Abstract
Sensor histidine kinases regulate adaptive cellular responses to changes in the chemical or physical state of the environment. HWE/HisKA2-family kinases comprise a subset of histidine kinases that is defined by unique sequence motifs in both the catalytic and non-catalytic regions. Recent crystal structures have defined conserved intramolecular interactions that inform models of kinase regulation that are unique to the HWE/HisKA2 superfamily. Emerging genetic, biochemical and genomic data indicate that, unlike typical histidine kinases, HWE/HisKA2 kinases do not generally signal via classical DNA-binding response regulators. Rather, these unusual kinases are often part of atypical regulatory pathways that control changes in gene expression via modulation of protein-protein interactions or transcription anti-termination.
Introduction
Sensor histidine kinases (HKs) are important regulators of environmental adaptation that are conserved across bacteria, archaea, fungi, and plants. Thousands of members of this diverse group of protein sensors have been annotated in genomes and classified into sequence families [1–5]. Among these is the HWE family, which has several unusual features that distinguish its primary structure from other sensor HKs. HWE was initially described as an atypical histidine kinase family often located on the same polypeptide as light-responsive phytochrome sensory domains, and that is enriched in the Rhizobiaceae family of Alphaproteobacteria [6]. Like typical histidine kinases, HWE family kinases bear a two-helix DHp (dimerization and histidine phosphotransfer) domain followed by a CA (catalytic ATP-binding) domain, and have demonstrated ATP hydrolysis and autophosphorylation activities [6–15]. However, several conserved sequence motifs are unique to this family, while motifs present in most other HKs are missing in HWE kinases. As a result, HWE family members were historically not classified as histidine kinases in several sequence databases. Even today, HK domain models may fail to identify the CA domain of many HWE kinases.
Recent biochemical analyses provide evidence that HWE kinases may exist in unusual oligomeric states, and high-resolution crystal structures have shed light on structural roles of sequence features specific to the HWE family. Histidine kinases are typically postulated to participate in two-component phosphoryltransfer pathways with cognate DNA-binding response regulators that regulate transcription initiation. This canonical signaling modality appears to be the exception for HWE kinases. Specifically, there is increasing evidence that HWE kinases are often part of signaling systems with multiple sensory kinase inputs that converge to regulate a common gene expression output by controlling protein-protein interactions, such as σ/anti-σ, or by directly regulating proteins that control transcription termination. Here, we highlight distinguishing features of this atypical class of histidine kinases, with respect to their primary structures, taxonomic distribution and genomic context. We further review recent data on the structure and functional roles of HWE and related HisKA2 kinases.
Distinguishing features of HWE/HisKA2 histidine kinase primary structure
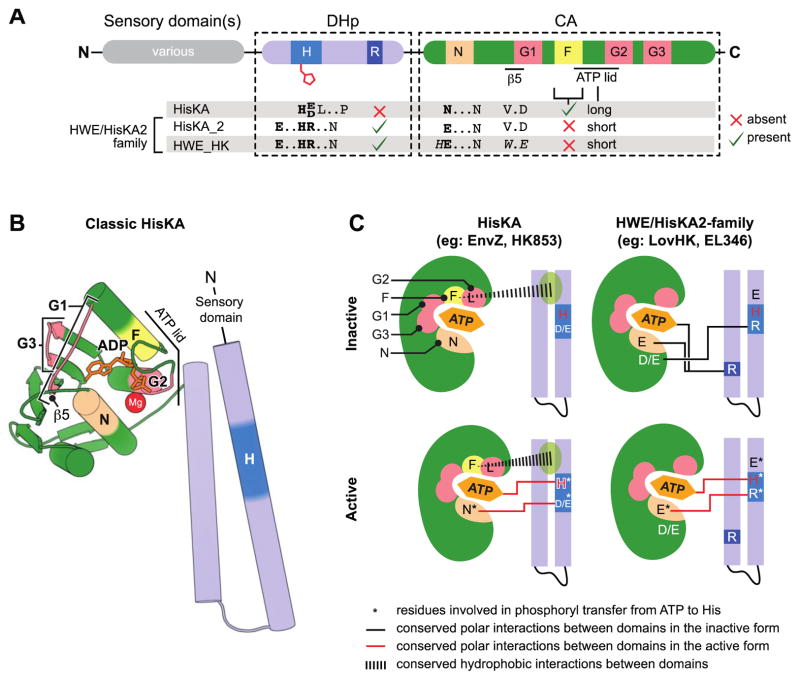
Given that classification of HWE kinases is based on amino acid sequence, it is worth discussing the key features of primary structure that distinguish this group. The nomenclature “H-W-E” refers to three residues in the catalytic domain, namely a histidine in the N-box and a WxE motif near the G1-box [6] (Figure 1A). However, the Pfam and SMART sequence models bearing the HWE name are based on motifs in the DHp domain, not the catalytic domain (Pfam: PF07536 and SMART: SM00911) [1,4]. Two key features in the DHp domain distinguish HWE-family kinases from typical HisKA kinases (Pfam: PF00512 and SMART: SM00388): 1) a unique set of residues that surround the phosphoryl-accepting histidine (H-box), and 2) a conserved arginine present in the second helix that is not found in other histidine kinases (R-box)(Figure 1A). The DHp domain model known as HisKA2 (Pfam: PF07568) shares both of these features and, together with HWE, form a superfamily (CDD: Cl06527) [16]. The shared H-box motif has resulted in the moniker, HRxxN kinases [9,17]. Indeed this is the most easily identified sequence characteristic of this kinase superfamily. We collectively refer to this group as HWE/HisKA2 kinases.
Figure 1. Characteristic features which distinguish HWE/HisKA2 histidine kinases.
(A) Primary structure features. Domain organization (top) highlights conserved histidine kinase boxes (H, R, N, G1, F, G2, G3). Distinguishing sequence motifs in Pfam:HisKA, Pfam:HisKA_2 or Pfam:HWE_HK kinases are highlighted below the domains. HisKA2 and HWE_HKs motifs are closely related. (B) Structural organization of a classical HisKA-family histidine kinase based on the crystal structure of Thermotoga maritima HK853 (PDB: 2C2A)[24]. The DHp domain (blue) forms two alpha helices that mediate dimerization and contain the phosphorylated histidine (dark blue). The catalytic ATPase (CA) domain (green) adopts the same basic globular fold as other GHKL ATPase superfamily domains [54]. Highlighted regions correspond to conserved boxes colored as in panel A. Sensory input domains are N-terminal to the DHp domain. (C) Conserved interdomain interactions in active and inactive conformations in HisKA and HWE/HisKA2 kinases. The CA domain is cartooned as a shell around the ATP binding pocket. The approximate positions of the conserved boxes are colored as in A and B. Key regulatory residues are marked with letters within the structure models. Residues involved in catalyzing histidine phosphorylation are highlighted with an asterisk. Conserved polar interactions between DHp and CA domains are shown as black lines in the inactive conformations and red lines in the active conformations.
In addition to unique features in the DHp domain, HWE/HisKA2 kinases bear several notable features in the catalytic domain. These kinases lack the F-box found in HisKA kinases and have a short ATP lid. In addition, the first asparagine in the N-box is replaced by a glutamate (Figure 1A) [6]. Two recent structures of HWE/HisKA2 kinases provide insight into the function of these distinguishing primary structure features [18,19].
Biochemical properties of HWE/HisKA2 kinases
Oligomeric state
Histidine kinases function almost exclusively as homodimers that can autophosphorylate either in cis or in trans depending on the tertiary and quaternary organization of the dimerized DHp domains [20,21]. Typical HK dimers are specific and stable: homodimer affinities range from mid-nanomolar to low micromolar [22]. While the oligomeric state of HWE/HisKA2 kinases has not been systematically investigated, studies point to diversity in intermolecular interactions in this family. For example, EsxG of Rhizobium, Agrobacterium and related genera is reported to form a stable hexameric structure composed of a trimer of dimers [13]. The kinase domain of Brucella LovhK appears to be dimeric in a crystal lattice, but these dimers are unstable in solution [18]. Finally, multiple lines of evidence show that Erythrobacter litoralis kinase EL346 is monomeric in crystals and in solution [19]. The monomeric nature of this particular kinase is enforced by a structure in which the sensory domain occludes the typical HK homodimer interface.
Catalytic residues
The fundamental chemistry required to transfer ATP γ-phosphate to a nitrogen on the imidazole ring of histidine is certainly similar between HisKA-family kinases and HWE/HisKA2-family kinases. However, the residues that participate in this catalysis are apparently not shared between these families. In HisKA kinases, a conserved acidic residue (E or D) directly adjacent the site of histidine phosphorylation (H+1 in the H-box) is proposed to act as a catalytic base that deprotonates histidine and enhances its nucleophilicity. This proton abstraction step promotes histidine nucleophilic attack on the γ-phosphate of ATP. Furthermore, the first asparagine in the N-box interacts with the H+1 acidic residue to both enhance histidine nucleophilicity and facilitate close positioning of the ATP to the H-box [21,23–25].
In HWE/HisKA2-family kinases, an arginine rather than an acidic residue is found at the H+1 position. While there is evidence for arginine serving as a general acid/base catalyst in some proteins [26,27], it is rare. Recent structures of the kinase domain of Brucella abortus LovHK [18] and full length Erythrobacter litoralis EL346 [19] provide testable models with regard to residues that participate in phosphoryltransfer chemistry. These structures suggest that either a conserved glutamic acid at the H minus 3 position of the H-box, or the conserved glutamic acid in the N-box could function as a general base to deprotonate histidine [18,19]. It remains unclear whether differences in the residues that catalyze phosphoryltransfer chemistry reflect higher order structural differences between HWE/HisKA2 and other HKs such as oligomeric state. Differences in catalytic residues may also be shaped by unique interactions between HWE/HisKA2 kinases and associated receiver domain substrates, such as those containing the so-called FATGY motif [28,29].
Structural control of kinase activity
A fundamental feature of sensory histidine kinases is their ability to regulate phosphoryltransfer activity in response to signal detection. Structural mechanisms by which signals may be transduced from sensory domain(s) to kinase domain in the more studied HisKA family proteins have been recently reviewed [30,31]. The HWE/HisKA2 kinase family, which exhibits atypical oligomeric states, catalytic residues and interdomain interactions, may utilize different kinase control mechanisms.
HK crystal structures show conformational flexibility in the orientation of the CA domain with respect to the DHp domain. The relative orientation of these domains determines kinase activity. Specifically, stabilization of conformations that position the ATP binding pocket of the CA domain proximal to the H-box of the DHp domain are predicted to promote histidine phosphorylation, while conformations that separate these domains likely reduce kinase activity.
In HisKA-family kinases two conserved hydrophobic residues, a phenylalanine in the F-box and a leucine in the G2 box are buried in the hydrophobic pocket between α1 and α2 of the DHp domain (see Figure 1C) [23,24,32]. HisKA crystal structures provide evidence that these conserved residues function as a pivot point that affects CA-to-DHp interdomain orientation. Structures of inactive conformations reveal variable polar and non-polar DHp-to-CA interdomain interactions [23,25,33]. Active forms of these kinases, in which the CA ATP binding pocket is proximal to DHp, are apparently stabilized by conserved interactions between the N-box asparagine and the acidic residue at position H+1 (Figure 1C) [21,23–25].
HWE/HisKA2 kinases lack the F-box. However, conserved hydrophobic residues present in the G2 box may fulfill the same function. Although only two crystal structures for this family are available, both support a model in which the N-box glutamic acid toggles between the conserved arginine at H+1 in the active conformation (Figure 1C), and a conserved arginine on the opposing helix of the DHp domain (i.e., the R-box [18]) in the inactive conformation [18,19]. The inactive conformation is further stabilized by a polar interaction between the H+1 arginine and an acidic residue outside of the N-box (Figure 1C). This regulatory switching model is consistent with data showing that mutation of the R-box of E. litoralis EL346 increases kinase activity, presumably by destabilizing the inactive conformation. Mutation of the H+1 arginine to alanine impairs kinase activity, consistent with a role for this residue in stabilizing the active conformation [19].
What about the WxE motif?
Based on their high conservation, it was initially hypothesized that the W and E residues may be involved in catalysis. Indeed, mutation of either residue to alanine abolished auto-phosphorylation in the kinase BphP2 [6]. However, these residues facilitate packing of β5 against α4 and thereby stabilize the fold of the CA domain [18]. In HisKA kinases, these positions are occupied by less bulky V and D residues, which enables closer β5- α4 packing. The functional role of these particular conserved residues remains undefined.
The genomic context of HWE kinases
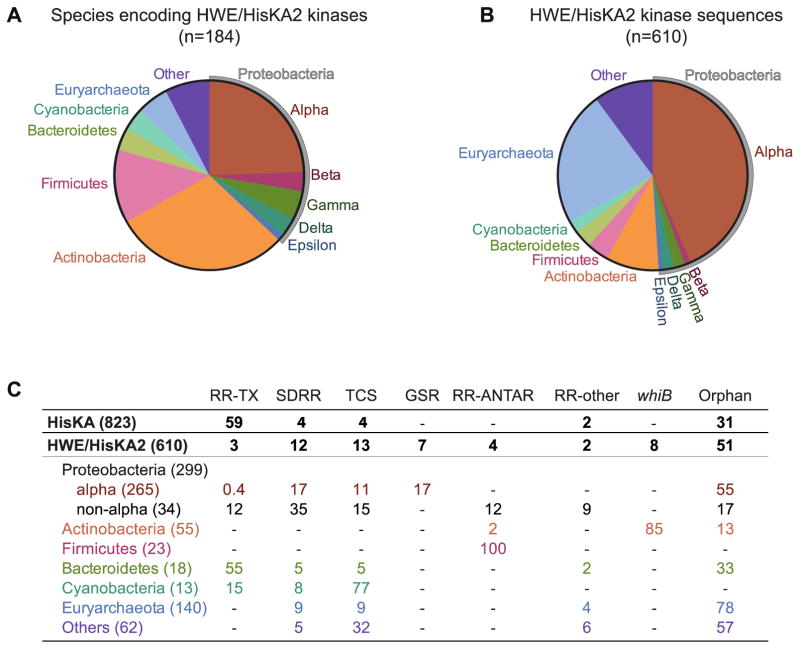
HisKA-family HKs are often encoded adjacently to DNA-binding response regulators, which directly regulate transcription (Figure 2) [5]. Such adjacent genes typically comprise classical two-component systems, whereby a sensor kinase phosphorylates a single, cognate response regulator to regulate its DNA-binding activity. Examination of the genes surrounding HWE/HisKA2 kinases, on the other hand, reveals that HWE/HisKA2 is rarely adjacent to DNA-binding response regulators (Figure 2C). Rather, these genes are most likely found a) as orphans (i.e. no adjacent response regulator gene), b) adjacent to genes encoding a single domain response regulator (SDRR) that has no ‘output’ domain, or c) at chromosomal loci that contain multiple two-component signaling genes including other histidine kinases - often hybrid histidine kinases (Figure 2C). This hints at a model in which HWE/HisKA2 kinases function in a manner that is distinct from archetypal two-component systems.
Figure 2. Phylogenetic distribution and genomic context of HWE/HisKA2 histidine kinases.
(A) Distribution of species encoding at least one HWE/HisKA2 kinase, grouped by phylum. (B) Distribution of HWE/HisKA2 kinase sequences, grouped by phylum. Pie charts are comprised of sequences containing either HisKA2 or HWE_HK domains in the Microbial Signal Transduction (MiST) Database version 2.2 (mistdb.com) [5]. Given that these domain models are highly similar, queries with either domain returned sets of sequences that overlapped by ≈70%. The distributions in panels A and B reflect the first 610 sequences in the Mist2.2 database that match either or both domain models. Supplemental File 1 lists the species included in this analysis. (C) Genomic neighborhoods of HWE/HisKA2 kinase genes. We identified the genes adjacent to each HWE/HisKA2 kinase gene tallied in panels A and B. The number in parentheses is the number of kinase genes identified in each phylogenetic group. Numbers in the table are the percentage of sequences identified in each genomic context (e.g. 100% of 23 tallied Firmicute HWE/HisKA2 kinase genes are adjacent to an RR-ANTAR gene). RR-TX: traditional response regulator with receiver domain and a DNA-binding transcriptional regulatory ‘output’ domain; SDRR: single domain response regulator; TCS: two-component locus encoding at least one other histidine kinase; RR-ANTAR: response regulator fused to a RNA-binding antiterminator domain[55,56]; RR-other: receiver domain fused to other ‘output’ domains such as a diguanylate cyclase, phosphodiesterase, or uncharacterized domains; Orphan: no adjacent genes with two-component receiver or kinase domains; whiB: a subset of orphan kinases adjacent to whiB-family transcriptional regulators. In this analysis, hybrid kinases (with a C-terminal receiver domain) were counted with together with non-hybrid kinases. For comparison, we conducted the same analysis on 823 HisKA kinases. Of the classical non-hybrid HisKA kinase genes, 75% are adjacent to a RR-TX gene.
Phylogenetic distribution and function
Alphaproteobacteria
Classical HisKA-family sensor histidine kinases are broadly distributed among bacteria, eukarya, and archaea [1], while HWE/HisKA2-family kinases are confined to a subset of bacteria and archaea [1,5,6,34] (Figure 2A and B). The majority of HWE/HisKA2 kinases are found in Alphaproteobacteria, which typically encode at least one and often ten or more members of this superfamily [17,34,35]. Emerging studies of alphaproteobacterial cell physiology have defined a central role for HWE/HisKA2 kinases as environmental sensors in the general stress response (GSR) transcriptional regulatory pathway. GSR transcription is triggered by phosphorylation of an unusual response regulator known as PhyR, which in turn activates an EcfG-family alternative sigma factor (reviewed in [17,35]). Staron and Mascher [34] noted that genomes containing PhyR and EcfG also contain HWE/HisKA2 kinases, and suggested this family of kinases as candidate regulators of GSR. Indeed, all histidine kinases demonstrated to phosphorylate PhyR and/or regulate GSR transcription to date are HWE/HisKA2 kinases [7,9–11,36–40]. The only histidine kinase known to signal to the GSR pathway that is not HWE/HisKA2 does so indirectly via an HWE/HisKA2 kinase that directly phosphorylates PhyR [9,41].
Studies of GSR regulation in Alphaproteobacteria have also revealed an important role for SDRRs, which interact biochemically and/or genetically with HWE/HisKA2 kinases to modulate GSR transcription [7,9,36,41–43]. Though the mechanistic basis by which SDRRs execute this function remains undefined, they have been hypothesized to function as allosteric regulators of HWE/HisKA2, competitive kinase inhibitors, phospho-sinks, or as components of parallel regulatory pathways. We note that data focused on SDRRs in regulation of GSR transcription do not support linear phosphorelay models. Instead, the data are more consistent with branched signaling pathways whereby HWE/HisKA2 kinases phosphorylate multiple receiver domains to control PhyR activity and GSR transcription.
Firmicutes and Actinobacteria
In Gram-positive phyla, Firmicutes and Actinobacteria, many species encode a single HWE/HisKA2 kinase with a conserved domain structure. These proteins are comprised of cytoplasmic GAF-PAS sensory domains at the N-terminus and a C-terminal HWE/HisKA2 kinase domain [44]. The genomic context of these GAF-PAS-kinase genes in each phylum is highly conserved, implying a specialized function that has not expanded or diversified within either group.
Among Firmicutes that encode ethanolamine utilization (eut) genes, the eutW HWE/HisKA2 kinase is always encoded adjacent to eutV, a response regulator that contains an RNA-binding output domain (ANTAR) rather than a DNA-binding output domain (Figure 2C). EutW and EutV regulate expression of ethanolamine utilization genes via phosphorylation-dependent control of RNA binding and transcription termination ([8,45]; reviewed in [46]).
In both sporulating and non-sporulating Actinobacteria, a single HWE/HisKA2 kinase gene is very often (85%) encoded opposite a WhiB-like (Wbl) family transcriptional regulator (Figure 2C). Wbl-family transcription factors do not contain a receiver domain. These small (~100 aa) proteins are unique to this phylum [47], regulate transcription in response to redox changes via an iron-sulfur cluster, and control cellular development [48,49]. The Wbl family protein adjacent to HWE/HisKA2 kinase gene is called WblE, and is widely distributed throughout Actinobacteria [50]. The WblE homolog in Mycobacterium tuberculosis H37RV (Rv3219), also named WhiB1 or WhmE [50], is an essential, nitric oxide-responsive transcription factor [51,52]. To our knowledge, a functional relationship between the HWE/HisKA2 kinase and WblE has not been reported. Nonetheless, co-occurance and co-location of these genes across diverse Actinobacteria suggests a functional connection.
We note that PdtaS, the HWE/HisKA2 kinase encoded adjacently to wblE in M. tuberculosis, also phosphorylates an RR-ANTAR protein, PdtaR, though pdtaS and pdtaR are not adjacent on the chomosome [53]. The sensor domains of PdtaS are related to EutW, but there is no evidence that this system responds to ethanolamine [44]. The transcripts controlled by PdtaR are unknown. Nevertheless, the connection between HWE/HisKA2 and RR-ANTAR proteins in Firmicutes and Actinobacteria demonstrate a conserved mode of regulation in these phyla in which HWE/HisKA2 kinases exert control on gene expression via phosphorylation of RNA-binding anti-termination proteins.
Archaea
HWE/HisKA2 kinases are not common in the Archaea, though two classes of Euryarchaeota (Methanobacteria and Methanomicrobia) contain species with a large number of these kinases. Some species in these classes have no histidine kinases (e.g. Methanobrevibacter spp.), while others have dozens of histidine kinases, the majority of which are HWE/HisKA2 [5]. Enrichment of this superfamily among select Archaea in these classes may have occurred via lateral gene transfer followed by duplication. Indeed, many HWE/HisKA2 kinases in these species have similar domain structures and are located in tandem on the chromosome, which is consistent with recent duplication events. The vast majority of archaeal HWE/HisKA2 kinase genes are not located adjacent to other two-component signaling genes. This is reminiscent of the Alphaproteobacteria, in which multiple orphan kinases appear to converge on and control a single transcriptional regulatory pathway. It is tempting to speculate that HWE/HisKA2 kinases function in a similar manner in these select species of the Euryarchaeota. Such a model is consistent with the observation that kinases outnumber receiver proteins at least 2:1 in archaeal species encoding many HWE/HisKA2 kinases [5]. The vast majority of receiver domains in these species are either SDRR or are located on the same polypeptide as the kinase (i.e. hybrid HWE/HisKA2 kinases).
HWE/HisKA2 as protein photosensors
Like all histidine kinases, HWE/HisKA2 kinases are appended to a diverse array of N-terminal signal input domains, and can exist as either membrane bound sensors or as soluble cytoplasmic sensors. There is a clear connection between HWE/HisKA2 kinases and bacterial photoperception. Indeed, much of the research on this kinase superfamily has been driven by an interest in bacterial photobiology and protein photosensors. The HWE motif was first described in a subset of kinases with red light-sensing bacteriophytochrome domains [6,15]. Though the hexameric EsxG HWE/HisKA2 kinase does not itself sense photons, phosphorylation of its N-terminal receiver domain is controlled by a bacteriophytochrome [13]. Furthermore, interest in blue light-sensing LOV domains has fueled many biochemical and physiological studies on LOV HWE/HisKA2 kinases in Alphaproteobacteria [7,10–12,14,36]. Indeed, the only crystal structures of HWE/HisKA2 domains to date are from LOV kinases [18,19]. Emerging data provide evidence that HWE/HisKA2 kinases containing photosensory LOV domains serve to regulate transcription of the general stress response (GSR) regulon in Alphaproteobacteria [7,9–11,36]. The functional connection between LOV domains, blue light, and GSR transcription remains largely undefined, though data point to a model in which light serves as an environmental signal that modulates the alphaproteobacterial general stress response.
Conclusion
HWE histidine kinases were described as an atypical sensor histidine kinase family over a decade ago. Features of HWE primary structure are largely shared with the HisKA2 family. Together, HWE and HisKA2 are classified as a superfamily. Recent studies have revealed other unusual features of this understudied group of sensor kinases including: 1) HWE/HisKA2 kinases are not typically involved in signaling pathways with classical DNA-binding response regulators. Rather, these kinases are associated with response regulators that have unusual output domains such as anti-anti-σ domains or RNA-binding anti-termination (ANTAR) domains. 2) In the Alphaproteobacteria, multiple HWE/HisKA2 kinases can coordinately control phosphorylation of a single receiver domain to regulate transcription of a stress response regulon; single domain response regulators both activate and repress this response. 3) There is evidence that HWE/HisKA2 kinases can adopt unusual oligomeric states (e.g. monomer or hexamer). 4) Recent crystal structures of HWE/HisKA2 kinases suggest unique conserved residues in the H- and N- boxes are involved in catalysis. Specifically, these structures inform a regulatory model in which a conserved glutamate in the N-box stabilizes either active or inactive conformations via interactions with conserved arginines in the H-box and R-box of the DHp helices.
Supplementary Material
Highlights.
HWE and HisKA2 domain families comprise an unusual subset of sensor histidine kinases
HWE/HisKA2 possess unique sequence motifs and structural features
This kinase superfamily is involved in atypical two-component signaling processes
HWE/HisKA2 are found in Alphaproteobacteria, select Gram-positives and archaea
Acknowledgments
This work was supported by grants from the National Institutes of Health (1R01GM087353 and 1R01AI107159).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 3.Kim D, Forst S. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology. 2001;147:1197–1212. doi: 10.1099/00221287-147-5-1197. [DOI] [PubMed] [Google Scholar]
- 4.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38:D401–407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karniol B, Vierstra RD. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correa F, Ko WH, Ocasio V, Bogomolni RA, Gardner KH. Blue light regulated two-component systems: enzymatic and functional analyses of light-oxygen-voltage (LOV)-histidine kinases and downstream response regulators. Biochemistry. 2013;52:4656–4666. doi: 10.1021/bi400617y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Papa MF, Perego M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol. 2008;190:7147–7156. doi: 10.1128/JB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •9.Kaczmarczyk A, Hochstrasser R, Vorholt JA, Francez-Charlot A. Complex two-component signaling regulates the general stress response in Alphaproteobacteria. Proc Natl Acad Sci U S A. 2014;111:E5196–5204. doi: 10.1073/pnas.1410095111. These authors systematically demonstrate that the majority of HWE/HisKA2 kinases in Sphingomonas coordinately phosphorylate the general stress response regulator, PhyR. In addition, authors provide evidence that four SDRRs coordinately regulate signalling with HWE/HisKA2 kinases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HS, Willett JW, Jain-Gupta N, Fiebig A, Crosson S. The Brucella abortus virulence regulator, LovhK, is a sensor kinase in the general stress response signalling pathway. Mol Microbiol. 2014;94:913–925. doi: 10.1111/mmi.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sycz G, Carrica MC, Tseng TS, Bogomolni RA, Briggs WR, Goldbaum FA, Paris G. LOV Histidine Kinase Modulates the General Stress Response System and Affects the virB Operon Expression in Brucella abortus. PLoS One. 2015;10:e0124058. doi: 10.1371/journal.pone.0124058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wojnowska M, Yan J, Sivalingam GN, Cryar A, Gor J, Thalassinos K, Djordjevic S. Autophosphorylation activity of a soluble hexameric histidine kinase correlates with the shift in protein conformational equilibrium. Chem Biol. 2013;20:1411–1420. doi: 10.1016/j.chembiol.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, et al. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 15.Karniol B, Vierstra RD. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc Natl Acad Sci U S A. 2003;100:2807–2812. doi: 10.1073/pnas.0437914100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francez-Charlot A, Kaczmarczyk A, Fischer HM, Vorholt JA. The general stress response in Alphaproteobacteria. Trends Microbiol. 2015;23:164–171. doi: 10.1016/j.tim.2014.12.006. [DOI] [PubMed] [Google Scholar]
- ••18.Rinaldi J, Arrar M, Sycz G, Cerutti ML, Berguer PM, Paris G, Estrin DA, Marti MA, Klinke S, Goldbaum FA. Structural Insights into the HWE Histidine Kinase Family: The Brucella Blue Light-Activated Histidine Kinase Domain. J Mol Biol. 2016;428:1165–1179. doi: 10.1016/j.jmb.2016.01.026. The first crystal structure of an HWE kinase. Together with 19 provides understanding of how conserved residues mediate intradomain interactions. [DOI] [PubMed] [Google Scholar]
- ••19.Rivera-Cancel G, Ko WH, Tomchick DR, Correa F, Gardner KH. Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation. Proc Natl Acad Sci U S A. 2014;111:17839–17844. doi: 10.1073/pnas.1413983111. The first crystal structure of a HisKA2 kinase. Together with 18 provides understanding of how conserved residues mediate interdomain interactions. Authors provide evidence that this kinase is monomeric in solution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashenberg O, Keating AE, Laub MT. Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. J Mol Biol. 2013;425:1198–1209. doi: 10.1016/j.jmb.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casino P, Miguel-Romero L, Marina A. Visualizing autophosphorylation in histidine kinases. Nat Commun. 2014;5:3258. doi: 10.1038/ncomms4258. [DOI] [PubMed] [Google Scholar]
- 22.Ashenberg O, Rozen-Gagnon K, Laub MT, Keating AE. Determinants of homodimerization specificity in histidine kinases. J Mol Biol. 2011;413:222–235. doi: 10.1016/j.jmb.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mechaly AE, Sassoon N, Betton JM, Alzari PM. Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol. 2014;12:e1001776. doi: 10.1371/journal.pbio.1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marina A, Mott C, Auyzenberg A, Hendrickson WA, Waldburger CD. Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism. J Biol Chem. 2001;276:41182–41190. doi: 10.1074/jbc.M106080200. [DOI] [PubMed] [Google Scholar]
- 25.Dago AE, Schug A, Procaccini A, Hoch JA, Weigt M, Szurmant H. Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proc Natl Acad Sci U S A. 2012;109:E1733–1742. doi: 10.1073/pnas.1201301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guillen Schlippe YV, Hedstrom L. Is Arg418 the catalytic base required for the hydrolysis step of the IMP dehydrogenase reaction? Biochemistry. 2005;44:11700–11707. doi: 10.1021/bi048342v. [DOI] [PubMed] [Google Scholar]
- 27.Keenholtz RA, Mouw KW, Boocock MR, Li NS, Piccirilli JA, Rice PA. Arginine as a general acid catalyst in serine recombinase-mediated DNA cleavage. J Biol Chem. 2013;288:29206–29214. doi: 10.1074/jbc.M113.508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campagne S, Dintner S, Gottschlich L, Thibault M, Bortfeld-Miller M, Kaczmarczyk A, Francez-Charlot A, Allain FH, Vorholt JA. Role of the PFXFATG[G/Y] Motif in the Activation of SdrG, a Response Regulator Involved in the Alphaproteobacterial General Stress Response. Structure. 2016;24:1237–1247. doi: 10.1016/j.str.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Sheftic SR, Garcia PP, White E, Robinson VL, Gage DJ, Alexandrescu AT. Nuclear magnetic resonance structure and dynamics of the response regulator Sma0114 from Sinorhizobium meliloti. Biochemistry. 2012;51:6932–6941. doi: 10.1021/bi300922z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••30.Bhate MP, Molnar KS, Goulian M, DeGrado WF. Signal transduction in histidine kinases: insights from new structures. Structure. 2015;23:981–994. doi: 10.1016/j.str.2015.04.002. A thorough review of histidine kinase conformational dynamics and mechanisms of intramolecular regulation of kinase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 32.Wang C, Sang J, Wang J, Su M, Downey JS, Wu Q, Wang S, Cai Y, Xu X, Wu J, et al. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 2013;11:e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albanesi D, Martin M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, Buschiazzo A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci U S A. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staron A, Mascher T. General stress response in alpha-proteobacteria: PhyR and beyond. Mol Microbiol. 2010;78:271–277. doi: 10.1111/j.1365-2958.2010.07336.x. [DOI] [PubMed] [Google Scholar]
- 35.Fiebig A, Herrou J, Willett J, Crosson S. General Stress Signaling in the Alphaproteobacteria. Annu Rev Genet. 2015;49:603–625. doi: 10.1146/annurev-genet-112414-054813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foreman R, Fiebig A, Crosson S. The LovK-LovR two-component system is a regulator of the general stress pathway in Caulobacter crescentus. J Bacteriol. 2012;194:3038–3049. doi: 10.1128/JB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaczmarczyk A, Campagne S, Danza F, Metzger LC, Vorholt JA, Francez-Charlot A. Role of Sphingomonas sp. strain Fr1 PhyR-NepR-sigmaEcfG cascade in general stress response and identification of a negative regulator of PhyR. J Bacteriol. 2011;193:6629–6638. doi: 10.1128/JB.06006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lourenco RF, Kohler C, Gomes SL. A two-component system, an anti-sigma factor and two paralogous ECF sigma factors are involved in the control of general stress response in Caulobacter crescentus. Mol Microbiol. 2011;80:1598–1612. doi: 10.1111/j.1365-2958.2011.07668.x. [DOI] [PubMed] [Google Scholar]
- 39.Sauviac L, Bruand C. A putative bifunctional histidine kinase/phosphatase of the HWE family exerts positive and negative control on the Sinorhizobium meliloti general stress response. J Bacteriol. 2014;196:2526–2535. doi: 10.1128/JB.01623-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu N, Lima A, Bandeali Z, Anderson B. Characterization of the general stress response in Bartonella henselae. Microb Pathog. 2016;92:1–10. doi: 10.1016/j.micpath.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaczmarczyk A, Hochstrasser R, Vorholt JA, Francez-Charlot A. Two-tiered histidine kinase pathway involved in heat shock and salt sensing in the general stress response of Sphingomonas melonis Fr1. J Bacteriol. 2015;197:1466–1477. doi: 10.1128/JB.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzger LC, Francez-Charlot A, Vorholt JA. Single-domain response regulator involved in the general stress response of Methylobacterium extorquens. Microbiology. 2013;159:1067–1076. doi: 10.1099/mic.0.066068-0. [DOI] [PubMed] [Google Scholar]
- 43.Ocasio VJ, Correa F, Gardner KH. Ligand-induced folding of a two-component signaling receiver domain. Biochemistry. 2015;54:1353–1363. doi: 10.1021/bi501143b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preu J, Panjikar S, Morth P, Jaiswal R, Karunakar P, Tucker PA. The sensor region of the ubiquitous cytosolic sensor kinase, PdtaS, contains PAS and GAF domain sensing modules. J Struct Biol. 2012;177:498–505. doi: 10.1016/j.jsb.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao B, Paramanathan R, Gupta RS. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Van Leeuwenhoek. 2006;90:69–91. doi: 10.1007/s10482-006-9061-2. [DOI] [PubMed] [Google Scholar]
- 48.Chim N, Johnson PM, Goulding CW. Insights into redox sensing metalloproteins in Mycobacterium tuberculosis. J Inorg Biochem. 2014;133:118–126. doi: 10.1016/j.jinorgbio.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.den Hengst CD, Buttner MJ. Redox control in actinobacteria. Biochim Biophys Acta. 2008;1780:1201–1216. doi: 10.1016/j.bbagen.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Soliveri JA, Gomez J, Bishai WR, Chater KF. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000;146( Pt 2):333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 51.Smith LJ, Stapleton MR, Buxton RS, Green J. Structure-function relationships of the Mycobacterium tuberculosis transcription factor WhiB1. PLoS One. 2012;7:e40407. doi: 10.1371/journal.pone.0040407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith LJ, Stapleton MR, Fullstone GJ, Crack JC, Thomson AJ, Le Brun NE, Hunt DM, Harvey E, Adinolfi S, Buxton RS, et al. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem J. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morth JP, Gosmann S, Nowak E, Tucker PA. A novel two-component system found in Mycobacterium tuberculosis. FEBS Lett. 2005;579:4145–4148. doi: 10.1016/j.febslet.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 54.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 55.Shu CJ, Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem Sci. 2002;27:3–5. doi: 10.1016/s0968-0004(01)02036-9. [DOI] [PubMed] [Google Scholar]
- 56.Ramesh A, DebRoy S, Goodson JR, Fox KA, Faz H, Garsin DA, Winkler WC. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet. 2012;8:e1002666. doi: 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.