in sigma(S) by the proteolytic targeting factor RssB and ClpX. EMBO J. 2003;22:4111–4120. doi: 10.1093/emboj/cdg411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Molecular microbiology. 2013;88:1083–1092. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai J-W, Alley MRK. Proteolysis of the Caulobacter McpA Chemoreceptor Is Cell Cycle Regulated by a ClpX-Dependent Pathway. Journal of Bacteriology. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gora KG, Cantin A, Wohlever M, Joshi KK, Perchuk BS, Chien P, Laub MT. Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Molecular microbiology. 2013;87:1277–1289. doi: 10.1111/mmi.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. The EMBO Journal. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 50.Beaufay F, Coppine J, Mayard A, Laloux G, De Bolle X, Hallez R. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J. 2015;34:1786–1800. doi: 10.15252/embj.201490730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radhakrishnan SK, Pritchard S, Viollier PH. Coupling Prokaryotic Cell Fate and Division Control with a Bifunctional and Oscillating Oxidoreductase Homolog. Developmental Cell. 2010;18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 52.Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Molecular Microbiology. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 53.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 54.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes & Development. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proceedings of the National Academy of Sciences. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A Dynamically Localized Protease Complex and a Polar Specificity Factor Control a Cell Cycle Master Regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 57.Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Molecular Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. Dynamics of Two Phosphorelays Controlling Cell Cycle Progression in Caulobacter crescentus. Journal of Bacteriology. 2009;191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith SC, Joshi KK, Zik JJ, Trinh K, Kamajaya A, Chien P, Ryan KR. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14229–14234. doi: 10.1073/pnas.1407862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan IS, Weiss CA, Popham DL, Ramamurthi KS. A Quality-Control Mechanism Removes Unfit Cells from a Population of Sporulating Bacteria. Dev Cell. 2015;34:682–693. doi: 10.1016/j.devcel.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Bivalent Tethering of SspB to ClpXP Is Required for Efficient Substrate Delivery: A Protein-Design Study. Molecular Cell. 2004;13:443–449. doi: 10.1016/s1097-2765(04)00027-9. [DOI] [PubMed] [Google Scholar]
- 62.Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol. 2015;13:285–297. doi: 10.1038/nrmicro3438. [DOI] [PubMed] [Google Scholar]
- 63.Bonissone S, Gupta N, Romine M, Bradshaw RA, Pevzner PA. N-terminal protein processing: a comparative proteogenomic analysis. Mol Cell Proteomics. 2013;12:14–28. doi: 10.1074/mcp.M112.019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt R, Zahn R, Bukau B, Mogk A. ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol Microbiol. 2009;72:506–517. doi: 10.1111/j.1365-2958.2009.06666.x. [DOI] [PubMed] [Google Scholar]
- 65.Ninnis RL, Spall SK, Talbo GH, Truscott KN, Dougan DA. Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. EMBO J. 2009;28:1732–1744. doi: 10.1038/emboj.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Humbard MA, Surkov S, De Donatis GM, Jenkins LM, Maurizi MR. The N-degradome of Escherichia coli: limited proteolysis in vivo generates a large pool of proteins bearing N-degrons. J Biol Chem. 2013;288:28913–28924. doi: 10.1074/jbc.M113.492108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ravid T, Hochstrasser M. Degradation signal diversity in the ubiquitin-proteasome system. Nature reviews Molecular cell biology. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shang F, Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free radical biology & medicine. 2011;51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strieter ER, Korasick DA. Unraveling the complexity of ubiquitin signaling. ACS Chem Biol. 2012;7:52–63. doi: 10.1021/cb2004059. [DOI] [PMC free article] [PubMed] [Google Scholar]