Abstract
The bacterial chromosome is compacted in a manner optimal for DNA transactions to occur. The degree of compaction results from the level of DNA-supercoiling and the presence of nucleoid-binding proteins. DNA-supercoiling is homeostatically maintained by the opposing activities of relaxing DNA topoisomerases and negative supercoil-inducing DNA gyrase. DNA-supercoiling acts as a general cis regulator of transcription, which can be superimposed upon other types of more specific trans regulatory mechanism. Transcriptomic studies on the human pathogen Streptococcus pneumoniae, which has a relatively small genome (∼2 Mb) and few nucleoid-binding proteins, have been performed under conditions of local and global changes in supercoiling. The response to local changes induced by fluoroquinolone antibiotics, which target DNA gyrase subunit A and/or topoisomerase IV, involves an increase in oxygen radicals which reduces cell viability, while the induction of global supercoiling changes by novobiocin (a DNA gyrase subunit B inhibitor), or by seconeolitsine (a topoisomerase I inhibitor), has revealed the existence of topological domains that specifically respond to such changes. The control of DNA-supercoiling in S. pneumoniae occurs mainly via the regulation of topoisomerase gene transcription: relaxation triggers the up-regulation of gyrase and the down-regulation of topoisomerases I and IV, while hypernegative supercoiling down-regulates the expression of topoisomerase I. Relaxation affects 13% of the genome, with the majority of the genes affected located in 15 domains. Hypernegative supercoiling affects 10% of the genome, with one quarter of the genes affected located in 12 domains. However, all the above domains overlap, suggesting that the chromosome is organized into topological domains with fixed locations. Based on its response to relaxation, the pneumococcal chromosome can be said to be organized into five types of domain: up-regulated, down-regulated, position-conserved non-regulated, position-variable non-regulated, and AT-rich. The AT content is higher in the up-regulated than in the down-regulated domains. Genes within the different domains share structural and functional characteristics. It would seem that a topology-driven selection pressure has defined the chromosomal location of the metabolism, virulence and competence genes, which suggests the existence of topological rules that aim to improve bacterial fitness.
Keywords: DNA supercoiling, DNA topoisomerases, fluoroquinolones, global transcription, interactome, novobiocin, seconeolitsine, topological domains
Introduction
The compaction of DNA by up to 1000-fold (Holmes and Cozzarelli, 2000) in the bacterial chromosome, or nucleoid, achieves the optimal condition under which its essential functions – replication, segregation and gene expression (reviewed by Dorman, 2013) – can be reconciled. This compaction is mediated by both the natural supercoiling of the DNA, and by the binding of nucleoid-associated proteins (NAPs) (Wang et al., 2013). NAPs form a functional network that maintains DNA topology by bending, wrapping, bridging and constraining supercoils. Although several NAPs have been characterized in the Gram-negative bacterium Escherichia coli, very few have been detected in Gram-positive bacteria, including the human pathogen Streptococcus pneumoniae (Dillon and Dorman, 2010). In bacteria, gene transcription is regulated by DNA-supercoiling. This functions as a general cis regulator of transcription, and can be superimposed upon other types of more specific trans regulatory mechanisms. cis regulation can also occur via promoter DNA sequences. Factors acting in trans include structural and regulatory proteins. NAPs (structural proteins) target a number of genes (Dillon and Dorman, 2010), while specific regulatory proteins facilitate or inhibit the interaction of RNA polymerase with specific promoter regions (Browning and Busby, 2004). The precision balance of DNA supercoiling is thus modulated by a network of self-regulating factors.
DNA topoisomerases, which are present in all bacteria, are responsible for the maintenance of DNA-supercoiling. These enzymes are classified into two types based on their DNA cleavage pattern: type I, which cleaves only one DNA strand, and type II, which cleaves both. The type II topoisomerases, gyrase and topoisomerase IV (Topo IV), are tetrameric proteins with two subunits: GyrA2GyrB2 in gyrase, and ParC2ParE2 in Topo IV. Supercoiling homeostasis is achieved by the competing activities of gyrase and topoisomerase I (Topo I, a type I isomerase) plus IV (Champoux, 2001); gyrase introduces negative supercoils into DNA (Gellert et al., 1976), Topo I relaxes DNA, and Topo IV both relaxes DNA and participates in chromosome partitioning (Kato et al., 1990). S. pneumoniae (the pneumococcus) has a relatively small genome (∼2 Mb compared to ∼4.6 Mb for E. coli) rich in AT (60%), that carries genes for all three of the above enzymes. These characteristics are shared by other pathogens of the genus Streptococcus, including S. pyogenes and S. suis.
Streptococcus pneumoniae is the primary cause of community-acquired pneumonia, meningitis, bacteremia, and otitis media in children. Worldwide, 1 million children under 5 years of age die every year of pneumococcal infections (World Health Organization, 2007). The use of the pneumococcal 7-valent conjugate vaccine, which covers the serotypes most often associated with resistance to antibiotics, has achieved a decline in the incidence of invasive pneumococcal disease (Whitney et al., 2003; Kyaw et al., 2006) and a reduction in penicillin resistance rates (Kyaw et al., 2006; Pilishvili et al., 2010). However, serotypes not included in the vaccine soon emerged, highlighting the limitations of anti-pneumococcal prophylaxis (Moore et al., 2008; Fenoll et al., 2009).
The post-genomic age is beginning to provide answers to questions regarding how chromosomes are topologically organized, and how this organization influences bacterial evolution. Several degrees of organization in bacterial chromosomes have been observed, based on size (for a recent review see Badrinarayanan et al., 2015). Macrodomains are found at the megabase-size range. E. coli, for example, has four macrodomains: Ori (origin of replication), Ter (terminus of replication), Left, and Right, plus two less-structured regions flanking the Ori macrodomain (Espeli et al., 2008). Macrodomains may be maintained by specific proteins, such as the macrodomain Ter proteins (MatPs) that bind, as the name suggests, to specific sites in the Ter macrodomain (Dupaigne et al., 2012). However, no such proteins stabilizing the other macrodomains have been identified, and MatP proteins are found only in enteric bacteria. Non-homologous proteins may therefore take on similar roles in other bacteria. Supercoiling domains are found at the kilobase range. These are isolated loops that coil up around themselves; proteins at their bases help to topologically isolate the looped DNA. These loops were initially detected in electron micrographs of lysed E. coli cells (Kavenoff and Bowen, 1976). Later studies estimated the number of supercoil domains by assessing the numbers of nicks required to fully relax the chromosome. From these experiments it was estimated that the E. coli chromosome contains about 40 domains of around 100 kb (Worcel and Burgi, 1972; Sinden and Pettijohn, 1981). Studies in Caulobacter crescentus suggested domains ranging in length from 30 to 420 kb (Le et al., 2013). In Salmonella enterica, these domains were estimated to be 20 kb long by taking into account the site-specific recombination events that occurred between chromosomal sites distant from one another (Higgins et al., 1996). Later, transcriptional data predicted sizes of ∼10 kb for E. coli (Postow et al., 2004). Controversy regarding the size and definition of domains remains, perhaps as a consequence of the different methods being used in their calculation.
The availability of drugs against all the topoisomerases of S. pneumoniae (Figure 1) has helped in determining the existence of chromosomal domains. This review summarizes the transcriptomic alterations induced by these agents, and how these changes can be interpreted to provide definitions of the chromosome domains in this bacterium. Changes induced by the clinically used fluoroquinolones (FQs) levofloxacin (LVX), and moxifloxacin (MOX) are first considered, followed by those that occur concomitantly with a global change in supercoiling, as induced by novobiocin (NOV, an inhibitor of the gyrase B subunit) and seconeolitsine (SCN, an inhibitor of Topo I). Overall, these studies reveal the S. pneumoniae genome to be organized into topology-reacting gene clusters, or supercoiling domains. The conservation of the location of these domains in the Streptococcus genus, and their enrichment for specific functions, suggests the existence of topological rules that aim to improve fitness via tight physiological feedback.
FIGURE 1.
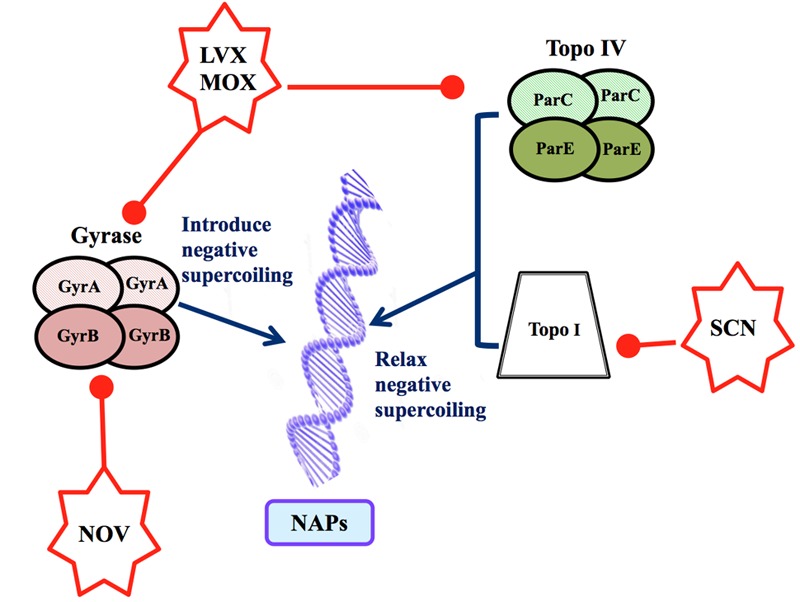
Factors determining the topology of the Streptococcus pneumoniae chromosome. The level of supercoiling is controlled by three DNA topoisomerases. Gyrase (GyrA2GyrB2) is inhibited by LVX, MOX (which inhibit GyrA) and NOV (which inhibits GyrB). Topo IV (ParC2ParE2) is inhibited by LVX and MOX (which inhibit ParC). Topo I, a monomer, is inhibited by SCN. The topological organization of the chromosome depends on the level of DNA supercoiling and on the presence of NAPs.
Control of Transcription by Local Changes in Supercoiling
Strains of S. pneumoniae resistant to antibiotics that act on the cell wall (beta-lactams) and on protein synthesis (macrolides) have proliferated in the last 30 years (Jacobs et al., 2003; Liñares et al., 2010). Consequently, pneumococcal infections are nowadays fought with LVX and MOX, which inhibit DNA topoisomerases. FQs target the type II DNA topoisomerases gyrase and Topo IV. Their mechanism of action involves the formation of DNA-FQ-topoisomerase complexes, which sterically inhibit replication and transcription and the subsequent generation of detrimental double-stranded DNA breaks (Drlica et al., 2008). Bacterial survival depends on the resolution of these breaks. Reactive oxygen species (ROS), such as superoxide anions, hydrogen peroxide and hydroxyl radicals contribute to FQ-mediated cell death via a protein synthesis-dependent pathway (Wang et al., 2010). This observation is consistent with the general model explaining the lethality of bactericidal antibiotics, which attributes a role to ROS generated via the Fenton reaction. The original reports supporting this model based their conclusions on the use of microarrays to study the transcriptional response to the inhibition of E. coli GyrA by an FQ or the peptide toxin CcdB. Under these conditions, global transcription was altered. In addition to the up-regulation of SOS damage response genes, genes related to superoxide stress, iron-sulfur cluster synthesis and iron uptake were up-regulated too (Dwyer et al., 2007). ROS production was also observed with a variety of bactericidal antibiotic families, in addition to FQs, each with a different intracellular target (reviewed by Dwyer et al., 2015). However, the intervening pathways lying between the initial antibiotic-target interaction and ROS formation have yet to be fully characterized.
The treatment of S. pneumoniae with FQs involves causing double-stranded breaks in the bacterial chromosome (Ferrándiz et al., 2016b), and as in other bacteria this requires active protein synthesis (Brito et al., 2017). Treatment with LVX or MOX (Ferrándiz and de la Campa, 2014; Ferrándiz et al., 2016b) is reported not to alter the level of global supercoiling. Nor are changes in supercoiling observed in E. coli exposed to oxolinic acid (Snyder and Drlica, 1979), although changes have been observed in the latter after treatment with the FQ norfloxacin (Peter et al., 2004). These differences might be attributable to species-dependent affinities of each drug for Topo IV or gyrase. For instance, Topo IV is the primary target of most FQs in Gram-positive bacteria, including S. pneumoniae, with gyrase a secondary target (Janoir et al., 1996; Muñoz and de la Campa, 1996; Tankovic et al., 1996; Fernández-Moreira et al., 2000). In contrast, in Gram-negative bacteria, including E. coli, gyrase is the primary target. At the LVX concentrations used in S. pneumoniae experiments, only Topo IV would have been inhibited, and no global change in supercoiling would be expected. However, at the MOX concentrations used, both gyrase and Topo IV would have been inhibited, suggesting that the inhibition of their opposing activities preserved the net level of supercoiling. Nevertheless, local topological changes are predictable in both cases and these would produce alterations in the transcriptome. Indeed, FQs induce a transcriptional response in S. pneumoniae, in which the differentially expressed genes (DEGs) account for 5.2 and 6.5% of the genome for LVX and MOX, respectively. In this bacterium, which lacks a proper SOS-like system, activation of the competence regulon has been reported with both FQs (Ferrándiz and de la Campa, 2014; Ferrándiz et al., 2016b), supporting the idea that competence is a general stress response in S. pneumoniae (Prudhomme et al., 2006). In addition, both LVX and MOX induce transcriptional alterations, which, although different, ultimately stimulate the Fenton reaction, increasing ROS accumulation and contributing to cell death (Ferrándiz and de la Campa, 2014; Ferrándiz et al., 2016b). Although S. pneumoniae is a facultative anaerobe, the increased lethality of FQs mediated by an increase in ROS fits with the antibiotic lethality model proposed for aerobic bacteria (Dwyer et al., 2007, 2014; Kohanski et al., 2007; Wang and Zhao, 2009). Via local supercoiling changes, the response to LVX specifically triggers the up-regulation of the fatDCEB operon. This causes an increase in intracellular iron, and in turn, a shift in the Fenton reaction toward the production of hydroxyl radicals. With MOX, the response leads to the up-regulation of the glycolytic pathway, with a noticeable increase in pyruvate and a subsequent increase in hydrogen peroxide (Figure 2). The different alterations in the patterns of gene expression induced by LVX and MOX are due to local changes in supercoiling, which are dependent on whether Topo IV (LVX) or both Topo IV and gyrase (MOX) are inhibited.
FIGURE 2.
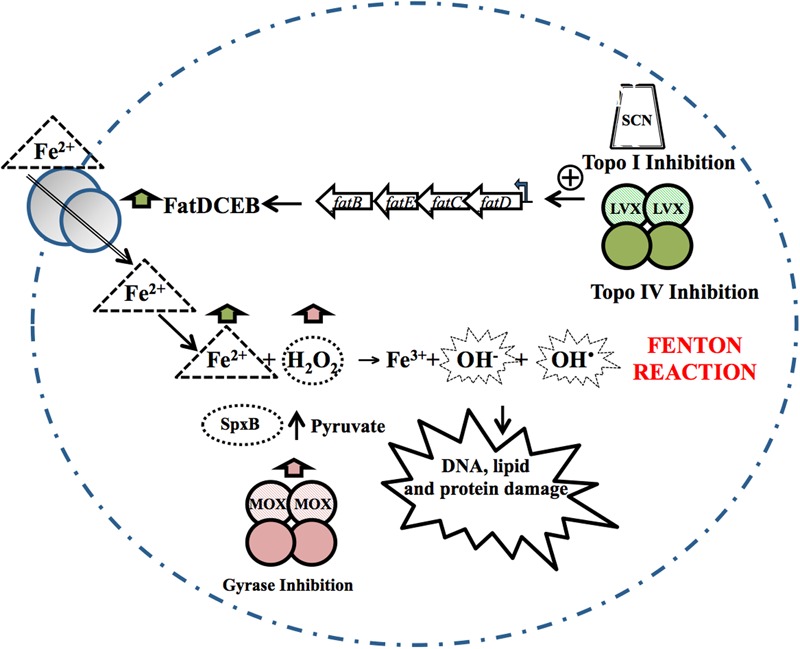
Oxidative damage cell death pathway. The inhibition of Topo IV by LVX, or of Topo I by SCN, causes a local increase in supercoiling, resulting in the up-regulation of the fatDCEB operon. The consequent increase in the iron transporter it encodes causes an increase in intracellular Fe2+. MOX alters the transcriptome, up-regulating genes from several metabolic pathways involved in the production of pyruvate. Pyruvate can then be converted by pyruvate oxidase (SpxB) into hydrogen peroxide (H2O2). Fe2+ and H2O2 are the substrates of the Fenton reaction. The Fenton reaction renders hydroxyl radicals, which oxidatively damage DNA, proteins and lipids. Taken from Ferrándiz and de la Campa (2014), with modifications.
Since both Topo IV and gyrase produce double-stranded breaks in the DNA when bound to FQs, the differential transcriptional alterations caused by these drugs might also be related to subtle, yet important, differences in sequence recognition (Leo et al., 2005), which are themselves affected by DNA supercoiling and bending (Arnoldi et al., 2013). Sequence recognition mediated by local supercoiling levels might explain the unique distribution of genes affected by LVX or MOX. In addition, the location of FQ-topoisomerase complexes relative to the replication forks, which is different for gyrase and Topo IV (Postow et al., 2001), may be involved in their different transcriptional outcomes.
Control of Transcription by Global Changes in Supercoiling
Response to Relaxation Caused by the Inhibition of Gyrase
The homeostatic control of supercoiling was first described in E. coli. In this bacterium, the transcription of topA (which codes for Topo I) was found to decrease under DNA relaxation (Tse-Dinh, 1985), while that of gyrA, and gyrB (which code for the two gyrase subunits) were found to increase (Menzel and Gellert, 1983, 1987a,b). An increase in gyrase expression in response to relaxation has also been observed in Streptomyces and Mycobacterium (Thiara and Cundliffe, 1989; Unniraman et al., 2002). However, in Staphylococcus aureus, treatment with NOV affects the transcription of the gyrase genes but not of topA (Schroder et al., 2014). In S. pneumoniae, treatment with NOV was also found to increase the transcription of gyrase genes, and diminish the expression of Topo I and Topo IV. In addition, global relaxation followed by a recovery of the native level of supercoiling was observed at low drug concentrations (Ferrándiz et al., 2010). The distribution of topoisomers in plasmid pLS1 (Stassi et al., 1981) was used to estimate the chromosomal superhelical density (σ), and returned a mean value of about -0.06 (Figure 3), which is within the range reported for the E. coli chromosome (Deng et al., 2005). At subinhibitory NOV concentrations (0.5× MIC), a transcriptomic response allowed the restoration of the native level of supercoiling after an initial relaxation causing a σ variation of 23%. A similar effect was observed at 1× MIC. However, higher concentrations of NOV increased the degree of relaxation with no further restoration of supercoiling, compatible with the saturation of the homeostatic capacity that results in the inhibition of cell division. The range of σ variation permitting homeostatic recovery of the supercoiling observed in S. pneumoniae is in agreement with the estimated ±20% variation compatible with normal cell growth in E. coli (Drlica, 1992). Supercoiling recovery in the pneumococcus occurred after the up-regulation of the gyrase genes gyrA and gyrB and the down-regulation of the Topo I (topA) and Topo IV (parEC) genes (Ferrándiz et al., 2010). In E. coli, the expression of the gyrase and Topo I genes is also mediated by NAPs, which affect DNA supercoiling (Travers and Muskhelishvili, 2005; Vora et al., 2009). However, these regulatory mechanisms may not function in S. pneumoniae for which NAP scarcity is predicted, and which certainly lacks most of the NAPs found in E. coli. Thus, supercoiling maintenance in S. pneumoniae appears to depend mainly on the regulation of topoisomerase transcription.
FIGURE 3.
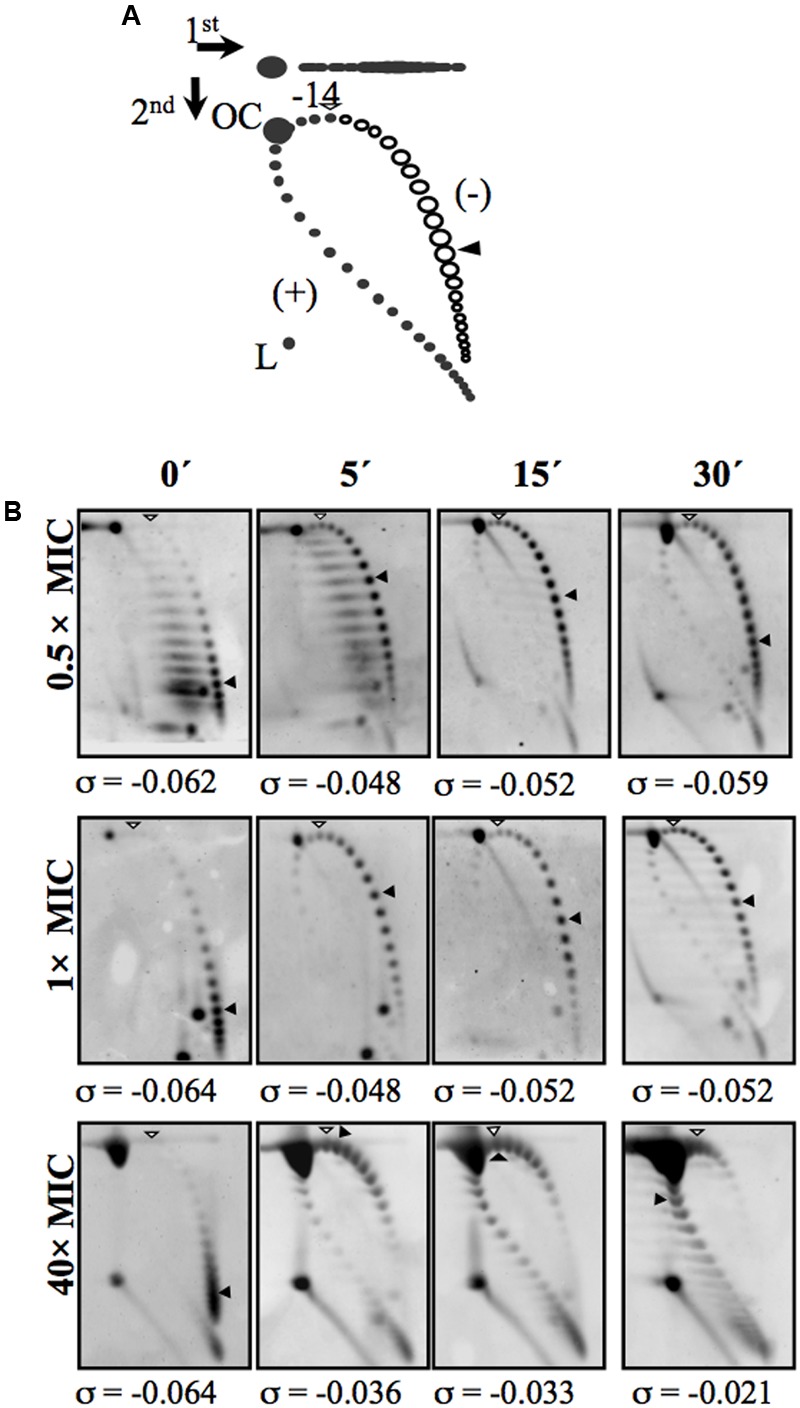
Treatment with the GyrB inhibitor NOV causes relaxation and subsequent recovery of supercoiling levels. (A) Diagram showing plasmid pLS1 topoisomer distribution after two-dimensional electrophoresis in agarose gels run in the presence of 1 and 2 μg/ml chloroquine in the first and second dimensions, respectively. Arrows at the top left corner indicate the running direction of the first and second dimensions, respectively. OC, open circle; L, linear form. Negative supercoiled topoisomers are in white and positive supercoiled topoisomers in black. 2 μg/ml chloroquine introduces 14 positive supercoils. A white arrowhead indicates the topoisomer that migrated with ΔLk of 0 in the second dimension; it migrated with a ΔWr of –14 in the first dimension. A black arrowhead indicates the most abundant topoisomer. (B) pLS1 topoisomer distribution after different NOV treatments. Samples were taken before the addition of the drug (time 0 min) and at the times indicated. The corresponding supercoiling density (σ) value is indicated below each autoradiogram. Taken from Ferrándiz et al. (2010), with modifications.
The Transcriptional Response to DNA Relaxation Involves Topology-Reactive Gene Clusters
The modulation of the expression of topoisomerase genes in S. pneumoniae is part of a global genome response (Ferrándiz et al., 2010). At subinhibitory concentrations, i.e., under physiological conditions, and short treatment times (5 and 15 min), DEGs were found to account for about 13% of the genome. An attenuation in the response at 30 min was observed, the number of DEGs being reduced to account for just 5.7% of the genome (Figure 4A), reflecting the recovery of supercoiling (Figure 3). Some 13% of the pneumococcal genome was therefore involved in the cellular response to moderate relaxation, allowing the recovery of the initial level of supercoiling. At fully inhibitory concentrations, the proportion of the genome covered by DEGs increased with time, from 14.4% at 5 min to 24% over longer periods (Figure 4). This agrees with the inhibition of cell division and with the continuous relaxation of the DNA (Figure 3). This proportion of the genome covered by DEGs upon relaxation is larger than in other bacteria. In Gram-negative bacteria, DEGs were found to account for 7% of the genome in E. coli [as determined using both gyrase inhibitors and gyrase thermosensitive mutants (Peter et al., 2004)], and for 8% in Haemophilus influenzae [as determined using NOV (Gmüender et al., 2001)]. In Gram-positive Staphylococcus aureus, treatment with NOV affected the transcription of 11% of the genome (Schroder et al., 2014).
FIGURE 4.
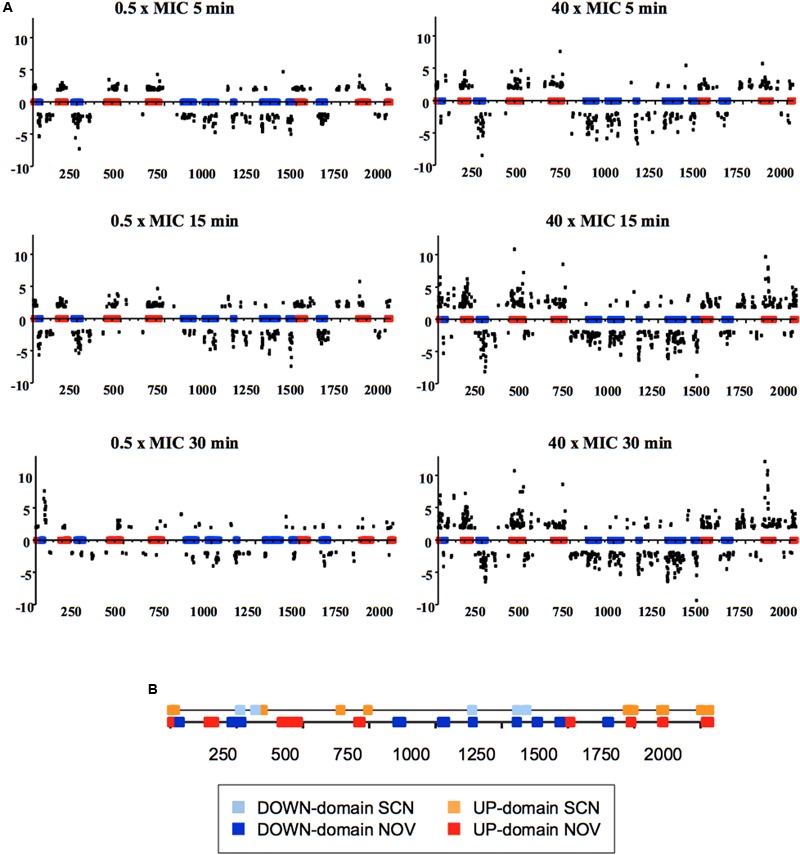
Global transcriptomic responses of S. pneumoniae to relaxation with NOV. (A) The relative fold variation of each gene is represented against the 3′ location of each open reading frame in the S. pneumoniae R6 chromosome (bases 1 to 2,038,615). Boxes indicate the transcriptional clusters: up-regulated in red, down-regulated in blue. (B) Localization of topological clusters detected as a result of treatment with either SCN or NOV. Taken from Ferrándiz et al. (2010, 2016a), with modifications.
It should be noted that the transcriptomic response to relaxation in S. pneumoniae involves topology-reactive gene clusters, or domains, that show coordinated up- or down-regulation. A total of 15 clusters have been detected, corresponding to 37% of the genome (Figure 4) (Ferrándiz et al., 2010). The sizes of these clusters varies from 14.6 to 85.6 kb (mean ± SD: 51.8 ± 21.8) and they contain 15–43 responsive genes (mean ± SD: 28 ± 9). They also include more than 68% of the DEGs. This has allowed topological clusters to be identified in which gene co-regulation is clearly more complex than would be expected simply from the number of genes in operons. In addition, the direction of transcription of the DEGs showed no preference for leading or lagging strands, providing additional evidence that topological control is structurally dependent.
The AT content over the genome correlates with domain location, and is higher in up-regulated (UP) than in down-regulated (DOWN) domains. These results suggest that the relaxation of DNA in AT-rich (ATr) regions favors the access of RNA polymerase to their promoters. On the contrary, a low AT content in DOWN clusters obstructs the access of RNA polymerase. Enrichment in the AT content of the region from positions -800 to +200 of genes up-regulated under relaxation has been reported in E. coli (Peter et al., 2004).
The organization of the S. pneumoniae chromosome into domains was further confirmed by the introduction of a cat heterologous gene cassette into the different types of domain (Figure 5A) (Ferrándiz et al., 2014). In response to relaxation with NOV, the transcription of cat was dependent on its chromosomal location, being up-regulated when located in UP domains, down-regulated when located in DOWN domains, and showing almost no changes when located in the non-regulated (NR) domains (Figure 5B). This all supports the idea that the chromosome is organized into topological domains that are reactive to interference in the supercoiling status. These results contrast, however, with those obtained in E. coli, in which the 306 DEGs were not only functionally diverse but widely dispersed throughout the chromosome (Peter et al., 2004), and with results obtained for Staphylococcus aureus, in which NOV-responsive genes were randomly distributed throughout the chromosome (Schroder et al., 2014).
FIGURE 5.
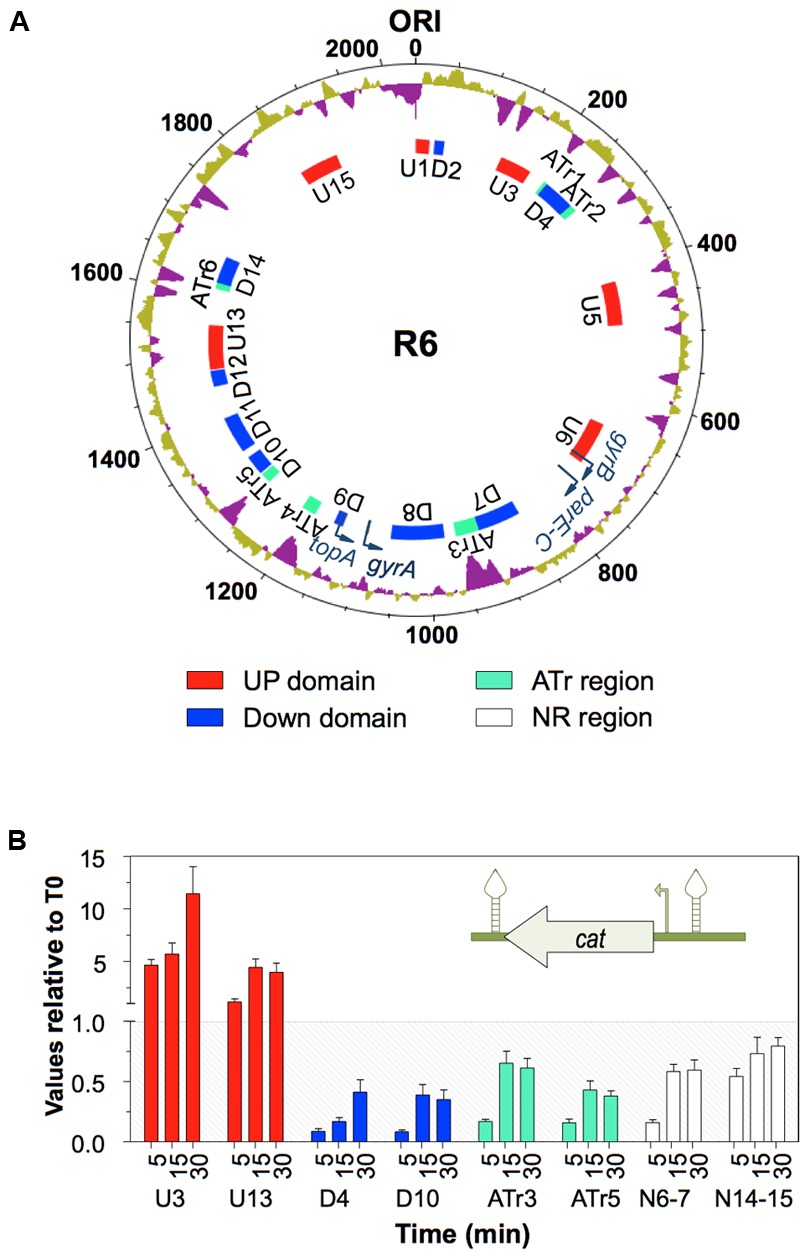
The topology-dependent transcription of Pccat is dependent on its chromosomal location. (A) Organization of the S. pneumoniae R6 chromosome in topological domains. Circles, from outside to inside, represent: % GC (values above the average in purple); DNA topoisomerase genes (dark blue curved arrows); topology-responsive domains. The chromosome is organized into domains up-regulated (U, red boxes) or down-regulated (D, blue boxes) in response to DNA relaxation, and ATr domains (green boxes). (B) Transcriptional response to DNA relaxation by NOV measured by qRT–PCR. A Ptccat cassette, coding for chloramphenicol-acetyl-transferase, which carries its own promoter (curved arrow) and is flanked by two transcriptional terminators (stem and loop structures), was inserted into different supercoiling domains. Cultures of the R6-CAT strains were treated with NOV and the transcription of cat analyzed by qRT–PCR. Taken from Ferrándiz et al. (2014), with modifications.
Response to Hypernegative Supercoiling Caused by the Inhibition of Topo I
The negative supercoiled state is the natural state of DNA homeostatic equilibrium in many bacteria. However, hypernegative supercoiling has been reported in E. coli topA mutants. With the exception of the topA10 mutant, all have acquired compensatory mutations in the gyrase genes (DiNardo et al., 1982). The topA10 mutant shows a notable 22% increase in negative supercoiling (Pruss et al., 1982), which probably represents the limit viable cells can afford in the long term. The inhibition of Topo I would produce greater hyper-supercoiling. Topo I plays an essential role in transcription, given its physical interaction with RNA polymerase (Cheng et al., 2003). During transcription, hypernegative supercoiling occurs behind the RNA polymerase, leading to RNA-DNA hybrid (R-loop) stabilization (Drolet, 2006). Topo I relaxes this supercoiling and prevents R-loop formation (Drolet et al., 1994; Phoenix et al., 1997; Masse and Drolet, 1999), allowing transcription to continue. Thus, the effects of hypernegative supercoiling in transcription depend directly on the activity of Topo I.
However, Topo I-targeting compounds are extremely scarce. Cheng et al. (2007) identified an alkaloid, which, although it inhibits the activity of E. coli Topo I, did not inhibit cell growth significantly. Our group discovered a new inhibitor of S. pneumoniae Topo I, SCN, which inhibits its relaxation activity at concentrations equivalent to those that inhibit cell growth. The modeling of pneumococcal Topo I, based on the crystal structure of the E. coli enzyme (Figure 6), and docking to SCN, revealed strong interactions between the drug and the DNA-binding site of Topo I to correlate with the inhibitory effect observed (García et al., 2011).
FIGURE 6.
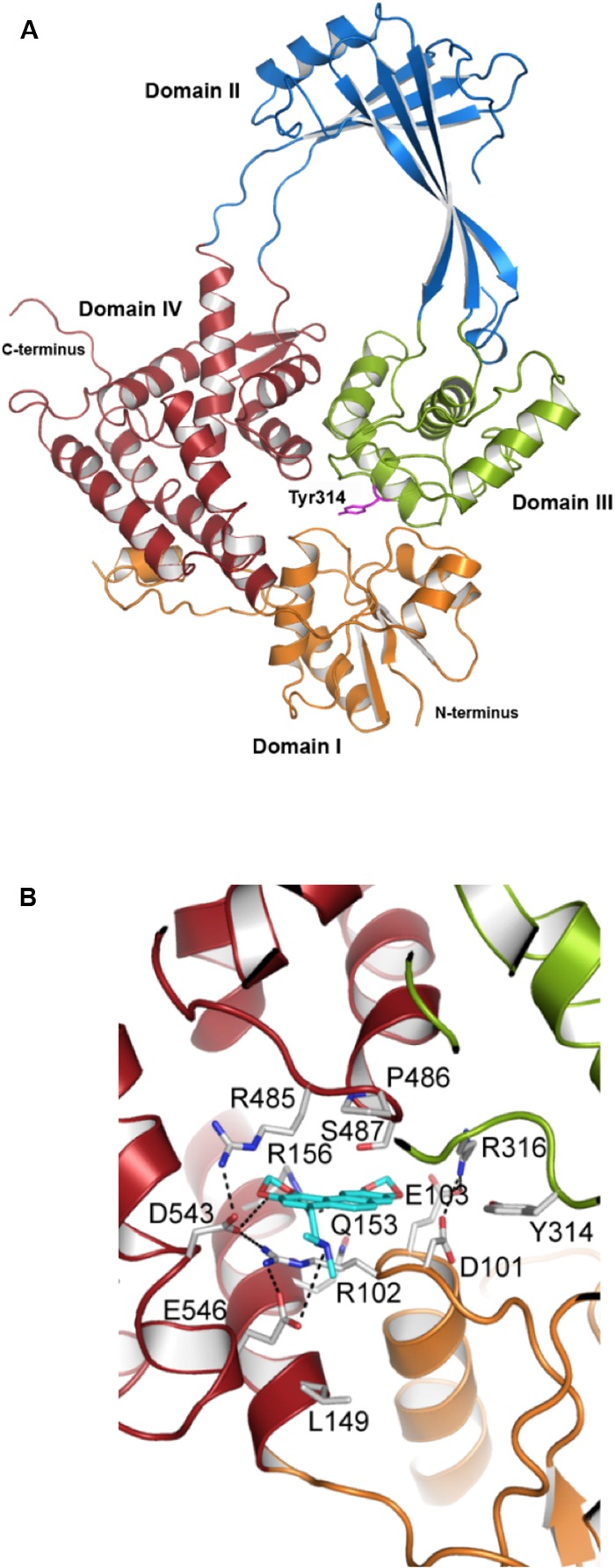
Structural modeling of the interaction of N-methyl SCN with S. pneumoniae topoisomerase I. (A) Modeling of the 67 kDa fragment of Topo I, showing domains I–IV and the catalytic Tyr314. (B) N-methyl-SCN (in blue) bound to the nucleotide-binding site of Topo I. Hydrogen bonds and salt-bridge interactions are indicated by dashed lines. Taken from García et al. (2011), with modifications.
Our group was the first to use SCN in studies of the transcriptomic response to hypernegative supercoiling in bacteria (Ferrándiz et al., 2016a). The viability of S. pneumoniae and the increase in supercoiling is affected by SCN in a concentration-dependent manner (Figure 7). Treatment with 6 μM SCN produced a peak σ increase of 41% at 5 min, which later recovered. Treatment with 8 μM SCN resulted in higher and longer lasting increases in the σ value, with partial recovery after 120 min. These results show that treatment with subinhibitory SCN concentrations permit the recovery of peak σ increases of up to 41% without affecting cell viability. This tolerance to increases in supercoiling levels is greater than the 25% observed for DNA relaxation upon NOV treatment (Figure 8A) (Ferrándiz et al., 2010), and indicates that S. pneumoniae, and very likely genetically related bacteria, are naturally more tolerant to hyper-negative supercoiling than to hyper-relaxation. Similarly, the results of experimental evolution assays with E. coli revealed increasing supercoiling (associated with mutations in topA) to increase bacterial fitness (Crozat et al., 2005). A similar homeostatic mechanism allowing increased negative supercoiling might also exist in bacteria with reverse gyrase. These bacteria keep DNA in a slightly overwound state to protect their genome from heat damage (Ogawa et al., 2015).
FIGURE 7.
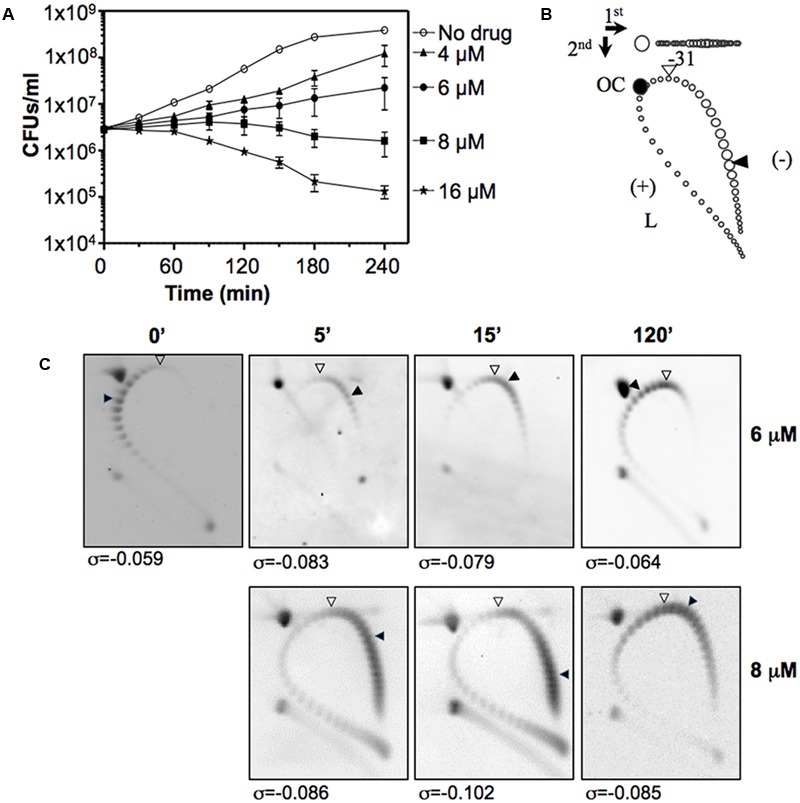
Seconeolitsine affects cell viability and induces hypersupercoiling. (A) Viability of R6 (pLS1) in a medium containing different SCN concentrations. Samples were taken before the addition of the drug (zero time), and at the indicated times were plated on drug-free agar medium. (B) Diagram showing topoisomer distribution in plasmid pLS1 subjected to 2D-agarose gel electrophoresis run in the presence of 5 and 15 μg/ ml chloroquine in the first and second dimensions, respectively. OC, open circle; L, linear form. A white arrowhead points to the topoisomer migrating with a ΔLk of 0 in the second dimension; it had a ΔWr of –31 in the first dimension. A black arrowhead points to the more abundant topoisomer. (C) Distribution of pLS1 topoisomers in 2D-gels after treatment with the indicated SCN concentrations. The supercoiling density (σ) values are indicated. Taken from Ferrándiz et al. (2016a), with modifications.
FIGURE 8.
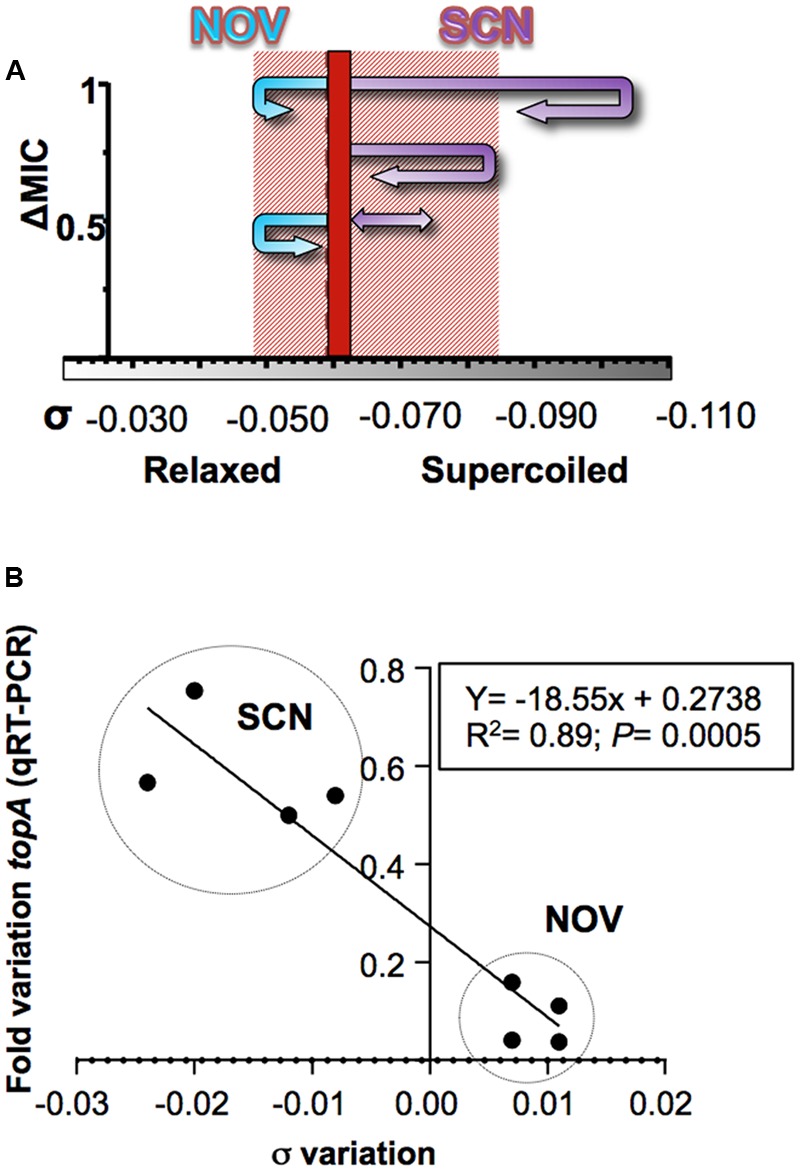
Responses to changes in supercoiling are conducted by Topo I. (A) Diagram showing the trajectory of the supercoiling density at the indicated NOV and SCN concentrations. A dashed line indicates the supercoiling density of DNA of non-treated cells. The shaded area represents the supercoiling density interval in which cells can survive. (B) Correlation between changes in supercoiling level and the transcription of topA. The data correspond to samples treated with either SCN or NOV at concentrations that allowed cell growth and the recovery of DNA supercoiling. Taken from Ferrándiz et al. (2016a), with modifications.
The transcription levels of topA in S. pneumoniae at subinhibitory concentrations of SCN or NOV (which allow for cell growth and the recovery of supercoiling) show a good correlation with the induced variation in σ (Figure 8B). The regulation of topA therefore plays a fundamental role in the recovery of supercoiling levels. The variations seen in topA expression were, however, only part of a global transcriptomic response. Treatment with subinhibitory concentrations of SCN (8 μM, 0.5× MIC) generated a two-stage transcriptomic response: (i) early response and (ii) recovery. The former, which represents an active response against sharply increased supercoiling, was observed at 5 and 15 min of treatment, and involved about 11% of the genome. During recovery, only about 2% of the genome was involved at 30 min. In the early response, transcriptional variations also occurred in clusters, with DEGs grouping into topologically sensitive domains. The average size of a SCN cluster is 14.0 ± 7.6, similar to the 10 kb E. coli domains predicted using transcriptional data (Postow et al., 2004). Although the NOV and SCN clusters are not identical, their position in the chromosome nearly overlap (Figure 4B) – an unexpected finding given the opposing nature of DNA relaxation and supercoiling. These results support the idea that the chromosome is divided into topological domains with fixed locations.
Regulation of DNA Topoisomerase Gene Transcription
In E. coli, several NAPs are involved in the regulation of topoisomerases. One such NAP is the FIS protein, which regulates the expression of genes coding for the subunits of gyrase (Schneider et al., 1999), Topo I (Weinstein-Fischer and Altuvia, 2007), and the genes coding for other NAPs involved in DNA supercoiling (Claret and Rouviere-Yaniv, 1996; Falconi et al., 1996; Grainger et al., 2008). In addition, two further NAPs, FIS, and H-NS proteins control both the level of supercoiling and global transcription (Blot et al., 2006; Marr et al., 2008). The corresponding situation in S. pneumoniae, which lacks these NAPs, seems to be much simpler.
The transcription of gyrB and topA in S. pneumoniae is regulated by their strategic chromosomal location in topological domains, since the expression driven by their promoters differs whether they are located in their natural chromosomal locations or in a replicating plasmid (Ferrándiz et al., 2014). Transcriptional fusions of these promoters to a reporter gene in plasmid pLS1 have been measured after DNA relaxation induced by NOV. As expected, relaxation caused down-regulation of topA and up-regulation of gyrB when the genes were located in their native chromosomal sites (DOWN9 for topA and UP6 for gyrB in Figure 5A). However, transcription from both promoters in the plasmid fusions was down-regulated. These results indicate that both topA and gyrB are under supercoil-mediated regulation, and that the plasmid behaves as a DOWN domain. This may serve to neutralize the high copy number of the plasmid genes and/or favor their replication.
In contrast, the Topo IV genes (parE and parC) and gyrA are located in NR domains, and their expression depends on specific regulatory signals located in the promoter region. The expression of the Topo IV genes from their common promoter (Balsalobre and de la Campa, 2008) is equivalent in their natural chromosomal location and in plasmids (Ferrándiz et al., 2014). With respect to the gyrA gene, its upstream region (PgyrA126, nt -126 to +1 in Figure 9A) shows an intrinsic DNA curvature (Balas et al., 1998). This was fused to cat and cloned into plasmid pLS1, and the curvature either eliminated by a 5 bp insertion (PgyrA126Pae) or by a 5 bp deletion (PgyrA121Pae), and a direct correlation observed between cat expression and the curvature under basal conditions (the specific activity of the PgyrA126 fusion was ∼3-fold higher than that recorded for plasmids lacking the curvature). This shows that the curvature behaves as an activator per se, providing better recruitment of either the RNA polymerase complex or specific regulatory proteins. The role of curvatures as regulators of transcription has previously been established in bacteria (Pérez-Martín et al., 1994), including S. pneumoniae (Pérez-Martín and Espinosa, 1991). In addition, the transcription levels from the chromosomal PgyrA and the PgyrAcat fusions in plasmids in the presence of NOV have been determined. While in the plasmid carrying the wild-type promoter (PgyrA126) the up-regulation of cat was similar to that of the chromosomal gyrA, down-regulation of cat was observed in the plasmids lacking the curvature (Figure 9B). These results suggest that the signals regulating gyrA transcription are included within the above-mentioned 126 nt region, and that bending is a key element for its regulation under relaxation by acting as a sensor of the supercoiling level.
FIGURE 9.
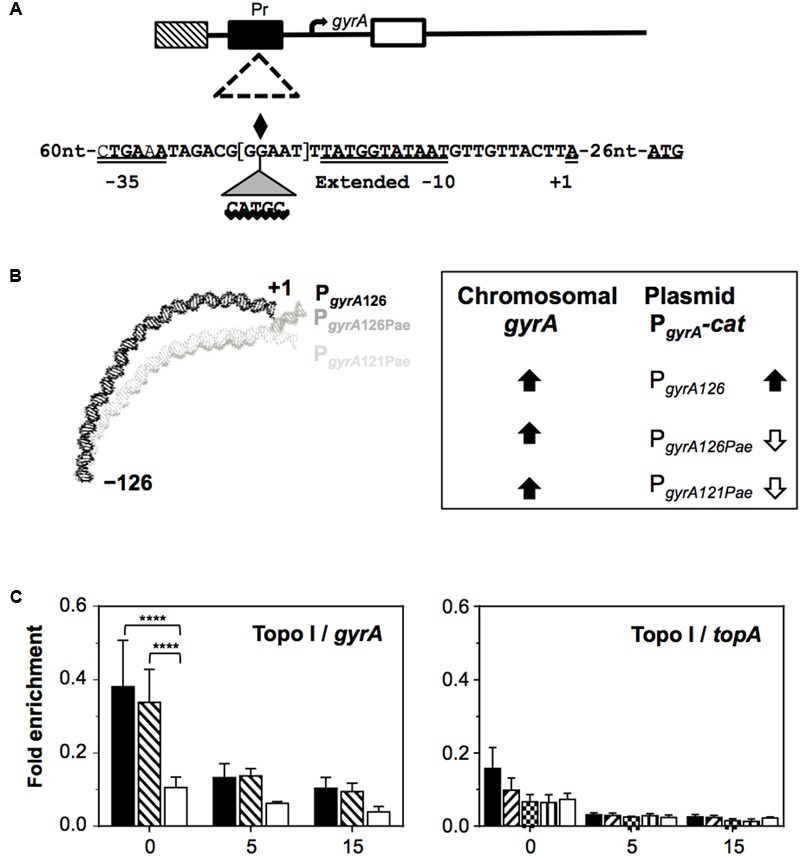
Control of the transcription of DNA topoisomerase genes by supercoiling. (A) Representation of the gyrA coding region and of the regions tested in chromatin immunoprecipitation, showing the sequences of the wild-type PgyrA126, PgyrA126Pae, and PgyrA121Pae derivatives. The –35 and extended –10 boxes, the nucleotide at which transcription is initiated (+1), the center of the intrinsic DNA curvature (diamond), and the location of the inserted CATGC sequence that creates a PaeI restriction site, are all indicated. The five nucleotides deleted in PgyrA121Pae are in brackets. (B) The relaxation-induced up-regulation of PgyrA depends on intrinsic bending: curvature prediction and results obtained from qRT-PCR analysis. (C) Recruitment of Topo I to topA and gyrA upstream sequences. Exponentially growing cells were subjected to chromatin immunoprecipitation using anti-Topo I antibodies; the pulled-down DNA was subsequently analyzed by qPCR. The graphs show the pulldown efficiency (ChIP-DNA/input DNA) for each primer pair. Values are the average ± SD of three independent replicates. ∗∗∗∗P < 0.0001. Taken from Ferrándiz et al. (2014, 2016a), with modifications.
Chromatin immunoprecipitation experiments using antibodies directed against the pneumococcal GyrA subunit and Topo I (Ferrándiz et al., 2016a) have shown PgyrA to recruit Topo I, but not gyrase (Figure 9C). The region to which Topo I binds includes the -35 and extended -10 boxes on PgyrA, plus the DNA curvature (Balas et al., 1998). Thus, Topo I, the transcription of which is regulated by supercoiling levels, appears to be the key factor regulating gyrA expression.
Evolutionary Pressure Drives the Organization of the Chromosome into Domains
Domain Conservation in Streptococci
Gene order in bacterial chromosomes surpasses the level of the operon (Lathe et al., 2000; Reams and Neidle, 2004). As explained above, and based on its transcriptome under DNA relaxation, the chromosome of S. pneumoniae R6 appears to be organized into four types of topological domains: UP, DOWN, NR, and ATr. The analysis of 12 S. pneumoniae complete genome sequences has revealed the conservation of the UP and DOWN domains (Figure 10). The gene-lack index (number of genomes in which a gene is absent divided by the total number of genomes) revealed lower values for the UP (1.51) and DOWN (1.65) domains than the genome average (1.91). However, ATr domains have high gene-lack indices (average 4.66), suggesting extensive gene interchange in these domains. To study the conservation of domains, normalized location dispersion indices (nLDI: values that quantify the position deviation of a given gene with respect to the Ori, and relative to homologs in several genomes (Martín-Galiano et al., 2017)) were calculated across S. pneumoniae genomes; the values returned were very small since synteny is highly conserved in this species. The same was then calculated for representative strains of 25 species of Streptococcus in order to detect distinguishing differences. The conservation of S. pneumoniae domains across these Streptococcus representatives was then determined. Two assumptions were made: (i) that the gene order is relatively conserved, as seen in gamma-proteobacteria (Sobetzko et al., 2012), and (ii) that chromosomal topology is conserved, given that species share core gene pools (Lefebure and Stanhope, 2007), similar genome lengths, and a similar AT content. Similar approaches have been followed to examine chromosomal patterning in other bacteria (Wright et al., 2007; Khedkar and Seshasayee, 2016). In S. pneumoniae, 571 genes (28.0%) had nLDI values of <1, which indicates they tend to locate to positions more stable than the average for maintained homologs (Martín-Galiano et al., 2017). Several genes from the UP and DOWN domains were present in most streptococci at equivalent positions. The greatest position conservation was observed in 40 genes near the Ori, indicating strong topological pressure to maintain functionalities in this region. Genes near the Ori have high copy numbers (Slager and Veening, 2016) and show a peculiar pattern of NAP binding (Sobetzko et al., 2012). Moreover, seven clusters with conserved positions were detected for NR genes, and named pcNR domains (position-conserved Non-Regulated domains). Most of the remaining NR genes were organized into 14 domains (≥10 genes) termed pvNR domains (position-variable Non-Regulated). ATr regions accounted for 13 domains (Figure 11). Strikingly, the pcNR domains appeared symmetrically located at regular intervals (∼200, 400, and 800 kb) on both sides of the Ori and were interleaved between UP, DOWN, and pvNR domains (Figure 11A). The size of these domains appeared compatible with the 100 kb lengths estimated for them using different techniques (Worcel and Burgi, 1972; Sinden and Pettijohn, 1981; Le et al., 2013). This suggests a potential higher-order macrostructural unit above the domain level controlling the genetic stability and plasticity required to face new environments (Rocha, 2004a).
FIGURE 10.
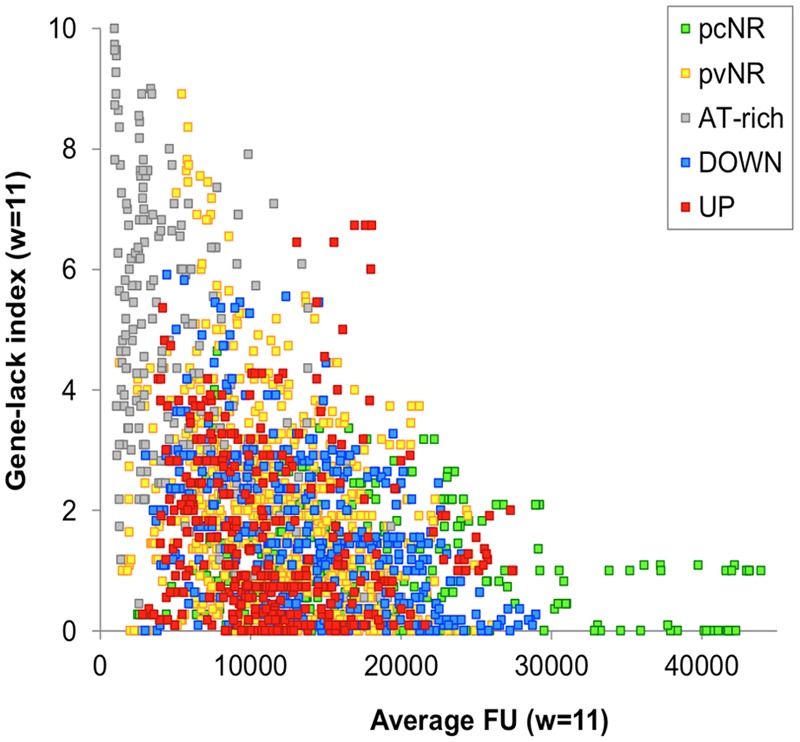
Evidence for conservation of UP and DOWN domains in S. pneumoniae. Relationship between the gene-lack index and gene expression (fluorescent units, FU) in S. pneumoniae R6 as detected in high density microarrays. A total of 12 genome sequences (from 11 clinical isolates and R6) were analyzed. Genes of clinical isolates were considered equivalent to those of R6 when their products shared ≥80% similarity over ≥80% of the sequence length. An 11-gene window (about 10 kb) was contemplated.
FIGURE 11.
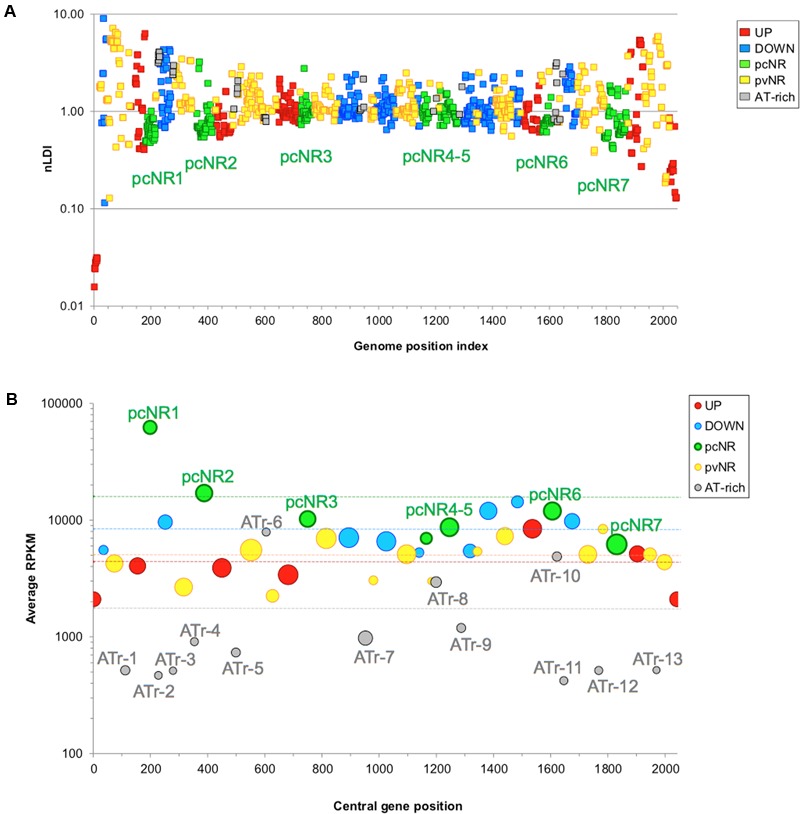
Different domains have different locations and transcription levels. (A) Location of NOV-reactive and pcNR domains. Dots indicate nLDI values for S. pneumoniae R6 genes for the whole Streptococcus genus. A total of 1216 genes coding for proteins in S. pneumoniae R6, with homologs in ≥10 species, were contemplated in the analysis. Gene indexing starts from the replication origin. (B) Abundance of RNA transcripts according to domain type. RNA-Seq data were normalized by gene size (kb) using the reads per kb per million mapped reads (RPKM). The positions of the domains in the genome are represented by the position of the central gene (central gene position, X-axis). The size of the circles is proportional to the number of genes in the domain. Only pvNR clusters with at least 10 members are shown. Horizontal lines in the corresponding color indicate the average RPKM value for each domain class. Taken from Martín-Galiano et al. (2017).
Levels of Protein Expression and Essentiality of the Domains
The transcriptomes of exponentially growing cultures (Ferrándiz et al., 2016a,b) showed the pcNR domain transcription level to be higher than that of the ATr domains (Figure 11B). Two factors contribute to these transcriptional differences. First, long repeat sequences (BOX, RUP, and SPRITE) (Croucher et al., 2011), which are associated with the repression of transcription, are few in pcNR domains, and second, the codon adaptation index (CAI), which is related to the translation rate and mRNA levels (Martín-Galiano et al., 2004), is high in pcNR domains (Martín-Galiano et al., 2017). Gene location also affects protein levels (Ochman et al., 2000; Rocha, 2004b), a pattern associated with the distance to the Ori. Genes at the Ori are doubly represented with respect to genes at the Ter in E. coli during exponential growth (Chandler and Pritchard, 1975). Accordingly, the relocation of genes coding for ribosomal proteins and the RNA polymerase alpha subunit to positions distant to the Ori, reduces their transcription rates, which was associated with slower growth in Vibrio cholerae (Soler-Bistue et al., 2015). Similarly, in Salmonella typhimurium, genes relocated near the Ori are expressed more strongly than those relocated near the Ter (Schmid and Roth, 1987). The regular positioning of strongly expressed genes may mark the limits of domains, as reported for Caulobacter crescentus (Le et al., 2013).
The fraction of essential genes, as determined by Tn-seq (van Opijnen and Camilli, 2012), is notably higher in pcNR domains than in the other domains (Figure 12A). The co-localization of essential genes beyond randomness has also been reported for Bacillus subtilis and E. coli (Fang et al., 2005), perhaps because clustering makes genomes more resistant to deletions (Fang et al., 2008). The number of pcNR genes in the lagging strand was 15.6%, significantly lower than the average in the remaining S. pneumoniae genome (22.3%). This would reduce the chances of collision between DNA and RNA polymerases, resulting in the discontinuation of transcription (French, 1992). Essential genes also tend to be more strongly expressed (Rocha and Danchin, 2003), as confirmed for pneumococcal pcNR genes. Essential gene clustering at regular intervals, and not affected by topological stress as defined for pcNR, appears to reflect a favorable “supercoiling environment” for protein expression.
FIGURE 12.
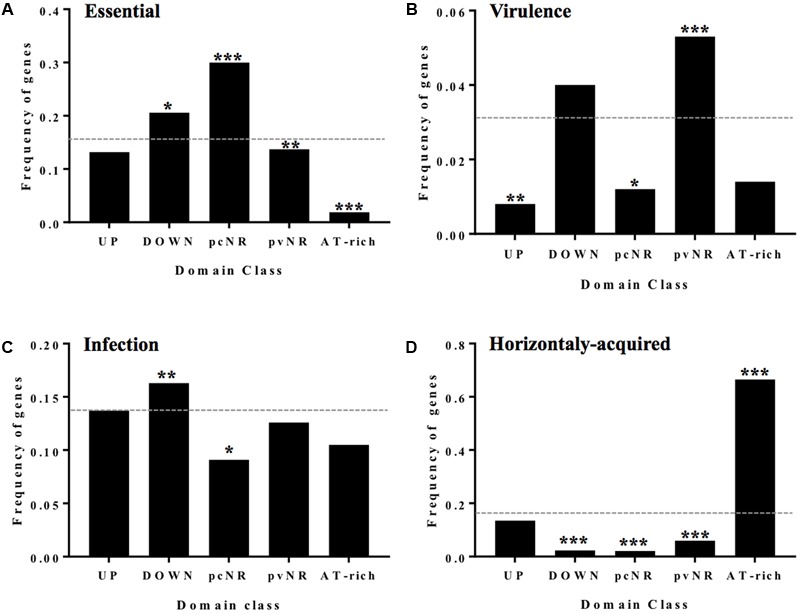
Genes of the different domains have different functionalities. (A) Fraction of essential genes in different domains. (B) Virulence factors. (C) Horizontally acquired genes abundance and domain class. (D) Essential genes for infection by STM. Statistical significance with respect to the genome average (dashed line): ∗P ≤0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001. Taken from Martín-Galiano et al. (2017), with modifications.
The Different Domains Contain Genes with Different Functions
Importance of the Protein Interaction Network
A significant fraction of the pcNR genes codes for proteins with important roles in central metabolism and that have a high number of protein–protein interactions (PPIs). PPIs provide a rough estimate of a protein’s importance in cell physiology. The estimated amounts of protein produced, and their functions, support the idea that the genes of pcNR domains are more involved in the central metabolic network than are those of the pvNR domains. In stark contrast, ATr genes appear to play little or no role in central metabolism; their PPI values are at most only about one third of the average for the remaining genome. As mentioned above, changes in the location of genes could lead to alterations in cell physiology, which holds true for both central metabolic (Soler-Bistue et al., 2015) and regulatory genes (Gerganova et al., 2015). The physical positioning of specific supercoiling-favorable regions in the chromosome is also related to the ability to gain access to cytoplasmic regions rich in ribosomes (Soler-Bistue et al., 2015).
Overall, the evidence supports the idea that the function, expression, essentiality and stability of genomic positions are interconnected, as reported for Dickeya dadantii and E. coli (Sobetzko et al., 2012; Jiang et al., 2015). Altogether, the pcNR genes reflect a multistep adaptation in the transcription-translation-interaction cascade that facilitates the activity of these genes’ products, thereby increasing bacterial fitness.
Pathogenesis and Immunogenicity
DNA topology regulates the expression of virulence factors in several bacteria (Dorman and Porter, 1998; Cameron and Dorman, 2012; Reverchon and Nasser, 2013; Jiang et al., 2015). In S. pneumoniae, three types of virulence genes show differences in their distribution among domains. Widely accepted virulence factors are more abundant in pvNR domains (Figure 12B), while genes contributing [as estimated by signature-tagged mutagenesis (Hensel et al., 1995)] to intranasal colonization, meningitis or otitis (Chen et al., 2008; Molzen et al., 2011) are more abundant in DOWN domains (Figure 12C). Finally, genes coding for proteins that trigger an immune response in humans (Giefing et al., 2009), and which are therefore candidate targets for a serotype-independent protein-based vaccine against pneumococcus, are predominant in the pvNR domains. The pvNR domains also contain more genes coding for extracellular proteins or proteins anchored in the cell wall than do pcNR domains. All in all, pvNR domains show strong allelic variation by being subjected to selective pressure during adhesion, cytotoxic challenge and immune system evasion. This variation also increases the genome pool of the species via gene duplication/paralogs in which one copy is not subject to immediate pressure (Mira et al., 2010). The link between supercoiling stress and virulence enhancement does not seem to be the rule for S. pneumoniae, the canonical virulence and accessory factors of which are preferentially encoded in the pvNR or DOWN domains.
Genes Involved in Competence
Gene transfer is a primary driver of evolution in bacteria, but the introduction of new genetic material at random can perturb chromosomal topology. S. pneumoniae is a naturally transformable bacterium (Claverys et al., 2006; Martin et al., 2006), the evolution of which (including its antibiotic-resistance and virulence factors) depends on both intra-species and inter-species chromosomal transformation (Dowson et al., 1989, 1990; Balsalobre et al., 2003; Ferrándiz et al., 2005). Competence involves the transient transcriptional modulation of ∼10% of the genome with strict timing (Peterson et al., 2004). When under stress (the X state), the competence system -which bears some resemblance to the SOS repair system of E. coli and other bacteria – is activated (Claverys et al., 2006). In fact, FQs induce the SOS response since they cause double-strand breaks in chromosomes (Drlica et al., 2008). As described above, local supercoiling changes triggered by FQs activate pneumococcal competence, but global supercoiling changes do so too. The early and delayed up-regulated competence genes (those activated during stress) are mainly located in UP domains. Many pcNR genes are, however, down-regulated, indicating that during the X-state the topology of the chromosome is perturbed to a degree that threatens cell viability via effects on the central metabolic machinery. This explains why growth is slowed during competence (Oggioni et al., 2004) and why several mechanisms have been acquired, including the use of small untranslated RNAs and proteases to actively terminate the X-state and promptly recover the normal topological situation (Echenique et al., 2000; Cassone et al., 2012).
Horizontally Acquired Genes
In S. pneumoniae R6, up to 12.1% of the genome is thought to have been acquired by horizontal gene transfer. The distribution of these acquired genes among domains is uneven, with a clear bias toward ATr domains (Figure 12D). This suggests that these domains act as structural or parasitic DNA hotspots, which agrees with their low transcriptional level and annotated functions (Ferrándiz et al., 2010, 2014, 2016a). It remains open the possibility that the ATr regions influence the organization of topological dynamics, or that they are involved in the acquisition of foreign genes.
Conclusion and Perspectives
The transcriptome of S. pneumoniae alters with local or global changes in supercoiling. Local changes induced by the clinically used FQs LVX, and MOX, which target GyrA and/or Topo IV, trigger a transcriptional response. Both FQs up-regulate the competence regulon in response to stress, and, respectively, cause an increase in intracellular ROS by increasing the uptake of iron (through up-regulation of the fatDCEB transporter) and hydrogen peroxide (through up-regulation of the glycolytic pathway), both of which are involved in the Fenton reaction.
Changes in global supercoiling induced by NOV (which targets GyrB), or by SCN (which targets Topo I), have revealed the existence of topological domains that react in a coordinated fashion. In S. pneumoniae, the control of DNA-supercoiling occurs mainly via the regulation of transcription of the topoisomerase genes: relaxation triggers the up-regulation of gyrA and gyrB and the down-regulation of the Topo I (topA) and Topo IV (parEC) genes, while hypernegative supercoiling triggers the down-regulation of topA. The transcription of gyrB and topA is regulated by their strategic chromosomal location in the topological domains, while the expression of parEC and gyrA depends on the specific regulation of their promoters. Although the regulators of parEC are unknown, the promoter of gyrA shows an intrinsic curvature that acts as a sensor of the supercoiling level. In addition, chromatin immunoprecipitation experiments have revealed Topo I to bind to the gyrA promoter. Therefore, Topo I, the transcription of which is regulated by the supercoiling level, appears to regulate gyrA expression.
The regulation of topoisomerase genes is part of a global response to changes in supercoiling. Relaxation affects >13% of the genome (from 13 to 24%), while hypernegative supercoiling affects 10%. In both cases, responsive genes are grouped into domains that essentially overlap, suggesting that they have a fixed chromosomal location. Based on their structural and functional characteristics, and the change in the domains detected under relaxation, the following types can be defined: UP, DOWN, pcNR, pvNR, and ATr. The genes of the UP, DOWN, and pcNR domains have been found at equivalent positions present in most streptococci, especially near the Ori. pcNR domains are interleaved between UP, DOWN, and pvNR domains, which suggests a higher-order macrostructural unit. The pcNRs genes show the highest level of transcription, and contain most of the essential genes plus those involved in the central metabolic network. In stark contrast, the ATr domains show the lowest transcriptional levels, and the genes they contain appear to have little to do with the central metabolic network. This explains the tropism of pcNR genes for topologically secure areas, helping to maintain the constant provision of central proteins.
The genes coding for the classical virulence factors, plus those coding for immunogenic proteins, are more common in the pvNR domains, while genes contributing toward the establishment of infection are more common in the DOWN domains. The distribution of horizontally acquired genes is clearly biased toward ATr domains, suggesting these to be hotspots for the acquisition of foreign genes.
In general, UP gene expression is favored by topological stress; DOWN genes are highly expressed under favorable conditions and less so during such stress. ATr domains may sense topological stress and modify supercoiling in their area to reduce the transcription of adjacent genes, preferentially those in the DOWN domains. The chromosome supercoiling structure may act as a multi-sensor with homeostatic capacity, adapted to react to unfavorable conditions.
Pneumococcal genes appear to be subject to topology-driven selection that defines the chromosomal location of genes involved in metabolism, virulence and competence. Together, these organizational features reveal the genome of S. pneumoniae to be influenced by physiology-related topological rules. A global topology theory might be envisaged in which gene positioning is far from random. Many aspects of the importance of gene location – such as the idiosyncrasy of the domains and how this affects fundamental aspects of bacterial biology – are only now becoming understood.
Topological genomics – topogenomics – provides an alternative paradigm of genome analysis. Certainly, genome architecture plays an important role in the pathobiology and evolution of S. pneumoniae, and it is tempting to speculate that in other species too, the genes are subjected to topology-driven selection pressure that defines their chromosomal locations. Data from many species will, however, be needed before a full understanding of all the rules underlying topogenomics are known and understood.
Author Contributions
All authors made intellectual contributions to the work and approved it for publication. AdC supervised all the studies and wrote the manuscript. MF performed most of the experiments related to determinations of supercoiling densities and transcriptomic studies. AM-G performed the bioinformatic studies. MG performed the characterization of topoisomerase I and its inhibition by seconeolitsine. JT-V contributed to the experiments of chromatin immunoprecipitation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Monica Amblar (Centro Nacional de Microbiología, ISCIII, Madrid, Spain), Pablo Hernández (Centro de Investigaciones Biológicas, CSIC, Madrid, Spain), and Pedro A. Lazo-Zbikowski (Instituto de Biología Molecular y Celular del Cáncer, CSIC, Salamanca, Spain), and for their critical reading of the manuscript.
Footnotes
Funding. AM-G is funded by a Miguel Servet contract from the Instituto de Salud Carlos III-MINECO. This work was supported by the Ministerio de Economía y Competitividad (BIO2014-55462-R).
References
- Arnoldi E., Pan X. S., Fisher L. M. (2013). Functional determinants of gate-DNA selection and cleavage by bacterial type II topoisomerases. Nucleic Acids Res. 41 9411–9423. 10.1093/nar/gkt696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayanan A., Le T. B., Laub M. T. (2015). Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 31 171–199. 10.1146/annurev-cellbio-100814-125211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balas D., Fernández-Moreira E., de la Campa A. G. (1998). Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J. Bacteriol. 180 2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre L., de la Campa A. G. (2008). Fitness of Streptococcus pneumoniae fluoroquinolone-resistant strains with topoisomerase IV recombinant genes. Antimicrob. Agents Chemother. 52 822–830. 10.1128/AAC.00731-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre L., Ferrándiz M. J., Liñares J., Tubau F., de la Campa A. G. (2003). Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47 2072–2081. 10.1128/AAC.47.7.2072-2081.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blot N., Mavathur R., Geertz M., Travers A., Muskhelishvili G. (2006). Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 7 710–715. 10.1038/sj.embor.7400729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito L., Wilton J., Ferrándiz M. J., Gómez-Sanz A., de la Campa A. G., Amblar M. (2017). Absence of tmRNA has a protective effect against fluoroquinolones in Streptococcus pneumoniae. Front. Microbiol. 7:2164 10.3389/fmicb.2016.02164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning D. F., Busby S. J. (2004). The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2 57–65. 10.1038/nrmicro787 [DOI] [PubMed] [Google Scholar]
- Cameron A. D., Dorman C. J. (2012). A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet. 8:e1002615 10.1371/journal.pgen.1002615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone M., Gagne A. L., Spruce L. A., Seeholzer S. H., Sebert M. E. (2012). The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J. Biol. Chem. 287 38449–38459. 10.1074/jbc.M112.391482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. (2001). DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70 369–413. 10.1146/annurev.biochem.70.1.369 [DOI] [PubMed] [Google Scholar]
- Chandler M. G., Pritchard R. H. (1975). The effect of gene concentration and relative gene dosage on gene output in Escherichia coli. Mol. Gen. Genet. 138 127–141. 10.1007/BF02428117 [DOI] [PubMed] [Google Scholar]
- Chen H., Ma Y., Yang J., O’Brien C. J., Lee S. L., Mazurkiewicz J. E., et al. (2008). Genetic requirement for pneumococcal ear infection. PLoS ONE 3:e2950 10.1371/journal.pone.0002950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B., Liu I. F., Tse-Dinh Y. C. (2007). Compounds with antibacterial activity that enhance DNA cleavage by bacterial DNA topoisomerase I. J. Antimicrob. Chemother. 59 640–645. 10.1093/jac/dkl556 [DOI] [PubMed] [Google Scholar]
- Cheng B., Zhu C. X., Ji C., Ahumada A., Tse-Dinh Y. C. (2003). Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J. Biol. Chem. 278 30705–30710. 10.1074/jbc.M303403200 [DOI] [PubMed] [Google Scholar]
- Claret L., Rouviere-Yaniv J. (1996). Regulation of HU alpha and HU beta by CRP and FIS in Escherichia coli. J. Mol. Biol. 263 126–139. 10.1006/jmbi.1996.0564 [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Prudhomme M., Martin B. (2006). Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60 451–475. 10.1146/annurev.micro.60.080805.142139 [DOI] [PubMed] [Google Scholar]
- Croucher N. J., Vernikos G. S., Parkhill J., Bentley S. D. (2011). Identification, variation and transcription of pneumococcal repeat sequences. BMC Genomics 12:120 10.1186/1471-2164-12-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat E., Philippe N., Lenski R. E., Geiselmann J., Schneider D. (2005). Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics 169 523–532. 10.1534/genetics.104.035717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S., Stein R. A., Higgins N. P. (2005). Organization of supercoil domains and their reorganization by transcription. Mol. Microbiol. 57 1511–1521. 10.1111/j.1365-2958.2005.04796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S. C., Dorman C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8 185–195. 10.1038/nrmicro2261 [DOI] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. (1982). Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell 31 43–51. 10.1016/0092-8674(82)90403-2 [DOI] [PubMed] [Google Scholar]
- Dorman C. J. (2013). Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat. Rev. Microbiol. 11 349–355. 10.1038/nrmicro3007 [DOI] [PubMed] [Google Scholar]
- Dorman C. J., Porter M. E. (1998). The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29 677–684. 10.1046/j.1365-2958.1998.00902.x [DOI] [PubMed] [Google Scholar]
- Dowson C. G., Hutchison A., Brannigan J. A., George R. C., Hansman D., Liñares J., et al. (1989). Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 86 8842–8846. 10.1073/pnas.86.22.8842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson C. G., Hutchinson A., Woodford N., Johnson A. P., George R. C., Spratt B. G. (1990). Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 87 5858–5862. 10.1073/pnas.87.15.5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K. (1992). Control of bacterial DNA supercoiling. Mol. Microbiol. 6 425–433. 10.1111/j.1365-2958.1992.tb01486.x [DOI] [PubMed] [Google Scholar]
- Drlica K., Malik M., Kerns R. J., Zhao X. (2008). Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52 385–392. 10.1128/AAC.01617-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M. (2006). Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 59 723–730. 10.1111/j.1365-2958.2005.05006.x [DOI] [PubMed] [Google Scholar]
- Drolet M., Bi X., Liu L. F. (1994). Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J. Biol. Chem. 269 2068–2074. [PubMed] [Google Scholar]
- Dupaigne P., Tonthat N. K., Espeli O., Whitfill T., Boccard F., Schumacher M. A. (2012). Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome. Mol. Cell 48 560–571. 10.1016/j.molcel.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer D. J., Belenky P. A., Yang J. H., MacDonald I. C., Martell J. D., Takahashi N., et al. (2014). Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A. 111 E2100–E2109. 10.1073/pnas.1401876111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer D. J., Collins J. J., Walker G. C. (2015). Unraveling the physiological complexities of antibiotic lethality. Annu. Rev. Pharmacol. Toxicol. 55 313–332. 10.1146/annurev-pharmtox-010814-124712 [DOI] [PubMed] [Google Scholar]
- Dwyer D. J., Kohanski M. A., Hayete B., Collins J. J. (2007). Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3 91 10.1038/msb4100135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echenique J. R., Chapuy-Regaud S., Trombe M. C. (2000). Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36 688–696. 10.1046/j.1365-2958.2000.01891.x [DOI] [PubMed] [Google Scholar]
- Espeli O., Mercier R., Boccard F. (2008). DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol. Microbiol. 68 1418–1427. 10.1111/j.1365-2958.2008.06239.x [DOI] [PubMed] [Google Scholar]
- Falconi M., Brandi A., La Teana A., Gualerzi C. O., Pon C. L. (1996). Antagonistic involvement of FIS and H-NS proteins in the transcriptional control of hns expression. Mol. Microbiol. 19 965–975. 10.1046/j.1365-2958.1996.436961.x [DOI] [PubMed] [Google Scholar]
- Fang G., Rocha E., Danchin A. (2005). How essential are nonessential genes? Mol. Biol. Evol. 22 2147–2156. 10.1093/molbev/msi211 [DOI] [PubMed] [Google Scholar]
- Fang G., Rocha E. P., Danchin A. (2008). Persistence drives gene clustering in bacterial genomes. BMC Genomics 9:4 10.1186/1471-2164-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoll A., Gimenez M. J., Vicioso M. D., Granizo J. J., Robledo O., Aguilar L. (2009). Susceptibility of pneumococci causing meningitis in Spain and prevalence among such isolates of serotypes contained in the 7-valent pneumococcal conjugate vaccine. J. Antimicrob. Chemother. 64 1338–1340. 10.1093/jac/dkp376 [DOI] [PubMed] [Google Scholar]
- Fernández-Moreira E., Balas D., González I., de la Campa A. G. (2000). Fluoroquinolones inhibit preferentially Streptococcus pneumoniae DNA topoisomerase IV than DNA gyrase native proteins. Microb. Drug Resist. 6 259–267. 10.1089/mdr.2000.6.259 [DOI] [PubMed] [Google Scholar]
- Ferrándiz M. J., Ardanuy C., Liñares J., García-Arenzana J. M., Cercenado E., Fleites A., et al. (2005). New mutations and horizontal transfer of rpoB among rifampin-resistant Streptococcus pneumoniae from four Spanish hospitals. Antimicrob. Agents Chemother. 49 2237–2245. 10.1128/AAC.49.6.2237-2245.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz M. J., Arnanz C., Martín-Galiano A. J., Rodríguez-Martin C., de la Campa A. G. (2014). Role of global and local topology in the regulation of gene expression in Streptococcus pneumoniae. PLoS ONE 9:e101574 10.1371/journal.pone.0101574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz M. J., de la Campa A. G. (2014). The fluoroquinolone levofloxacin triggers the transcriptional activation of iron transport genes that contribute to cell death in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58 247–257. 10.1128/AAC.01706-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz M. J., Martín-Galiano A. J., Arnanz C., Camacho-Soguero I., Tirado-Vélez J. M., de la Campa A. G. (2016a). An increase in negative supercoiling in bacteria reveals topology-reacting gene clusters and a homeostatic response mediated by the DNA topoisomerase I gene. Nucleic Acids Res. 44 7292–7303. 10.1093/nar/gkw602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz M. J., Martín-Galiano A. J., Arnanz C., Zimmerman T., de la Campa A. G. (2016b). Reactive oxygen species contribute to the bactericidal effects of the fluoroquinolone moxifloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 60 409–417. 10.1128/AAC.02299-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz M. J., Martín-Galiano A. J., Schvartzman J. B., de la Campa A. G. (2010). The genome of Streptococcus pneumoniae is organized in topology-reacting gene clusters. Nucleic Acids Res. 38 3570–3581. 10.1093/nar/gkq106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S. (1992). Consequences of replication fork movement through transcription units in vivo. Science 258 1362–1365. 10.1126/science.1455232 [DOI] [PubMed] [Google Scholar]
- García M. T., Blázquez M. A., Ferrándiz M. J., Sanz M. J., Silva-Martín N., Hermoso J. A., et al. (2011). New alkaloid antibiotics that target the DNA topoisomerase I of Streptococcus pneumoniae. J. Biol. Chem. 286 6402–6413. 10.1074/jbc.M110.148148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., ODea H., Nash H. A. (1976). DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U.S.A. 73 3872–3876. 10.1073/pnas.73.11.3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerganova V., Berger M., Zaldastanishvili E., Sobetzko P., Lafon C., Mourez M., et al. (2015). Chromosomal position shift of a regulatory gene alters the bacterial phenotype. Nucleic Acids Res. 43 8215–8226. 10.1093/nar/gkv709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giefing C., Nagy E., von Gabain A. (2009). The antigenome: from protein subunit vaccines to antibody treatments of bacterial infections? Adv. Exp. Med. Biol. 655 90–117. 10.1007/978-1-4419-1132-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmüender H., Kuratli K., Di Padova K, Gray C. P., Keck W., Evers S. (2001). Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11 28–42. 10.1101/gr.157701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger D. C., Goldberg M. D., Lee D. J., Busby S. J. (2008). Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol. Microbiol. 68 1366–1377. 10.1111/j.1365-2958.2008.06253.x [DOI] [PubMed] [Google Scholar]
- Hensel M., Shea J. E., Gleeson C., Jones M. D., Dalton E., Holden D. W. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science 269 400–403. 10.1126/science.7618105 [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Yang X., Fu Q., Roth J. R. (1996). Surveying a supercoil domain by using the gamma delta resolution system in Salmonella typhimurium. J. Bacteriol. 178 2825–2835. 10.1128/jb.178.10.2825-2835.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes V. F., Cozzarelli N. R. (2000). Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl. Acad. Sci. U.S.A. 97 1322–1324. 10.1073/pnas.040576797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M. R., Felmingham D., Appelbaum P. C., Grüneberg R. N., Group T. A. P. (2003). The Alexander project 1998-200: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52 229–246. 10.1093/jac/dkg321 [DOI] [PubMed] [Google Scholar]
- Janoir C., Zeller V., Kitzis M.-D., Moreau N. J., Gutmann L. (1996). High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob. Agents Chemother. 40 2760–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Sobetzko P., Nasser W., Reverchon S., Muskhelishvili G. (2015). Chromosomal “stress-response” domains govern the spatiotemporal expression of the bacterial virulence program. MBio 6 e00353–15. 10.1128/mBio.00353-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. (1990). New topoisomerase essential for chromosome segregation in E. coli. Cell 63 393–404. 10.1016/0092-8674(90)90172-B [DOI] [PubMed] [Google Scholar]
- Kavenoff R., Bowen B. C. (1976). Electron microscopy of membrane-free folded chromosomes from Escherichia coli. Chromosoma 59 89–101. 10.1007/BF00328479 [DOI] [PubMed] [Google Scholar]
- Khedkar S., Seshasayee A. S. (2016). Comparative genomics of interreplichore translocations in bacteria: a measure of chromosome topology? G3 (Bethesda) 6 1597–1606. 10.1534/g3.116.028274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. (2007). A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130 797–810. 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- Kyaw M. H., Lynfield R., Schaffner W., Craig A. S., Hadler J., Reingold A., et al. (2006). Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 354 1455–1463. 10.1056/NEJMoa051642 [DOI] [PubMed] [Google Scholar]
- Lathe W. C., III, Snel B., Bork P. (2000). Gene context conservation of a higher order than operons. Trends. Biochem. Sci. 25 474–479. 10.1016/S0968-0004(00)01663-7 [DOI] [PubMed] [Google Scholar]
- Le T. B., Imakaev M. V., Mirny L. A., Laub M. T. (2013). High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342 731–734. 10.1126/science.1242059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebure T., Stanhope M. J. (2007). Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 8:R71 10.1186/gb-2007-8-5-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo E., Gould K. A., Pan X. S., Capranico G., Sanderson M. R., Palumbo M., et al. (2005). Novel symmetric and asymmetric DNA scission determinants for Streptococcus pneumoniae topoisomerase IV and gyrase are clustered at the DNA breakage site. J. Biol. Chem. 280 14252–14263. 10.1074/jbc.M500156200 [DOI] [PubMed] [Google Scholar]
- Liñares J., Ardanuy C., Pallarés R., Fenoll A. (2010). Changes in antimicrobial resistance, serotypes and genotypes in Streptococcus pneumoniae over a 30-year period. Clin. Microbiol. Infect. 16 402–410. 10.1111/j.1469-0691.2010.03182.x [DOI] [PubMed] [Google Scholar]
- Marr C., Geertz M., Hutt M. T., Muskhelishvili G. (2008). Dissecting the logical types of network control in gene expression profiles. BMC Syst. Biol. 2:18 10.1186/1752-0509-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B., Quentin Y., Fichant G., Claverys J. P. (2006). Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 14 339–345. 10.1016/j.tim.2006.06.007 [DOI] [PubMed] [Google Scholar]
- Martín-Galiano A. J., Ferrándiz M. J., de la Campa A. G. (2017). Bridging chromosomal architecture and pathophysiology of Streptococcus pneumoniae. Genome Biol. Evol. 9 350–361. 10.1093/gbe/evw299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Galiano A. J., Wells J. M., de la Campa A. G. (2004). Relationship between codon biased genes, microarray expression values and physiological characteristics of Streptococcus pneumoniae. Microbiology 150 2313–2325. 10.1099/mic.0.27097-0 [DOI] [PubMed] [Google Scholar]
- Masse E., Drolet M. (1999). Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 274 16659–16664. 10.1074/jbc.274.23.16659 [DOI] [PubMed] [Google Scholar]
- Menzel R., Gellert M. (1983). Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell 34 105–113. 10.1016/0092-8674(83)90140-X [DOI] [PubMed] [Google Scholar]
- Menzel R., Gellert M. (1987a). Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J. Bacteriol. 169 1272–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. (1987b). Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc. Natl. Acad. Sci. U.S.A. 84 4185–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A., Martín-Cuadrado A. B., D’Auria G., Rodríguez-Valera F. (2010). The bacterial pan-genome:a new paradigm in microbiology. Int. Microbiol. 13 45–57. 10.2436/20.1501.01.110 [DOI] [PubMed] [Google Scholar]
- Molzen T. E., Burghout P., Bootsma H. J., Brandt C. T., van der Gaast-de Jongh C. E., Eleveld M. J., et al. (2011). Genome-wide identification of Streptococcus pneumoniae genes essential for bacterial replication during experimental meningitis. Infect. Immun. 79 288–297. 10.1128/IAI.00631-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. R., Gertz J. R. E., Woodbury R. L., Barkocy-Gallagher G. A., Schaffner W., Lexau C., et al. (2008). Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197 1016–1027. 10.1086/528996 [DOI] [PubMed] [Google Scholar]
- Muñoz R., de la Campa A. G. (1996). ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40 2252–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G., Groisman E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405 299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yogo K., Furuike S., Sutoh K., Kikuchi A., Kinosita K., Jr. (2015). Direct observation of DNA overwinding by reverse gyrase. Proc. Natl. Acad. Sci. U.S.A. 112 7495–7500. 10.1073/pnas.1422203112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oggioni M. R., Iannelli F., Ricci S., Chiavolini D., Parigi R., Trappetti C., et al. (2004). Antibacterial activity of a competence-stimulating peptide in experimental sepsis caused by Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48 4725–4732. 10.1128/AAC.48.12.4725-4732.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J., Espinosa M. (1991). The RepA repressor can act as a transcriptional activator by inducing DNA bends. EMBO J. 10 1375–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J., Rojo F., de Lorenzo V. (1994). Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58 268–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter B. J., Arsuaga J., Breier A. M., Khodursky A. B., Brown P. O., Cozzarelli N. R. (2004). Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 5:R87 10.1186/gb-2004-5-11-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S. N., Sung C. K., Cline R., Desai B. V., Snesrud E. C., Luo P., et al. (2004). Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51 1051–1070. 10.1046/j.1365-2958.2003.03907.x [DOI] [PubMed] [Google Scholar]
- Phoenix P., Raymond M. A., Masse E., Drolet M. (1997). Roles of DNA topoisomerases in the regulation of R-loop formation in vitro. J. Biol. Chem. 272 1473–1479. 10.1074/jbc.272.3.1473 [DOI] [PubMed] [Google Scholar]
- Pilishvili T., Lexau C., Farley M. M., Hadler J., Harrison L. H., Bennett N. M., et al. (2010). Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J. Infect. Dis. 201 32–41. 10.1086/648593 [DOI] [PubMed] [Google Scholar]
- Postow L., Crisona N. J., Peter B. J., Hardy C. D., Cozzarelli N. R. (2001). Topological challenges to DNA replication: conformations at the fork. Proc. Natl. Acad. Sci. U.S.A. 98 8219–8226. 10.1073/pnas.111006998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L., Hardy C. D., Arsuaga J., Cozzarelli N. R. (2004). Topological domain structure of the Escherichia coli chromosome. Genes Dev. 18 1766–1779. 10.1101/gad.1207504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M., Attaiech L., Sanchez G., Martin B., Claverys J. P. (2006). Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313 89–92. 10.1126/science.1127912 [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Manes S. H., Drlica K. (1982). Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell 31 35–42. 10.1016/0092-8674(82)90402-0 [DOI] [PubMed] [Google Scholar]
- Reams A. B., Neidle E. L. (2004). Selection for gene clustering by tandem duplication. Annu. Rev. Microbiol. 58 119–142. 10.1146/annurev.micro.58.030603.123806 [DOI] [PubMed] [Google Scholar]
- Reverchon S., Nasser W. (2013). Dickeya ecology, environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 5 622–636. 10.1111/1758-2229.12073 [DOI] [PubMed] [Google Scholar]
- Rocha E. P. (2004a). Order and disorder in bacterial genomes. Curr. Opin. Microbiol. 7 519–527. 10.1016/j.mib.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Rocha E. P. (2004b). The replication-related organization of bacterial genomes. Microbiology 150 1609–1627. 10.1099/mic.0.26974-0 [DOI] [PubMed] [Google Scholar]
- Rocha E. P., Danchin A. (2003). Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat. Genet. 34 377–378. 10.1038/ng1209 [DOI] [PubMed] [Google Scholar]
- Schmid M. B., Roth J. R. (1987). Gene location affects expression level in Salmonella typhimurium. J. Bacteriol. 169 2872–2875. 10.1128/jb.169.6.2872-2875.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Travers A., Kutateladze T., Muskhelishvili G. (1999). A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol. 34 953–964. 10.1046/j.1365-2958.1999.01656.x [DOI] [PubMed] [Google Scholar]
- Schroder W., Bernhardt J., Marincola G., Klein-Hitpass L., Herbig A., Krupp G., et al. (2014). Altering gene expression by aminocoumarins: the role of DNA supercoiling in Staphylococcus aureus. BMC Genomics 15:291 10.1186/1471-2164-15-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. (1981). Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl. Acad. Sci. U.S.A. 78 224–228. 10.1073/pnas.78.1.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager J., Veening J. W. (2016). Hard-wired control of bacterial processes by chromosomal gene location. Trends Microbiol. 24 788–800. 10.1016/j.tim.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Drlica K. (1979). DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J. Mol. Biol. 131 287–302. 10.1016/0022-2836(79)90077-9 [DOI] [PubMed] [Google Scholar]
- Sobetzko P., Travers A., Muskhelishvili G. (2012). Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. U.S.A. 109 E42–E50. 10.1073/pnas.1108229109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Bistue A., Mondotte J. A., Bland M. J., Val M. E., Saleh M. C., Mazel D. (2015). Genomic location of the major ribosomal protein gene locus determines Vibrio cholerae global growth and infectivity. PLoS Genet. 11:e1005156 10.1371/journal.pgen.1005156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassi D., López P., Espinosa M., Lacks S. A. (1981). Cloning of chromosomal genes in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 78 7028–7032. 10.1073/pnas.78.11.7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tankovic J., Perichon B., Duval J., Courvalin P. (1996). Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob. Agents Chemother. 40 2505–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiara A. S., Cundliffe E. (1989). Interplay of novobiocinresistant and –sensitive DNA gyrase activities in selfprotection of the novobiocin producer Streptomyces sphaeroides. Gene 81 65–72. 10.1016/0378-1119(89)90337-5 [DOI] [PubMed] [Google Scholar]
- Travers A., Muskhelishvili G. (2005). DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 3 157–169. 10.1038/nrmicro1088 [DOI] [PubMed] [Google Scholar]
- Tse-Dinh Y.-C. (1985). Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling. Nucleic Acids Res. 13 4751–4763. 10.1093/nar/13.13.4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unniraman S., Chatterji M., Nagaraja V. (2002). DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J. Bacteriol. 184 5449–5456. 10.1128/JB.184.19.5449-5456.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T., Camilli A. (2012). A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 22 2541–2551. 10.1101/gr.137430.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora T., Hottes A. K., Tavazoie S. (2009). Protein occupancy landscape of a bacterial genome. Mol. Cell 35 247–253. 10.1016/j.molcel.2009.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Montero Llopis P., Rudner D. Z. (2013). Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14 191–203. 10.1038/nrg3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao X. (2009). Contribution of oxidative damage to antimicrobial lethality. Antimicrob. Agents Chemother. 53 1395–1402. 10.1128/AAC.01087-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao X., Malik M., Drlica K. (2010). Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J. Antimicrob. Chemother. 65 520–524. 10.1093/jac/dkp486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein-Fischer D., Altuvia S. (2007). Differential regulation of Escherichia coli topoisomerase I by Fis. Mol. Microbiol. 63 1131–1144. 10.1111/j.1365-2958.2006.05569.x [DOI] [PubMed] [Google Scholar]
- Whitney C. G., Farley M. M., Hadler J., Harrison L. H., Bennett N. M., Lynfield R., et al. (2003). Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348 1737–1746. 10.1056/NEJMoa022823 [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. (1972). On the structure of the folded chromosome of Escherichia coli. J. Mol. Biol. 71 127–147. 10.1016/0022-2836(72)90342-7 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2007). Pneumococcal conjugate vaccine for childhood immunization-WHO position paper. Wkly. Epidemiol. Rec. 82 93–104. [PubMed] [Google Scholar]
- Wright M. A., Kharchenko P., Church G. M., Segre D. (2007). Chromosomal periodicity of evolutionarily conserved gene pairs. Proc. Natl. Acad. Sci. U.S.A. 104 10559–10564. 10.1073/pnas.0610776104 [DOI] [PMC free article] [PubMed] [Google Scholar]


