Abstract
At present, most of released wheat cultivars or breeding lines in China are susceptible to powdery mildew (Pm) (caused by Blumeria graminis f. sp. tritici, Bgt), so there is an urgent need to rapidly transfer effective and broad-spectrum Pm resistance genes into elite cultivars/lines. Near-isogenic lines (NILs) with short target gene region are very important in molecular breeding and map-based cloning and can be developed by combining marker-assisted selection and conventional phenotypic identification. However, no Pm gene NILs were reported by using this method in the previous studies. A new broad-spectrum dominant resistance gene Pm2b, derived from the Chinese wheat breeding line KM2939, conferred high resistance to Pm at both the seedling and adult stages. In this study, with the aid of forward and background selection (FS and BS) using molecular markers, the Pm2b gene was introgressed into three elite susceptible commercial cultivars Shimai 15, Shixin 828, and Kenong 199 through the back-crossing procedure. With the appropriate backcrossing generations, selected population sizes and marker number for BS, the homozygous resistant BC3F2:3 NILs of Pm2b gene in the three genetic backgrounds with the highest recipient genome composition of about 99%, confirmed by simple sequence repeat markers and 660K single nucleotide polymorphic array, were developed and evaluated for the powdery mildew resistance and agronomic traits. The different resistance and similar or improved agronomic performance between Pm2b NILs and their corresponding recurrent parents indicated their potential value in the marker-assisted breeding of the Pm2b gene. Moreover, the development of four flanked diagnostic markers (CFD81, BWM25, BWM20, and BWM21) of the Pm2 gene can effectively assist the forward selection and accelerate the transfer and use of this resistance gene.
Keywords: Pm2b, near-isogenic line, Blumeria graminis, marker-assisted selection, wheat
Introduction
Common wheat (Triticum aestivum L.) is one of the most important staple food crop in China and the yield of wheat can seriously affect food safety. Of the several wheat production constraints, diseases are the most important stress, which can cause significant yield losses. Among the various wheat foliar diseases, Pm, caused by B. graminis f. sp. tritici (Bgt), is one of the most prevalent diseases occurring throughout the wheat growing regions of the world, which caused severe yield damage (Bennett, 1984; Sun et al., 2015). In China, most of the wheat cultivars released in recent years are susceptible to Pm and account for 83.1% of the total acreage (Li et al., 2011). In 2017, up to 8 million hectares of wheat production area are vulnerable to this disease (The Occurrence Tendency Prediction of Plant Diseases and Insect Pests in China, 2017). The use of resistant cultivars and resistance genes is the most efficient, economical and environmentally safe approach to curb this disease and reduce yield losses (Wang et al., 2015).
In wheat, two kinds of genes confers resistance against Pm: mlo genes and Pm resistance genes. The three MLO homoeologs in bread wheat (TaMLO-A1, TaMLO-B1, and TaMLO-D1) encode proteins to repress defenses against Pm diseases. Loss-of-function mlo mutant in all three MLO copies lead to broad-spectrum and durable resistance to Bgt (Wang et al., 2014). Different from mlo genes, most of reported Pm resistance genes with only one copy encode R proteins, which interacted with avirulent proteins of the pathogens to confer resistance to the diseases. At present, more than 70 formally designated Pm resistance genes Pm1 – Pm58 at 53 loci (Pm8 is allelic to Pm17, Pm18 = Pm1c, Pm22 = Pm1e, Pm23 = Pm4c, and Pm31 = Pm21) and more than 20 temporarily named Pm genes have been reported. These genes are distributed on all chromosomes (Hao et al., 2015; Petersen et al., 2015; Zhang et al., 2016; Li et al., 2017; McIntosh et al., 2017). Most of these resistance genes are major genes conferring race-specific resistance and are easily used in disease-resistance breeding. However, relatively few Pm genes including Pm2a, Pm4a, Pm6, Pm8, and Pm21 have been successfully used in breeding or in the development of resistant cultivars/lines in China (Zhang et al., 2010; Huang et al., 2012). There is an urgent need to rapidly transfer effective and broad-spectrum Pm resistance genes into elite released cultivars.
However, the transfer and introgression of resistance genes into released cultivars is time-consuming and inefficient when based on conventional selection strategies alone. Target resistance genes of donors may be lost due to uncertain phenotype identification. The superior agronomic traits of recurrent parents are not completely restored because of recombination and difficulties in phenotypic evaluation. Furthermore, diluted or suppressed resistance and adverse linkage drag or negative associations often occur unpredictably (Friebe et al., 1994; Zeller and Hsam, 1996; Li et al., 2017). Instead, combining marker-assisted forward and background selection (FS and BS) with conventional phenotypic selection, the target resistance gene can be precisely introgressed into the wheat cultivar background in a short time pairing with few negative association. Generally the tightly linked markers to the target gene were used to trace this gene (forward selection) and the whole genome markers were used to detect the genetic similarity between the progeny plants with their recurrent parents (BS). With the help of MAS, more and more resistance genes in wheat have been transferred into recurrent parent backgrounds (Vida et al., 2009; Kumar et al., 2010; Xue et al., 2010; Elkot et al., 2015; Guo et al., 2015; Yaniv et al., 2015).
A better way to utilize target resistance gene is to develop its NILs in the genetic backgrounds of elite cultivars and use those NILs in the molecular MAB (Tanksley et al., 1996; Zhou et al., 2005). In addition, NILs of resistance genes are valuable for their validation and determination of gene effects, differential expression profiling, fine mapping and map-based cloning (Xue et al., 2010). With the help of some closely linked markers, NILs for some resistance genes in wheat have been developed (Xue et al., 2010; Khanna et al., 2015; Habib et al., 2016; Zheng et al., 2017). However, in wheat, no NIL for Pm resistance gene has been developed through MAS.
Chinese wheat breeding line KM2939 confers high resistance to Pm at both the seedling and adult stages. It carried a broad-spectrum dominant resistance gene Pm2b, which was mapped on chromosome 5DS (Ma et al., 2015a). In this study, the Pm2b gene was introgressed into the backgrounds of commercial cultivars SM15, SX828 and KN199 with high yield and susceptibility to Pm, which are widely grown on the North China Plain (Ma et al., 2015a), and the NILs of Pm2b from different genetic backgrounds were developed and evaluated for their Pm resistance and agronomic traits.
Materials and Methods
Plant Materials
Chinese winter wheat breeding line KM2939 was characterized by high resistance to Pm at both the seedling and adult stages and was crossed as the Pm2b gene donor to three high yield and Pm susceptible commercial cultivars SM15 (Jimai38/92R137//Jimai 38), SX828 (422/Shixin63//612), and KN199 (Shi4185/Kenong 9204), as the recurrent parents. All the progeny plants/lines, derived from the combinations of KM2939/SM15, SX828/KM2939, and KN199/KM2939, were used to develop Pm2b NILs in different genetic backgrounds. The BC2F1 population of KM2939/SM15 was also used to genotype Pm2b gene and develop diagnostic markers (Ma et al., 2015a).
Marker Analysis
Marker CFD81 was regarded as a co-dominant diagnostic marker for the FS of Pm2b (Ma et al., 2015a). Furthermore, three tightly linked SSR markers Xbwm20, Xbwm21, and Xbwm25 of PmPB3558, a novel Pm2 allele (Lu et al., 2015), were used as co-dominant diagnostic markers for tracing Pm2b gene. Genotyping and marker map construction were as described by Ma et al. (2015a). A total of 182 SSR markers evenly distributed on all 21 wheat chromosomes (Somers et al., 2004) were chosen to assess the genetic similarity between the progeny plants and their corresponding recurrent parents (Xue et al., 2010). The wheat 660K SNP array (designed at the Chinese Academy of Agricultural Sciences and synthesized by Affymetrix1) analysis also was used to determine the RGC of the Pm2b NIL in the genetic background of SM15. PCR amplification was performed in a Veriti® thermal cycler (Applied Biosystems, Foster, CA, United States) following the procedures described by Xu et al. (2015). PCR products were separated in 8% non-denaturing polyacrylamide gels with 25:1 ratios of acrylamide and bis-acrylamide, and visualized by silver-staining as described in Santos et al. (1993).
Phenotyping
During the transfer of Pm2b gene, all the progeny plants were phenotyped by Bgt isolate E09. To verify the resistance of Pm2b NILs, they were tested singly by 25 single-pustule-derived Pm isolates (E01, E02, E05, E06, E07, E09, E11, E13, E15, E16, E17, E18, E20, E23-1, E23-2, E26, E30-1, E30-2, E31, E49, E50, Bg01, Bg02, Bg03, and Bg04) that were avirulent to Pm2b gene (Ma et al., 2015a) at the seedling stage and by their composite mixture at the adult stage (Zadoks et al., 1974). Reactions of plants/lines to Bgt isolates and infection types (ITs) on each plant were assessed on a 0–4 scale as described by Xu et al. (2015). Plants with one of the four IT 0, 0;, 1, or 2 were regarded as resistant, and those with one of the IT 3 or 4 as susceptible. All tests and identification were repeated to assure the reliability of the data and only resistant progeny plants were retained after phenotyping.
To evaluate the agronomic performance of Pm2b NILs, they were planted at Luancheng Agro-ecosystem Experimental Station (37°53′15″N, 114°40′47″E) together with their respective recurrent parents and donor parent during 2015–2016 in a randomized complete block design with two replicates. Each parent/line was grown 5 row plots (20 kernels/row) with 1.5 m-long, 1.0 m-width and 0.25 m apart with two replicates. In each plot, 10 plants in the middle three internal rows were sampled to investigate the following traits: PH, SNPP, and GY determined from the mean of the ten plants; SL, TSS, SSS, KNS determined from the mean of the main spikes of the ten plants; TKW evaluated after harvest by weighing three samples of 500 kernels.
Data Analysis
Chi-squared (χ2) tests for goodness-of-fit in each backcross generations were used to evaluate deviations of observed data from expected segregation ratios. The software MAPMAKER/Exp (version 3.0b) was used to determine linkage with a LOD score of 3.0 as the threshold for declaration of linkage (Lander et al., 1987). Genetic distances were estimated from the recombination values using the Kosambi mapping function (Kosambi, 1943). T-tests were conducted between the Pm2b NILs and their recurrent parents for agronomic traits.
Using the formula described by Xue et al. (2010) and Ma et al. (2015a), the RGC of progeny plants was estimated as %RGC = 100%∗1/2(2BB + AB)/(AA+AB+BB). AA, AB and BB represented the number of genotyped marker loci of homozygous donor, heterozygous and homozygous recipient genomes, respectively.
Results
The Introgression of Pm2b Gene
Wheat breeding line KM2939, used as the Pm2b gene donor, was crossed with the three wheat cultivars SM15, SX828, and KN199. Their F1 progeny plants were then backcrossed three times with their corresponding recurrent parents. After selfing, the BC3F2:3 lines were developed. During the transfer of Pm2b into the recurrent parents, the resistant donor parent KM2939, recurrent parents SM15, SX828, and KN199, and all their derived generation plants were tested against Bgt isolate E09, which is avirulent to Pm2b gene at the seedling stage and their inheritance of resistance was analyzed, and only the resistant target progeny plants after FS and BS were selected for further backcrossing or selfing (Table 1).
Table 1.
The seedling resistance identification with E09 isolate and marker-assisted selection for lines of Pm2b gene in the genetic backgrounds of Shimai 15 (SM15), Shixin828 (SX828), and Kenong 199 (KN199).
| Generations | Resistant | Susceptible | Chi-c2 | P-value | Target plants after FS | Selected plant | Markers for BS | RGC% |
|---|---|---|---|---|---|---|---|---|
| KM2939 × SM15 BC1F1 | 36 | 29 | c21:1 = 0.55 | 0.46 | 33 | KS-6 | 101 | 89.1 |
| KM2939 × SM15 BC2F1 | 236 | 255 | c21:1 = 0.66 | 0.42 | 209 | KS-6-88 | 101 | 96.5 |
| KM2939 × SM15 BC3F1 | 21 | 30 | c21:1 = 1.25 | 0.26 | 20 | KS-6-88-11 | 101 | 98.0 |
| KM2939 × SM15 BC3F2 | 68 | 20 | c23:1 = 0.14 | 0.71 | 22 | KS-6-88-11-2 | 101 | 99.0 |
| KM2939 × SM15 BC3F2:3 | 23RR+45Rr | 20rr | c21:2:1 = 0.25 | 0.88 | – | – | – | – |
| SX828 × KM2939 BC1F1 | 13 | 15 | c21:1 = 0.036 | 0.85 | 13 | KSX-2 | 94 | 87.2 |
| SX828 × KM2939 BC2F1 | 11 | 13 | c21:1 = 0.042 | 0.84 | 11 | KSX-2-3 | 94 | 93.6 |
| SX828 × KM2939 BC3F1 | 26 | 28 | c21:1 = 0.074 | 0.79 | 24 | KSX-2-3-15 | 94 | 96.2 |
| SX828 × KM2939 BC3F2 | 71 | 25 | c23:1 = 0.056 | 0.81 | 23 | KSX-2-3-15-22 | 94 | 98.9 |
| SX828 × KM2939 BC3F2:3 | 24RR+47Rr | 25rr | c21:2:1 = 0.063 | 0.97 | – | – | – | – |
| KN199 × KM2939 BC1F1 | 12 | 10 | c21:1 = 0.045 | 0.83 | 12 | KK-3 | 99 | 87.8 |
| KN199 × KM2939 BC2F1 | 10 | 11 | c21:1 = 0.048 | 0.83 | 10 | KK-3-4 | 99 | 93.9 |
| KN199 × KM2939 BC3F1 | 72 | 70 | c21:1 = 0.028 | 0.87 | 69 | KK-3-4-31 | 99 | 98.5 |
| KN199 × KM2939 BC3F2 | 78 | 27 | c23:1 = 0.029 | 0.87 | 24 | KK-3-4-31-54 | 99 | 99.0 |
| KN199 × KM2939 BC3F2:3 | 25RR+53Rr | 27rr | c21:2:1 = 0.086 | 0.96 | – | – | – | – |
BS, background selection; FS, forward ground selection; RGC, recipient genome composition.
Development of Pm2b Diagnostic Markers
In a previous study, Pm2b gene was reported tightly linked to four marker loci, namely Xscar112, Xscar203, Xmag6176, and Xcfd81 (Ma et al., 2015a). However, only CFD81 was a co-dominant marker and could be regarded as diagnostic marker for tracing Pm2b gene. To identify more flanking co-dominant diagnostic markers of Pm2b gene, three tightly linked co-dominant SSR markers BWM20, BWM21, and BWM25 of PmPB3558, a novel Pm2 allele (Lu et al., 2015), were used to map Pm2b gene. The results indicated that Pm2b gene co-segregated with Xbwm25 and was flanked by Xbwm20 and Xcfd81 at genetic distances of 2.6 and 1.4 cM, respectively (Figure 1). In addition, Xbwm21 was also tightly linked to Pm2b with a genetic distance of 3.9 cM (Figure 1). Thus, BWM20, BWM21, and BWM25 together with CFD81 were regarded as the diagnostic markers and were used to trace the Pm2b gene in this study.
FIGURE 1.
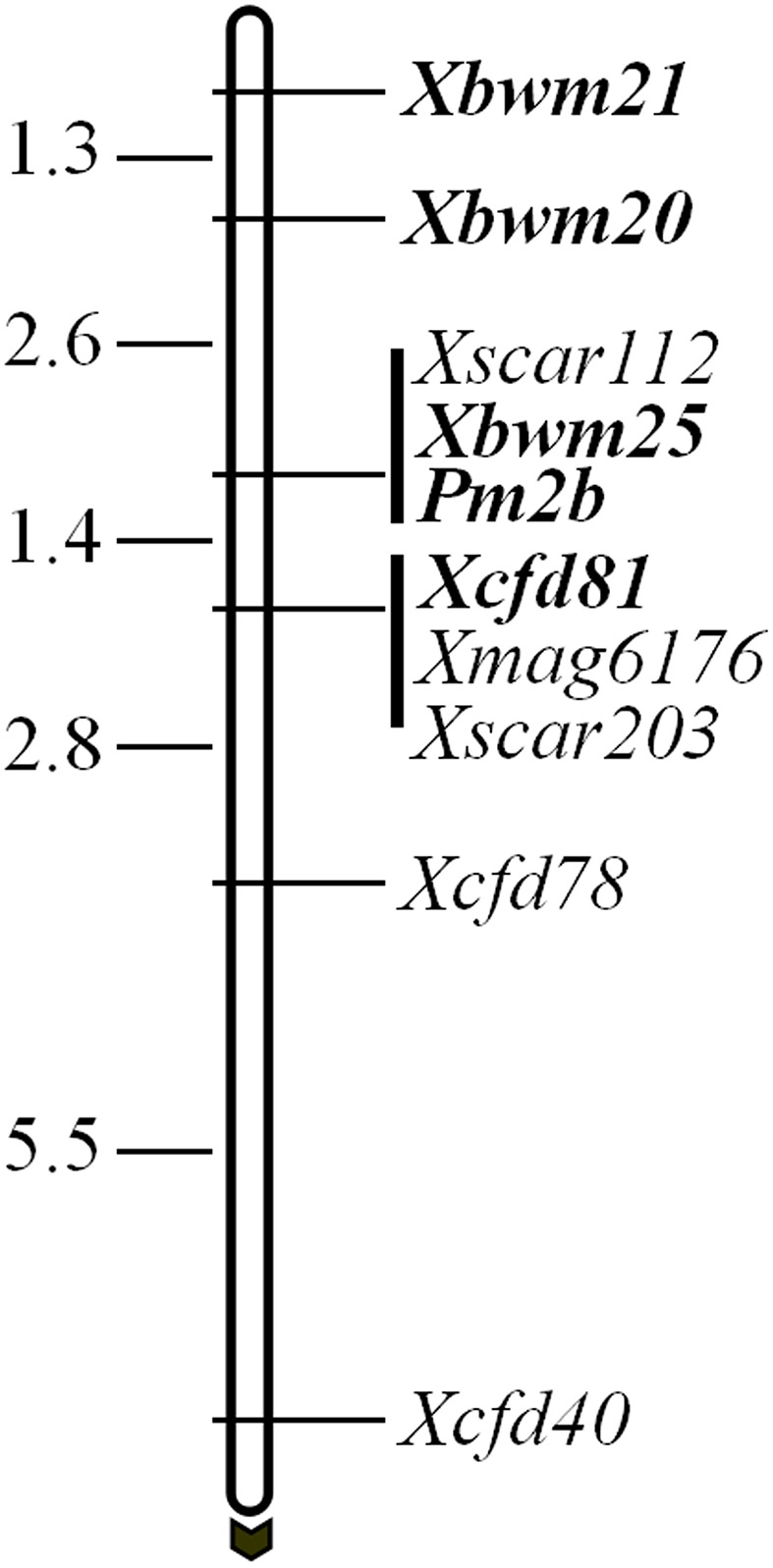
Genetic maps and diagnostic markers for Pm2b gene. Genetic distances are shown on the left and the black arrow indicated the centromere direction. The diagnostic markers and Pm2b gene are shown with bold.
Marker-Assisted Selection for Pm2b Gene
During the backcross transfer of Pm2b gene into the genetic backgrounds of SM15, SX828 and KN199, most of the resistant progeny plants were genotyped with the four Pm2b diagnostic markers CFD81, BWM20, BWM21, and BWM25. As shown in the Table 1, all BC1F1, BC2F1, and BC3F1 progeny plants of KM2939/SM15, SX828/KM2939, and KN199/KM2939 combinations, conferring high resistance to Bgt isolate E09, were used in the FS by using the four Pm2b diagnostic markers CFD81, BWM20, BWM21, and BWM25. After FS, all the resistant backcrossed progeny plants with heterozygous genotypes for Xcfd81, Xbwm20, Xbwm21, and Xbwm25, were regarded as the target plants and chosen for assessing genetic similarity (i.e., %RGC) with their corresponding recurrent parents (Table 1). Then, target progeny plants with the highest %RGC were selected for further backcrossing or selfing. Finally, the homozygous resistant BC3F2 progeny plants after genotyping with the four Pm2b diagnostic markers CFD81, BWM20, BWM21, and BWM25, were assessed genetic similarity to their corresponding recurrent parents (Table 1). The homozygous resistant BC3F2 progeny plants with the highest %RGC, derived from different combinations, were regarded as the Pm2b NILs of their corresponding genetic backgrounds. For example, after the 36 BC1F1 resistant progeny plants of the KM2939/SM15 were genotyped with the four Pm2b diagnostic markers CFD81, BWM20, BWM21, and BWM25, 33 target plants were assessed for genetic similarity to SM15 (Table 1). After the first FS and BS, the target BC1F1 progeny plant KS-6 with the highest %RGC was further backcrossed with the recurrent parent SM15. Using continuous FS and BS, the target BC3F2 progeny plant KS-6-88-11-2 with the highest %RGC was regarded as the Pm2b NIL of SM15 (Table 1).
Based on the genetic marker map (Somers et al., 2004), a total of 182 SSR markers distributed on all 21 wheat chromosomes were chosen to assess the genetic similarity between the progeny plants and their corresponding recurrent parents. A polymorphic survey indicated that 101, 94, and 99 markers out of these 182 SSR markers were polymorphic between KM2939 and SM15, KM2939 and SX828, and KM2939 and KN199, respectively. In the BC1F1, the number of selected plants of KM2939/SM15, SX828/KM2939, and KN199/KM2939 combinations was 33, 13, and 12, and the highest %RGC were correspondingly 89.1, 87.2, and 87.8%, all much higher than 75%, which is the average expected value when no selection was used (Xue et al., 2010) (Table 1). In BC2F1 and BC3F1, the highest %RGC of KM2939/SM15, SX828/KM2939, and KN199/KM2939 combinations were also higher than the average expect value 87.5 and 93.75% without selection. In BC3F2, through marker-assisted FS and BS, the progeny plants KS-6-88-11-2, KSX-2-3-15-22, and KK-3-4-31-54 in the genetic backgrounds of SM15, SX828, and KN199, respectively, were selected for the homozygous Pm2b gene and 99% RGC (Table 1).
To verify the reliability of BS with genome SSR markers, the BC3F2 progeny plant KS-6-88-11-2, derived from the combination KM2939/SM15, together with its recurrent parent SM15, were genotyped with 660K wheat SNP array. Among a total of 606,470 SNPs identified, 587,587 SNPs were polymorphic between KM2939 and SM15, and 573,739, 12,488 and 1,360 were homozygous SM15/SM15, heterozygous KM2939/SM15 and homozygous KM2939/KM2939 genotypes, respectively, in KS-6-88-11-2. Thus, the %RGC of KS-6-88-11-2 was 98.7% assessed by 660K wheat SNP array, which nearly equal to the 99.0% RGC identified by genome SSR markers for BS (Table 1).
Phenotype and Genotype Identification of Pm2b NILs
Pm2b NILs (Pm2b-SM, Pm2b-SX, and Pm2b-KN) in the genetic background of SM15, SX828, and KN199 were developed by selfing the BC3F2 progeny plants KS-6-88-11-2, KSX-2-3-15-22, and KK-3-4-31-54, respectively. Firstly, these Pm2b NILs were identified with tightly linked diagnostic markers Xcfd81, Xbwm20, Xbwm21, and Xbwm25. Figure 2 showed the polymorphic bands about 258/274 bp (Figure 2A), 189/263 bp (Figure 2B), 222/235 and 160/167 bp (Figure 2C), and 236/256 and 183/202 bp (Figure 2D) in the Pm2b-SM, Pm2b-SX, and Pm2b-KN NILs amplified by Xcfd81, Xbwm20, Xbwm21, and Xbwm25, respectively. The results indicated that all Pm2b NILs contained the homozygous Pm2b gene. To verify the resistance to Pm, the Pm2b-SM, Pm2b-SX, and Pm2b-KN, together with the donor and recurrent parents, were separately inoculated with 25 Bgt isolates avirulent to Pm2b gene at the seedling stage and with mixtures of above isolates at the adult stage. All three Pm2b NILs resultly high resistance to Pm at both the seedling and adult stages (Figure 3).
FIGURE 2.
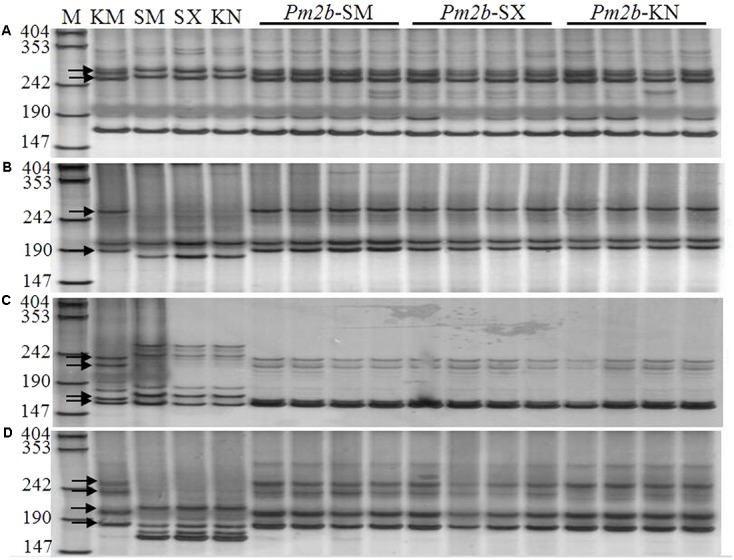
Amplification profiles generated by primers CFD81 (A), BWM20 (B), BWM21 (C), and BWM25 (D) on KM2939, Shimai15, Shixin 828, Kenong 199 and their respective NILs in the backgrounds of Shimai15 (Pm2b-SM), Shixin 828 (Pm2b-SX), and Kenong 199 (Pm2b-KN) (Lanes 1–16). M: pUC18/MspI, numbers to the left are band sizes (bp), and black arrows indicate the polymorphic bands in KM2939.
FIGURE 3.

Powdery mildew reactions of KM2939, Shimai 15, Shixin 828, Kenong 199 and their respective NILs in the backgrounds of Shimai 15 (Pm2b-SM), Shixin 828 (Pm2b-SX), and Kenong 199 (Pm2b-KN) to Bgt isolate E09 at the seedling stage (A) and Bgt mixture at the adult stage (B).
Evaluation of Agronomic Traits of Pm2b NILs
The Pm2b donor breeding line KM2939 had a higher PH, SL, and KNS but a lower SNPP than the three recurrent parents. Compared with the donor and the recurrent parents, the three developed Pm2b NILs showed improved PH and KNS and similar SSS and TKW (Table 2). Moreover, there was no significant difference apart from a few exceptions for SL, TSS, and GY between the Pm2b NILs and their respective recurrent parents. Pm2b-SM showed higher SL and TSS than its recurrent parent SM15, while Pm2b-SX had higher GY than SX828 (Table 2). These results indicated that the developed Pm2b NILs had greater KNS than their recurrent parents and PH than their donor parents. They showed the elite agronomic traits of both their donor and recurrent parents.
Table 2.
The agronomic evaluation of near isogenic lines of Pm2b gene in the genetic backgrounds of Shimai 15 (SM15), Shixin828 (SX828), and Kenong 199 (KN199).
| Cultivars/Lines | PH (cM) | SNPP | SL | TSS | SSS | KNS | TKW (g) | GY (g) |
|---|---|---|---|---|---|---|---|---|
| KM2939 | 91.3 | 6.3 | 10.9 | 20.8 | 0 | 89.0 | 53.9 | 22.9 |
| Pm2b-SM | 73.8∗∗ | 11.4 | 8.1∗∗ | 23.0∗∗ | 0.4 | 69.4∗ | 50.7 | 24.9 |
| Shimai 15 | 69.3 | 10.0 | 7.2 | 21.3 | 0.8 | 62.5 | 51.7 | 21.8 |
| Pm2b-SX | 79.3∗∗ | 11.5 | 7.7 | 21.0 | 0.8 | 71.3∗ | 52.2 | 34.1∗∗ |
| Shixin 828 | 64.3 | 10.6 | 7.7 | 21.0 | 0 | 61.0 | 48.8 | 17.5 |
| Pm2b-KN | 77.0∗∗ | 10.3 | 7.5 | 21.3 | 0.7 | 69.7∗ | 55.7 | 23.1 |
| Kenong 199 | 63.0 | 10.8 | 7.2 | 21.2 | 0 | 60.8 | 51.4 | 22.3 |
∗Significance between the Pm2b NILs and their respective recurrent parent at P ≤ 0.05.
∗∗Significance between the Pm2b NILs and their respective recurrent parent at P ≤ 0.01.
GY, grain yield per plant; KN199, Kenong 199; KNS, kernel number per spike; PH, plant height; RGC, recipient genome composition; SL, spike length; SM15, Shimai 15; SNPP, spike number per plant; SSS, sterile spikelet number per spike; SX828, Shixin 828; TKW, thousand kernel weight; TSS, total spikelet number per spike.
Discussion
In China, only a few Pm genes, which include Pm2, Pm4a, Pm6, Pm8, and Pm21, have been successfully used in resistant breeding in China (Zhang et al., 2010; Huang et al., 2012). Recently only Pm2a was still extensively used by breeders due to the lost (Pm8) or reduced (Pm4a and Pm6) resistance or negative linkages (Pm21) of the other four genes (Wang et al., 2005; Zhu et al., 2005; Ren et al., 2011). However, more and more virulent isolates against Pm2a have been reported in succession (Huang et al., 2012; Ma et al., 2015a; Xu et al., 2015). At the same time, at least 13 new Pm2 alleles or closely linked genes have also been documented, such as PmD57-5D (Ma et al., 2011), PmLX66 (Huang et al., 2012), Pm48 (Gao et al., 2012), PmX3986-2 (Ma et al., 2014), PmW14 (Song et al., 2014), PmPB3558 (Lu et al., 2015), Pm2b (Ma et al., 2015a), PmWFJ34 (Ma et al., 2015b), PmYB (Ma et al., 2015c), PmZ155 (Sun et al., 2015), Pm2c (Xu et al., 2015), PmPB74 (Lu et al., 2016), PmFG (Ma et al., 2016). These resistance genes can make up or replace Pm2a for resistance breeding. However, so far few genes have been transferred into susceptible elite cultivars (Ma et al., 2015a).
Near-isogenic lines with shortest target gene region and highest %RGC are very important in molecular breeding, which can be developed by combining MAS and phenotypic selection. However, no Pm gene NILs have been created using this method in any previous study. In this study, the Pm2b gene conferring high Pm resistance at both the seedling and adult stages was introgressed into the susceptible elite cultivars SM15, SX828, and KN199, which were developed into the corresponding NILs by combining MAS with phenotypic evaluation. Multiple Bgt isolates tests for the Pm2b NILs demonstrated that they carried Pm resistance unlike the recurrent parents SM15, SX828, and KN199. These results not only validated the previous target gene mapping studies, but also further demonstrated the feasibility of MAB for Pm resistance gene Pm2b.
During the backcross transfers of Pm2b gene into the genetic backgrounds of the recurrent parents, it was essential to combine phenotypic identification with MAS. The strategy ensured the successful development of Pm2b NILs in different genetic backgrounds. All the progeny plants were phenotyped for their resistance to E09 isolate and genotyped. Then, resistant progeny plants were implemented by both marker-assisted FS and BS. To avoid the loss of target resistance gene because of double crossover, which occurs with increasing probability in successive backcrosses, multiple markers covering the target regions were used for FS. In addition, large target intervals are often accompanied by deleterious linkage drag (Friebe et al., 1994; Zeller and Hsam, 1996; Li et al., 2017), so tightly linked co-dominant markers of the Pm2b gene for FS minimized negative associations and ensured the precise transfer of target gene. Through both FS and BS, homozygous Pm2b NILs were obtained in the target regions and characterized as about 99% RGC after only three generations of backcrosses and one generation of selfing (Table 1). Generally two to four more generations are needed to achieve a similar goal with the conventional selection alone (Xue et al., 2010). Furthermore, the appropriate backcrossing generations, selected population sizes and marker number for BS assisted in the development of Pm2b NILs with the highest %RGC of 99%. During the BS procedure, selected progeny plants of every generation had a RGC significantly higher than the expected value (Table 1). Generally, through backcrossing three generations and selfing with a population of about 100 selected progeny plants, the NILs with the highest %RGC of 99% could be obtained with the BS for more than 100 genome SSR markers. In addition, 660K SNP array played an important role in precisely assessing the %RGC of Pm2b NIL compared with genome SSR markers. These results indicated that compared with conventional selection, MAS generation by generation was a significantly faster approach for recovery of the recurrent genetic backgrounds than conventional breeding.
This study compared the recurrent parents with the developed Pm2b NILs and showed improved agronomic traits, especially for KNS. Therefore, significantly improved resistance to Pm was combined with improved agronomic traits in the Pm2b NILs compared with their respective recurrent parents, which shows that the Pm2b gene can be easily used in the resistant breeding programs with the help of MAS. Moreover, these NILs will be valuable for the molecular design breeding, high-resolution mapping and forward map-based cloning of this gene.
Conclusion
This study showed: (1) By combining MAS and phenotypic selection, a new broad-spectrum Pm resistance gene Pm2b was introgressed into the genetic backgrounds of three elite susceptible commercial cultivars and three Pm2b NILs with the RGC about 99% were developed; (2) the appropriate backcrossing generations (three generations), selected population sizes (about 100 progeny plants) and marker number for BS (more than 100 markers) could be used to develop the Pm2b NILs with the highest %RGC of 99%; (3) the %RGC of Pm2b NILs could be precisely assessed combining genome SSR markers and SNP array; (4) the four diagnostic markers of Pm2b gene can effectively accelerate the transfer and use of this resistance gene.
Author Contributions
HX: Data analysis and manuscript preparation. YC: MAS for Pm2b gene. YX: Agronomic traits evaluation. PM: Mapping of Pm2b gene. FM: Development of Pm2b NILs. LS: Resistance identification. LL and DA: Experimental design and manuscript revised.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Professor Yilin Zhou, Institute of Plant Protection, Chinese Academy of Agricultural Sciences for his help on powdery mildew tests.
Abbreviations
- Bgt
Blumeria graminis f. sp. tritici
- BS
background selection
- FS
forward ground selection
- GY
grain yield per plant
- ITs
infection types
- KN199
Kenong 199
- KNS
kernel number per spike
- MAB
marker-assisted breeding
- MAS
marker-assisted selection
- NIL
near-isogenic line
- PH
plant height
- Pm
powdery mildew
- RGC
recipient genome composition
- SL
spike length
- SM15
Shimai 15
- SNP
single nucleotide polymorphic
- SNPP
spike number per plant
- SSR
simple sequence repeat
- SSS
sterile spikelet number per spike
- SX828
Shixin 828
- TKW
thousand kernel weight
- TSS
total spikelet number per spike
Funding. This research was financially supported by the National Key Research and Development Program of China (2016YFD0100102), the Chinese Academy of Sciences (#XDA08030107), the National Natural Science Foundation (31671771) and Natural Science Foundation of Hebei Province (C2016503016).
References
- Bennett F. G. A. (1984). Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes. Plant Pathol. 33 279–300. 10.1111/j.1365-3059.1984.tb01324.x [DOI] [Google Scholar]
- Elkot A. F., Chhuneja P., Kaur S., Saluja M., Keller B., Singh K. (2015). Marker assisted transfer of two powdery mildew resistance genes PmTb7A.1 and PmTb7A.2 from Triticum boeoticum (Boiss.) to Triticum aestivum (L.). PLoS ONE 10:e0128297 10.1371/journal.pone.0128297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe B., Heun M., Tuleen N., Zeller F. J., Gill B. S. (1994). Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci. 34 621–625. 10.2135/cropsci1994.0011183X003400030003x [DOI] [Google Scholar]
- Gao H., Zhu F., Jiang Y., Wu J., Yan W., Zhang Q., et al. (2012). Genetic analysis and molecular mapping of a new powdery mildew resistant gene Pm46 in common wheat. Theor. Appl. Genet. 125 967–973. 10.1007/s00122-012-1886-7 [DOI] [PubMed] [Google Scholar]
- Guo J., Zhang X., Hou Y., Cai J., Shen X., Zhou T., et al. (2015). High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor. Appl. Genet. 128 2301–2316. 10.1007/s00122-015-2586-x [DOI] [PubMed] [Google Scholar]
- Habib A., Shabala S., Shabala L., Zhou M., Liu C. (2016). Near-isogenic lines developed for a major QTL on chromosome arm 4HL conferring Fusarium crown rot resistance in barley. Euphytica 209 555–563. 10.1007/s10681-015-1623-9 [DOI] [Google Scholar]
- Hao Y., Parks R., Cowger C., Chen Z., Wang Y., Bland D., et al. (2015). Molecular characterization of a new powdery mildew resistance gene Pm54 in soft red winter wheat. Theor. Appl. Genet. 128 465–476. 10.1007/s00122-014-2445-1 [DOI] [PubMed] [Google Scholar]
- Huang J., Zhao Z., Song F., Wang X., Xu H., Huang Y., et al. (2012). Molecular detection of a gene effective against powdery mildew in the wheat cultivar Liangxing 66. Mol. Breed. 30 1737–1745. 10.1007/s11032-012-9757-0 [DOI] [Google Scholar]
- Khanna A., Sharma V., Ellur R. K., Shikari A. B., Krishnan S., Singh U. D., et al. (2015). Development and evaluation of near-isogenic lines for major blast resistance gene(s) in Basmati rice. Theor. Appl. Genet. 128 1243–1259. 10.1007/s00122-015-2502-4 [DOI] [PubMed] [Google Scholar]
- Kosambi D. D. (1943). The estimation of map distances from recombination values. Ann. Eugen. 12 172–175. 10.1111/j.1469-1809.1943.tb02321.x [DOI] [Google Scholar]
- Kumar J., Mir R. R., Kumar N., Kumar A., Mohan A., Prabhu K. V., et al. (2010). Marker-assisted selection for pre-harvest sprouting tolerance and leaf rust resistance in bread wheat. Plant Breed. 129 617–621. 10.1111/j.1439-0523.2009.01758.x [DOI] [Google Scholar]
- Lander E. S., Green P., Abrahamson J., Barlow A., Daly M., Lincoln S., et al. (1987). Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1 174–181. 10.1016/0888-7543(87)90010-3 [DOI] [PubMed] [Google Scholar]
- Li H., Jiang B., Wang J., Lu Y., Zhang J., Pan C., et al. (2017). Mapping of novel powdery mildew resistance gene(s) from Agropyron cristatum chromosome 2P. Theor. Appl. Genet. 130 109–121. 10.1007/s00122-016-2797-9 [DOI] [PubMed] [Google Scholar]
- Li H. J., Wang X. M., Song F. J., Wu C. P., Wu X. F., Zhang N., et al. (2011). Response to powdery mildew and detection of resistance genes in wheat cultivars from China. Acta Agron. Sin. 37 943–954. 10.3724/sp.j.1006.2011.00943 [DOI] [Google Scholar]
- Lu Y., Wu X., Yao M., Zhang J., Liu W., Yang X., et al. (2015). Genetic mapping of a putative Agropyron cristatum-derived powdery mildew resistance gene by a combination of bulked segregant analysis and single nucleotide polymorphism array. Mol. Breed. 35:96 10.1007/s11032-015-0292-7 [DOI] [Google Scholar]
- Lu Y., Yao M., Zhang J., Song L., Liu W., Yang X., et al. (2016). Genetic analysis of a novel broad-spectrum powdery mildew resistance gene from the wheat-Agropyron cristatum introgression line Pubing 74. Planta 244 713–723. 10.1007/s00425-016-2538-y [DOI] [PubMed] [Google Scholar]
- Ma H., Kong Z., Fu B., Li N., Zhang L., Jia H., et al. (2011). Identification and mapping of a new powdery mildew resistance gene on chromosome 6D of common wheat. Theor. Appl. Genet. 123 1099–1106. 10.1007/s00122-011-1651-3 [DOI] [PubMed] [Google Scholar]
- Ma P., Xu H., Li L., Zhang H., Han G., Xu Y., et al. (2016). Characterization of a new Pm2 allele conferring powdery mildew resistance in the wheat germplasm line FG-1. Front. Plant Sci. 7:546 10.3389/fpls.2016.00546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Xu H., Luo Q., Qie Y., Zhou Y., Xu Y., et al. (2014). Inheritance and genetic mapping of a gene for seedling resistance to powdery mildew in wheat line X3986-2. Euphytica 200 149–157. 10.1007/s10681-014-1178-1 [DOI] [Google Scholar]
- Ma P., Xu H., Xu Y., Li L., Qie Y., Luo Q., et al. (2015a). Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor. Appl. Genet. 128 613–622. 10.1007/s00122-015-2457-5 [DOI] [PubMed] [Google Scholar]
- Ma P., Xu H., Zhang H., Li L., Xu Y., Zhang X., et al. (2015b). The gene PmWFJ is a new member of the complex Pm2 locus conferring unique powdery mildew resistance in wheat breeding line Wanfengjian 34. Mol. Breed. 35:210 10.1007/s11032-015-0403-5 [DOI] [Google Scholar]
- Ma P., Zhang H., Xu H., Xu Y., Cao Y., Zhang X., et al. (2015c). The gene PmYB confers broad-spectrum powdery mildew resistance in the multi-allelic Pm2 chromosome region of the Chinese wheat cultivar YingBo 700. Mol. Breed. 35:124 10.1007/s11032-015-0320-7 [DOI] [Google Scholar]
- McIntosh R. A., Dubcovsky J., Rogers W. J., Morris C., Xia X. C. (2017). Catalogue of Gene Symbols for Wheat: 2017 Supplement. Available at: http://www.shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp [Google Scholar]
- Petersen S., Lyerly J. H., Worthington M. L., Parks W. R., Cowger C., Marshall D. S., et al. (2015). Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theor. Appl. Genet. 128 303–312. 10.1007/s00122-014-2430-8 [DOI] [PubMed] [Google Scholar]
- Ren T. H., Chen F., Zhang H. Q., Yan B. J., Ren Z. L. (2011). Genetic suppression of the powdery mildew resistance gene Pm21 in common wheat. Acta Phytopathol. Sin. 42 57–64. 10.13926/j.cnki.apps.2012.01.017 [DOI] [Google Scholar]
- Santos F. R., Pena S. D. J., Epplen J. T. (1993). Genetic and population study of a Y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic technique. Hum. Genet. 90 655–656. 10.1007/bf00202486 [DOI] [PubMed] [Google Scholar]
- Somers D. J., Isaac P., Edwards K. (2004). A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109 1105–1114. 10.1007/s00122-004-1740-7 [DOI] [PubMed] [Google Scholar]
- Song W., Sun H. G., Sun Y. L., Zhao Z. H., Wang X. O., Wu X. F., et al. (2014). Chromosomal localization of the gene for resistance to powdery mildew in the wheat cultivar Wennong14. Acta Agron. Sin. 40 798–804. 10.3724/sp.j.1006.2014.00798 [DOI] [Google Scholar]
- Sun H., Song W., Sun Y., Chen X., Liu J., Zou J., et al. (2015). Resistance of ‘Zhongmai 155’ wheat to powdery mildew: effectiveness and detection of the resistance gene. Crop Sci. 55 1017–1025. 10.2135/cropsci2014.05.0355 [DOI] [Google Scholar]
- Tanksley S. D., Grandillo S., Fulton T. M., Zamir D., Eshed Y., Petiard V., et al. (1996). Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 92 213–224. 10.1007/bf00223378 [DOI] [PubMed] [Google Scholar]
- The Occurrence Tendency Prediction of Plant Diseases and Insect Pests in China (2017). Intelligence of Plant Diseases and Insect Pests. Available at: http://cb.natesc.gov.cn/Html/2017_01_11/28092_151760_2017_01_11_436738.html [Google Scholar]
- Vida G., Gál M., Uhrin A., Veisz O., Syed N. H., Flavell A. J., et al. (2009). Molecular markers for the identification of resistance genes and marker-assisted selection in breeding wheat for leaf rust resistance. Euphytica 170 67–76. 10.1007/s10681-009-9945-0 [DOI] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32 947–951. 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- Wang Z., Li H., Zhang D., Guo L., Chen J., Chen Y., et al. (2015). Genetic and physical mapping of powdery mildew resistance gene MlHLT in Chinese wheat landrace Hulutou. Theor. Appl. Genet. 128 365–373. 10.1007/s00122-014-2436-2 [DOI] [PubMed] [Google Scholar]
- Wang Z. L., Li L. H., He Z. H., Duan X. Y., Zhou Y. L., Chen X. M., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89 457–463. 10.1094/pd-89-0457 [DOI] [PubMed] [Google Scholar]
- Xu H., Yi Y., Ma P., Qie Y., Fu X., Xu Y., et al. (2015). Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor. Appl. Genet. 128 2077–2084. 10.1007/s00122-015-2568-z [DOI] [PubMed] [Google Scholar]
- Xue S., Li G., Jia H., Lin F., Cao Y., Xu F., et al. (2010). Marker-assisted development and evaluation of near-isogenic lines for scab resistance QTLs of wheat. Mol. Breed. 25 397–405. 10.1007/s11032-009-9339-y [DOI] [Google Scholar]
- Yaniv E., Raats D., Ronin Y., Korol A. B., Grama A., Bariana H., et al. (2015). Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol. Breed. 35:43 10.1007/s11032-015-0238-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks J. C., Chang T. T., Konzak C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14 415–421. 10.1111/j.1365-3180.1974.tb01084.x [DOI] [Google Scholar]
- Zeller F. J., Hsam S. L. K. (1996). Chromosomal location of a gene suppressing powdery mildew resistance genes Pm8 and Pm17 in common wheat (Triticum aestivum L. em. Thell.). Theor. Appl. Genet. 93 38–40. 10.1007/bf00225724 [DOI] [PubMed] [Google Scholar]
- Zhang H., Guan H., Li J., Zhu J., Xie C., Zhou Y., et al. (2010). Genetic comparative genomics mapping reveals that a powdery mildew resistance gene Ml3D232 originating from wild emmer co-segregates with an NBS-LRR analog in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 121 1613–1621. 10.1007/s00122-010-1414-6 [DOI] [PubMed] [Google Scholar]
- Zhang R., Sun B., Chen J., Cao A., Xing L., Feng Y., et al. (2016). Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 129 1975–1984. 10.1007/s00122-016-2753-8 [DOI] [PubMed] [Google Scholar]
- Zheng S., Li Y., Lu L., Liu Z., Zhang C., Ao D., et al. (2017). Evaluating the contribution of Yr genes to stripe rust resistance breeding through marker-assisted detection in wheat. Euphytica 213:50 10.1007/s10681-016-1828-6 [DOI] [Google Scholar]
- Zhou R., Zhu Z., Kong X., Huo N., Tian Q., Li P., et al. (2005). Development of wheat near-isogenic lines for powdery mildew resistance. Theor. Appl. Genet. 110 640–648. 10.1007/s00122-004-1889-0 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Zhou R., Kong X., Dong Y., Jia J. (2005). Microsatellite markers linked to 2 powdery mildew resistance genes introgressed from Triticum carthlicum accession PS5 into common wheat. Genome 48 585–590. 10.1139/g05-016 [DOI] [PubMed] [Google Scholar]


