Abstract
The ECF sigma family was identified 23 years ago as a distinct group of σ70-like factors. ECF sigma factors have since emerged as a major form of bacterial signal transduction that can be grouped into over 50 phylogenetically distinct subfamilies. Advances in our understanding of these sigma factors and the signaling pathways governing their activity have elucidated conserved features as well as aspects that have evolved over time. All ECF sigma factors are predicted to share a common streamlined domain structure and mode of promoter interaction. The activity of most ECF sigma factors is controlled by an anti-sigma factor. The nature of the anti-sigma factor and the activating signaling pathways appear to be conserved within ECF families, while considerable diversity exists between different families.
Introduction
Nearly all cells, whether a bacterium invading a plant leaf or a cell in a developing human liver, must be able to sense their surroundings and adapt gene expression accordingly. In bacteria, a major form of transcriptional regulation is through the exchange of the primary sigma subunit of RNA polymerase (RNAP) with an alternative sigma factor redirecting RNAP to specific promoters and rapidly reprogramming gene expression. Most bacterial species have an array of alternative sigma factors, the largest group of which is the ExtraCytoplasmic Function (ECF) family.
From the founding members to the ECF sigma factor family of today
The ECF sigma factors (also known as Group 4) were first recognized as a distinct subgroup of σ70-like factors in 1994 [1]. The founding members of the group were σE from Escherichia coli, AlgU from Pseudomonas aeruginosa, σE from Streptomyces coelicolor, CarQ from Myxococcus xanthus, CnrH from Alcaligenes eurotrophas, HrpL from Pseudomonas syringae, SigX from Bacillus subtilis, and FecI from E. coli. All the proteins, with the exception of SigX, which was an ORF of unknown function, positively regulated gene expression, and E. coli and S. coelicolor σE had been shown to act as sigma factors in biochemical assays [2–8]. Sequence comparisons with the primary sigma factor indicated that these proteins were essentially streamlined versions of sigma factors [1]. They contained conserved domains 2 and 4, which are responsible for the fundamental properties of sigma factors, core RNAP binding and specific promoter recognition, but lacked conserved domains found in other sigma factors (Figure 1) [9]. Not only were the primary sequences of this new group of sigma factors conserved, but the promoters they recognized, which had been identified for AlgU, σEEc, σESc, CarQ, and FecI, were also similar, particularly in the −35 element [1,6]. This group of proteins shared other features as well. All of the proteins regulated genes with extracytoplasmic functions and responded to extracytoplasmic signals, inspiring the name of the group [1]. Four of the family members, AlgU, σEEc, CarQ, and CnrH, were known or predicted to be negatively regulated by downstream genes in the same operon [1,7,8,10]. However, it was clear that other regulatory mechanisms existed because HrpL was shown to be activated at the level of transcription by HrpR and HrpS, two NtrC-like proteins [11]. In this review, we will explore how many of the core themes in ECF sigma factor biology identified with the founding eight members still hold true today. Ironically, a major exception is the name itself. Although many ECF sigma factors do have extracytoplasmic functions, there are members of the family that sense intra-cellular signals and govern responses that primarily influence the cytoplasm.
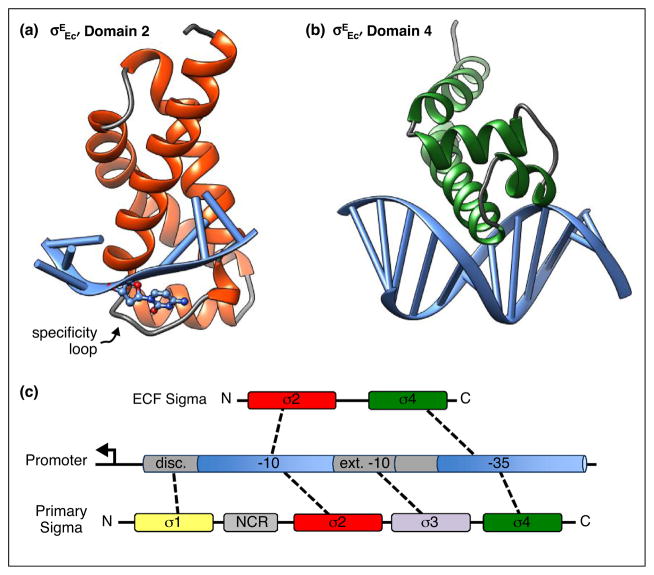
Figure 1.
Promoter recognition by ECF sigma factors. (a) Domain 2 from σEEc bound to the nontemplate strand of the −10 promoter element is shown. Protein helices are in red and loops are in gray, while promoter DNA is in blue. The loop that determines binding specificity is indicated. It forms a pocket for the cytosine base, shown in ball and stick format, that is flipped out from the ssDNA helix. (b) Domain 4 from σEEc bound to the −35 promoter element is shown. Protein helices are in green and loops in gray. (c) Schematic of conserved regions of ECF and primary sigma factors are diagrammed. Interactions between individual sigma factor domains and the promoter are indicated by dashed lines. NCR is the nonconserved region of primary sigma factors. Disc. is the discriminator motif and ext. −10 is the extended −10 motif. Structural representations were generated from PDB 4LUP [17] and 2H27 [16] using Chimera [44].
The ECF sigma factor family now comprises approximately 38 000 genes from 7679 bacterial genomes and constitutes one of the major signal transduction systems in bacteria [12••,13]. Phylogenetic analyses have defined 56 major and 32 minor ECF sigma factor families based on sequence similarity and genomic context [12••,13,14]. Predicted and confirmed ECF sigma factors and their family affiliation, where known, can be found in the Microbial Signal Transduction (MiST2) database (http://mistdb.com/). Most bacteria possess multiple ECF sigma factors, the number of which is roughly correlated with the size of the genome and the lifestyle of the organism [12••,13]. In general, bacteria with large genomes, complex developmental programs, and that live in niches subject to fluctuating environmental conditions have larger numbers of ECF sigma factors, whereas species with smaller genomes that inhabit relatively stable environments have few or no ECF sigma factors.
Promoter recognition and transcription initiation
The hallmark of the ECF family is the reduced domain structure. ECF sigma factors lack most of domain 1, which in the primary sigma factor binds to the discriminator sequence downstream of the −10 promoter element, and domain 3, which recognizes the extended −10 sequence upstream of the −10 promoter element (Figure 1) [9]. Domain 3 has also been proposed to stabilize the incoming nucleotides during initial transcript formation [9,15]. The implications of not having these domains are not fully understood and await detailed biochemical studies of transcription initiation directed by ECF sigma factors and structural studies with an ECF sigma factor-RNAP holoenzyme bound to promoter DNA.
Although a structure of an ECF holoenzyme has yet to be determined, structures of the isolated sigma factor domains 2 (σ2) and 4 (σ4) from E. coli σE bound to the −10 and −35 promoter elements, respectively, are available and provide a wealth of information [16,17••]. The structures illustrate both similarities and differences in the ways that ECF sigma factors recognize and melt promoter DNA compared to primary sigma factors. The overall docking on the DNA is likely to be conserved among ECF sigma factors given their sequence conservation, although the details of any given specific interaction will vary.
Sequence alignments of the founding group of ECF sigma factors clearly showed that the ECF σ2 was divergent from that of other σ70 family members [1]. σ2 stabilizes DNA strand separation by binding to the non-template strand of the −10 promoter element in the open complex [9,15]. The primary sigma factors have a series of conserved hydrophobic and basic amino acids that form a pocket that cradles two bases flipped out from the base stack of the non-template strand during promoter melting [9,15]. These amino acids are not conserved in the ECF σ2 domains [1,17••]. Recent structural studies with σEEc domain 2 demonstrate that ECF sigmas have solved the problem of promoter melting with a related, but different mechanism [17••]. Only one base is flipped out of the ssDNA base stack and it is stabilized by a flexible loop connecting helices 2.3 and 2.4 of σ2 (Figure 1a) [17••]. In an interesting twist, sequences of the loop vary among ECF sigma factors such that different loops recognize different flipped-out bases making this region a specificity determinant [17••].
The sequence of domain 4 of ECF sigma factors more closely resembles that of the primary sigma factor [1]. However, the structure of σEEc domain 4 bound to the −35 promoter element revealed that the ECF σ4 sits further in the major groove and makes more extensive contacts with the DNA than σ70 domain 4 (Figure 1b) [16]. Most of the residues that are critical for promoter recognition are conserved and structures of σ4 from different ECF sigma factors align well with each other indicating that recognition of the −35 promoter element is likely to be conserved [16,18,19,20•]. These findings are consistent with observations made with the founding members that the promoters they recognized were related, particularly in the −35 element [1].
Studies on promoter specificity suggest that within an ECF family, individual sigma factors often recognize overlapping sets of the promoters, whereas little crosstalk is found for ECF sigma factors from different families [13,21]. Bacteria with multiple ECF sigma factors tend to have more ECF sigma factors from non-crosstalking families than from families that have a broader promoter specificities, indicating that there is evolutionary pressure on bacteria to insulate individual ECF signaling systems [21]. However, variations on this theme do occur. B. subtilis has a series of ECF sigma factors that respond to partially overlapping types of cell envelope stresses and have partially overlapping promoter specificities [22–24]. This arrangement enables a complex response to envelope stress in which a core set of genes is induced by most stresses, while other genes are induced in a stress-specific manner [21,22,25]. Finally, the ECF sigma factors appear to operate in a modular manner in which recognition of the −35 promoter element is independent of the −10 element. Functional chimeric sigma factors with novel binding specificities can be constructed by combining σ2 from one ECF sigma factor with σ4 from a different sigma factor along with the respective −35 and −10 promoter sequences [21].
Antisigma factors
Anti-sigma domains
When the founding ECF sigma factors were first described, it was noted that several were negatively regulated by co-transcribed genes encoding anti-sigma factors [1]. With more ECF sequences available, this model of co-transcription of the ECF sigma factor with an anti-sigma factor has become a defining characteristic for many of the ECF families [13]. Anti-sigma factors share little primary sequence homology (~25% conservation over 75 amino acids) making them difficult to identify from sequence information alone [26]. Seminal structural studies of E. coli σE and R. sphaeroides σE bound to their cognate anti-sigma factors, RseA and ChrR respectively, revealed that the two anti-sigma factors shared a structural motif that was termed the anti-sigma domain (ASD) [26,27]. This domain consists of a three-helix bundle and is followed by a fourth helix whose position varies (Figure 2) [28]. Currently, bioinformatic analyses that combine structural and sequence information have identified ASD-like domains in genes co-transcribed with ECF sigma factors for approximately 50% of the ECF families [12••,13,26]. The majority of these proteins are predicted to be single-pass transmembrane proteins with the ASD in the N-terminal cytoplasmic domain [13]. The periplasmic domains of the predicted anti-sigma factors vary considerably among different ECF families suggesting that they have diverged to recognize disparate signals [12••,13].
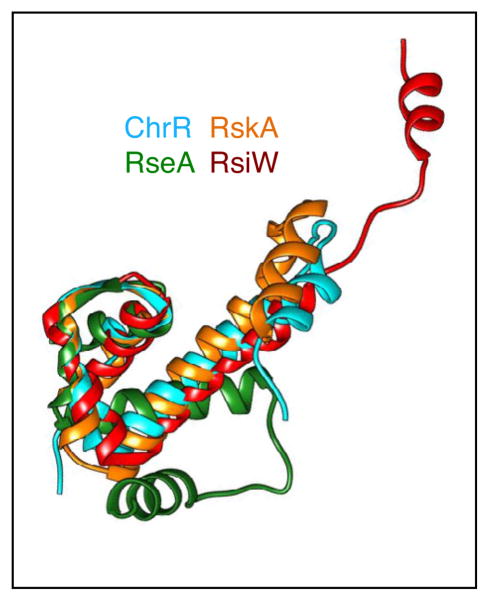
Figure 2.
Structural conservation of the ASD from ChrR, RseA, RskA, and RsiW. Structural alignments were performed using Swiss-PDBViewer 4.0 and displayed using Chimera [71]. The three-helix bundle is superimposable for all the anti-sigma factors, while the position of the fourth helix is variable. ChrR is shown in cyan, RseA in green, RskA in orange, and RsiW in red.
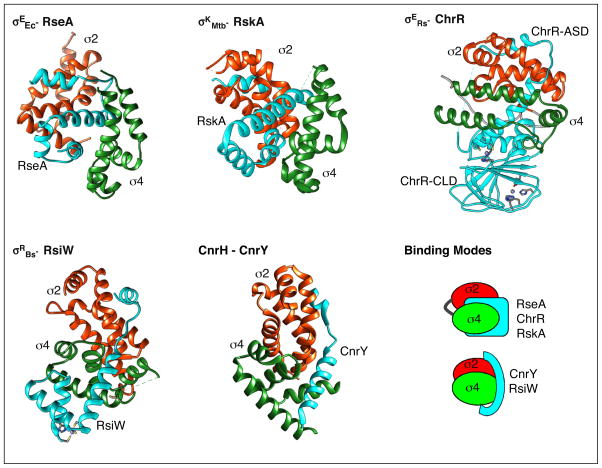
Co-crystal structures have been solved for four ECF sigma factors from different families bound to their cognate anti-sigma factors, E. coli σE-RseA, R. sphaeroides σE-ChrR, B.subtilis σW-RsiW, and M. tuberculosis σK-RskA [26,27,29,30•]. RseA, RskA, and RsiW are all inner membrane proteins and the cytoplasmic domain was used for structural studies, while ChrR is a soluble cytoplasmic protein and a proteolytically stable fragment lacking the C-terminal 10 amino acids was used [26,27,29,30•]. The ASD fold formed by the first three helices is highly conserved, although the fourth helix assumes a different position in each structure (Figure 2). Interestingly, the way in which each anti-sigma factor binds to the ECF sigma factor varies, despite conservation of the ASD fold (Figure 3) [26,27,29,30•]. RseA, ChrR, and RskA all bind between the σ2 and σ4 domains, but they dock on the cognate sigma factor in different manners (Figure 3) [26,27,30•]. RskA blocks DNA binding and RNAP binding surfaces on σK. RseA and ChrR block RNAP binding surfaces in a similar fashion on σ2, but interact differently with σ4 [26,27,30•]. Remarkably, the structure of RsiW bound to σW reveals a completely different mode of inhibition (Figure 3) [29]. Instead of the two sigma factor domains sandwiching the anti-sigma factor, σW forms a compact structure in which σW2 and σW4 interact with each other in a way that masks the DNA binding surface of σW2 [29]. RsiW wraps around the outside of σW with the ASD occluding the promoter-binding surface of σW4 and the fourth helix reaching around σW2 [29]. Therefore, although the conformation of the ASD has been highly conserved among anti-sigma factors, the mechanism of inhibition of sigma factor activity is unique for each cognate sigma/anti-sigma factor pair examined thus far.
Figure 3.
Two modes of sigma factor/anti-sigma factor interactions. Structures of σEEc bound to RseA (top left), σKMtb bound to RskA (top center), σERs bound to ChrR (top right), σWBs bound to RsiW (bottom left), and CnrH bound to CnrY (bottom center) are shown. Domains 2 and 4 of the sigma are in red and green, respectively. The anti-sigma factors are shown in turquoise. For ChrR, ChrR-ASD is the N-terminal anti-sigma domain and ChrR-CLD is the C-terminal cupin-like domain. A cartoon representation of the two binding modes is shown (bottom right). Structural representations were generated from PDB 1OR7 [27], 2Q1Z [26], 4NQW [30•], 5WUQ [29] and 4CXF [35••] using Chimera [71].
Zinc binding anti-sigma (ZAS) factors
Over 30% of anti-sigma factors with an ASD contain a Zn-binding motif (C/H-X23–26-H-X3-C-X2-C) within the ASD [12••,13,26]. The motif was first identified in the S. coelicolor anti-sigma factor RsrA, which regulates σR in response to disulfide stress, and then visualized structurally in ChrR, which responds to singlet oxygen [26,31,32]. Structural and functional studies reveal at least three major classes of ZAS factors whose Zn-binding properties vary in their redox sensitivity and role in ECF sigma factor regulation. ChrR binds two Zn atoms, one via the Zn binding motif in the amino terminal ZAS domain and the other in the carboxy-terminal cupin-like domain (CLD) [26]. Zn stabilizes the fold of the ASD of ChrR and is therefore required for σE binding [26]. However, the Zn ion in the ASD is not easily dissociated and does not appear to be involved in redox sensing [26]. The ASD of RsiW contains a ZAS motif, although σW has not been found to respond to oxidative stress [25]. A comparison of structures of the reduced Zn-bound state and oxidized Zn-free state of RsiW in complex with σW revealed only minor rearrangements [29]. The overall structure and σW binding remain unperturbed by oxidation, leaving the role of Zn unclear [29]. In contrast, structural studies of reduced and oxidized RsrA along with a homology model of the RsrA-σR complex demonstrate how Zn binding plays critical roles for both structure and redox sensing by RsrA [33••]. In solution structures of the reduced Zn-bound form of RsrA, the protein folds into a 4-helix bundle that does not resemble the ASD fold [33••]. Neither cocrystal nor solution structures of a RsrA-σR complex could be obtained, therefore a homology model of the complex was built based on structures of anti-sigma factor/sigma factor complexes and constrained by cross-linking data with RsrA and σR [33••]. In the resulting model, RsrA undergoes extensive structural rearrangements upon binding σR and adopts a classical ASD fold [33••]. In structures of the oxidized form of RsrA, a Zn atom is no longer bound and two of the cysteine residues that were involved in zinc binding form a disulfide bond. The disulfide bond helps stabilize a compact helical structure in which the σR binding surfaces, Zn binding site, and ASD fold are disrupted [33••].
Short extended anti-sigma domains
In addition to the ASD family, a second class of putative anti-sigma factors has been proposed in which the ECF sigma is co-transcribed with a predicted transmembrane protein containing a short, <50 amino acid, amino-terminal cytoplasmic domain that may act as an inhibitor of the sigma factor [12••,13]. Approximately 25% of the ECF sigma factor families have these candidate anti-sigma sequences [12••,13]. CnrH, one of the founding ECF sigma factors, is regulated by an anti-sigma factor belonging to this class, CnrY [34,35••]. CnrY is single-pass transmembrane protein with a 45 amino acid cytoplasmic domain that acts as the anti-sigma factor [34]. In the cocrystal structure of the CnrH-CnrY complex, the σ2 and σ4 domains of CnrH bind to each other concealing the DNA binding surfaces [35••]. CnrY wraps across CnrH and blocks sites where the beta subunit of RNAP binds (Figure 3) [35••]. A similar mechanism of binding was seen between the anti-sigma NepR and the ECF sigma mimic PhyR from Alphaproteobacteria, indicating that this mode of inhibition is not unique to CnrH [36,37]. There is no detectable sequence homology between NepR and CnrY making bioinformatic identification of these short anti-sigma factors difficult. Interestingly the overall mechanism of inhibition of CnrH by CnrY, in which the anti-sigma factor wraps around the ECF sigma factor, is analogous to inhibition of σW by RsiW [29].
Fused regulatory domains
A putative anti-sigma factor could not be identified in 20% of the ECF sigma factor families [12••,13]. Some of these sigma factors, including two of the founding ECF sigma factors, HrpL and σESc, are primarily regulated at the level of expression and not by anti-sigma factors [2,38]. In other families, the ECF sigma factor itself contains additional domains beyond σ2 and σ4 that could be regulatory [12••,13]. Once such family is the ECF41 group of sigma factors whose members have been identified in bacterial genomes from 10 different phyla [13]. These ECF sigma factors appear to be regulated by a C-terminal domain, partial truncation of which leads to increased sigma factor activity in vivo, but lower RNAP binding in vitro compared to the full-length protein [39]. Surprisingly, complete truncation of the C-terminal extension eliminated sigma factor activity in vivo and RNAP binding, even though the σ2 and σ4 domains were intact [39]. The details about how the regulatory domain senses signals and controls RNAP interaction remain to be elucidated.
Signal sensing
Most ECF sigma factors, with the exception of those that are primarily regulated at the level of transcription, are held in an inactive state by an anti-sigma factor or by a cis-acting inhibitory domain that is disrupted in response to a stimulus. The activating signals, sensed indirectly via a signal transduction pathway or directly by the anti-sigma factor itself, trigger release of the sigma factor so that it can associate with RNAP and initiate the response [28]. Several major classes of activation mechanisms have been identified and include regulated proteolysis, partner switching, and conformational changes [28,40]. A general theme that is emerging from these studies is that the overall framework of the signal transduction pathway is often conserved, especially within an ECF family, while the inducing signals and accessory factors that modulate the signal transduction pathways vary. Here we focus on two of the most well understood activation mechanisms, sensing of cell envelope signals by regulated proteolysis and redox sensing by conformational changes, to illustrate both the similarities and variations in how these system have evolved.
Indirect sensing-regulated proteolysis
Regulated proteolysis of the anti-sigma factors is one of the best understood mechanisms of ECF sigma factor activation particularly for transmembrane anti-sigma factors that sense signals in the cell envelope [41]. The anti-sigma factor is completely degraded in response to the inducing signal by the sequential action of two transmembrane proteases followed by cytoplasmic pro-teases (Figure 4a–c). The initiating signal triggers the first protease to cleave the periplasmic domain from the anti-sigma factor [41]. Release of the periplasmic domain removes inhibitory interactions allowing the second pro-tease, usually a member of the RIP family of intramembrane proteases, to cleave the anti-sigma factor in the transmembrane region thereby releasing the amino terminal domain bound to the sigma factor into the cytoplasm [41]. The remaining fragment of the anti-sigma factor is then degraded to completion by cytoplasmic proteases, mainly ClpXP, freeing the sigma factor to bind to RNAP and initiate transcription [41].
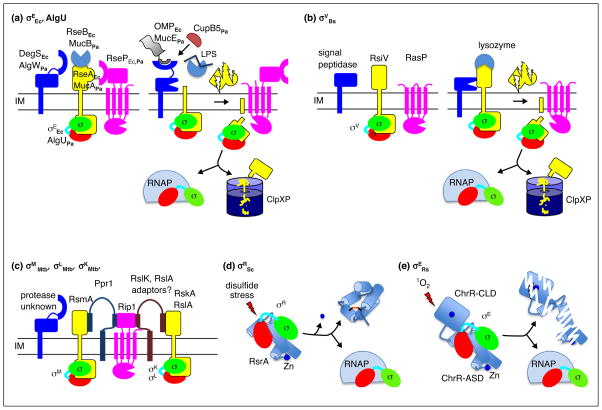
Figure 4.
Envelope sensing by regulated proteolysis and redox sensing by conformational changes. (a) Roles of the key players in the pathways regulating σEEc and AlgU are depicted as the system transitions from the inactive to the active state. Proteolysis of RseA and MucA is triggered by binding of OMP peptides to DegS and AlgW in conjunction with release RseB and MucB from the anti-sigma factor upon LPS binding. For P. aeruginosa, CupB5 can activate AlgW bypassing the need for LPS binding to MucB. (b) Activation of σV by degradation of RsiV following lysozyme binding is depicted. In the inactive state, the first protease to act, signal peptidase, cannot access RsiV. Binding of lysozyme to RsiV exposes the cleavage site in RsiV to signal peptidase initiating the proteolytic cascade. (c) A related proteolytic pathway regulates degradation of the RsmA, RskA, and RslA anti-sigma factors from M. tuberculosis. The initiating protease and inducing signals have yet to be identified. Rip1 is responsible for the second cleavage in the proteolytic cascade for all three anti-sigma factors. The adapter protein, Ppr1, bridges Rip1 and RsmA and prevents cleavage. Similar adapter proteins have been postulated for RskA and RslA. (d) Redoxing sensing by RsrA is depicted. In the inactive state, σR is sequestered by the reduced Zn bound form of RsrA. Exposure to reactive electrophile species results in loss of Zn, formation of an intramolecular disulfide bond, structural rearrangement of RsrA, and release of σR. (e) Activation of σE by singlet oxygen (1O2). In the inactive state, σE is bound to ChrR. Singlet oxygen sensing involves ligands of the Zn atom in the ChrR-CLD, although loss of Zn is not absolutely required, and leads to degradation of ChrR by an unknown mechanism.
The overall layout of the proteolytic cascade is conserved in diverse bacteria from E. coli to B. subtilis to M. tuberculosis. However, the details in how the pathway is implemented have been adapted to fit the needs of the organism. The E. coli σE response is induced by stresses that disrupt the outer membrane and its regulon members include genes that facilitate delivery of proteins and LPS to the outer membrane [42–44]. Two signals, unfolded outer membrane porins (OMPs) and intermediates in LPS biosynthesis that do not reach the outer membrane, are required to trigger the proteolytic pathway responsible for degradation of RseA [45,46]. Exposed C-terminal peptides from unfolded OMPs bind to and allosterically activate DegS, the first protease in the cascade [45]. The LPS intermediates bind to RseB, an accessory protein that protects RseA from proteolysis, and dissociate it from RseA (Figure 4a) [46–48]. Each signal reports on a different aspect of outer membrane integrity, tying σE activity directly to outer membrane homeostasis. A remarkably similar proteolytic pathway regulates activation of P. aeruginosa AlgU via degradation of its anti-sigma factor, MucA (Figure 4a). OMP peptides, although from a smaller subset of porins than in E. coli, activate the first protease [49] and LPS intermediates bind MucB, the RseB homologue, to relieve inhibition of the first cleavage of MucA (Figure 4a) [46]. The P. aeruginosa system can also be activated by OMP peptides in combination with the CupB5 usher protein, when it is expressed without its cognate chaperone [50]. CupB5 is proposed to enhance the ability of the initiating protease, AlgW, to compete with MucB for access to MucA [50].
B. subtilis σW and σV are also activated in response to cell envelope stresses by regulated proteolysis of their cognate transmembrane anti-sigma factors, RsiW and RsiV respectively [51•,52–54]. The signals to initiate proteoly-sis of RsiW are not known, but the inducing signal and sensing mechanism have recently been elucidated for the RsiV-σV system (Figure 4b). Instead of the inducing signal directly activating the first protease in the cascade, degradation of RsiV is triggered by binding of lysozyme to RsiV, which makes RsiV accessible to the initiating protease [55••,56•]. In addition, the active site of lysozyme is occluded by RsiV such that binding not only activates σV but also inactivates lysozyme [55••]. Once released from RsiV, σV transcribes a set of genes that render the cells resistant to lysozyme [57,58].
M. tuberculosis, presents yet another variation on the proteolytic cascade. The same RIP protease, Rip1, acts as the second protease in the proteolytic cascades that degrade three different anti-sigma factors, RslA, RsmA, and RskA (Figure 4c) [59]. The initiating protease(s) has not yet been identified. Recently it was shown that an adaptor protein, Ppr1, binds to both RsmA and the substrate binding domain of Rip1 and prevents signal-independent degradation of RsmA (Figure 4c) [60]. The adaptor is proposed to provide specificity to the system by tethering RsmA to the protease. This model predicts that additional adaptor proteins exist to connect Rip1 with each of the remaining anti-sigma factors [60].
Direct sensing-conformational changes
The best characterized system for direct sensing of an inducing signal is disulfide stress sensing by RsrA. RsrA is activated by reactive electrophiles such as diamide and is less sensitive to reactive oxygen species like H2O2 [61,62•]. The rate-limiting step in activation is loss of Zn from RsrA, which is followed by release of σR and formation of a disulfide bond in RsrA (Figure 4d) [33••].
The disulfide bond stabilizes a conformation, described above, that does not bind σR [33••]. Once released, σR transcribes genes required to combat thiol stress [63]. A similar mechanism has been proposed for RslA, a M. tuberculosis ZAS-containing anti-sigma factor. H2O2 treatment of RslA results in loss of bound Zn and formation of a disulfide bond [18]. The affinity of Zn-free RslA for σL, its cognate ECF sigma factor, is less than that of the Zn-bound form [18]. However, treatment with DTT to prevent disulfide bond formation did not affect σL binding, indicating that the activation mechanism is not entirely the same as that governing RsrA–σR interactions [18,33••]. Unlike RsrA, which is cytoplasmic, RslA is a transmembrane anti-sigma factor and is also regulated by a proteolytic cascade [59]. How these two activating mechanisms interact to modulate σL activity in vivo is not yet understood.
An entirely different redox sensing mechanism is employed by the ChrR-σE system of R. sphaeroides (Figure 4e). ChrR binds two Zn ions, one in the N-terminal ZAS domain and the other in a C-terminal cupin-like domain (Figure 3) [26]. Although the N-terminal ZAS domain requires Zn to bind σE, the N-terminal domain is not involved in sensing singlet oxygen [26]. Instead the C-terminal cupin-like domain is required for signal sensing. Interestingly, the conserved amino acids that coordinate the Zn atom in the cupin-like domain are required for the response to singlet oxygen, but release of the Zn ion is not necessary [26,64]. Mutations of these residues render ChrR signal blind, even though Zn binding is relatively unperturbed [64]. Singlet oxygen also leads to degradation of ChrR, suggesting that regulated proteolysis is involved in the signaling pathway [64,65]. The protease(s) responsible for degrading ChrR have yet to be identified [65].
Interactions with other regulatory networks
The ECF sigma factors are often regarded as stand-alone pathways that are activated solely by a dedicated signal transduction pathway. However, several ECF sigma factors have been shown to respond to additional regulators, indicating that they are more fully integrated into cellular regulatory networks than originally thought. E. coli σE can be activated independently of RseA and the envelope stress signaling pathway by the alarmone ppGpp, a global regulator that mediates the cellular response to nutrient limitation and other stresses [66,67•,68]. The stress signaling pathway that controls the degradation rate of RseA sets the overall amount of σE that is free to interact with RNAP, while ppGpp activates σE-dependent transcription initiation [66,69]. B. subtilis σX and σM were recently shown to be activated independently of their anti-sigma factors in response to glucose [70•]. In these cases, anti-sigma factor-independent activation provides a way for the bacterium to increase expression of genes controlled by these sigma factors in the absence of envelope stress, perhaps preparing the cell in advance should such stresses arise. As more ECF sigma factors are studied, it will be important to determine whether integration into cellular regulatory networks is prevalent, or whether the primary role of these pathways is as dedicated, stand-alone signaling modules.
Future directions
From their start as a small group of sigma factors, the ECF family now constitutes a central pillar of bacterial signal transduction. Research into these regulators and the pathways that control their activity has illuminated many general principles about their mechanisms of action and roles in cellular signaling. However, we have barely scratched the surface in exploring this large and diverse family. Clearly the themes and variations outlined here will continue to evolve as more ECF systems are investigated.
Acknowledgments
This work was supported by the National Institutes of Health [Grant number R01 GM097365].
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Lonetto MA, Brown KL, Rudd KE, Buttner MJ. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci U S A. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao YX, Hutcheson SW. A single promoter sequence recognized by a newly identified alternate sigma-factor directs expression of pathogenicity and host-range determinants in Pseudomonas syringae. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buttner MJ, Smith AM, Bibb MJ. At least three different RNA polymerase holoenzymes direct transcription of the agarase gene (dagA) of Streptomyces coelicolor A3(2) Cell. 1988;52:599–607. doi: 10.1016/0092-8674(88)90472-2. [DOI] [PubMed] [Google Scholar]
- 4.Erickson JW, Gross CA. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 5.Deretic V, Gill JF, Chakrabarty AM. Pseudomonas aeruginosa infection in cystic fibrosis—nucleotide-sequence and transcriptional regulation of the algd gene. Nucleic Acids Res. 1987;15:4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enz S, Braun V, Crosa JH. Transcription of the region encoding the ferric dicitrate-transport system in Escherichia coli: similarity between promoters for fecA and for extracytoplasmic function sigma factors. Gene. 1995;163:13–18. doi: 10.1016/0378-1119(95)00380-o. [DOI] [PubMed] [Google Scholar]
- 7.Liesegang H, Lemke K, Siddiqui RA, Schlegel HG. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes-eutrophus CH34. J Bacteriol. 1993;175:767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGowan SJ, Gorham HC, Hodgson DA. Light-induced carotenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol Microbiol. 1993;10:713–735. doi: 10.1111/j.1365-2958.1993.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 9.Feklístov A, Sharon BD, Darst SA, Gross CA. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol. 2014;68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 10.Martin DW, Schurr MJ, Mudd MH, Deretic V. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol Microbiol. 1993;9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson SW. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Huang X, Pinto D, Fritz G, Mascher T. Environmental sensing in Actinobacteria: a comprehensive survey on the signaling capacity of this phylum. J Bacteriol. 2015;197:2517–2535. doi: 10.1128/JB.00176-15. In this work, the authors expand and refine the ECF family through a comprehensive analysis of predicted ECF sigma factors and anti-sigma factors in the Actinobacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol. 2009;74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 14.Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, Kolter R. Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics. J Bacteriol. 2012;194:6419–6430. doi: 10.1128/JB.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Borukhov S. Bacterial RNA polymerase-DNA interaction-the driving force of gene expression and the target for drug action. Front Mol Biosci. 2016;3:73. doi: 10.3389/fmolb.2016.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane WJ, Darst SA. The structural basis for promoter −35 element recognition by the group IV sigma factors. PLoS Biol. 2006;4:e269. doi: 10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Campagne S, Marsh ME, Capitani G, Vorholt JA, Allain FH-T. Structural basis for −10 promoter element melting by environmentally induced sigma factors. Nat Struct Mol Biol. 2014;21:269–276. doi: 10.1038/nsmb.2777. NMR and crystallographic structures of domain 2 from an ECF sigma factor bound to the nontemplate strand of promoter DNA provide important insights into how these proteins recognize and initiate open complex formation. [DOI] [PubMed] [Google Scholar]
- 18.Thakur KG, Praveena T, Gopal B. Structural and biochemical bases for the redox sensitivity of Mycobacterium tuberculosis RslA. J Mol Biol. 2010;397:1199–1208. doi: 10.1016/j.jmb.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thakur KG, Joshi AM, Gopal B. Structural and biophysical studies on two promoter recognition domains of the extra-cytoplasmic function sigma factor sigma(C) from Mycobacterium tuberculosis. J Biol Chem. 2007;282:4711–4718. doi: 10.1074/jbc.M606283200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Kim KY, Park JK, Park S. In Streptomyces coelicolor SigR, methionine at the +35 element interacting region 4 confers the −31′-adenine base selectivity. Biochem Biophys Res Commun. 2016;470:257–262. doi: 10.1016/j.bbrc.2016.01.075. The authors present a structure of domain 4 of an ECF sigma factor and identify a specificity determining protein-DNA contact. [DOI] [PubMed] [Google Scholar]
- 21.Rhodius VA, Segall-Shapiro TH, Sharon BD, Ghodasara A, Orlova E, Tabakh H, Burkhardt DH, Clancy K, Peterson TC, Gross CA, et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol Syst Biol. 2013;9:702. doi: 10.1038/msb.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mascher T, Hachmann A-B, Helmann JD. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J Bacteriol. 2007;189:6919–6927. doi: 10.1128/JB.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Y, Asai K, Sadaie Y, Helmann JD. Transcriptomic and phenotypic characterization of a Bacillus subtilis strain without extracytoplasmic function σ factors. J Bacteriol. 2010;192:5736–5745. doi: 10.1128/JB.00826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu J, Helmann JD. The −10 region is a key promoter specificity determinant for the Bacillus subtilis extracytoplasmic-function sigma factors sigma(X) and sigma(W) J Bacteriol. 2001;183:1921–1927. doi: 10.1128/JB.183.6.1921-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmann JD. Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr Opin Microbiol. 2016;30:122–132. doi: 10.1016/j.mib.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell EA, Greenwell R, Anthony JR, Wang S, Lim L, Das K, Sofia HJ, Donohue TJ, Darst SA. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol Cell. 2007;27:793–805. doi: 10.1016/j.molcel.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 28.Paget MS. Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules. 2015;5:1245–1265. doi: 10.3390/biom5031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devkota SR, Kwon E, Ha SC, Chang HW, Kim DY. Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW. PLoS One. 2017;12:e0174284. doi: 10.1371/journal.pone.0174284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Shukla J, Gupta R, Thakur KG, Gokhale R, Gopal B. Structural basis for the redox sensitivity of the Mycobacterium tuberculosis SigK–RskA σ–anti-σ complex. Acta Crystallogr Sect D Biol Crystallogr. 2014;70:1026–1036. doi: 10.1107/S1399004714000121. Crystallographic studies of the RskA-SigK complex demonstrate the mechanism of inhibition of SigK by RskA. Biochemical experiments explore the redox sensitivity of Zn binding by RskA and its implications for anti-sigma factor activity. [DOI] [PubMed] [Google Scholar]
- 31.Anthony JR, Warczak KL, Donohue TJ. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci U S A. 2005;102:6502–6507. doi: 10.1073/pnas.0502225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paget MS, Bae JB, Hahn MY, Li W, Kleanthous C, Roe JH, Buttner MJ. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol Microbiol. 2001;39:1036–1047. doi: 10.1046/j.1365-2958.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- 33••.Rajasekar KV, Zdanowski K, Yan J, Hopper JTS, Francis M-LR, Seepersad C, Sharp C, Pecqueur L, Werner JM, Robinson CV, et al. The anti-sigma factor RsrA responds to oxidative stress by reburying its hydrophobic core. Nat Commun. 2016;7:12194. doi: 10.1038/ncomms12194. Structural and biochemical studies demonstrate the basis for redox sensing by RsrA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grass G, Fricke B, Nies DH. Control of expression of a periplasmic nickel efflux pump by periplasmic nickel concentrations. Biometals. 2005;18:437–448. doi: 10.1007/s10534-005-3718-6. [DOI] [PubMed] [Google Scholar]
- 35••.Maillard AP, Girard E, Ziani W, Petit-Härtlein I, Kahn R, Covès J. The crystal structure of the anti-σ factor CnrY in complex with the σ factor CnrH shows a new structural class of anti-σ factors targeting extracytoplasmic function σ factors. J Mol Biol. 2014;426:2313–2327. doi: 10.1016/j.jmb.2014.04.003. The structure of the CnrY anti-sigma factor bound to CnrH reveals a novel form of inhibition in which the anti-sigma factor wraps around the ECF sigma factor. [DOI] [PubMed] [Google Scholar]
- 36.Campagne S, Damberger FF, Kaczmarczyk A, Francez-Charlot A, Allain FH-T, Vorholt JA. Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria. Proc Natl Acad Sci U S A. 2012;109:E1405–E1414. doi: 10.1073/pnas.1117003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrou J, Rotskoff G, Luo Y, Roux B, Crosson S. Structural basis of a protein partner switch that regulates the general stress response of α-proteobacteria. Proc Natl Acad Sci U S A. 2012;109:E1415–E1423. doi: 10.1073/pnas.1116887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong HJ, Paget MSB, Buttner MJ. A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol Microbiol. 2002;44:1199–1211. doi: 10.1046/j.1365-2958.2002.02960.x. [DOI] [PubMed] [Google Scholar]
- 39.Wecke T, Halang P, Staroń A, Dufour YS, Donohue TJ, Mascher T. Extracytoplasmic function σ factors of the widely distributed group ECF41 contain a fused regulatory domain. Microbiologyopen. 2012;1:194–213. doi: 10.1002/mbo3.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mascher T. Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr Opin Microbiol. 2013;16:148–155. doi: 10.1016/j.mib.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Barchinger SE, Ades SE. Regulated proteolysis: control of the Escherichia coli σ(E)-dependent cell envelope stress response. Subcell Biochem. 2013;66:129–160. doi: 10.1007/978-94-007-5940-4_6. [DOI] [PubMed] [Google Scholar]
- 42.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 43.Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- 44.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 46.Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science. 2013;340:837–841. doi: 10.1126/science.1235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaba R, Alba BM, Guo MS, Sohn J, Ahuja N, Sauer RT, Gross CA. Signal integration by DegS and RseB governs the sigmaE-mediated envelope stress response in Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:2106–2111. doi: 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cezairliyan BO, Sauer RT. Inhibition of regulated proteolysis by RseB. Proc Natl Acad Sci U S A. 2007;104:3771–3776. doi: 10.1073/pnas.0611567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Regt AK, Yin Y, Withers TR, Wang X, Baker TA, Sauer RT, Yu HD. Overexpression of CupB5 activates alginate overproduction in Pseudomonas aeruginosa by a novel AlgW-dependent mechanism. Mol Microbiol. 2014;93:415–425. doi: 10.1111/mmi.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Ellermeier CD, Losick R. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 2006;20:1911–1922. doi: 10.1101/gad.1440606. This work identifies a novel participant in the proteolytic pathway governing AlgU activity in Psuedomonas aeruginosa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinrich J, Hein K, Wiegert T. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol Microbiol. 2009;74:1412–1426. doi: 10.1111/j.1365-2958.2009.06940.x. [DOI] [PubMed] [Google Scholar]
- 53.Heinrich J, Wiegert T. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol Microbiol. 2006;62:566–579. doi: 10.1111/j.1365-2958.2006.05391.x. [DOI] [PubMed] [Google Scholar]
- 54.Hastie JL, Williams KB, Ellermeier CD. The activity of σV, an extracytoplasmic function σ factor of Bacillus subtilis, is controlled by regulated proteolysis of the anti-σ factor RsiV. J Bacteriol. 2013;195:3135–3144. doi: 10.1128/JB.00292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Hastie JL, Williams KB, Bohr LL, Houtman JC, Gakhar L, Ellermeier CD. The anti-sigma factor RsiV is a bacterial receptor for lysozyme: co-crystal structure determination and demonstration that binding of lysozyme to RsiV is required for σV activation. PLoS Genet. 2016;12:e1006287. doi: 10.1371/journal.pgen.1006287. The structure of the periplasmic domain of RsiV bound to lysozyme demonstrates a novel mechanism to induce the regulated proteolysis of an anti-sigma factor. Lysozyme binding to RsiV triggers cleavage of RsiV and also inhibits lysozyme activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Hastie JL, Williams KB, Sepúlveda C, Houtman JC, Forest KT, Ellermeier CD. Evidence of a bacterial receptor for lysozyme: binding of lysozyme to the anti-σ factor RsiV controls activation of the ECF σ factor σV. PLoS Genet. 2014;10:e1004643. doi: 10.1371/journal.pgen.1004643. This paper identifies lysozyme binding to RsiV as the molecular signal required for activation of σV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guariglia-Oropeza V, Helmann JD. Bacillus subtilis σ(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J Bacteriol. 2011;193:6223–6232. doi: 10.1128/JB.06023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho TD, Hastie JL, Intile PJ, Ellermeier CD. The Bacillus subtilis extracytoplasmic function σ factor σ(V) is induced by lysozyme and provides resistance to lysozyme. J Bacteriol. 2011;193:6215–6222. doi: 10.1128/JB.05467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sklar JG, Makinoshima H, Schneider JS, Glickman MS. M. tuberculosis intramembrane protease Rip1 controls transcription through three anti-sigma factor substrates. Mol Microbiol. 2010;77:605–617. doi: 10.1111/j.1365-2958.2010.07232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider JS, Reddy SP, Hock YE, Evans HW, Glickman MS. Site-2 protease substrate specificity and coupling in trans by a PDZ-substrate adapter protein. Proc Natl Acad Sci U S A. 2013;110:19543–19548. doi: 10.1073/pnas.1305934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paget MS, Kang JG, Roe JH, Buttner MJ. SigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2) EMBO J. 1998;17:5776–5782. doi: 10.1093/emboj/17.19.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.Lee K-L, Yoo J-S, Oh G-S, Singh AK, Roe J-H. Simultaneous activation of iron- and thiol-based sensor-regulator systems by redox-active compounds. Front Microbiol. 2017;8:139. doi: 10.3389/fmicb.2017.00139. In this article the authors investigate the effects of redox-active compounds in vivo on key S. coelicolor redox stress-sensing systems, including RsrA. RsrA is primarily induced via reactive eletrophile species (RES) and intramolecular disulfide bonds form in RsrA in vivo in response to RES, concomitant with release of σR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim M-S, Dufour YS, Yoo J-S, Cho Y-B, Park J-H, Nam G-B, Kim HM, Lee K-L, Donohue TJ, Roe J-H. Conservation of thiol-oxidative stress responses regulated by SigR orthologues in actinomycetes. Mol Microbiol. 2012;85:326–344. doi: 10.1111/j.1365-2958.2012.08115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greenwell R, Nam T-W, Donohue TJ. Features of Rhodobacter sphaeroides ChrR required for stimuli to promote the dissociation of ((E)/ChrR complexes. J Mol Biol. 2011;407:477–491. doi: 10.1016/j.jmb.2011.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nam T-W, Ziegelhoffer EC, Lemke RAS, Donohue TJ. Proteins needed to activate a transcriptional response to the reactive oxygen species singlet oxygen. MBio. 2013;4:e00541–12. doi: 10.1128/mBio.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol. 2008;67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 67•.Gopalkrishnan S, Nicoloff H, Ades SE. Co-ordinated regulation of the extracytoplasmic stress factor, sigmaE, with other Escherichia coli sigma factors by (p)ppGpp and DksA may be achieved by specific regulation of individual holoenzymes. Mol Microbiol. 2014;93:479–493. doi: 10.1111/mmi.12674. In this work, the authors detail how E. coli σE is regulated independently of the stress signaling pathway in response to nutrient limitation by the alarmone ppGpp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3″,5-″ bispyrophosphate (ppGpp) J Bacteriol. 2006;188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli sigmaE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70•.Ogura M, Asai K. Glucose induces ECF sigma factor genes, sigX and sigM, independent of cognate anti-sigma factors through acetylation of CshA in Bacillus subtilis. Front Microbiol. 2016;7:1918. doi: 10.3389/fmicb.2016.01918. This paper is the first to demonstrate that ECF sigma factors from B. subtilis can be regulated by an alternative pathway that does not involve the anti-sigma factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]